Abstract
The inhibition of the proliferation and apoptosis of bone marrow-derived mesenchymal stem cells (BMSCs) triggered by the excessive use of glucocorticoids, is considered a potential mechanism for the pathogenesis of steroid-induced osteonecrosis of the femoral head (SONFH). Long non-coding RNAs (lncRNAs) have been proven to influence the proliferation, apoptosis and differentiation of BMSCs by regulating the expression of critical genes. A previous microarray analysis by the authors confirmed the significant downregulation of LINC00473 in human BMSCs (hBMSCs) from patients with SONFH. However, the underlying role and molecular mechanisms of LINC00473 on dexamethasone (Dex)-stimulated hBMSCs remains unknown. In the present study, the expression of LINC00473 was determined in the hBMSCs of patients with SONFH and control patients. In addition, the protective effects and underlying molecular mechanisms of LINC00473 in Dex-stimulated hBMSCs were investigated. The results revealed that LINC00473 expression was significantly down-regulated in hBMSCs from patients with SONFH compared with the controls, and that the upregulation of LINC00473 attenuated the inhibitory effects exerted by 1 µM Dex on the proliferation and apoptosis of hBMSCs. Moreover, the upregulation of LINC00473 significantly promoted the protein expression of phosphorylated (p-)Akt, p-Bcl-2-associated death promoter (p-Bad) and B-cell lymphoma 2 (Bcl-2), whereas it decreased the cleavage of caspase-3, thus preventing the Dex-induced apoptosis of hBMSCs. Of note, the regulatory effects of LINC00473 on the Akt/Bad/Bcl-2 signaling pathway and its anti-apoptotic effects were similar to those of SC79 (an Akt activator), and were inhibited by MK-2206 (an Akt inhibitor). In further experiments, it was found that the upregulation of LINC00473 markedly promoted the phosphorylation of Akt in Dex-stimulated hBMSCs, and increased the protein level of phosphatidylethanolamine-binding protein 1 (PEBP1). Alternatively, the promoting effect on Akt phosphorylation induced by LINC00473 was significantly attenuated following the knockdown of PEBP1. Furthermore, the upregulation of PEBP1 triggered a marked increase in the levels of Akt phosphorylation in Dex-stimulated hBMSCs, which was line with the upregulation of LINC00473. Taken together, the results of the present study demonstrate that LINC00473 has the ability to rescue hBMSCs from Dex-induced apoptosis through the PEBP1-mediated activation of the Akt/Bad/Bcl-2 signaling pathway.
Keywords: lncRNA LINC00473, human bone marrow mesenchymal stem cells, dexamethasone, apoptosis, phosphatidylethanolamine-binding protein 1, Akt/Bad/Bcl-2 signaling pathway
Introduction
Steroid-induced osteonecrosis of the femoral head (SONFH) is a common and progressive musculoskeletal disorder characterized by severe pain and dysfunction of the hip joint, caused by the excessive use of glucocorticoids (1-3). Although the explicit mechanisms responsible for the development of SONFH remain unclear, a growing body of evidence has recognized an association between abnormal cellular behaviors of bone marrow-derived mesenchymal stem cells (BMSCs) and the development of SONFH (4-6). Emerging evidence has also demonstrated that long-term exposure to high doses of glucocorticoids, such as dexamethasone (Dex), can inhibit the proliferation and induce the apoptosis of BMSCs (7,8), which is considered a potential mechanism for the pathogenesis of SONFH.
Long noncoding RNAs (lncRNAs) are RNA transcripts that are >200 nucleotides in length, which are implicated in various pathophysiological processes by regulating the expression of critical genes to influence cell differentiation, proliferation and apoptosis, although they have no coding potential (9-11). LINC00473, a lncRNA located on human chromosome 6q27 (12), has been shown to be significantly downregulated in human BMSCs (hBMSCs) obtained from patients with SONFH, based on a previous microarray analysis by the authors (13). Several studies have confirmed the role of LINC00473 in the proliferation and apoptosis of various tumor cells (14-17); however, the underlying effects and molecular mechanisms of LINC00473 on Dex-stimulated hBMSCs remain unknown.
Protein kinase B (Akt) is a primary mediator of the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, which has been shown to affect cell apoptosis, proliferation and migration (18-20). Increases in the levels of B-cell lymphoma 2 (Bcl-2), an anti-apoptotic protein, can lead to the inhibition of apoptosis through the decrease of cleaved caspase-3. Moreover, the phosphorylation of Akt increases the expression of Bcl-2 through the inactivation of the Bcl-2-associated death promoter (Bad) (21,22). Taken together, these results indicate that the Akt/Bad/Bcl-2 signaling cascade plays a key role in cell apoptosis, proliferation and survival. Moreover, Dex has been reported to regulate cellular behavior, including the inhibition of cancer cell migration and osteoblast osteogenesis, by suppressing the Akt signaling pathway (23,24), which prob-ably is the molecular mechanism responsible for the apoptosis of BMSCs induced by Dex.
Phosphatidylethanolamine-binding protein 1 (PEBP1), which is also referred to as Raf1 kinase inhibitory protein (RKIP), has been found to be potentially involved in diverse biological functions through the regulation of various protein kinases in several signaling pathways, such as the RAF/MEK/mitogen-activated protein kinase (MAPK)/extra-cellular regulated kinase (ERK), nuclear factor (NF)-κB and PI3K/Akt/mammalian target of rapamycin (mTOR) pathways (25,26). In addition, the influence of PEBP1 on cell apoptosis, proliferation, migration and autophagy has been demonstrated. For instance, PEBP1 has been reported to inhibit starvation-induced autophagy through the activation of the Akt/MTORC1 pathway (27).
Surprisingly, it was found that PEBP1 is involved in the competitive endogenous RNA (ceRNA) network of LINC00473, and there may be a potential correlation between LINC00473 and PEBP1, based on the statistics obtained from the Encyclopedia of RNA Interactomes (ENCORI; http://starbase.sysu.edu.cn/index.php) (28). Notably, a majority of these data have been obtained from cancer-related studies, but almost none are from studies on BMSCs. Due to this disparity, the present study aimed to investigate whether LINC00473 can rescue hBMSC from apoptosis induced by Dex via the Akt/Bad/Bcl-2 signaling pathway. In addition, the present study aimed to further determine whether the activation of the Akt/Bad/Bcl-2 signaling pathway induced by LINC00473 is mediated by PEBP1, in order to obtain evidence to identify potential targets of LINC00473.
Materials and methods
Clinical tissue sample extraction
The present study was approved by the Ethics Committee of Qingdao University Affiliated Hospital (Qingdao, China). Bone marrow tissue of the proximal femur was obtained from 6 patients with SONFH (4 males and 2 females, aged 52 to 67 years) and 6 patients (3 males and 3 females, aged 61 to 71 years) with femoral neck fractures as the controls, during total hip arthroplasty (THA) surgery at the Department of Orthopedics at Qingdao University Affiliated Hospital. All donors were Chinese and provided informed consent for participation.
hBMSC isolation and culture
Based on previously described methods, hBMSCs were isolated from the bone marrow tissue and cultured (29). Briefly, the bone marrow was mixed with an equal volume of phosphate-buffered saline (PBS), and then spread on lymphocyte separation medium (LSM; Beijing Solarbio Science & Technology Co., Ltd.; 1:1, v/v). After washing with PBS, the mononuclear cell layer was collected following centrifugation at 500 × g for 30 min at room temperature. The cells were then cultured in low-glucose DMEM (Beijing Solarbio Science & Technology Co., Ltd.) containing 10% (v/v) fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) and 100 units/ml penicillin-streptomycin (Beijing Solarbio Science & Technology Co., Ltd.), and maintained in a humidified incubator (HERAcell vios 160i, Thermo Fisher Scientific, Inc.) at 5% CO2 and 37°C. When 80-90% confluency was reached, the hBMSCs were detached using 0.05% trypsin-EDTA (Beijing Solarbio Science & Technology Co., Ltd.) and sub-cultured at a ratio of 1:2 in new culture flasks. The cells at passage 3 were used in subsequent experiments.
Phenotyping of hBMSCs
Flow cytometric analysis for surface markers was performed following each extraction of primary cells, and 3 replicates were made in the experiment. In brief, after the 3rd passage hBMSCs were collected, rinsed and blocked with pre-cold PBS containing 1% FBS 3 times, the cell suspension was incubated for 30 min in the dark at 37°C with the following 2 pairs of mouse anti-human antibodies (30): 20 µl of CD34PE (cat. no. 560941) and 20 µl of CD45-FITC (cat. no. 555482); 20 µl of CD73PE (cat. no. 561014) and 2 µl of CD90FITC (cat. no. 555595), through double-staining based on previous descriptions (31). The cells were incu-bated with isotype control antibodies for PE Mouse IgG1, κ (cat. no. 555749) and FITC Mouse IgG1, κ (cat. no. 554679) served as a negative control as recommended by the manufacturer. The expression levels of hBMSC surface markers were investigated using an Apogee A50-MICRO flow cytometer (Apogee Corporation). All antibodies were purchased from BD Biosciences.
Osteogenic and adipogenic differentiation of hBMSCs
The capacity of the hBMSCs to undergo osteogenic and adipogenic differentiation was assessed using a differentiation medium (Biogene Biotechnology, Inc.), as per the manufacturer's instructions. Osteogenic induction medium was composed of complete medium supplemented with 1% glutamine, 1% penicillin-streptomycin, 1% β-glycerophosphate, 0.2% ascorbate acid and 0.01% Dex. Adipogenic induction medium was composed of complete medium supplemented with 0.1% insulin, 0.1% isobutylmethylxanthine, 0.1% rosiglitazone and 0.1% dexamethasone.
For osteogenic differentiation, the cells were treated with the osteogenic differentiation medium after the confluency reached 60%, and were fixed with 0.2 ml of 4% paraformaldehyde solution for 15 min, and incubated with alkaline phosphatase (Beijing Solarbio Science & Technology Co., Ltd.) for 20 min in dark at 37°C after 14 days. For adipogenic differentiation, the cells were treated with the adipogenic differentiation medium after confluency reached 90%, and were then fixed with 0.2 ml of 4% paraformaldehyde solution for 15 min, and stained with Oil Red O (Beijing Solarbio Science & Technology Co., Ltd.) for 15 min after 21 days. The activity part of ALP and lipid droplet formation in cells were observed under an inverted phase-contrast microscope (CKX41, Olympus Corporation).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cultured cells using a RNAiso plus kit (Takara Bio Inc.), and was then reverse transcribed into cDNA using a PrimeScript RT reagent kit (Takara Bio Inc.). qPCR was performed using a SYBR Premix Ex Taq II kit (Takara Bio Inc.) along with a Roche LightCycler 480 Detection System (Roche Diagnostics) as instructed by the manufacturer. The RT-qPCR thermocycling conditions were as follows: Denaturation at 95°C for 30 sec; 40 cycles at 95°C for 5 sec and 60°C for 30 sec; dissociation at 95°C for 5 sec and 60°C for 1 min; and cooling at 50°C for 30 sec. The primers used in the present study were provided by Guangzhou RiboBio Co., Ltd. The sequences of the primers were as follows: Human LINC00473 forward, 5′-GGC AGC CTC AGG TTA CAA AT-3′ and reverse, 5′-AGG AGC AGG TAG GGA AAT GA-3′; human PEBP1 forward, 5′-CTC GCG ATG CTG GTG TAC C-3′ and reverse, 5′-GGA TCC CTG CTT CCC ACA CAG C-3′; and human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward, 5′-CCC ACG CCT CCT CCG TTG AC-3′ and reverse, 5′-ATA CCT GGA AAT CAG CTT TAC AA-3′. The relative expression levels of LINC00473 and PEBP1 were evaluated using the 2−ΔΔCq method (32), and were normalized to the GAPDH levels. The experiment was performed in triplicate.
Lentivirus vector construction and infection of hBMSCs
The pRLenti-EF1a-EGFP-CMV-LINC00473-up lentivirus (H12785) and negative control lentivirus (GL103) were provided by OBiO Technology (Shanghai) Corp., Ltd. The hBMSCs were infected with the LINC00473-up lentivirus at a final multiplicity of infection (MOI) of 100 containing 5 µg/ml polybrene, and were observed for the expression of green fluorescent protein (GFP) under an inverted fluorescence microscope (EVOS FL, Invitrogen; Thermo Fisher Scientific, Inc.) after 24 h. The efficiency of LINC00473-up and the negative control were determined by RT-qPCR after 3 days.
Treatment of hBMSCs
The hBMSCs obtained from the patients with femoral neck fractures were used in the following experiments. The cells treated with 1 µM Dex were named as the Dex group (Dex), while the cells infected with LINC00473-up lentivirus following exposure to 1 µM Dex were named as the Dex + LINC00473-up group (Dex + LINC00473-up), and cells infected with a random sequence following exposure to 1 µM Dex were named as the Dex + Vector group (Dex + Vector). Cells cultured in complete medium were regarded as the normal group (Normal).
In addition, MK-2206 (Selleckchem), an Akt inhibitor, and SC79 (Selleckchem), an Akt activator, were used in the antagonist and activation experiments, respectively. Cells in the Dex + LINC00473-up group were treated with MK-2206 (5 µmol/l) to create the Dex + LINC00473-up + MK-2206 group (Dex + LINC00473-up + MK-2206), while cells in the Dex group were treated with SC79 (8 µg/ml) to create the Dex + SC79 group (Dex + SC79).
Cell proliferation analysis
In brief, 5×103 cells/well were seeded into 96-well plates, and were treated according to their grouping after 24 h. Subsequently, a Cell Counting Kit-8 (CCK-8 assay kit; Beijing Solarbio Science & Technology Co., Ltd.) was used to analyze cell proliferation from day 1 to 7, as recommended by the manufacturer. A microplate reader (5082, Tecan Austria GmbH) was used to measure the absorbance at 450 nm. The experiment was performed in triplicate.
Assessment of the morphology of apoptotic cells
In brief, 5×104 cells/well were seeded into 24-well plates, and were then treated based on their grouping. After 7 days, the morphology of the apoptotic cells was assessed using a chromatin dye Hoechst 33342 kit (Beijing Solarbio Science & Technology Co., Ltd.), as recommended by the manufacturer. Apoptotic cells were observed through fluorescence microscopy (EVOS FL, Invitrogen; Thermo Fisher Scientific, Inc.) and identified as the cells with blue hyper-fluorescence, and with one of the following morphological alterations: Chromatic agglutination, karyopyknosis, nuclear fragmentation (33,34), and were then counted through 6 randomly selected fields. The experiment was performed in triplicate.
Flow cytometric analysis of apoptosis
In brief, 2×105 cells/well were seeded into 6-well plates, and were then treated based on their grouping. After 7 days, the apoptotic cells were analyzed using an Apogee A50-MICRO flow cytometer (Apogee Corporation) along with an Annexin V-PE/7-AAD apoptosis detection kit (BD Biosciences) as instructed by the manufacturer, and at least 104 cells in each sample were analyzed. The emission max wavelength of PE and 7-AAD was 578 nm and 655 nm, respectively. Both of their excitation wavelengths were 488 nm (35). The experiment was performed in triplicate.
Western blot analysis
A pre-cold RIPA Lysis Buffer (Beijing Solarbio Science & Technology Co., Ltd.) containing 1% protease inhibitor cocktail (MedChemExpress) was used to extract total protein from the hBMSCs treated based on their grouping. After the protein concentration was determined by a bicinchoninic acid (BCA) protein assay kit (Beijing Solarbio Science & Technology Co., Ltd.), the cell lysates were mixed with a protein loading buffer (EpiZyme) at a ratio of 1:5, and heated to 95°C for 5 min. The separation of the protein sample (50 µg) in each group was conducted by 12.5% SDS-PAGE (EpiZyme), and the protein samples were electrotransferred onto a PVDF membrane (MilliporeSigma) following separation. Subsequently, 5% non-fat milk in TBST (Beijing Solarbio Science & Technology Co., Ltd.) was used to block the PVDF membranes for 2 h at 37°C. Following incubation with primary antibodies at 4°C overnight, the PVDF membranes were incubated with corresponding secondary antibodies (HRP-conjugated) at 37°C for 1 h. The target bands were visualized using ECL-PLUS reagents (MilliporeSigma), and were scanned using a BioSpectrum Imaging System (UVP, Thermo Fisher Scientific, Inc.). The results of western blot analysis were quantified through integrated density using ImageJ software (version 1.52u), and were normalized to GAPDH levels.
The primary antibodies, including rabbit anti-human Akt (cat. no. 4685S), phosphorylated (p-)Akt (cat. no. 13038S), Bad (cat. no. 9239S), p-Bad (cat. no. 4366S), Bcl-2 (cat. no. 4223S), cleaved caspase-3 (cat. no. 9664S), caspase-3 (cat. no. 14220S), PEBP1 (cat. no. 13006S) antibodies, were provided by Cell Signaling Technology, Inc. and were diluted in antibody dilution buffer (Boster Biological Technology, Inc.) at ratio of 1:1,000 as recommended by the manufacturer. The primary antibody for GAPDH (cat. no. E-AB-20072) and all the secondary antibodies (cat. nos. : E-AB-1003) were purchased from Elabscience Biotechnology Inc. and were diluted in antibody dilution buffer (Boster Biological Technology, Inc.) at ratio of 1:5,000 and 1:1,000, respectively.
ceRNA interation network of LINC00473 in humans
The data of the ceRNA interation network of LINC00473 in humans were obtained from ENCORI. Briefly, on the home page of the ENCORI website (http://starbase.sysu.edu.cn/index.php), 'ceRNA-Network' was selected from the navigation bar, and 'lncRNA-ceRNA' was then selected. Subsequently, 'human' was selected in the section of 'Genome', and a search for 'LINC00473' was made in the section of 'ceRNA Gene' to complete the ceRNA interaction network of LINC00473 in humans. 'Pan-Cancer' was then selected to obtain the coefficient-R value in various types of cancer, which were created as a statistics chart using GraphPad Prism 8 software (GraphPad, Inc.).
PEBP1 recombinant plasmid and transfection of hBMSCs with shRNA
To further determine whether the activation of the Akt/Bad/Bcl-2 signaling pathway by LINC00473 is mediated by PEBP1, the shRNA and overexpression plasmid of PEBP1 were transfected into Dex-induced hBMSCs using a Lipofectamine 2000 system (Thermo Fisher Scientific, Inc.), as recommended by the manufacturer. Three shRNAs of PEBP1, including sh-PEBP1-1 (target sequence, tgGGA TGA CTA TGT GCC CAA A), sh-PEBP1-2 (target sequence, caAAT TCA AGG TGG CGT CCT T) and sh-PEBP1-3 (target sequence, cgAGC AGG ACA GGC CGC TAA A), and the overexpression plasmid (vector name, GV657) for PEBP1 (PEBP1-up) were provided by Genechem Corporation. Moreover, a random sequence (TTC TCC GAA CGT GTC ACG T) was selected as the negative control of shRNA (sh-Control), and a blank vector was used as the negative control of the overexpression plasmid for PEBP1 (PEBP1-Control).
Following infection with the LINC00473-up lentivirus, the Dex-induced hBMSCs were transfected with a shRNA of PEBP1 to create the Dex + LINC00473-up + sh-PEBP1 group (Dex + LINC00473-up + sh-PEBP1). The Dex-stimulated hBMSCs were transfected with the recombinant plasmid of PEBP1 to create the Dex + PEBP1-up group (Dex + PEBP1-up). Following transfection for 24 h, the transfection efficiency of PEBP1 was detected by RT-qPCR and western blot analysis.
Statistical analysis
SPSS 19.0 software (IBM Corporation) was used to perform one-way analysis of variance (ANOVA) to compare the results between >3 groups, and an unpaired t-test to compare the data of 2 groups. Parametric data are presented as the means ± standard deviation (SD). Furthermore, the homogeneity test for variance was also performed. When the statistics exhibited heteroscedasticity, Tamhane's T2 test was used. A P-value <0.05 was considered to indicate a statistically significant result. GraphPad Prism 8 software (GraphPad, Inc.) was used to create the statistics charts.
Results
Identification of hBMSCs
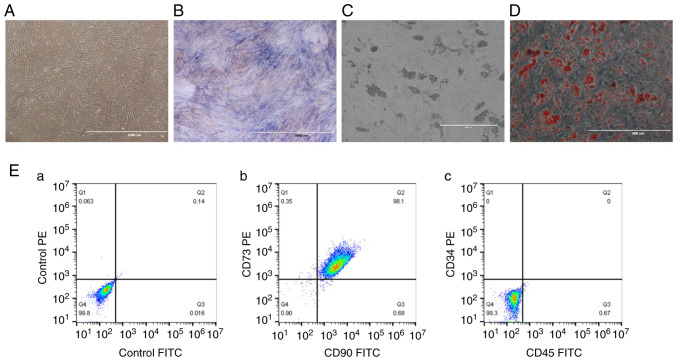
The third passage of cells was observed to have a homogeneous fibroblast-like, spindle-shaped morphology (Fig. 1A). Furthermore, their ability to differentiate into osteogenic and adipogenic lineages was confirmed using ALP and Oil Red O staining (Fig. 1B-D). The phenotyping of the hBMSCs detected using flow cytometry further suggested that the majority of the isolated cells expressed the two typical surface markers of marrow-derived stem cells: CD73 (98.45%) and CD90 (98.78%) (Fig. 1Eb), and few cells expressed the two specific cell surface markers of hematopoietic cells: CD34 (0%) and CD45 (0.67%) (Fig. 1Ec).
Figure 1.
Identification of hBMSCs. (A) Representative images showing morphology of hBMSCs under an inverted phase contrast microscope (scale bar, 1,000 µm). (B) ALP staining (scale bar, 1,000 µm). (C) Lipid droplets formation (scale bar, 400 µm). (D) Oil Red O staining (scale bar, 400 µm). (E) Phenotypic analysis of hBMSCs by flow cytometry (CD34, CD45, CD73 and CD90): (E-a) Cells were incubated with isotype control antibodies for PE mouse IgG1, κ and FITC mouse IgG1, κ served as a control; (E-b) Most of the isolated cells expressed the two typical surface markers of MSCs: CD73 (98.45%) and CD90 (98.78%); (E-c) Few cells expressed the two specific cell surface markers of hematopoietic cells: CD34 (0%) and CD45 (0.67%). hBMSCs, human bone marrow-derived mesenchymal stem cells; ALP, alkaline phosphatase.
LINC00473 is downregulated in the hBMSCs of patients with SONFH
To determine the role of LINC00473 in SONFH, the expression of LINC00473 in hBMSCs of the proximal femur obtained from patients with SONFH and femoral neck fractures were assessed by RT-qPCR. The relative LINC00473 expression in hBMSCs obtained from the patients with SONFH was significantly lower than that of the cells from the control patients (Fig. 2), which is in line with the previous microarray analysis (13) that indicated that this may be caused by glucocorticoids.
Figure 2.
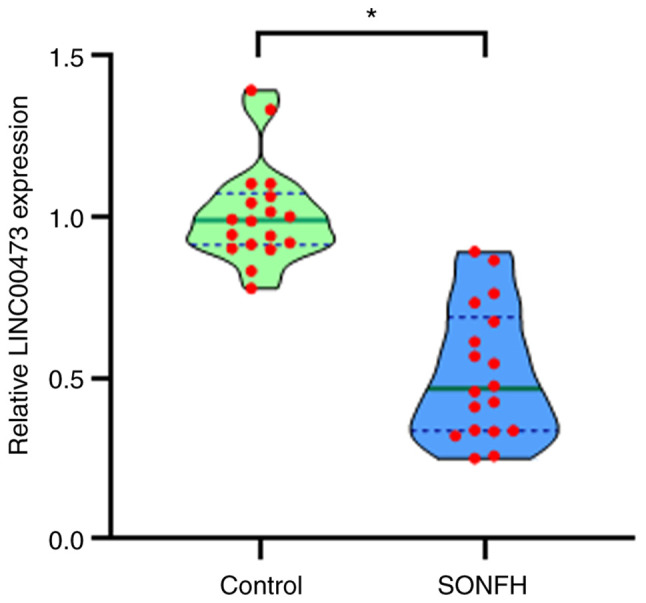
Relative LINC00473 expression examined by RT-qPCR in hBMSCs from patients with SONFH compared to the controls. All data are presented as the means relative LINC00473 expression ± standard deviation, n=18 (n=6×3; the experiment was performed in triplicate), *P<0.05 compared with the control group. LINC00473, lncRNA-00473; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; hBMSCs, human bone marrow-derived mesenchymal stem cells; SONFH, steroid-induced osteonecrosis of the femoral head.
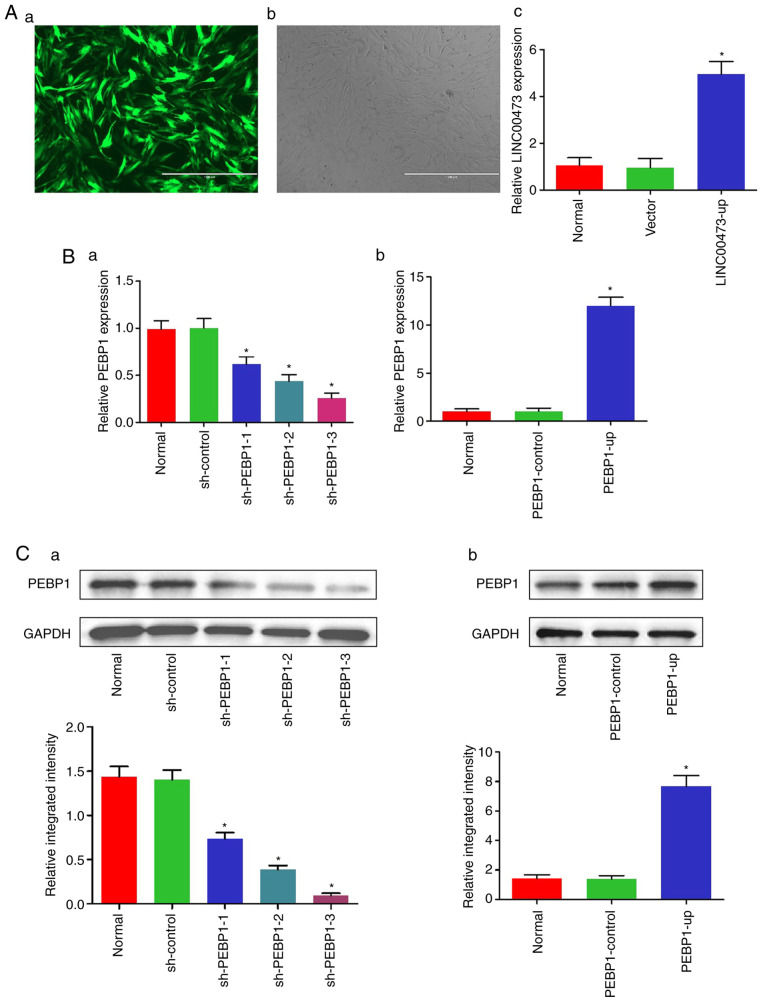
Efficiency of transfection of LINC00473 and PEBP1 into hBMSCs
Following infection with LINC00473-up lentivirus, >90% of the hBMSCs were positive for GFP (Fig. 3Aa and b). Moreover, the relative expression of LINC00473 in the LINC00473-up group was significantly upregulated by LINC00473-up lentivirus (494%), compared with the level observed in the normal group and vector group (Fig. 3Ac).
In addition, the relative expression of PEBP1 mRNA in the transfected hBMSCs was downregulated by all 3 types of sh-PEBP1 at varying degrees (37.7% by sh-PEBP1-1, 55.9% by sh-PEBP1-2 and 74% by sh-PEBP1-3), compared with the normal and sh-Control cells (Fig. 3Ba). The relative mRNA expression of PEBP1 in the hBMSCs was significantly upregulated by PEBP1-up (1,169%), compared with the normal and PEBP1-control cells (Fig. 3Bb). Correspondingly, the protein level of PEBP1 in the transfected hBMSCs was significantly downregulated by sh-PEBP1-1 (48.7%), sh-PEBP1-2 (73.1%) and sh-PEBP1-3 (93.5%) (Fig. 3Ca), and upregulated by PEBP1-up (441%) (Fig. 3Cb), compared with the normal, sh-Control and PEBP1-control cells.
The above-mentioned results demonstrated that the hBMSCs were successfully transfected with the LINC00473-up lentivirus, shRNA and recombinant plasmids of PEBP1 using the present method. sh-PEBP1-3 was found to exert the most significant effect on the knockdown of PEBP1 mRNA and protein expression levels and was thus selected for use in the following experiments.
LINC00473 reverses the inhibitory effect induced by Dex on the proliferation of hBMSCs through the activation of the Akt signaling pathway
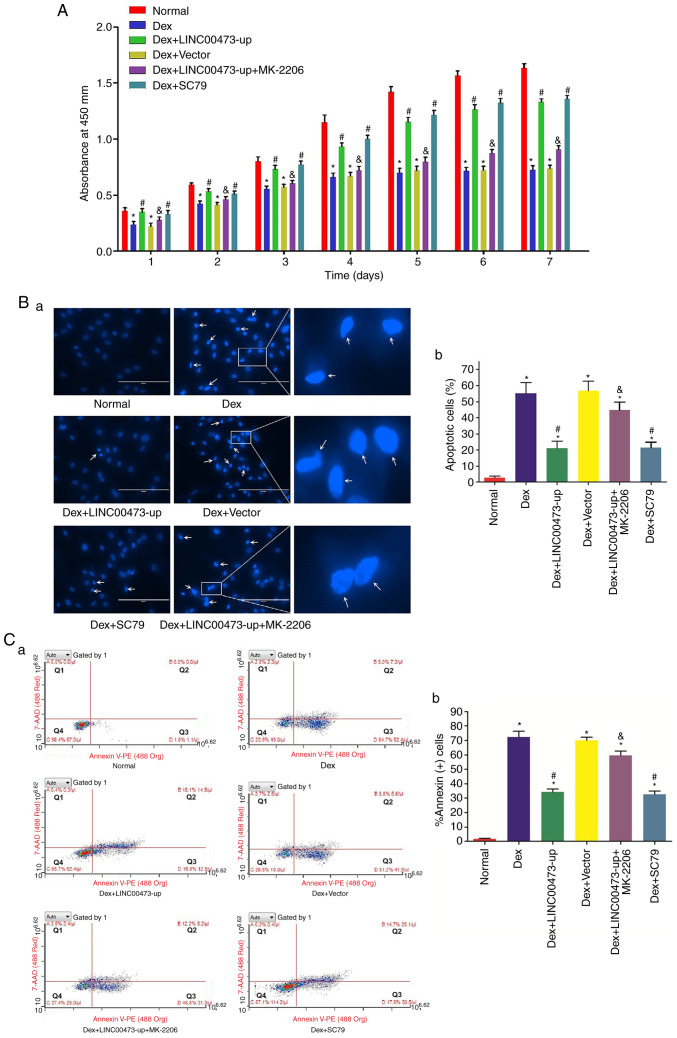
The effect of LINC00473 on the proliferation of hBMSCs in the presence of Dex (1 µM) was evaluated by CCK-8 assay. As also previously reported (13), the proliferation of hBMSCs was markedly inhibited following exposure to 1 µM Dex, whereas the inhibitory effect of Dex on hBMSCs was significantly reversed by the upregulation of LINC00473. Notably, the effect of LINC00473 on the proliferation of hBMSCs was similar to that of SC79 (a unique specific activator of Akt signaling pathway). However, this effect was weakened by MK-2206 (a selective inhibitor of Akt signaling pathway) (Fig. 4A). The results revealed that LINC00473 attenuated the inhibitory effects exerted by a high concentration of Dex (1 µM) on the proliferation of hBMSCs through the activation of the Akt signaling pathway.
Figure 4.
LINC00473 attenuates the Dex-induced inhibition of the viability of hBMSCs by activating the Akt signaling pathway. (A) The viability of hBMSCs in each group was evaluated by CCK-8 assay from 1 to 7 days. (B-a) The apoptotic cells were investigated by a fluorescence microscope through Hoechst 33342 staining (scale bar, 200 µm). White arrows indicate the cells with karyopyknosis. (B-b) The percentage of apoptotic cells was calculated by cell count. A total of 6 randomly selected fields were quantified, and a total of 300 cells in each group were counted. (C-a) Flow cytometric analysis revealed the apoptotic cells with Annexin V-PE and 7-AAD staining: Cells in Q1 represent necrotic cells, cells in Q2 represent late apoptotic cells, cells in Q3 represent early apoptotic cells, cells in Q4 represent normal cells. (C-b) Percentage of Annexin+ cells in Q2 and Q3 in each group. All data are presented as the mean value ± standard deviation of 3 independent experiments. *P<0.05 compared with the normal group, #P<0.05 compared with the Dex group and the Dex + Vector group, &P<0.05 compared with the Dex + LINC00473-up group. LINC00473, lncCRNA-00473; hBMSCs, human bone marrow-derived mesenchymal stem cells; Dex, dexamethasone; Akt, protein kinase B; CCK-8, Cell Counting Kit-8; Q, quadrant.
LINC00473 attenuates the Dex-induced apoptosis of hBMSCs through the activation of the Akt signaling pathway
From the assessment of apoptosis morphology, characteristics of apoptotic cells, such as chromatic agglutination, karyopyknosis and nuclear fragmentation, were observed in the hBMSCs stained with Hoechst 33342 following continuous exposure to 1 µM Dex for 7 days, as reported in a previous study (13). By contrast, the upregulation of LINC00473 significantly decreased the number of apoptotic hBMSCs observed in response to Dex, which resembled the effects of SC79 under the same conditions. Nevertheless, the effect of LINC00473 on Dex-induced hBMSCs were inhibited by MK-2206 (Fig. 4Ba and b).
Likewise, the flow cytometry assay conducted through Annexin V-PE/7-AAD double-staining revealed that both LINC00473 and SC79 markedly reversed the inhibitory effects on hBMSC viability induced by Dex. Moreover, supplementation with MK-2206 suppressed the anti-apoptotic effects of LINC00473 on Dex-stimulated hBMSCs (Fig. 4Ca and b). Taken together, these data demonstrated that LINC00473 reversed the Dex-induced apoptosis of hBMSCs through the activation of the Akt signaling pathway.
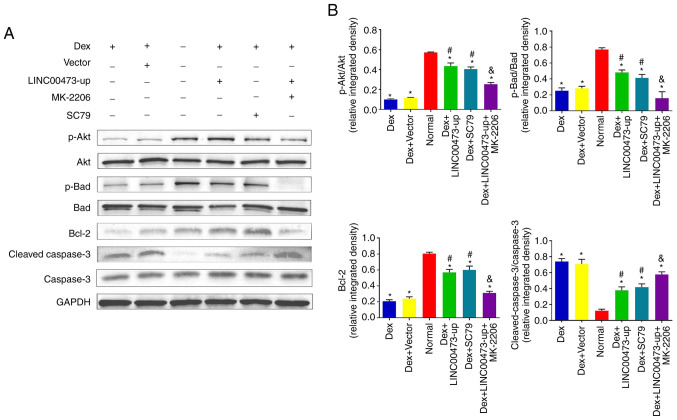
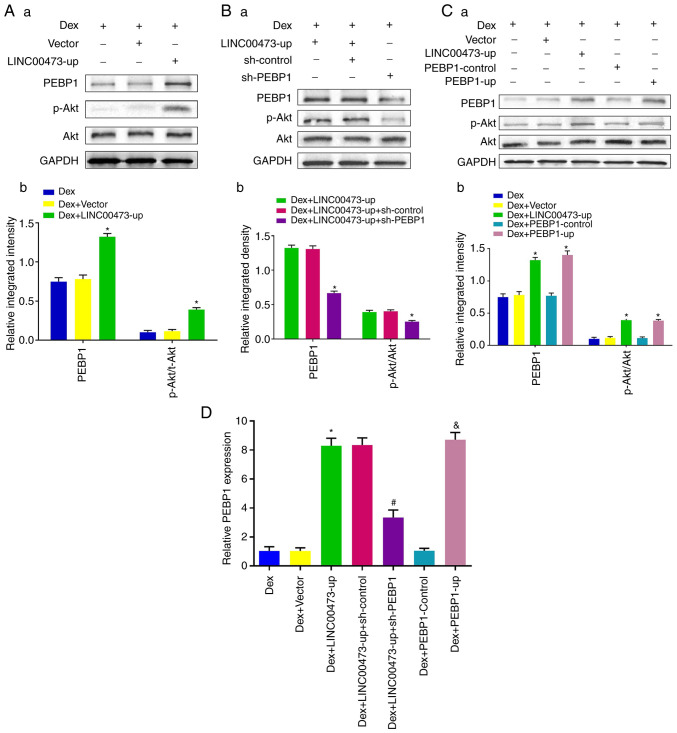
LINC00473 activates the Akt/Bad/Bcl-2 signaling pathway in Dex-stimulated hBMSCs
To further explore the role of LINC00473 in the Akt/Bad/Bcl-2 signaling cascade, western blot analysis was conducted to detect the phosphorylation of Akt and Bad, in addition to the protein expression of Bcl-2 and cleaved caspase-3, after the cells were treated based on their grouping for 24 h. The results demonstrated that expo-sure to 1 µM Dex not only inhibited the phosphorylation of Akt and Bad, but also decreased the protein expression of Bcl-2 and increased the cleavage of caspase-3, which is in line with the results of previous research studies (23,24). However, the upregulation of LINC00473 significantly attenuated the negative effects of Dex by promoting the protein level of p-Akt, p-Bad and Bcl-2, while inhibiting the cleavage of caspase-3, which is consistent with the effect observed with SC79 (an Akt activator). Notably, the regulatory effects of LINC00473 on p-Akt, p-Bad, Bcl-2 levels and the cleavage of caspase-3 in the Dex-stimulated hBMSCs were antagonized by MK-2206 (an Akt inhibitor) (Fig. 5A and B). Collectively, these findings demonstrated that LINC00473 attenuated the negative effects on hBMSCs induced Dex by activating the Akt/Bad/Bcl-2 signaling pathway.
Figure 5.
LINC00473 activates the Akt/Bad/Bcl-2 signaling pathway in Dex-stimulated hBMSCs. The results of western blot analysis revealed (A) the protein expression levels of p-Akt, Akt, p-Bad, Bad, Bcl-2 and cleaved caspase-3 in each group, and (B) the quantification of integrated density in target bands by normalization to GAPDH. All data are presented as the mean value ± standard deviation of 3 independent experiments. *P<0.05 compared with the normal group, #P<0.05 compared with the Dex group and the Dex + Vector group, &P<0.05 compared with the Dex + LINC00473-up group. LINC00473, lncRNA-00473; Akt, protein kinase B; Bad, Bcl-2-associated death promoter; Bcl-2, B-cell lymphoma 2; hBMSCs, human bone marrow-derived mesenchymal stem cells; Dex, dexamethasone.
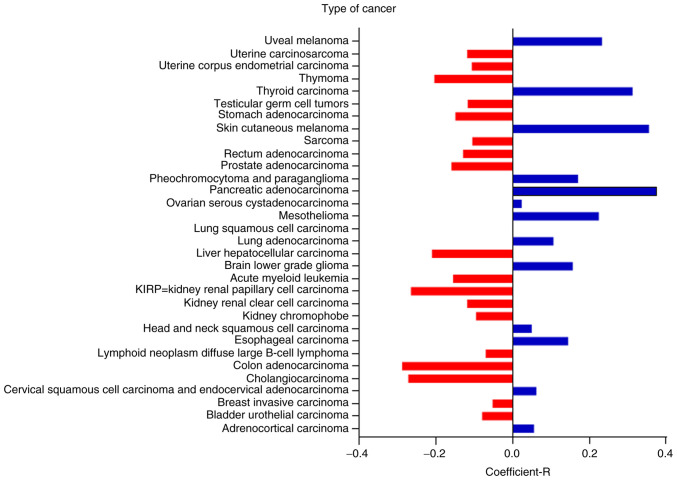
PEBP1 mediates the activation of the Akt/Bad/Bcl-2 signaling pathway through LINC00473 in Dex-stimulated hBMSCs
To further reveal the underlying mechanisms of LINC00473 in the regulation of the Akt/Bad/Bcl-2 signaling pathway, ENCORI (http://starbase.sysu.edu.cn/index.php) was searched to deter-mine the ceRNA network of LINC00473 (28), and it was found that there was a potential correlation between LINC00473 and PEBP1 in 32 types of cancer (Fig. 6). Furthermore, PEBP1 has been reported to inhibit starvation-induced autophagy by promoting the phosphorylation of Akt (27). Therefore, the present study investigated the role of PEBP1 in the activation of the Akt/Bad/Bcl-2 signaling pathway by LINC00473 in Dex-stimulated hBMSCs. As shown in Fig. 7Aa and b, the upregulation of LINC00473 markedly promoted the phosphorylation of Akt in Dex-stimulated hBMSCs, while increasing the protein level of PEBP1. However, the elevation of Akt phosphorylation by LINC00473 was significantly suppressed following the knockdown of PEBP1 (Fig. 7Ba and b). Furthermore, the upregulation of LINC00473 and PEBP1 resulted in an analogous and marked increase in Akt phosphorylation in the Dex-stimulated hBMSCs (Fig. 7Ca and b). In addition, the results of RT-qPCR suggested that the changes in the PEBP1 mRNA expression levels were consistent with those observed at the protein level for PEBP1 (Fig. 7D). Taken together, these results demonstrated that PEBP1 mediated the activation of the Akt/Bad/Bcl-2 signaling pathway induced by LINC00473 in Dex-stimulated hBMSCs.
Figure 6.
Correlation between LINC00473 and PEBP1 in 32 types of cancer. All data were obtained from ENCORI. PEBP1, phosphatidylethanolamine-binding protein 1; LINC00473, lncRNA-00473; ENCORI, Encyclopedia of RNA Interactomes.
Figure 7.
PEBP1 mediates the activation of the Akt/Bad/Bcl-2 signaling pathway by LINC00473 in Dex-stimulated hBMSCs. The result of western blot analysis revealed (Aa and b) the protein expression levels of PEBP1, p-Akt and Akt in each groups following infection with LINC00473-up lentivirus in Dex-induced hBMSCs (*P<0.05 compared with the Dex group and Dex + Vector group), and (Ba and b) the protein expression levels of PEBP1 and p-Akt in each groups following co-transfection with LINC00473 and PEBP1 in Dex-stimulated hBMSCs (*P<0.05 compared with the Dex + LINC00473-up group and Dex + LINC00473-up + sh-Control group), and (Ca and b) the protein expression levels of PEBP1 and p-Akt in each group following transfection with LINC00473 and PEBP1 respectively in Dex-induced hBMSCs (*P<0.05 compared with the Dex group, Dex + Vector group and Dex + PEBP1-Control group). (D) The results of RT-qPCR revaled the PEBP1 mRNA expression in each groups (*P<0.05 compared with the Dex group and Dex + Vector group, #P<0.05 compared with the Dex + LINC00473-up group, &P<0.05 compared with the Dex group and Dex + PEBP1-Control group). All data are presented as the mean values ± standard deviation 3 three independent experiments. PEBP1, phosphatidylethanolamine-binding protein 1; Akt, protein kinase B; Bad, Bcl-2-associated death promoter; Bcl-2, B-cell lymphoma 2; LINC00473, lncRNA-00473; hBMSCs, human bone marrow-derived mesenchymal stem cells; Dex, dexamethasone; p-Akt, phosphorylated Akt; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
BMSCs have the ability of self-renewal, excellent proliferation and multipotential differentiation into an osteogenic lineage, adipogenic lineage and chondrogenic lineage, as well as other embryonic lineages (36). Therefore, the dysfunction of BMSCs can lead to a variety of skeletal muscle diseases (6,37). As a typical representative, the development of osteonecrosis of the femoral head (ONFH) is closely associated with the abnormal cellular behavior of BMSCs (6). The excessive use of glucocorticoids is the primary cause of the pathogeny of SONFH, and to a certain extent, is known to be due to glucocorticoid-induced inhibition of the proliferation and apoptosis of BMSCs (6,24).
It has been confirmed that lncRNAs play important roles in cell proliferation, apoptosis and differentiation by regulating the expression levels of critical genes (9-11). In the present study, the expression of LINC00473 was detected in hBMSCs obtained from patients with SONFH, and it was found that LINC00473 was significantly downregulated in these hBMSCs, compared with the controls, which was in agreement with the results of our previous microarray analysis (13). LINC00473 is an intergenic lncRNA located on human chromosome 6q27 that consists of 2 exons and 2 annotated transcript isoforms (12). The role of LINC00473 on the proliferation and apoptosis of various tumor cells has been elucidated in several studies (14-17); however, the effect of LINC00473 on BMSCs has not yet been fully elucidated.
Dex is a common glucocorticoid drug, and emerging studies have proven that Dex exerts variable effects on BMSCs based on concentrations and exposure times (7,8,38). Recent evidence has indicated that long-term exposure to a high concentration of Dex (1 µM) can significantly inhibit the proliferation and induce the apoptosis of BMSCs (8,13). In the present study, based on previous findings, the concentration of 1 µM Dex was selected for use as the condition that induces the apoptosis of hBMSCs (8,13), to mimic the cellular micro-environments of SONFH. Moreover, hBMSCs were infected with the LINC00473-up lentivirus in consideration of the decrease in LINC00473 expression in hBMSCs obtained from patients with SONFH to investigate the role of LINC00473 on hBMSCs induced by 1 µM Dex. As previously reported (8,13), 1 µM Dex significantly inhibited the proliferation and induced the apoptosis of hBMSCs. However, it was found that the upregulation of LINC00473 attenuated the inhibitory effects induced by 1 µM Dex on the proliferation and apoptosis of the hBMSCs, indicating the anti-apoptotic effects of LINC00473.
Akt is a primary mediator of the PI3K/Akt signaling pathway, which has been shown to be involved in cell apoptosis, proliferation and migration (18-20). In terms of the anti-apoptotic effect, the Akt/Bad/Bcl-2 signaling cascade is the focus of current research. In general, the phosphorylation-mediated activation of Akt leads to the inactivation of Bad, which increases the expression of Bcl-2 protein, which then inhibits apoptosis of cells by suppressing the cleavage of caspase-3 (a downstream key factor of apoptosis) (21,22). Although the explicit mechanisms of the Dex-induced apoptosis of BMSCs remain unclear, they may potentially involve the suppression of the Akt signaling pathway (23,24).
Therefore, the present investigated the effects of LINC00473 on the protein levels of the Akt/Bad/Bcl-2 signaling pathway, and performed antagonist and activation experiments to further elucidate the molecular mechanisms through which LINC00473 inhibits the Dex-induced apoptosis of hBMSCs. The results demonstrated that 1 µM Dex suppressed the phosphorylation of Akt and Bad, decreased the protein expression of Bcl-2, and increased the expression of cleaved caspase-3, leading to the apoptosis of hBMSCs, which was in agreement with previous research results (23,24). Moreover, the upregulation of LINC00473 significantly attenuated the negative effects induced by Dex by promoting the phosphorylation of Akt and Bad, and increasing the protein level of Bcl-2, while inhibiting the cleavage of caspase-3. Notably, MK-2206, a selective inhibitor of Akt, partially inhibited the upregulation of the protein expression levels of the Akt/Bad/Bcl-2 signaling pathway triggered by LINC00473, while attenuating the anti-apoptotic effect of LINC00473 on Dex-induced hBMSCs. Furthermore, the results of the activation experiment revealed that the anti-apoptotic effect of LINC00473 and the regulatory effect induced by LINC00473 on the Akt/Bad/Bcl-2 signaling pathway in Dex-stimulated hBMSCs, was similar to that of SC79 (a unique specific activator of Akt). Taken together, it is reasonable to speculate that LINC00473 rescued the hBMSCs from Dex-induced apoptosis by activating the Akt/Bad/Bcl-2 signaling pathway. However, the explicit mechanisms through which LINC00473 regulates the Akt/Bad/Bcl-2 signaling pathway warrant further investigation.
To the best of our knowledge, certain lncRNAs act as ceRNAs to regulate the expression of mRNAs that affect the biological function of cells (39). Surprisingly, ENCORI we searched to identify the ceRNA network of LINC00473 (28), and a potential correlation was found between LINC00473 and PEBP1 in 32 types of cancer. PEBP1 is also known as RKIP and is involved in diverse biological functions through its regulation of various signaling pathways, and has been reported to inhibit starvation-induced autophagy through the activation of the Akt/MTORC1 pathway. Consequently, PEBP1 was selected to explore the mechanisms through which LINC00473 regulates the Akt/Bad/Bcl-2 signaling pathway.
In further experiments, it was found that the upregulation of LINC00473 significantly promoted the phosphorylation of Akt in Dex-stimulated hBMSCs and increased the protein level of PEBP1. However, the promotion of Akt phosphorylation by LINC00473 was significantly suppressed following the knockdown of PEBP1. Furthermore, the upregulation of PEBP1 triggered a significant increase in Akt phosphorylation in Dex-stimulated hBMSCs, which corresponds with the results of the upregulation of LINC00473. Taken together, the results demonstrated that PEBP1 mediated the activation of the Akt/Bad/Bcl-2 signaling pathway induced by LINC00473 in Dex-stimulated hBMSCs.
In conclusion, the present study confirmed the downregulation of LINC00473 in the hBMSCs of patients with SONFH, compared with the controls. In addition, LINC00473 reserved the inhibitory effects induced by Dex on the proliferation of hBMSCs and attenuated the Dex-induced apoptosis of hBMSCs by activating the Akt/Bad/Bcl-2 signaling pathway. It is noteworthy that PEBP1 mediated this effect of LINC00473. Collectively, LINC00473 rescued the hBMSCs from apoptosis induced by Dex through the PEBP1-mediated Akt/Bad/Bcl-2 signaling pathway.
Figure 3.
Efficiency of transfection of hBMSCs with LINC00473 and PEBP1. (A-a) hBMSCs expressing GFP tagged LINC00473-up lentivirus under a fluorescence microscope (scale bar, 1,000 µm) and (A-b) under a light microscope (scale bar, 1,000 µm). The results of RT-qPCR revealed that (A-c) the relative expression of LINC00473 was significantly upregulated by LINC00473-up lentivirus (494%), *P<0.05 compared with the normal group and vector group, and (B-a) the relative expression of PEBP1 was significantly downregulated by 3 shRNAs for PEBP1 (sh-PEBP1), *P<0.05 compared with the normal group and sh-Control group, and (B-b) the relative expression of PEBP1 was a significantly upregulated by PEBP1 overexpression vector (PEBP1-up), *P<0.05 compared with the normal group and PEBP1-Control group. The results of western blot analysis revealed that (C-a) the relative expression of PEBP1 protein was significantly downregulated by the 3 sh-PEBP1, *P<0.05 compared with the normal group and sh-Control group, and (C-b) the relative expression of PEBP1 protein was a significantly upregulated by PEBP1-up, *P<0.05 compared with the normal group and PEBP1-Control group. All data were presented as the means ± standard deviation. hBMSCs, human bone marrow-derived mesenchymal stem cells; LINC00473, lncCRNA-00473; PEBP1, phosphatidylethanol-amine-binding protein 1; GFP, green fluorescent protein; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81802151), the Shandong Province Natural Science Foundation (grant nos. ZR2016HQ05, ZR2017BH089 and ZR2019MH012), the China Postdoctoral Science Foundation (grant no. 2018M642616), and the Qingdao Applied Foundational Research Youth Project (grant no. 19-6-2-55-cg).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YX, YJ and ZZ performed the experiments and analyzed the results. YX and YJ wrote and drafted the manuscript. TL wrote, reviewed and edited the manuscript. TL and YW conceived the methodology, while TL and YW designed the research study and were major contributors in recruiting the donors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Qingdao University Affiliated Hospital (Qingdao, China). All donors were Chinese and provided informed consent for participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang XS, Zhuang QY, Weng XS, Lin J, Jin J, Qian WW. Etiological and clinical analysis of osteonecrosis of the femoral head in Chinese patients. Chin Med J (Engl) 2013;126:290–295. [PubMed] [Google Scholar]
- 2.Hao C, Yang S, Xu W, Shen JK, Ye S, Liu X, Dong Z, Xiao B, Feng Y. MiR-708 promotes steroid-induced osteonecrosis of femoral head, suppresses osteogenic differentiation by targeting SMAD3. Sci Rep. 2016;6:22599. doi: 10.1038/srep22599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang YL, Yin JH, Ding H, Zhang W, Zhang CQ, Gao YS. Vitamin K2 prevents glucocorticoid-induced osteonecrosis of the femoral head in rats. Int J Biol Sci. 2016;12:347–358. doi: 10.7150/ijbs.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han L, Wang B, Wang R, Gong S, Chen G, Xu W. The shift in the balance between osteoblastogenesis and adipogenesis of mesenchymal stem cells mediated by glucocorticoid receptor. Stem Cell Res Ther. 2019;10:377. doi: 10.1186/s13287-019-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Wei Z, Li D, Yang Z, Tian M, Kang P. Glucocorticoid enhanced the expression of ski in osteonecrosis of femoral head: The effect on adipogenesis of rabbit BMSCs. Calcif Tissue Int. 2019;105:506–517. doi: 10.1007/s00223-019-00592-3. [DOI] [PubMed] [Google Scholar]
- 6.Houdek MT, Wyles CC, Packard BD, Terzic A, Behfar A, Sierra RJ. Decreased osteogenic activity of mesenchymal stem cells in patients with corticosteroid-induced osteonecrosis of the femoral head. J Arthroplasty. 2016;31:893–898. doi: 10.1016/j.arth.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Song IH, Caplan AI, Dennis JE. Dexamethasone inhibition of confluence-induced apoptosis in human mesenchymal stem cells. J Orthop Res. 2009;27:216–221. doi: 10.1002/jor.20726. [DOI] [PubMed] [Google Scholar]
- 8.Fan Q, Zhan X, Li X, Zhao J, Chen Y. Vanadate inhibits dexamethasone-induced apoptosis of rat bone marrow-derived mesenchymal stem cells. Ann Clin Lab Sci. 2015;45:173–180. [PubMed] [Google Scholar]
- 9.Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014;49:16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei B, Wei W, Zhao B, Guo X, Liu S. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of the femoral head. PLoS One. 2017;12:e0169097. doi: 10.1371/journal.pone.0169097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Zheng Y, Jia L, Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/smad3/HDAC signalinging pathway by deriving miR-675. Stem Cells. 2015;33:3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 12.Pruunsild P, Bengtson CP, Bading H. Networks of cultured ipsc-derived neurons reveal the human synaptic activity-regulated adaptive gene program. Cell Rep. 2017;18:122–135. doi: 10.1016/j.celrep.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Jiang Y, Wang Y, Ren Y, Zhao Z, Wang T, Li T. LINC00473 regulated apoptosis, proliferation and migration but could not reverse cell cycle arrest of human bone marrow mesenchymal stem cells induced by a high-dosage of dexamethasone. Stem Cell Res. 2020;48:101954. doi: 10.1016/j.scr.2020.101954. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Yang F, Li X, Gong ZJ, Wang LW. Long noncoding RNA lnc473 inhibits the ubiquitination of survivin via association with usp9x and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2018;99:702–710. doi: 10.1016/j.bbrc.2018.03.215. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Song Y. LINC00473 predicts poor prognosis and regulates cell migration and invasion in gastric cancer. Biomed Pharmacother. 2018;107:1–6. doi: 10.1016/j.biopha.2018.07.061. [DOI] [PubMed] [Google Scholar]
- 16.Shi C, Yang Y, Yu J, Meng F, Zhang T, Gao Y. The long noncoding rna linc00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ilf2 degradation in cervical cancer. Am J Cancer Res. 2017;7:2157–2168. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu SB, Fu W, Zhang L, Fu K, Hu JH, Jia W, Liu G. LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of wilms tumour via IKKα. Cell Prolif. 2018;51:e12416. doi: 10.1111/cpr.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Xia Y, Wang Z, Cao X, Da Z, Guo G, Qian J, Liu X, Fan Y, Sun L, et al. Suppression of the PI3K-akt pathway is involved in the decreased adhesion and migration of bone marrow-derived mesenchymal stem cells from non-obese diabetic mice. Cell Biol Int. 2011;35:961–966. doi: 10.1042/CBI20100544. [DOI] [PubMed] [Google Scholar]
- 20.Gu YX, Du J, Si MS, Mo JJ, Qiao SC, Lai HC. The roles of PI3K/akt signalinging pathway in regulating MC3T3-E1 preosteo-blast proliferation and differentiation on SLA and SLActive titanium surfaces. J Biomed Mater Res A. 2013;101:748–754. doi: 10.1002/jbm.a.34377. [DOI] [PubMed] [Google Scholar]
- 21.Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, Parsons MJ, Zheng JH, Brown SA, Pelletier S, et al. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell. 2016;165:421–433. doi: 10.1016/j.cell.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Wu G, Zhu G, Ma C, Zhao H. Chronic sleep restriction induces changes in the mandibular condylar cartilage of rats: Roles of akt, bad and caspase-3. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Int J Clin Exp Med. 2014;7:2585–2592. [PMC free article] [PubMed] [Google Scholar]
- 23.Pan JM, Wu LG, Cai JW, Wu LT, Liang M. Dexamethasone suppresses osteogenesis of osteoblast via the PI3K/akt signalinging pathway in vitro and in vivo. J Recept Signal Transduct Res. 2019;39:80–86. doi: 10.1080/10799893.2019.1625061. [DOI] [PubMed] [Google Scholar]
- 24.Tao SC, Yuan T, Rui BY, Zhu ZZ, Guo SC, Zhang CQ. Exosomes derived from human platelet-rich plasma prevent apoptosis induced by glucocorticoid-associated endoplasmic reticulum stress in rat osteonecrosis of the femoral head via the akt/bad/bcl-2 signaling pathway. Theranostics. 2017;7:733–750. doi: 10.7150/thno.17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoentgen F, Jonic S. PEBP1/RKIP behavior: A mirror of actin-membrane organization. Cell Mol Life Sci. 2020;77:859–874. doi: 10.1007/s00018-020-03455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W, Cho SB, Jung HY, Yoo DY, Oh JK, Choi GM, Cho TG, Kim DW, Hwang IK, Choi SY, Moon SM. Phosphatidylethanolamine-Binding protein 1 ameliorates ischemia-induced inflammation and neuronal damage in the rabbit spinal cord. Cells. 2019;8:1370. doi: 10.3390/cells8111370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noh HS, Hah YS, Zada S, Ha JH, Sim G, Hwang JS, Lai TH, Nguyen HQ, Park JY, Kim HJ, et al. PEBP1, a RAF kinase inhibitory protein, negatively regulates starvation induced autophagy by direct interaction with LC3. Autophagy. 2016;12:2183–2196. doi: 10.1080/15548627.2016.1219013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuru S, Hofmann TJ, Olson TS, Dominici M, Horwitz EM. Improved isolation and expansion of bone marrow mesenchymal stromal cells using a novel marrow filter device. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Cytotherapy. 2013;15:146–153. doi: 10.1016/j.jcyt.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini FC, Krause DS, Deans RJ, Keating A, Prockop DJ, Horwitz EM. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 31.Fu L, Tang T, Miao Y, Zhang S, Qu Z, Dai K. Stimulation of osteogenic differentiation and inhibition of adipogenic differentiation in bone marrow stromal cells by alendronate via ERK and JNK activation. Bone. 2008;43:40–47. doi: 10.1016/j.bone.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Chen J, Cong X, Hu S, Chen X. Hypoxia and serum deprivation-induced apoptosis in mesenchymal stem cells. Stem Cells. 2006;24:416–425. doi: 10.1634/stemcells.2005-0121. [DOI] [PubMed] [Google Scholar]
- 34.Wang XY, Fan XS, Cai L, Liu S, Cong XF, Chen X. Lysophosphatidic acid rescues bone mesenchymal stem cells from hydrogen peroxide-induced apoptosis. Apoptosis. 2015;20:273–284. doi: 10.1007/s10495-014-1074-0. [DOI] [PubMed] [Google Scholar]
- 35.Schmid I, Krall WJ, Uittenbogaart CH, Braun J, Giorgi JV. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13:204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 36.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 37.Yeung DK, Griffith JF, Antonio GE, Lee FK, Woo J, Leung PC. Osteoporosis is associated with increased marrow fat content and decreased marrow fat unsaturation: A proton MR spectroscopy study. Mesenchymal stem cells in health and disease. J Magn Reson Imaging. 2005;22:279–285. doi: 10.1002/jmri.20367. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Pang B, Li Y, Zhu D, Pang T, Liu Y. Dexamethasone has variable effects on mesenchymal stromal cells. Cytotherapy. 2012;14:423–430. doi: 10.3109/14653249.2011.652735. [DOI] [PubMed] [Google Scholar]
- 39.Song X, Cao G, Jing L, Lin S, Wang X, Zhang J, Wang M, Liu W, Lv C. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18:991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.