Abstract
The coronavirus disease 2019 (COVID-19) outbreak, which has caused >46 millions confirmed infections and >1.2 million coronavirus related deaths, is one of the most devastating worldwide crises in recent years. Infection with COVID-19 results in a fever, dry cough, general fatigue, respiratory symptoms, diarrhoea and a sore throat, similar to those of acute respiratory distress syndrome. The causative agent of COVID-19, SARS-CoV-2, is a novel coronavirus strain. To date, remdesivir has been granted emergency use authorization for use in the management of infection. Additionally, several efficient diagnostic tools are being actively developed, and novel drugs and vaccines are being evaluated for their efficacy as therapeutic agents against COVID-19, or in the prevention of infection. The present review highlights the prevalent clinical manifestations of COVID-19, characterizes the SARS-CoV-2 viral genome sequence and life cycle, highlights the optimal methods for preventing viral transmission, and discusses possible molecular pharmacological mechanisms and approaches in the development of anti-SARS-CoV-2 therapeutic agents. In addition, the use of traditional Chinese medicines for management of COVID-19 is discussed. It is expected that novel anti-viral agents, vaccines or an effective combination therapy for treatment/management of SARS-CoV-2 infection and spread therapy will be developed and implemented in 2021, and we would like to extend our best regards to the frontline health workers across the world in their fight against COVID-19.
Keywords: angiotensin-converting enzyme 2, acute respiratory distress syndrome, chloroquine, COVID-19, 3-chymotrypsin-like cysteine protease, hydroxychloroquine, traditional Chinese medicine, transmembrane protease serine 2
1. Introduction
The coronavirus disease 2019 (COVID-19) outbreak has spread worldwide with overwhelming speed, infecting >48.3 million individuals and causing >1.23 million deaths across ~200 countries as of 2nd of November, 2020 (1). COVID-19, caused by SARS-CoV-2 virus, hit China, the US and European countries considerably hard, with the aforementioned countries becoming the epicentres of the SARS-CoV-2 virus pandemic (2). Early prevention of transmission of SARS-CoV-2 via imposed lockdowns and social distancing have been the primary means of preventing the spread of COVID-19 (2,3).
Strategies aimed at interrupting interactions between the virus and host have been primarily utilised from the viewpoint of public epidemiology (4,5). To control the spread of the virus, several countries have limited or outright banned accesses to international flights, locked down the entire country or several cities, have instructed the public to follow social distancing measures, and made the wearing of masks either mandatory or recommended. Moreover, body temperatures are being measured wherever individuals congregate and social activities have been diminished in hopes of curbing peak prevalence and death (5-7). Remdesivir has received emergency use authorization by the US Food and Drug Administration (FDA) for the treatment of COVID-19 (8) for SARS-CoV-2 virus infections (1). However, the development of novel agents and vaccines against SARS-CoV-2 is now one of the most intensively researched subjects worldwide.
The current review summarises the clinical manifestations, SARS-CoV-2 viral genome structure and sequence, SARS-CoV-2 viral life cycle, diagnosis, preventative methods, and management measures of COVID-19. Finally, an over-view is provided of the possible molecular pharmacological mechanisms of anti-SARS-CoV-2 agents and the effectiveness of remdesivir (GS-5734), chloroquine, hydroxychloroquine, steroids and anti-coagulant agents as well as traditional Chinese medicines (TCM) for management of COVID-19.
2. Clinical characteristics of COVID-19
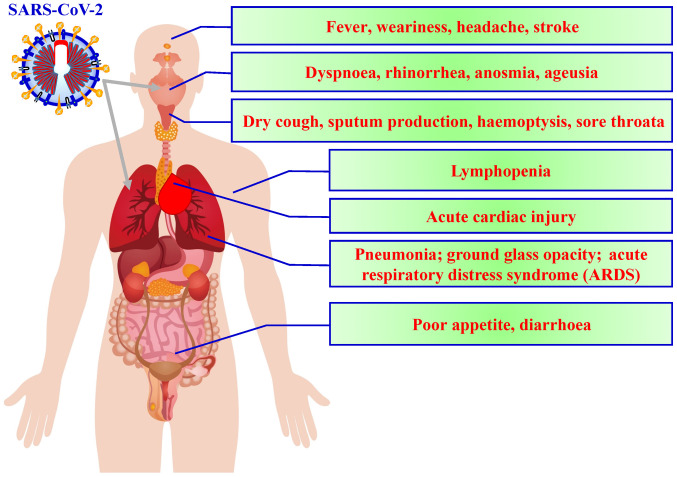
According to the current literature, fever, dry cough and fatigue are the most common symptoms observed at the onset of COVID-19, with other symptoms including muscle pain, productive cough, headache, diarrhoea, dyspnoea and haemoptysis developing later (Fig. 1) (9). Symptoms gener-ally appear ~5.2 days after COVID-19 (10). Although 50-75% of patients with COVID-19 remain asymptomatic, ~14% of infected individuals present with serious symptoms requiring hospitalisation and oxygen therapy, while 5% require intensive care. The median duration from symptom onset to intensive care unit admission is ~10 days, while the duration between symptom onset and death ranges from 2-8 weeks (10-13).
Figure 1.
Symptoms of SARS-CoV-2. Symptoms of SARS-CoV-2 include fever, dyspnoea, cough and loss of taste or smell.
Laboratory findings include elevated lactate dehydrogenase and ferritin levels (14). Moreover, although while white blood cell counts can vary, leucopoenia and lymphopenia have been the most commonly observed in individuals infected with SARS-CoV-2 infection (15). Chest radiography and computed tomography (CT) findings are diverse and nonspecific, commonly presenting as multiple ground-glass opacity lesions, bilateral patchy shadowing or local patchy shadowing (16). Severe cases tend to yield more prominent radiological findings (17). However, a few cases have presented with no notable imaging abnormalities (17.9% of non-severe cases and 2.9% of severe cases) (9). As the disease progresses, multiple ground-glass opacity lesions may progress into consolidation or superimposed interlobular/intralobular septal thickening (for example, crazy-paving pattern), which may expand consolidation (18). Several similarities (fever, cough and fatigue) exist between COVID-19 symptoms and those caused by other atypical pathogens such as Chlamydia, Legionella and Mycoplasma (19,20). However, COVID-19 exhibits some distinctive clinical characteristics as well, including targeting of the lower respiratory tract instead of the upper respiratory tract, which produces symptoms like sneezing, rhinorrhoea and a sore throat (21). Moreover, chest radiographs and CT scans upon patient admission revealed an infiltrate in the upper lobe of the lung that was associated with increasing dyspnoea with hypoxemia (22). Certain patients with COVID-19 also developed gastrointestinal distress, such as diarrhoea, whereas only a low percentage of patients with Middle East respiratory syndrome coronavirus (MERS-CoV) or severe acute respiratory syndrome coronavirus (SARS-CoV) experienced these symptoms (23). Finally, the majority of the patients with COVID-19 exhibited leucopoenia and lymphopenia on admission. Tan et al (24) demonstrated that patients with blood lymphocyte percentage (LYM%) >20% are in the process of recovery. In contrast, those with between 5 and 20% LYM% are still at risk, and those with LYM% <5% become critically ill with high mortality rates and require intensive care. Lymphopenia seems to be an effective and reliable indicator of severity and hospitalisation amongst patients with COVID-19 (25). Table I presents the classification of the clinical manifestations of COVID-19 (26).
Table I.
Classification of clinical manifestation of COVID-19.
| Symptoms of COVID-19 | Clinical manifestation |
|---|---|
| Asymptomatic | 25-50% of patients with SARS-CoV-2 infection are asymptomatic |
| Mild clinical manifestation without comorbidity | 1. Common symptoms of upper respiratory tract infection include cough, fever, sore throat, running nose, headache, malaise and muscle pain |
| 2. A few patients may have symptoms of diarrhea, nausea and vomiting | |
| Pneumonia | Cough, dyspnea and chest images presented as pneumonia patch or multiple ground-glass opacities, without manifestation of severe pneumonia or requirement for oxygen supply |
| Severe pneumonia | Pneumonia with requirement of oxygen therapy, plus respiratory rate >30 breaths/min, severe respiratory distress or SpO2 ≤93% on room air |
| ARDS | Chest images presented as pneumonia Oxygenation impairment: With the minimum level of PEEP 5 cm H2O, PaO2/FiO2 ratio at ≤300 and >200 is defined as mild ARDS; PaO2/FiO2 ratio at 100-200 is defined as moderate ARDS; PaO2/FiO2 ratio at <100 is defined as severe ARDS |
ARDS, Acute respiratory distress syndrome; PEEP, positive end-expiratory pressure; PaO2, arterial oxygen tension; FiO2, fraction of inspiration O2; SpO2, oxyhemoglobin saturation by pulse oximetry.
3. Structure, genome size and life cycle of SARS-CoV-2
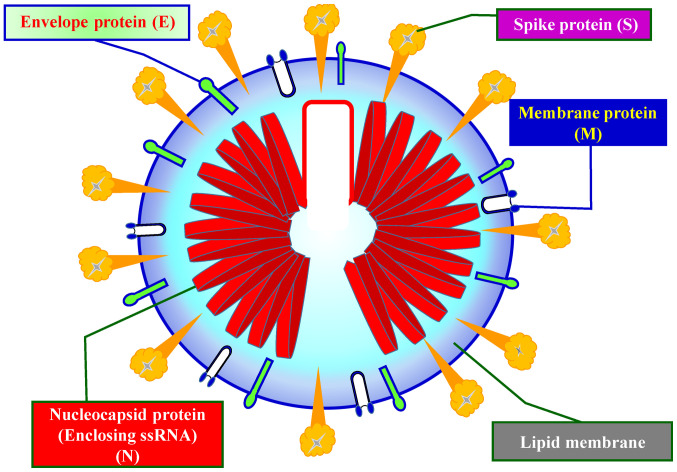
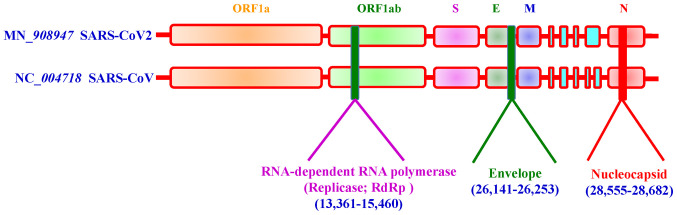
Coronaviruses primarily cause respiratory and gastrointestinal tract infections and are genetically classified into four major genera: α-coronavirus, β-coronavirus, γ-coronavirus and δ-coronavirus (27). Six types of human coronaviruses have been previously identified, which include HCoV-NL63 and HCoV-229E belonging to the α-coronavirus genus and HCoV-OC43, HCoV-HKU1, SARS-CoV and MERS-CoV belonging to the β-coronavirus genus (27). Coronaviruses had not attracted worldwide attention until the 2003 SARS pandemic, followed by the 2012 MERS and, most recently, the COVID-19 outbreaks (27). Both SARS-CoV-2 and MERS-CoV have been considered highly pathogenic (28). Fig. 2 shows the schematic structure of SARS-CoV-2 (29,30).
Figure 2.
Schematic structure of SARS-CoV-2. SARS-CoV-2 encodes four major structural proteins, including the envelope protein, membrane protein, nucleocapsid protein and spike protein. ssRNA, single stranded RNA.
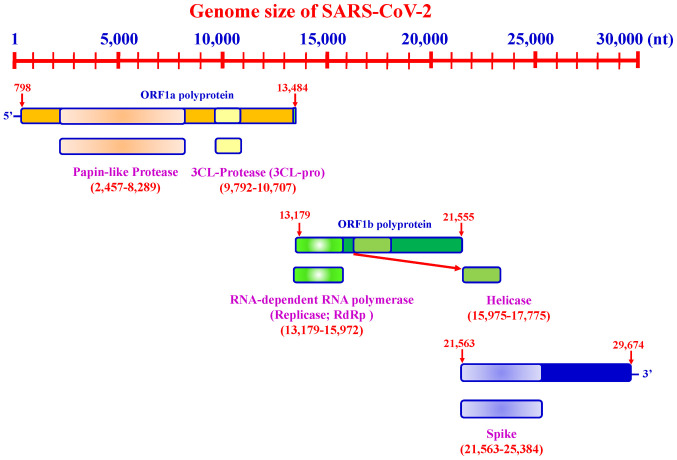
SARS-CoV-2 possesses a genome length of ~30 kb. Accordingly, SARS-CoV-2 genome sequences from NCBI (30), covering between ~798 and 29,674 bases, include a variable number of open reading frames (ORFs) (Fig. 3). The first ORF, representing ~67% of the entire genome, encodes two large polyproteins, PP1a and PP1ab, which are proteolytically cleaved into 16 non-structural proteins (NSPs), including papain-like protease, 3-chymotrypsin-like cysteine protease (3CLpro), RNA-dependent RNA poly-merase (RdRp), helicase and exonuclease (ExoN). The remaining ORFs encode accessory and structural proteins. The four major structural proteins include the spike surface glycoprotein (S) (31), envelope protein (E) (32), membrane (M) (33) and nucleocapsid protein (N) (34). Recent studies have revealed six major NSP subtypes, including nsp3 (35), nsp4 (36), nsp6 (37), nsp12 (38), nsp13 (39) and nsp14 (38) for SARS-CoV-2. Spike proteins of viruses bind to host cell receptors for entry. Accordingly, the spike proteins of SARS-CoV-2 and MERS-CoV bind to different host receptors through different receptor-binding domains. SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as one of the main receptors with CD209L as an alternative receptor, whereas MERS-CoV uses dipeptidyl peptidase 4 (DPP4, also known as CD26) as its primary receptor (40-44). The cleavage of trimer S protein is initiated by the cell surface-associated transmembrane protease serine 2 (TMPRSS2) and cathepsin (45,46).
Figure 3.
Genome size of SARS-CoV-2. The length of the SARS-CoV-2 genome is ~30 kb.
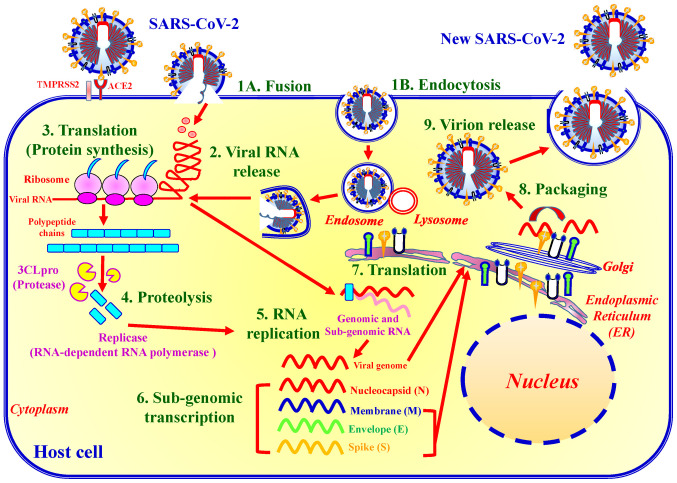
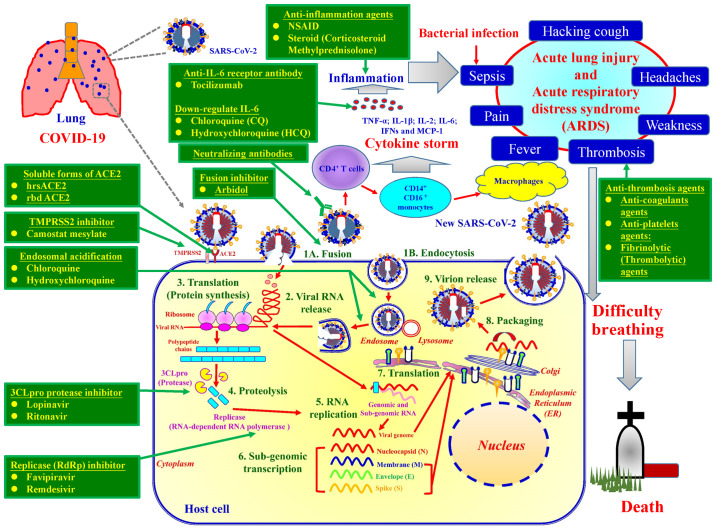
The life cycle of SARS-CoV-2 can be categorised into nine major steps (Fig. 4). Upon binding to ACE2 and TMPRSS2, SARS-CoV-2 enters host target cells through either fusion or endocytosis (step 1). In the endocytic pathway, the SARS-CoV-2 envelope fuses with the endosome membrane in the lysosomal acid environment, which promotes viral RNA genome release into the host cell cytoplasm (step 2). ORF1a/b encoding 3CLpro is then translated for the replication of genomic RNA (step 3). Thereafter, replicase polypeptide is cleaved (proteolysis), producing NSPs, such as RdRp and helicase (step 4). SARS-CoV-2 then undergoes viral RNA replication in the host cells (step 5). The viral sub-genome is transcribed (step 6). Viral N, M, E and S proteins are translated through the endoplasmic reticulum and Golgi apparatus (step 7). N protein and other structural proteins interact with viral genomic RNA to pack and form a novel virion (step 8). The assembled virion is then released via exocytosis into the extracellular compartment (step 9). The released viral particles are infectious and may begin a new life cycle (Fig. 4) (10,47).
Figure 4.
Life cycle of SARS-CoV-2. The SARS-CoV-2 life cycle consists of nine major stages: Step 1, virus entry either via fusion (1A) or endocytosis (1B); step 2, viral RNA release; step 3, translation of viral replication machinery protein; step 4, proteolysis; step 5, RNA replication; step 6, sub-genomic transcription; step 7, translation of viral structure protein; step 8, virion assembly; and step 9, virion release.
4. Diagnostic methods for COVID-19
Two approaches have generally been utilised for the diagnostic screening of SARS-CoV-2: i) Reverse transcription-quantitative PCR (RT-qPCR) and, ii) rapid screening (48,49). Detection time and duration until COVID-19 diagnosis are detailed in Table II.
Table II.
Primary means of diagnosis of COVID-19, including use timing, detection time, specificity and sensitivity.
| First author, year | Method | Use timing | Detection time | Specificity % | Sensitivity % | (Refs.) |
|---|---|---|---|---|---|---|
| Wang, 2020 | RT-qPCR | Early stage of clinical manifestation | 2-4 h | 100 | 98.7 | (58) |
| Porte, 2020 | Antigen method (rapid screening) | Early stage of clinical manifestation. Peak period of SARS-CoV-2 infection | 15 min | 100 | 93.9 | (161) |
| Wang, 2020 | Antibody method (Rapid screening) | After 7-day clinical manifestation | 15 min | 92.2 | 95.7 | (58) |
Specificity, ratio of non-sick individuals who get a negative reaction; sensitivity, ratio of sick individuals who get a positive reaction; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
RT-qPCR
RT-qPCR assay utilises viral RNA extracted from patient samples (for example, material collected through nasopharyngeal and oropharyngeal swabs), synthesises of complementary DNA (cDNA) through the action of a reverse transcriptase enzyme, and amplifies the target sequences of the viral genome from the cDNA template. RT-qPCR can be interpreted semi-quantitatively, with the target amplification speed dependent on the concentration and quality of the viral RNA in the initial sample, thereby allowing the amplification rate to be used as a proxy for the sample viral load (49). The three target screening assays include E gene, RdRp gene and N assaying (Fig. 5) (50). For a routine workflow, the Taiwan Centers for Disease Control recommends the E gene assay as the first-line screening tool, followed by confirmatory testing with the RdRp gene assay. Utilising the RdRp gene assay with dual colour technology can discriminate between SARS-CoV-2 (both probes positive) and SARS-CoV RNA provided that the latter is used as a positive control. Alternatively, laboratories may choose to run the RdRp assay with only the SARS-CoV-2-specific probe. Despite also performing well, the N gene assay has not been subjected to further intensive validation given its slightly inferior sensitivity (51).
Figure 5.
Three candidate diagnostic reverse transcription PCR assays for detection of infection with SARS-CoV-2. The relative genome positions of virions are used to assay for SARS-CoV-2. The three target screening assays include the E gene assay, RNA-dependent RNA polymerase gene assay and N gene assay. E, envelope protein; N, nucleocapsid protein; M, membrane protein.
Rapid screening
To date, five antibody-based tests have been used for detecting the presence of IgG and IgM in body fluids, such as whole blood, serum or plasma. The BioMedomics rapid test and Surescreen rapid test cassette utilise lateral flow immunoassays, which are diagnostic devices used to examine antibodies (12,52-55). Moreover, Goldsite diagnostics has developed a time-resolved fluorescence immunoassay kit, while the Assay Genie rapid POC kit and VivaDiag COVID-19 IgG-IgM tests are colloidal gold-based immunoassays for detecting viral infection (56). To perform the assay, a few drops of blood obtained from the individual using a finger-stick or vein are applied onto the immunoassay. A few drops of buffer solution are then added onto the assay, after which the results are obtained within 10-15 min at room temperature. RT-qPCR testing is used as the reference standard to which immunoassays are compared. Amongst the five rapid screening tests, the BioMedomics IgM-IgG rapid test has been widely used for detecting antibody production in the human body (57).
The results by Wang (58) indicated that diagnostic sensitivity and specificity were 95.7% (antibody-based tests) and 98.7% (RT-qPCR), 92.2% (antibody-based tests) and 100% (RT-qPCR) by total antibody tests and RT-qPCR, respectively.
5. Methods to prevent COVID-19
SARS-CoV-2 possesses several problematic properties, such as transmission from asymptomatic individuals and nonspecific features of COVID-19, and utilises the ACE2 and TMPRSS2 receptors for attachment and transmission (31,59). Both ACE2 and TMPRSS2 proteins are expressed in <10% of human respiratory and gastrointestinal tract cells, including nasal goblet secretory cells, lung type II pneumocytes and ileal absorptive enterocytes (60,61). At present, prevention of viral entry into the human body has been the best option for controlling viral spread. The TCDC has established technical guidelines for COVID-19 (1). The following are crucial steps for preventing viral spread: i) Stay at home, unless essential, the general public should avoid travelling to affected countries and regions, as well as avoiding contact with animals, dead or alive. The general public should make a habit of applying alcohol-based hand sanitisers after entering any public spaces. ii) Maintain decontamination: Rooms should be regularly decontaminated, preferably with 5 to 10% sodium hypo-chlorite. iii) Keep a safe social distance, the general public must avoid public gatherings. Individuals should preferably maintain a distance of at least 1.5 m (5 ft) between themselves and anyone who is coughing or sneezing indoors. Individuals should maintain a distance of at least 1 m (3 ft) distance between themselves and anyone else outdoors. iv) Regularly sanitize hands, individuals are advised to practice appropriate hygiene, such as frequently washing their hands with soap after sneezing or coughing. Avoid touching any secretions, such as stool or urine. In addition, individuals should refrain from touching their eyes, nose and mouth with unclean hands. v) Wear face masks, healthcare personnel must use personal protective equipment, such as medical masks (including surgical face masks and N95s), eye protection, gloves, gowns and protective gear. The general public must wear a face mask to help prevent viral transmission, particularly in public spaces. Given the supply shortages, each country has their own recommendations regarding wearing of face masks.
6. Current therapeutic modalities for COVID-19
Given the lack of clinical evidence supporting the efficacy of any existing anti-viral agents or the existence of vaccines which have completed Phase II clinical trials and have been approved by a regulatory body for COVID-19, supportive treatments for clinical conditions in the early stages is imperative. In addition, conservative fluid management should be employed among patients with COVID-19 when no evidence of shock is present. Details and targets of supportive treatments for clinical conditions are presented in Table III (26).
Table III.
Summary of anti-viral agents against COVID-19.
| Anti-viral agents for COVID-19 | Chemical formula | Molecular weight, g/mol g·mol−1 | Drug targets | Pharmacologic mechanisms | Inhibits stage | (Refs.) |
|---|---|---|---|---|---|---|
| Chloroquine (Aralan®) | C18H26ClN3 | 319.87 | 1. Endosomal acidification 2. Phosphatidylinositol binding clathrin assembly protein |
1. A lysosomotropic base 2. Increases endosomal and lysosomal pH (Alkalinizes vacuolar pH) 3. Disrupts intracellular trafficking. 4. Inhibits phosphatidylinositol binding clathrin assembly protein 5. Downregulates IL-6 |
Inhibits viral fusion events | (70, 90-95) |
| Hydroxychloroquine (Plaquenil®) | C18H26ClN3O | 335.87 | 1. Endosomal acidification 2. Phosphatidylinositol binding clathrin assembly protein |
1. A lysosomotropic base 2. Increases endosomal and lysosomal pH (Alkalinizes vacuolar pH) 3. Disrupts intracellular trafficking. 4. Inhibits phosphatidylinositol binding clathrin assembly protein 5. Downregulates IL-6 |
1. Inhibits viral fusion events | (70, 90-95) |
| Arbidol (Umifenovir®) | C22H25BrN2O3S | 477.41 | 1. Interacts with aromatic residues within the viral hemagglutinin glycoprotein | 1. Inhibition of viral entry and membrane fusion | 1. Inhibits viral entry and membrane fusion | (66,67) |
| Camostat mesylate (Foipan®) | C21H26N4O8S | 494.52 | Transmembrane protease serine 2 | 1. Inhibits transmembrane protease serine 2 | 1. Blocks entry of SARS-Cov-2 into lung cells | (62,63) |
| Remdesivir (GS-5734) | C27H35N6O8P | 602.58 | RNA-dependent RNA polymerase |
1. Inhibits RNA-dependent RNA polymerase 2. Evades proofreading by viral exoribonuclease (ExoN) |
1. Inhibits viral replication 2. Delays RNA chain termination |
(105, 106, 112) |
| Favipiravir (Avigan®) | C5H4FN3O2 | 157.10 | RNA-dependent RNA polymerase | 1. Inhibits RNA-dependent RNA polymerase | 1. Inhibits viral replication | (70, 104) |
| Ribavirin (Rebetol®) | C7H10N4O5 | 230.18 | RNA-dependent RNA polymerase | 1. Inhibits RNA-dependent RNA polymerase | 1. Inhibits viral RNA synthesis 2. Inhibits viral mRNA capping |
(109, 116-118) |
| Lopinavir | C37H48N4O5 | 628.80 | 3C-like protease | 1. Inhibits 3C-like protease | 1. Inhibits protease by proteolysis | (68, 119, 120) |
| Ritonavir | C37H48N6O5S2 | 720.94 | 3C-like protease | 1. Inhibits 3C-like protease | 1. Inhibits protease by proteolysis | (68, 119, 120) |
| Recombinant Interferon | Type 1 interferons (IFN-α; IFN-β) | 1. Induces the phosphorylation of transcriptional factors such as STAT1 2. Activates interferon-stimulated genes |
1. Immunomodulation 2. Activates both innate and adaptive immunity responses to against the virus |
(103, 115, 121, 127, 128, 131, 162) |
Several ongoing clinical trials have evaluated the following direct treatments for SARS-CoV-2: Chloroquine (Aralan®), hydroxychloroquine (Plaquenil®), arbidol (Umifenovir®), camostat mesylate (Foipan®), remdesivir (GS-5734), favipiravir (Avigan®), ribavirin (Rebetol®), lopinavir/ritonavir (Kaletra®) and interferon-α and interferon-β (47). The chemical structures of hydroxychloroquine (Plaquenil®), chloroquine (Aralan®), remdesivir (GS-5734), favipiravir (Avigan®), ribavirin (Rebetol®), lopinavir/ritonavir (Kaletra®), and camostat mesylate (Foipan®) are presented in Fig. 6. Table III and Fig. 7 summarise ongoing therapeutic agents being evaluated for management of COVID-19 and their molecular pharmacologic mechanisms. The mechanisms by which suitable therapeutic agents against SARS-CoV2 exhibit their effects are discussed below.
Figure 6.
Chemical structures of hydroxychloroquine, chloroquine, remdesivir, favipiravir, ribavirin, lopinavir/ritonavir, arbidol and camostat mesylate.
Figure 7.
Molecular pharmacological mechanisms of ongoing potential therapeutic agents for management of COVID-19.
Blocking coronavirus-host interactions and attachments
Camostat mesylate (Foipan®) is a serine protease inhibitor that inhibits TMPRSS2 and blocks virus entry into lung cells (62,63). In vitro studies have shown that Foipan® inhibits TMPRSS2 and blocks SARS-CoV and human coronavirus NL63 infection of HeLa cells (62,63). Hoffmann et al demonstrated that SARS-CoV-2 exploits ACE-2 for entry and serine protease TMPRSS2 for S protein priming (31). Moreover, reports have shown that Foipan® blocks SARS-CoV-2 infection of Calu-3 lung cells in vitro (63). Accordingly, four clinical trials on camostat mesylate for COVID-19 are currently ongoing worldwide (NCT 04353284, NCT 04321096, NCT 04338906 and NCT 04355052) (64).
Arbidol (Umifenovir®) is a small indole-derivative agent used for the treatment of respiratory viral infections (65-68). In vitro and in vivo studies have demonstrated that Umifenovir® inhibits a number of enveloped or non-enveloped RNA or DNA viruses, including influenza viruses A, B and C, SARS-CoV, adenovirus, poliovirus, rhinovirus, coxsackievirus, Hantaan virus, Chikungunya virus and Hepatitis B and C viruses (68-70). Umifenovir® interacts with aromatic residues within the viral hemagglutinin glycoprotein and inhibits viral entry (71-73). A total of eight clinical trials (NCT 04350684, NCT 04286503, NCT 04260594, NCT 04323345, NCT 04273763, NCT 04306497, NCT 04261907 and NCT 04333589) on Umifenovir® for COVID-19 are ongoing worldwide (64).
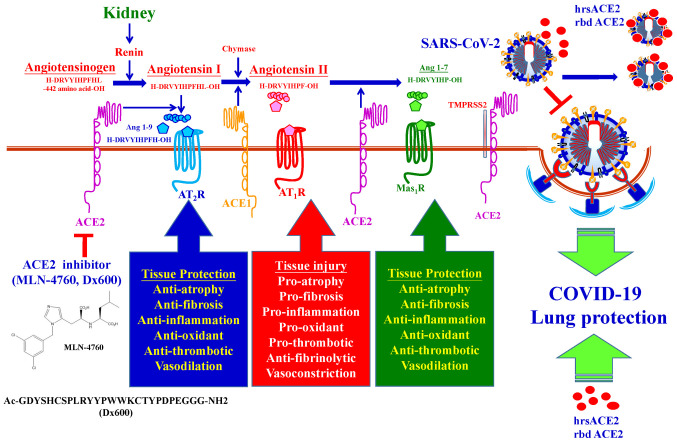
Recently, ACE2 has been considered as a target for the treatment of COVID-19 (74). ACE2 is abundantly expressed on vascular endothelial cells of the lung (75), heart (42), nervous system (76), intestine (77), kidneys (51), blood vessels (78) and muscles (75) on the cell surface. ACE2 possesses peptidyl dipeptidase activity by catalyzing the cleavage of Angiotensin II into Angiotensin, and is one of the means by which blood pressure and cardiovascular functions are regulated (1-7). SARS-CoV-2 binds ACE2 for entry into the host (40-44). The ACE2 specific inhibitors, including MLN-4760 and Dx600 are not used clinically (79,80). It was reported that administration of excessive soluble ACE2 may slow the entry of SARS-CoV-2 into the host cells (74). Soluble forms of ACE2 include recombinant human ACE2 protein (rhACE2) (81) and recombinant bacterial ACE2 receptors-like enzyme of B38-CAP (rbd ACE2) (82). Studies have suggested rhACE2 (83) or rbd ACE2 (82) competitively bind with SARS-CoV-2, neutralizing the virus and also rescuing host cellular ACE2 activity and protecting the lungs from injury. The molecular pharmacological mechanisms of ACE-2 inhibitors are summarized in Fig. 8. A soluble form of ACE2 (rhACE2 and rbd ACE2) on COVID-19 therapy may exert dual functions: i) Slowing viral entry into host cells and inhibiting viral spread; ii) protecting the lung from injury (84). A total of four clinical trials (NCT 04375046, NCT 04382950, NCT 04287686 and NCT 04335136) on ACE-2 inhibitors for management of COVID-19 are ongoing worldwide (64).
Figure 8.
Molecular pharmacological mechanisms of ACE2 inhibitors and soluble forms of ACE2. ACE2 possesses peptidyl dipeptidase activity by catalyzing the cleavage of Angiotensin II into Angiotensin. The ACE2 specific inhibitors including MLN-4760 and Dx600, but are not used clinically. Soluble forms of ACE2, including rhACE2 protein and rbd ACE2, competitively bind with SARS-CoV-2 to neutralize the virus and also rescue the hosts cellular ACE2 activity and thus protect the lungs from injury. ACE2, angiotensin-converting enzyme 2; rhACE2, recombinant human ACE2; rbd, ACE2, recombinant bacterial ACE2 receptors-like enzyme of B38-CAP.
Triggering lysosomal activation and disrupting intracellular trafficking
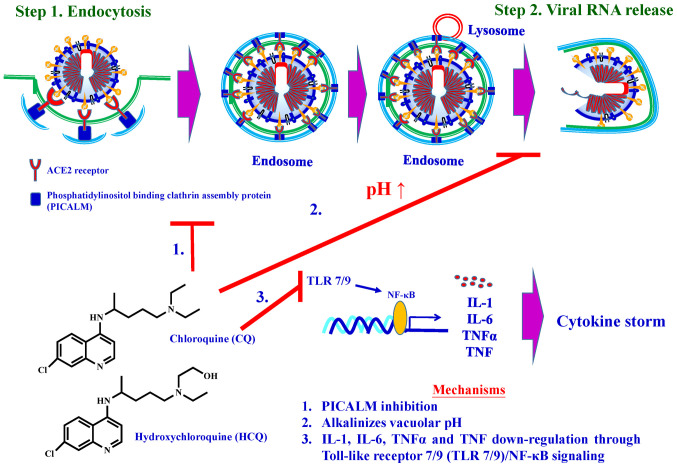
Chloroquine (Aralan®), a well-known anti-malarial and anti-autoimmune agent, has long been used to treat malaria (85) and autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis (86), Hydroxychloroquine (Plaquenil®) is synthesised by introducing a hydroxyl group into chloroquine. Animal studies have demonstrated that Plaquenil® is much less toxic than chloroquine (10,87-89).
Reports have shown that chloroquine and hydroxychlo-roquine increase endosomal and lysosomal pH (alkalinises vacuolar pH) and then disrupt intracellular trafficking (90-94). Recent studies have demonstrated that chloroquine reduces the expression of phosphatidylinositol binding clathrin assembly protein (PICALM), a cargo-selecting clathrin adaptor that senses and drives membrane curvature, which regulates endocytosis (95). In vitro studies have demonstrated that chloroquine significantly inhibits SARS-CoV-2 from infecting Vero E6 cells. One of the mechanisms for the chloroquine-mediated effects against SARS-CoV-2 is the decrease in the ability of cells to perform clathrin-mediated endocytosis of nanosized structures due to PICALM suppression (95).
Clinical investigations have shown that patients with COVID-19 had high concentrations of cytokines, such as IL-1β, IL-1β, IL-2, IL-6, IFNs and MCP-1 (96-98), in their plasma, subsequently causing a cytokine storm. In addition, hydroxychloroquine has been demonstrated to exhibit anti-inflammatory activity and can significantly decrease the IL-1, IL-6, TNF-α and TNF production through Toll-like receptor/NF-κB signalling (99,100).
The molecular pharmacological mechanisms of chloro-quine and hydroxychloroquine are summarised in Fig. 9. A total of 52 clinical trials on chloroquine and 150 clinical trials on hydroxychloroquine for the treatment of COVID-19 are ongoing (64). Given that chloroquine and hydroxychloroquine are longstanding therapeutic agents widely used for disease treatment in hospitals, several ongoing clinical trials on COVID-19 have focused on both. However, it has more recently been reported that hydroxychloroquine does exhibit beneficial effects in the management of infection with COVID-19, and may in fact result in increased deaths due to its side-effects, resulting in the early halting an Oxford-based study (101).
Figure 9.
Molecular mechanisms of chloroquine and hydroxychloroquine.
Inhibiting RdRp
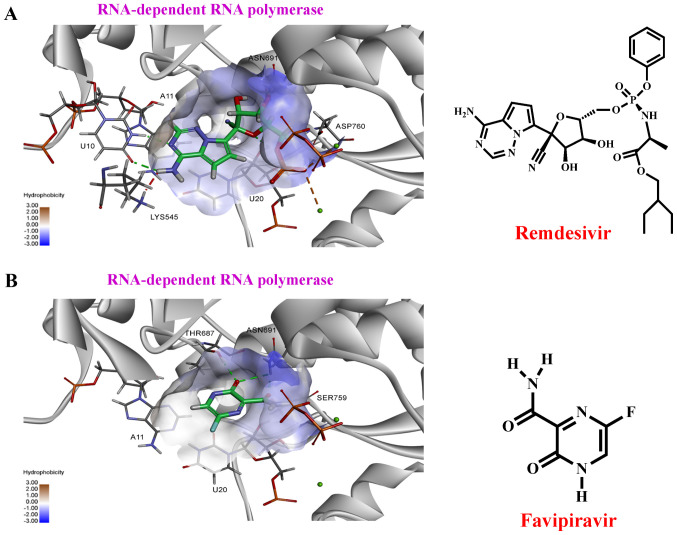
Remdesivir (GS-5734), favipiravir (Avigan®) (45,65,102-104). Remdesivir (GS-5734), a phosphoramidate prodrug of an adenine-derivative agent, was originally developed by Gilead Sciences (Gilead Sciences Inc.; patent holder) for the Ebola virus (105-108). Avigan®, a guanine-derived agent, has been approved for influenza for patients resistant to Tamiflu and Relenza treatment (10,109,110). Remdesivir and Avigan® are incorporated into nascent viral RNA and inhibit the RdRp (109,110). This results in the premature termination of the viral RNA chain and consequently halts the replication of the viral genome. Recent in vitro studies have reported that remdesivir and Avigan® possesses bioactive effects against SARS-CoV-2 (109,111,112). Our previous preliminary studies using Discovery Studio 2020 (DS 2020) software revealed that remdesivir and Avigan® exhibited strong binding potential to RdRp (Fig. 10 and Table SI). A total of 19 clinical trials on remdesivir and 12 clinical trials on Avigan® for the treatment of COVID-19 are ongoing (64). In April, 2020, a National Institutes of Health clinical trial reported that remdesivir accelerates recovery from COVID-19. On May 1, 2020, the US FDA issued an emergency authorisation for the use of investigational remdesivir in the treatment of suspected or laboratory-confirmed COVID-19 amongst adults and children hospitalised with severe disease (113). The US FDA has approved remdesivir for the treatment of hospitalized COVID-19 patients on October 23, 2020 (8). This is positive and exciting news for the treatment of COVID-19.
Figure 10.
Molecular docking of remdesivir and favipiravir binding to RdRp. (A) The right panel shows the structure of remdesivir, the left panel shows molecular docking simulation using Discovery Studio 2020. (B) The right panel shows the structure of favipiravir, the left panel shows molecular docking simulation. The structures of the drugs are presented using a stick model. Carbon atoms are coloured green. RdRp, RNA-dependent RNA polymerase.
Interfering with RNA metabolism required for viral replication
Ribavirin (Rebetol®) (109,114,115), a guano-sine-derived agent, had been approved for the treatment of Hepatitis C viral infection. Recent studies have demonstrated that ribavirin can be used to treat respiratory syncytial virus and SARS-CoV by inhibiting viral RNA synthesis, viral mRNA capping and RdRp (109,116-118). To date, five clinical trials on Rebetol® for the treatment of COVID-19 are ongoing worldwide (64).
Inhibiting 3CLpro
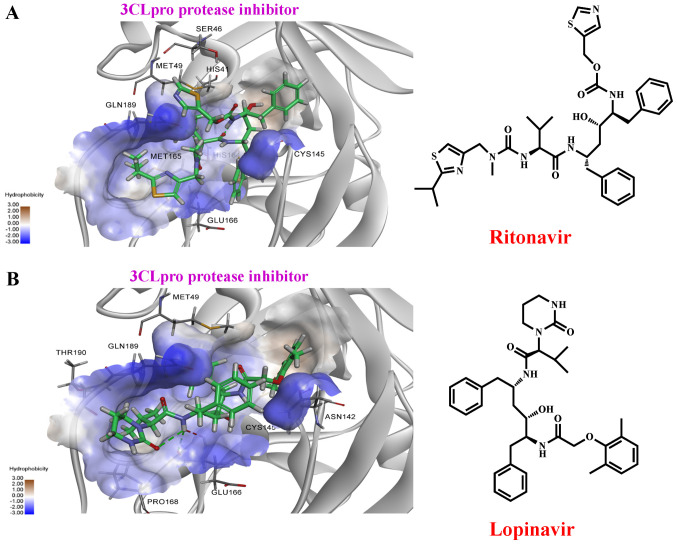
Lopinavir/Ritonavir (Kaletra®) (103,111,115) are widely used for treating HIV infection. However, early studies have demonstrated that lopinavir and ritonavir are active against SARS-CoV and MERS by inhibiting 3CLpro via proteolysis in SARS-CoV (68,119,120). In contrast, Wu et al (27) demonstrated that Kaletra® did not shorten the duration of SARS-CoV-2 infection amongst patients with mild pneumonia in Taiwan. Our previous preliminary study using DS 2020 software showed that lopinavir and ritonavir exhibits strong binding potential to 3CLpro (Fig. 11 and Table SI). A total of 45 clinical trials on Kaletra® for the treatment of COVID-19 are ongoing worldwide (64).
Figure 11.
Molecular docking of ritonavir and lopinavir binding to the 3CLpro. (A) The right panel shows the structure of ritonavir, the left panel shows molecular docking simulation using Discovery Studio 2020. (B) The right panel is the structure of lopinavir, the left panel shows molecular docking simulation. The structures of the drugs are presented using a stick model. Carbon atoms are coloured green. 3CLpro, 3-chymotrypsin-like cysteine protease.
Agents which exhibit immunotherapeutic properties
i) Type 1 IFN-α, pegylated IFNα-2a and α-2b and IFN-β (103,114,115,121); and ii) steroids (122,123). During viral infection, type I IFN synthesis is initially induced, which subsequently activates both the innate and adaptive immune response against the virus (124). The type I IFN family consists of IFN-α, IFN-β and other subtypes (121,125,126). When the virus infects target cells, RNA sensors induces IFN regulatory transcription factor translocation to the nucleus, which promotes type I IFN secretion. The secreted IFN inter-acts with IFN receptors on the cell membrane, which promotes phosphorylation of STAT1/2 transcriptional factors (127,128). The phosphorylated STAT1/2 localises to the nucleus, binds to IFN-stimulated response element responsible and activates IFN-stimulated genes, which then results in more production of type I IFN (129). Upon secretion of type I IFN, type I IFN-mediated innate immunity is initiated. Natural killer cells then become active and destroy infected cells. Type I IFN binds to the IFN receptors on cytotoxic T cells (CD8+ T cells), subsequently killing infected cells through cellular immunity (130). In addition, type I IFN stimulates B cells and induces production of neutralising antibodies, which serves a protective role by limiting later-phase infections and preventing future re-infections (128). Treatment with IFN-α2b significantly reduced the duration of SARS-CoV-2 in the upper respiratory tract and reduced the levs of the inflammatory cytokines IL-6 and CRP in COVID-19 patients (131).
Cells infected with SARS-CoV and MERS-CoV exhibit reduced type I IFN. As such, it is hypothesized that SARS-CoV-2 may utilise a similar manner for type I IFN reduction. It has previously been shown that type I IFN treatments improve anti-SARS-CoV and anti-MERS-CoV activity amongst infected mice and exhibits synergistic effects with ribavirin against SARS-CoV in vitro (132). Immunocompromised patients are at higher risk for severe COVID-19 than the general public. Type I IFN treatments can thus be a safe and efficient approach to manage SARS-CoV-2 infection (121,133). A total of 37 clinical trials on IFN for COVID-19 are ongoing worldwide (64). Fig. 12 presents a schematic overview of the type I IFN-mediated immune response mechanism following SARS-CoV, MERS-CoV and SARS-CoV-2 infection.
Figure 12.
Schematic overview of type I interferon-mediated immune response mechanism for SARS-CoV, MERS-CoV and SARS-CoV-2. IRF, interferon regulatory transcription factor; type 1 IFN, type 1 interferon; ISRE, IFN-stimulated response element; IFNR, interferon receptor; MHC, major histocompat-ibility complex; TCR, T cell receptor; CTL, cytotoxic T lymphocyte; CD40L, CD40 ligand.
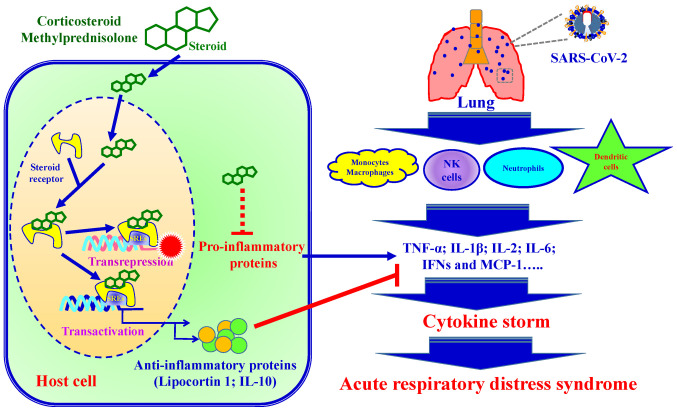
Combining anti-viral and anti-inflammatory agents is another attractive therapeutic option for the prevention and treatment of COVID-19. Upon infection, innate immune cells including macrophages, natural killer cells, neutrophils and dendritic cells produce large amounts of pro-inflammatory cytokines (TNF, Type 1 IFN, IL-6 and IL-12). Previous clinical studies have demonstrated that steroids (corticosteroid and methylprednisolone) modulate inflammatory responses, reducing the incidence of treatment failure and reducing cytokine storms (123,134). The anti-inflammatory mechanisms of steroids is involved in the presence of steroid receptors and regulates down-stream gene transcription processes (135). Steroid receptor signaling mechanisms regulate down-stream gene expression via transactivation and trans-repression. i) In the process of transactivation, steroid receptors bind steroid hormones and form dimers. The ligand bound steroid receptor dimer complex binds to specific DNA sequences (steroid response elements; SREs), increasing anti-inflammatory gene transcription (such as Lipocortin 1 and IL-10). ii) In the process of trans-repression, the ligand bound steroid receptor tethers to SREs and inters pro-inflammatory transcription factors, which leads to a reduction of pro-inflammatory cytokines (136). SARS-CoV-2 infection induces pro-inflammatory cytokine production, resulting in local tissue inflammation and a systemic inflammatory response, termed a cytokine storm (137). Cytokine storm injures host cells and causes an increased risk of respiratory failure such as acute respiratory distress syndrome (ARDS) and eventually death. Fig. 13 presents a schematic diagram of the steroid-mediated immune response following SARS-CoV-2 infection.
Figure 13.
Schematic diagram of the steroid-mediated immune response following SARS-CoV-2 infection. Steroid receptor signaling mechanisms regu-late down-stream gene expression via transactivation and transrepression. Steroids cause an increase in anti-inflammatory gene transcription and blocks pro-inflammatory cytokine production. SREs, steroid response elements.
7. Pharmacological agents targeting thrombosis
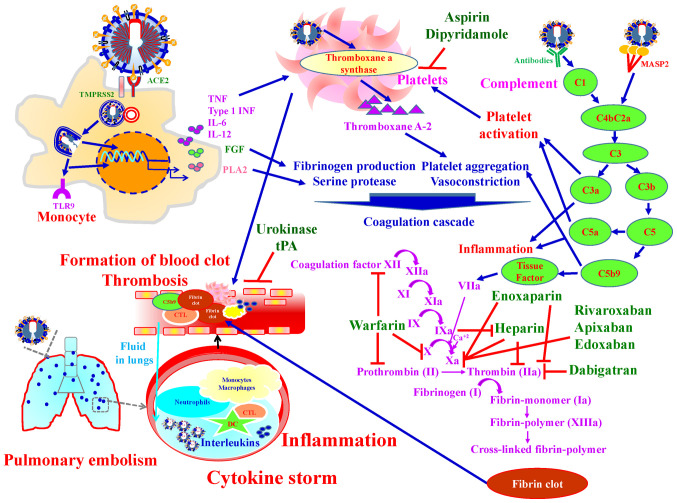
It was reported that SARS-CoV-2 infection increased the risk of thrombosis and up to 50% of severe COVID-19 patients developed coagulopathy (138). Coagulation and thrombosis are associated with pathogenesis of COVID-19 (139). SARS-CoV-2 results in direct injuries to the vascular endothelium cells or induces a cytokine storm, leading to systemic thrombus formation, thrombosis on pulmonary artery, and potentially lower limb arterial and cerebral infarction (138). Upon SARS-CoV-2 infection, the complement system, a bridge between innate and adaptive immune response, is activated and triggers inflammation. In addition, the complement system links the immune system with the coagulation system. C5a in the complement system induces tissue factor expression, and C5b-9 activates platelet (140) Tissue factor/factor VIIa complex converts prothrombin into thrombin. Thrombin catalyzes fibrinogen to fibrin and promotes fibrin formation by coagulation cascade activation and by activating platelets (140-144).
Pharmacological agents targeting thrombosis in COVID-19 are divided into three groups: i) Anti-coagulant agents; Unfractionated heparin, low-molecular-weight heparins (Enoxaparin), Danaparoid (a mixture of heparan sulfate, dermatan sulfate, and chondroitin sulfate) and Vitamin-K antagonists (warfarin). Heparin and Enoxaparin have strong anti-thrombotic activity, and anti-inflammatory properties via selectin blockade, bradykinin downregulation and thrombin generation. In addition, Heparins attenuate interactions between SARS-CoV-2 spike protein and ACE-2 (145). Enoxaparin is a first choice drug to prevent thromboembolic phenomena in COVID-19 patients (138). ii) Anti-platelets agents; Aspirin and Dipyridamole. Aspirin has been demonstrated with ARDS prevention and higher survival rates from acute lung injury in clinical studies (146,147). Dipyridamole is a phosphodiesterase inhibitor that inhibits platelet aggregation by increasing cyclic adenosine monophosphate concentrations. In vitro studies demonstrated that dipyridamole has anti-SARS-CoV-2 activity through binding with 3CLpro of SARS-CoV-2 (148). iii) Fibrinolytic (thrombolytic) agents; Urokinase, Streptokinase and Tissue-type plasminogen activator (tPA). The blood clots are broken down by plasmin call fibrinolysis. Fibrinolysis intermediate tPA and urokinase plasminogen activator convert plasminogen to plasmin. When tPA, a thrombolytic agent, is intravenously injected into the vasculature, increased fibrinolytic ability in the plasma and lyses of the thrombosis was observed in COVID-19 patients (149). Fig. 14 presents an over-view of the mechanisms by which anti-thrombotic agents exert their effects in the treatment of SARS-CoV-2 infection.
Figure 14.
Schematic overview of the mechanisms of anti-thrombotic agents used for the treatment of SARS-CoV-2 infection. Anti-thrombotic agents include: i) Anti-coagulants: Unfractionated heparin, Enoxaparin, Danaparoid and warfarin; ii) anti-platelet agents: Aspirin and Dipyridamole; and iii) Fibrinolytic (Thrombolytic) agents: Urokinase, Streptokinase and tPA. tPA, tissue-type plasminogen activator.
8. Neutralizing antibodies and vaccines against SARS-CoV-2
Two promising countermeasures for controlling the current COVID-19 pandemic are recombinant neutralizing anti-bodies (150) and vaccines (151) directed against SARS-CoV-2. Recombinant human or humanized monoclonal antibodies are proving to be safe, effective, and highly specific in their ability to target an invading pathogen. More than 70 recombinant monoclonal antibodies have now been approved by the FDA for use in the treatment of infectious, autoimmune and inflammatory, malignant, or cardiovascular diseases (152). Thus, recombinant neutralizing antibodies isolated from those infected with SARS-CoV-2 are the most rapid and readily manufacturable immune intervention for passive administration that may be developed to either prevent or treat COVID-19 disease. Of note, US President Donald Trump, who recently suffered from infection with COVID-19 was treated with monoclonal antibodies generated by Regeneron.
Vaccines are a time-honored method for establishing long-lived immune memory for controlling infectious diseases, and technologies have been developed such that vaccines can now be developed faster than previously (151). Over 100 companies or academic institutions are working on COVID-19 vaccines with strategies that include recombinant vectors, mRNA in lipid nanoparticles, DNA, inactivated virus, live attenuated virus, virus-like particles and protein subunits (153). Three vaccine candidates have already advanced to Phase II testing that include an mRNA vaccine encoding the viral spike protein from Moderna, an Adenotype 5 vector vaccine expressing the S protein from CanSino Biologicals, and a chimpanzee adenovirus encoding the spike protein from the Jenner Institute in Oxford, UK. There are several mRNA/LNP (for example, from Moderna/NIAID, BioNTech/Fosum, Pharma/Pfizer) or DNA (Inovio) vaccines as well as attenuated viruses, proteins, nanoparticles and viral vectors containing SARS-CoV-2 viral genes as vaccine candidates moving through safety and immunogenicity trials, and a smaller subset of vaccine candidates will be tested in Phase III or efficacy trials to better determine if they are safe, as well to determine their efficacy. In parallel now with Phase I and II trials, it is important to develop capacity for large-scale vaccine production, in the event of a successful efficacy trial (154). It is possible that genetic immunization strategies such as DNA or mRNA in LNPs can be manufactured more rapidly than proteins or viral vectors and can be more cost effective.
9. TCM and COVID-19
Based on >3,500 years of Chinese medical practice, TCM has spread to numerous countries worldwide, has profoundly influenced lives and has gradually merged with and complemented modern Western medicine and therapy (155). In recent decades, mounting evidence has suggested that TCM may be helpful in the prevention and treatment of human virus-related disorders, including influenza, liver diseases and acquired immune deficiency syndrome (114,156-158). Following the COVID-19 outbreak, TCM schemes have been included into the guidelines for the diagnosis and therapy of COVID-19 in China (114,159,160). Recently, in silico data showed that binding of curcuminoid derivatives to COVID-19 3CLpro is stronger than that of Lopinavir and curcumin (126). It is hypothesized that more convenient methods for the early detection of COVID-19 via genotyping will emerge in the near future. Even amongst severe/critical cases, TCMs can still serve as a complementary and integrative therapy to modern Western medicine to shorten the recovery period and relieve symptoms among patients with COVID-19.
10. Conclusions
This review describes several clinical manifestations of COVID-19, analyses the SARS-CoV-2 genome and outlines the life cycle of SARS-CoV-2. Several methods have been used to examine SARS-CoV-2 infections. For example, RT-qPCR has been widely applied for RNA detection, whereas rapid screening has been used for antibody or virus detection. Despite the lack of medications for COVID-19, several clinical trials have been proposed for its treatment. In addition, several TCMs have been discussed for the readers' reference.
Global interaction and cooperation amongst several countries is expected to underlie the development of rapid and accurate screening assays, produce vaccines, design novel agents against SARS-CoV-2 and reduce the side effects of therapeutic TCMs, with the ultimate, long-term goal of eradication of COVID-19.
Supplementary Data
Acknowledgments
We thank Kuan-Wen Chen and Tzu-Mao Hung in GGA Corporation, Molecular Science Center (Taiwan) for performing the molecular docking to RdRp and 3CLpro. We wish to acknowledge the work of Nian-Gu Chen, Pei-Jen Chung, Chien-Jung Huang, Yi-Chia Li and Chia-Wen Tsai for their assistance on this work. We also would also like thank Merck Ltd. Taiwan company for providing Synthia Organic Retrosynthesis Software.
Funding
This work was supported by the China Medical University Hospital (grant nos. DMR-109-147, CMU-103-S-16 and MOST 106-2314-B-039-046), and also by the Chinese Medicine Research Center, China Medical University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan.
Availability of data and materials
Not applicable.
Authors' contributions
SCT, FJT and JSY were involved in the conception of the study. CCL, DTB, YJC, YTY, YMH, CWF, SCK, YSL, HYC and YNJ were involved in the literature search and critical reviewing of the manuscript. SCT, CCL, DTB, YJC, YTY, YMH, SCK and JSY were involved in the preparation of the draft of the manuscript. SCT, FJT and JSY were involved in the revising and editing of the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.World Health Organization (WHO) Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed November 2, 2020.
- 2.Liu YC, Kuo RL, Shih SR. COVID-19: The first documented coronavirus pandemic in history. Biomed J. 2020;43:328–333. doi: 10.1016/j.bj.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsay SF, Kao CC, Wang HH, Lin CC. Nursing's response to COVID-19: Lessons learned from SARS in Taiwan. Int J Nurs Stud. 2020;108:103587. doi: 10.1016/j.ijnurstu.2020.103587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang CL, McAleer M. Alternative global health security indexes for risk analysis of COVID-19. Int J Environ Res Public Health. 2020;17:3161. doi: 10.3390/ijerph17093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu YC, Liu YA, Lin MH, Lee HW, Chen TJ, Chou LF, Hwang SJ. Visiting policies of hospice wards during the COVID-19 pandemic: An environmental scan in Taiwan. Int J Environ Res Public Health. 2020;17:2857. doi: 10.3390/ijerph17082857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz J, King CC, Yen MY. Protecting healthcare workers during the coronavirus disease 2019 (COVID-19) outbreak: lessons from Taiwan's severe acute respiratory syndrome response. Clin Infect Dis. 2020;71:858–860. doi: 10.1093/cid/ciaa255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu YC, Chen CS, Chan YJ. The outbreak of COVID-19: An overview. J Chin Med Assoc. 2020;83:217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.U.S. Food & Drug Administration (FDA) Coronavirus Disease 2019 (COVID-19) https://www.fda.gov/news-events/press-announcements/covid-19-update-fda-broadens-emergency-use-authorization-veklury-remdesivir-include-all-hospitalized. Accessed August 28, 2020. [PubMed]
- 9.Park SE. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19) Clin Exp Pediatr. 2020;63:119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day M. Covid-19: Identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;3:m1165. doi: 10.1136/bmj.m1165. [DOI] [PubMed] [Google Scholar]
- 12.Chang TH, Wu JL, Chang LY. Clinical characteristics and diagnostic challenges of pediatric COVID-19: A systematic review and meta-analysis. J Formos Med Assoc. 2020;119:982–989. doi: 10.1016/j.jfma.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CT, Bookman K, Sieja A, Markley K, Altman RL, Sippel J, Perica K, Reece L, Davis C, Horowitz E, et al. Clinical informatics accelerates health system adaptation to the COVID-19 pandemic: Examples from Colorado. J Am Med Inform Assoc. 2020 Jul 20; doi: 10.1093/jamia/ocaa171. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Zhang R, He G. Hematological findings in corona-virus disease 2019: Indications of progression of disease. Ann Hematol. 2020;99:1421–1428. doi: 10.1007/s00277-020-04103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ippolito D, Maino C, Pecorelli A, Allegranza P, Cangiotti C, Capodaglio C, Mariani I, Giandola T, Gandola D, Bianco I, et al. Chest X-ray features of SARS-CoV-2 in the emergency department: A multicenter experience from northern Italian hospitals. Respir Med. 2020;170:106036. doi: 10.1016/j.rmed.2020.106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, Lai WY, Yang DM, Chou SJ, Yang YP, et al. A review of SARS-CoV-2 and the Ongoing Clinical Trials. Int J Mol Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis B, Rothrock AN, Swetland S, Andris H, Davis P, Rothrock SG. Viral and atypical respiratory co-infections in COVID-19: A systematic review and meta-analysis. J Am Coll Emerg Physicians Open. 2020;1:533–548. doi: 10.1002/emp2.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliva A, Siccardi G, Migliarini A, Cancelli F, Carnevalini M, D'Andria M, Attilia I, Danese VC, Cecchetti V, Romiti R, et al. Co-infection of SARS-CoV-2 with Chlamydia or Mycoplasma pneumoniae: A case series and review of the literature. Infection. 2020 Jul 28; doi: 10.1007/s15010-020-01483-8. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. Lancet Infect Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, Nguyen TT, Cao TM, Pham QD. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ, Wang Q, Miao H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su YJ, Lai YC. Comparison of clinical characteristics of coronavirus disease (COVID-19) and severe acute respiratory syndrome (SARS) as experienced in Taiwan. Travel Med Infect Dis. 2020;36:101625. doi: 10.1016/j.tmaid.2020.101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung YY, Wu YC, Li CY, Hsu CY, Hsu CY, Liang ST, Huang WC, Pan KY, Tsai JH, Yen YH. Interim Guidelines for Clinical Management of SARS-CoV-2 Infection (5th edition) Ministry of Health and Welfare. Taiwan: Centers for Disease Control; 2020. https://www.cdc.gov.tw/File/Get/-ewtg9-RCAetCPKR4_rnCw. Accessed March 26, 2020. [Google Scholar]
- 27.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singhal T. A Review of coronavirus disease-2019 (COVID-19) Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 30.National Institutes of Health (NIH), National Center for Biotechnology Information (NCBI) NCBI SARS-CoV-2 Resources. https://www.ncbi.nlm.nih.gov/Structure/SARS-CoV-2.html. Accessed October 27, 2020.
- 31.Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoeman D, Fielding BC. Is there a link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front Microbiol. 2020;11:2086. doi: 10.3389/fmicb.2020.02086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik YA. Properties of coronavirus and SARS-CoV-2. Malays J Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 34.Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M, Ren Y, Li X, Geng Q, Xu Y, et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci. 2020;63:1–4. doi: 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J Virol. 2014;88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai Y, Kawachi K, Terada Y, Omori H, Matsuura Y, Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:e00524–13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rathnasinghe R, Karlicek RF, Schotsaert M, Koffas MA, Arduini B, Jangra S, Wang B, Davis JL, Alnaggar M, Costa A, et al. Scalable, effective, and rapid decontamination of SARS-CoV-2 contaminated N95 respirators using germicidal ultra-violet C (UVC) irradiation device. medRxiv. doi: 10.1101/2020.10.0520206953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boopathi S, Poma AB, Kolandaivel P. Novel 2019 corona-virus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2020 Apr 30; doi: 10.1080/07391102.2020.1758788. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen WH, Strych U, Hotez PJ, Bottazzi ME. The SARS-CoV-2 vaccine pipeline: An overview. Curr Trop Med Rep. 2020 Mar 3; doi: 10.1007/s40475-020-00201-6. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan VS, Chan KY, Chen Y, Poon LL, Cheung AN, Zheng B, Chan KH, Mak W, Ngan HY, Xu X, et al. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat Genet. 2006;38:38–46. doi: 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Jr, Thackray LB, Young MD, Mason RJ, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahlmann R, Lode H. Medication for COVID-19-an over-view of approaches currently under study. Dtsch Arztebl Int. 2020;117:213–219. doi: 10.3238/arztebl.2020.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kupferschmidt K, Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 48.Yip CC, Ho CC, Chan JF, To KK, Chan HS, Wong SC, Leung KH, Fung AY, Ng AC, Zou Z, et al. Development of a novel, genome subtraction-derived, SARS-CoV-2-specific COVID-19-nsp2 real-time RT-PCR assay and its evaluation using clinical specimens. Int J Mol Sci. 2020;21:2574. doi: 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan C, Cui J, Huang L, Du B, Chen L, Xue G, Li S, Zhang W, Zhao L, Sun Y, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahman H, Carter I, Basile K, Donovan L, Kumar S, Tran T, Ko D, Alderson S, Sivaruban T, Eden JS, et al. Interpret with caution: An evaluation of the commercial AusDiagnostics versus in-house developed assays for the detection of SARS-CoV-2 virus. J Clin Virol. 2020;127:104374. doi: 10.1016/j.jcv.2020.104374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pujadas E, Ibeh N, Hernandez MM, Waluszko A, Sidorenko T, Flores V, Shiffrin B, Chiu N, Young-Francois A, Nowak MD, et al. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas(R) 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J Med Virol. 2020 May 8; doi: 10.1002/jmv.25988. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montesinos I, Gruson D, Kabamba B, Dahma H, Van den Wijngaert S, Reza S, Carbone V, Vandenberg O, Gulbis B, Wolff F, Rodriguez-Villalobos H. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thabet L, Mhalla S, Naija H, Jaoua MA, Hannachi N, Fki-Berrajah L, Toumi A, Karray-Hakim H. SARS-CoV-2 infection virological diagnosis. Tunis Med. 2020;98:304–308. [PubMed] [Google Scholar]
- 54.Vásárhelyi B, Kristóf K, Ostorházi E, Szabó D, Prohászka Z, Merkely B. The diagnostic value of rapid anti IgM and IgG detecting tests in the identification of patients with SARS CoV-2 virus infection. Orv Hetil. 2020;161:807–812. doi: 10.1556/650.2020.31859. In Hungarian. [DOI] [PubMed] [Google Scholar]
- 55.Green K, Graziadio S, Turner P, Fanshawe T, Allen J. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Centre for Evidence- Based Medicine (CEBM); Oxford; 2020. https://www.cebm.net/covid-19/molecular-and-antibody-point-of-care-tests-to-support-the-screening-diagnosis-and-monitoring-of-covid-19/. Accessed April 7, 2020. [Google Scholar]
- 56.Xue X, Zhu C, Huang S, Pan L, Xu J, Li W. Effect of heat inactivation of blood samples on the efficacy of three detection methods of SARS-CoV-2 antibodies. Nan Fang Yi Ke Da Xue Xue Bao. 2020;40:316–320. doi: 10.12122/j.issn.1673-4254.2020.03.03. In Chinese. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang P, Liu T, Huang L, Liu H, Lei M, Xu W, Hu X, Chen J, Liu B. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295:22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P. Combination of serological total antibody and RT-PCR test for detection of SARS-COV-2 infections. J Virol Methods. 2020;283:113919. doi: 10.1016/j.jviromet.2020.113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lange C, Wolf J, Auw-Haedrich C, Schlecht A, Boneva S, Lapp T, Horres R, Agostini H, Martin G, Reinhard T, Schlunck G. Expression of the COVID-19 receptor ACE2 in the human conjunctiva. J Med Virol. 2020 May 6; doi: 10.1002/jmv.25981. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 61.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahman N, Basharat Z, Yousuf M, Castaldo G, Rastrelli L, Khan H. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020;25:2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang J, Song W, Huang H, Sun Q. Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: From mechanistic studies to clinical trials for COVID-19. J Clin Med. 2020;9:1131. doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. ClinicalTrials.gov NUnlom: COVID-19. https://clinicaltrials.gov/ct2/results?cond=covid-19. Accessed August 17, 2020.
- 65.Amawi H, Abu Deiab GI, A Aljabali AA, Dua K, Tambuwala MM. COVID-19 pandemic: An overview of epidemiology, pathogenesis, diagnostics and potential vaccines and therapeutics. Ther Deliv. 2020;11:245–268. doi: 10.4155/tde-2020-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents. 2020;56:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Z, Lu Z, Xu T, Chen C, Yang G, Zha T, Lu J, Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J Infect. 2020;81:e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng L, Li C, Zeng Q, Liu X, Li X, Zhang H, Hong Z, Xia J. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J Infect. 2020;81:e1–e5. doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, Li Y, Zhao L, Li W, Sun X, et al. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 71.Hulseberg CE, Fénéant L, Szymańska-de Wijs KM, Kessler NP, Nelson EA, Shoemaker CJ, Schmaljohn CS, Polyak SJ, White JM. Arbidol and other low-molecular-weight drugs that inhibit lassa and Ebola viruses. J Virol. 2019;93:e02185–18. doi: 10.1128/JVI.02185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci USA. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng LY, Yang J, Liu S. Investigational hemagglutinin-targeted influenza virus inhibitors. Expert Opin Investig Drugs. 2017;26:63–73. doi: 10.1080/13543784.2017.1269170. [DOI] [PubMed] [Google Scholar]
- 74.Roshanravan N, Ghaffari S, Hedayati M. Angiotensin converting enzyme-2 as therapeutic target in COVID-19. Diabetes Metab Syndr. 2020;14:637–639. doi: 10.1016/j.dsx.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu J, Lazartigues E. Expression of ACE2 in human neurons supports the neuro-invasive potential of COVID-19 virus. Cell Mol Neurobiol. 2020 Jul 4; doi: 10.1007/s10571-020-00915-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wedell J, Banzhaf G, Meier zu Eissen P, Schlageter M. Experiences with a subcutaneous, fully resorbable bridge in construction a double loop ileo- and colostomy. Chirurg. 1990;61:36–38. In German. [PubMed] [Google Scholar]
- 78.Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X, Zheng J, Yan Y, Ruan Z, Su Y, Wang J, Huang H, Zhang Y, Wang W, Gao J, et al. Angiotensin-converting enzyme 2 regulates autophagy in acute lung injury through AMPK/mTOR signaling. Arch Biochem Biophys. 2019;672:108061. doi: 10.1016/j.abb.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 80.de Moraes PL, Kangussu LM, Castro CH, Almeida AP, Santos RAS, Ferreira AJ. Vasodilator effect of angiotensin-(17) on vascular coronary bed of rats: Role of Mas, ACE and ACE2. Protein Pept Lett. 2017;24:869–875. doi: 10.2174/0929866524666170728154459. [DOI] [PubMed] [Google Scholar]
- 81.Basu R, Poglitsch M, Yogasundaram H, Thomas J, Rowe BH, Oudit GY. Roles of angiotensin peptides and recombinant human ACE2 in heart failure. J Am Coll Cardiol. 2017;69:805–819. doi: 10.1016/j.jacc.2016.11.064. [DOI] [PubMed] [Google Scholar]
- 82.Qiao B, Olvera de la Cruz M. Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano. 2020;14:10616–10623. doi: 10.1021/acsnano.0c04798. [DOI] [PubMed] [Google Scholar]
- 83.Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020;9:72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mejia Torres RE, Banegas EI, Mendoza M, Diaz C, Bucheli ST, Fontecha GA, Alam MT, Goldman I, Udhayakumar V, Zambrano JO. Efficacy of chloroquine for the treatment of uncomplicated Plasmodium falciparum malaria in Honduras. Am J Trop Med Hyg. 2013;88:850–854. doi: 10.4269/ajtmh.12-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taherian E, Rao A, Malemud CJ, Askari AD. The biological and clinical activity of anti-malarial drugs in autoimmune disorders. Curr Rheumatol Rev. 2013;9:45–62. doi: 10.2174/1573397111309010010. [DOI] [PubMed] [Google Scholar]
- 87.Pastick KA, Okafor EC, Wang F, Lofgren SM, Skipper CP, Nicol MR, Pullen MF, Rajasingham R, McDonald EG, Lee TC, et al. Review: Hydroxychloroquine and chloroquine for treatment of SARS-CoV-2 (COVID-19) Open Forum Infect Dis. 2020;7:ofaa130. doi: 10.1093/ofid/ofaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Piszczatoski CR, Powell J. Emergency approval of chloroquine and hydroxychloroquine for treatment of COVID-19. Ann Pharmacother. 2020;54:827–831. doi: 10.1177/1060028020925558. [DOI] [PubMed] [Google Scholar]
- 89.Shukla AM, Archibald LK, Shukla AW, Mehta HJ, Cherabuddi K. Chloroquine and hydroxychloroquine in the context of COVID-19. Drugs Context. 2020;9(2020) doi: 10.7573/dic.2020-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sturrock BR, Chevassut TJ. Chloroquine and COVID-19-a potential game changer? Clin Med (Lond) 2020;20:278–281. doi: 10.7861/clinmed.2020-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020;55:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Annangi S. Chloroquine and hydroxychloroquine for COVID-19: A word of caution. Respirology. 2020;25:683–684. doi: 10.1111/resp.13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferner RE, Aronson JK. Chloroquine and hydroxychloroquine in covid-19. BMJ. 2020;369:m1432. doi: 10.1136/bmj.m1432. [DOI] [PubMed] [Google Scholar]
- 94.Badyal DK, Mahajan R. Chloroquine: Can it be a novel drug for COVID-19. Int J Appl Basic Med Res. 2020;10:128–130. doi: 10.4103/ijabmr.IJABMR_141_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu TY, Frieman M, Wolfram J. Insights from nano-medicine into chloroquine efficacy against COVID-19. Nat Nanotechnol. 2020;15:247–249. doi: 10.1038/s41565-020-0674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ren Y, Yao MC, Huo XQ, Gu Y, Zhu WX, Qiao YJ, Zhang YL. Study on treatment of 'cytokine storm' by anti-2019-nCoV prescriptions based on arachidonic acid metabolic pathway. Zhongguo Zhong Yao Za Zhi. 2020;45:1225–1231. doi: 10.19540/j.cnki.cjcmm.20200224.405. In Chinese. [DOI] [PubMed] [Google Scholar]
- 97.McGonagle D, Sharif K, O'Regan A, Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev. 2020;19:102537. doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lagunas-Rangel FA, Chávez-Valencia V. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J Med Virol. 2020 Apr 16; doi: 10.1002/jmv.25900. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aizawa T, Imaizumi T, Hirono K, Watanabe S, Tsugawa K, Tanaka H. Chloroquine attenuates TLR3-mediated plasminogen activator inhibitor-1 expression in cultured human glomerular endothelial cells. Clin Exp Nephrol. 2019;23:448–454. doi: 10.1007/s10157-018-1659-y. [DOI] [PubMed] [Google Scholar]
- 100.Clancy RM, Markham AJ, Reed JH, Blumenberg M, Halushka MK, Buyon JP. Targeting downstream transcription factors and epigenetic modifications following Toll-like receptor 7/8 ligation to forestall tissue injury in anti-Ro60 associated heart block. J Autoimmun. 2016;67:36–45. doi: 10.1016/j.jaut.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mahase E. Hydroxychloroquine for covid-19: The end of the line? BMJ. 2020;369:m2378. doi: 10.1136/bmj.m2378. [DOI] [PubMed] [Google Scholar]
- 102.Wu R, Wang L, Kuo HD, Shannar A, Peter R, Chou PJ, Li S, Hudlikar R, Liu X, Liu Z, et al. An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Rep. 2020 May 11; doi: 10.1007/s40495-020-00216-7. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ahsan W, Javed S, Bratty MA, Alhazmi HA, Najmi A. Treatment of SARS-CoV-2: How far have we reached? Drug Discov Ther. 2020;14:67–72. doi: 10.5582/ddt.2020.03008. [DOI] [PubMed] [Google Scholar]
- 104.Du YX, Chen XP. Favipiravir: Pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin Pharmacol Ther. 2020;108:242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 105.Reina J. Remdesivir, the antiviral hope against SARS-CoV-2. Rev Esp Quimioter. 2020;33:176–179. doi: 10.37201/req/028.2020. In Spanish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cao YC, Deng QX, Dai SX. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med Infect Dis. 2020;35:101647. doi: 10.1016/j.tmaid.2020.101647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Augustin M, Hallek M, Nitschmann S. Remdesivir for patients with severe COVID-19. Internist (Berl) 2020;61:644–645. doi: 10.1007/s00108-020-00800-5. In German. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z, Wang X, Cao D, Sun R, Li C, Li G. Rapid review for the anti-coronavirus effect of remdesivir. Drug Discov Ther. 2020;14:73–76. doi: 10.5582/ddt.2020.01015. [DOI] [PubMed] [Google Scholar]
- 109.Jean SS, Lee PI, Hsueh PR. Treatment options for COVID-19: The reality and challenges. J Microbiol Immunol Infect. 2020;53:436–443. doi: 10.1016/j.jmii.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu CC, Chen MY, Lee WS, Chang YL. Potential therapeutic agents against COVID-19: What we know so far. J Chin Med Assoc. 2020;83:534–536. doi: 10.1097/JCMA.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Simsek Yavuz S, Ünal S. Antiviral treatment of COVID-19. Turk J Med Sci. 2020;50:611–619. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choy KT, Wong AY, Kaewpreedee P, Sia SF, Chen D, Hui KPY, Chu DKW, Chan MCW, Cheung PP, Huang X, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.U.S. Food & Drug Administration (FDA) Coronavirus (COVID-19) Update: FDA issues emergency use authorization for potential COVID-19 treatment. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment. Accessed May 1, 2020.
- 114.Chan KW, Wong VT, Tang SCW. COVID-19: An update on the epidemiological, clinical, preventive and therapeutic evidence and Guidelines of integrative Chinese-Western medicine for the management of 2019 novel coronavirus disease. Am J Chin Med. 2020;48:737–762. doi: 10.1142/S0192415X20500378. [DOI] [PubMed] [Google Scholar]
- 115.Martinez MA. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob Agents Chemother. 2020;64:e00399. doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guzik TJ, Mohiddin SA, Dimarco A, Patel V, Savvatis K, Marelli-Berg FM, Madhur MS, Tomaszewski M, Maffia P, D'Acquisto F, et al. COVID-19 and the cardiovascular system: Implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res. 2020;116:1666–1687. doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: Recent reports on antiviral therapies based on lopinavir/rito-navir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr Med Chem. 2020;27:4536–4541. doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- 120.Ye XT, Luo YL, Xia SC, Sun QF, Ding JG, Zhou Y, Chen W, Wang XF, Zhang WW, Du WJ, et al. Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. Eur Rev Med Pharmacol Sci. 2020;24:3390–3396. doi: 10.26355/eurrev_202003_20706. [DOI] [PubMed] [Google Scholar]
- 121.Sallard E, Lescure FX, Yazdanpanah Y, Mentre F, Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sauñe PM, Bryce-Alberti M, Portmann-Baracco AS, Accinelli RA. Methylprednisolone pulse therapy: An alternative management of severe COVID-19. Respir Med Case Rep. 2020;31:101221. doi: 10.1016/j.rmcr.2020.101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang JW, Yang L, Luo RG, Xu JF. Corticosteroid administration for viral pneumonia: COVID-19 and beyond. Clin Microbiol Infect. 2020;26:1171–1177. doi: 10.1016/j.cmi.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li SF, Gong MJ, Zhao FR, Shao JJ, Xie YL, Zhang YG, Chang HY. Type I interferons: Distinct biological activities and current applications for viral infection. Cell Physiol Biochem. 2018;51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- 125.Andreakos E, Tsiodras S. COVID-19: Lambda interferon against viral load and hyperinflammation. EMBO Mol Med. 2020;12:e12465. doi: 10.15252/emmm.202012465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Du B, Qiu HB, Zhan X, Wang YS, Kang HYJ, Li XY, Wang F, Sun B, Tong ZH. Pharmacotherapeutics for the New Coronavirus Pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:E012. doi: 10.3760/cma.j.issn.1001-0939.2020.0012. In Chinese. [DOI] [PubMed] [Google Scholar]
- 127.Vidal P. Interferon α in cancer immunoediting: From elimination to escape. Scand J Immunol. 2020;91:e12863. doi: 10.1111/sji.12863. [DOI] [PubMed] [Google Scholar]
- 128.Nelemans T, Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11:961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abdul-Sater AA, Majoros A, Plumlee CR, Perry S, Gu AD, Lee C, Shresta S, Decker T, Schindler C. Different STAT transcription complexes drive early and delayed responses to type I IFNs. J Immunol. 2015;195:210–216. doi: 10.4049/jimmunol.1401139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud JP, Boudinot P. The antiviral innate immune response in fish: Evolution and conservation of the IFN system. J Mol Biol. 2013;425:4904–4920. doi: 10.1016/j.jmb.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 131.Zhou Q, Wei XS, Xiang X, Wang X, Wang ZH, Chen V, Shannon CP, Tebbutt SJ, Kollmann TR, Fish EN. Interferon-a2b treatment for COVID-19. medRxiv. https://101101/2020040620042580.
- 132.Morgenstern B, Michaelis M, Baer PC, Doerr HW, Cinatl J., Jr Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Young MJ, Clyne CD, Chapman KE. Endocrine aspects of ACE2 regulation: RAAS, steroid hormones and SARS-CoV-2. J Endocrinol. 2020;247:R45–R62. doi: 10.1530/JOE-20-0260. [DOI] [PubMed] [Google Scholar]
- 135.Xavier AM, Anunciato AK, Rosenstock TR, Glezer I. Gene expression control by glucocorticoid receptors during innate immune responses. Front Endocrinol (Lausanne) 2016;7:31. doi: 10.3389/fendo.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hardy RS, Raza K, Cooper MS. Therapeutic glucocorticoids: Mechanisms of actions in rheumatic diseases. Nat Rev Rheumatol. 2020;16:133–144. doi: 10.1038/s41584-020-0371-y. [DOI] [PubMed] [Google Scholar]
- 137.Song P, Li W, Xie J, Hou Y, You C. Cytokine storm induced by SARS-CoV-2. Clin Chim Acta. 2020;509:280–287. doi: 10.1016/j.cca.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Komiyama M, Hasegawa K. Anticoagulant therapy for patients with coronavirus disease 2019: Urgent need for enhanced awareness. Eur Cardiol. 2020;15:e58. doi: 10.15420/ecr.2020.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Merrill JT, Erkan D, Winakur J, James JA. Emerging evidence of a COVID-19 thrombotic syndrome has treatment implications. Nat Rev Rheumatol. 2020;16:581–589. doi: 10.1038/s41584-020-0474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fletcher-Sandersjöö A, Bellander BM. Is COVID-19 associated thrombosis caused by overactivation of the complement cascade? A literature review. Thromb Res. 2020;194:36–41. doi: 10.1016/j.thromres.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Conway EM, Pryzdial ELG. Is the COVID-19 thrombotic catastrophe complement-connected? J Thromb Haemost. 2020 Aug 6; doi: 10.1111/jth.15050. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Furie B, Furie BC. Mechanisms of thrombus formation. N Engl J Med. 2008;359:938–949. doi: 10.1056/NEJMra0801082. [DOI] [PubMed] [Google Scholar]
- 143.Giannis D, Ziogas IA, Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J Clin Virol. 2020;127:104362. doi: 10.1016/j.jcv.2020.104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bikdeli B, Madhavan MV, Jimenez D, Chuich T, Dreyfus I, Driggin E, Nigoghossian C, Ageno W, Madjid M, Guo Y, et al. COVID-19 and thrombotic or thromboembolic disease: Implications for prevention, antithrombotic therapy, and follow-up: JACC State-of-the-Art review. J Am Coll Cardiol. 2020;75:2950–2973. doi: 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu J, Li J, Arnold K, Pawlinski R, Key NS. Using heparin molecules to manage COVID-2019. Res Pract Thromb Haemost. 2020;4:518–523. doi: 10.1002/rth2.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Panka BA, de Grooth HJ, Spoelstra-de Man AM, Looney MR, Tuinman PR. Prevention or treatment of ards with aspirin: A review of preclinical models and meta-analysis of clinical studies. Shock. 2017;47:13–21. doi: 10.1097/SHK.0000000000000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen W, Janz DR, Bastarache JA, May AK, O'Neal HR, Jr, Bernard GR, Ware LB. Prehospital aspirin use is associated with reduced risk of acute respiratory distress syndrome in critically ill patients: A propensity-adjusted analysis. Crit Care Med. 2015;43:801–807. doi: 10.1097/CCM.0000000000000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li Z, Li X, Huang YY, Wu Y, Liu R, Zhou L, Lin Y, Wu D, Zhang L, Liu H, et al. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc Natl Acad Sci USA. 2020;117:27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Whyte CS, Morrow GB, Mitchell JL, Chowdary P, Mutch NJ. Fibrinolytic abnormalities in acute respiratory distress syndrome (ARDS) and versatility of thrombolytic drugs to treat COVID-19. J Thromb Haemost. 2020;18:1548–1555. doi: 10.1111/jth.14872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 151.Graham BS. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 152.Shepard HM, Phillips GL, D Thanos C, Feldmann M. Developments in therapy with monoclonal antibodies and related proteins. Clin Med (Lond) 2017;17:220–232. doi: 10.7861/clinmedicine.17-3-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 154.Corey L, Mascola JR, Fauci AS, Collins FS. A strategic approach to COVID-19 vaccine R&D. Science. 2020;368:948–950. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- 155.Zhao Z, Li Y, Zhou L, Zhou X, Xie B, Zhang W, Sun J. Prevention and treatment of COVID-19 using Traditional Chinese Medicine: A review. Phytomedicine. 2020;153308 doi: 10.1016/j.phymed.2020.153308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang YS, Cong WH, Zhang JJ, Guo FF, Li HM. Research progress of intervention of Chinese herbal medicine and its active components on human coronavirus. Zhongguo Zhong Yao Za Zhi. 2020;45:1263–1271. doi: 10.19540/j.cnki.cjcmm.20200219.501. In Chinese. [DOI] [PubMed] [Google Scholar]
- 157.McKimm-Breschkin JL, Jiang S, Hui DS, Beigel JH, Govorkova EA, Lee N. Prevention and treatment of respiratory viral infections: Presentations on antivirals, traditional therapies and host-directed interventions at the 5th ISIRV Antiviral Group conference. Antiviral Res. 2018;149:118–142. doi: 10.1016/j.antiviral.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Teschke R, Larrey D, Melchart D, Danan G. Traditional Chinese Medicine (TCM) and herbal hepatotoxicity: RUCAM and the role of novel diagnostic biomarkers such as MicroRNAs. Medicines (Basel) 2016;3:18. doi: 10.3390/medicines3030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hu XY, Logue M, Robinson N. Antimicrobial resistance is a global problem - a UK perspective. Eur J Integr Med. 2020;36:101136. doi: 10.1016/j.eujim.2020.101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ho LTF, Chan KKH, Chung VCH, Leung TH. Highlights of traditional Chinese medicine frontline expert advice in the China national guideline for COVID-19. Eur J Integr Med. 2020;36:101116. doi: 10.1016/j.eujim.2020.101116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, Pizarro G, Vial P, Iruretagoyena M, Dittrich S, Weitzel T. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mirijello A, D'Errico MM, Lamarca A, Piscitelli P, De Cosmo S. Comment on Matricardi PM et al: The first, holistic immunological model of COVID-19: Implications for prevention, diagnosis, and public health measures. Pediatr Allergy Immunol. 2020 May 17; doi: 10.1111/pai.13277. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.