Abstract
Existing adenine and cytosine base editors induce only a single type of modification, limiting the range of DNA alterations that can be created. Here we describe a CRISPR-Cas9-based synchronous programmable adenine and cytosine editor (SPACE) that can concurrently introduce A-to-G and C-to-T substitutions with minimal RNA off-target edits. SPACE expands the range of possible DNA sequence alterations, broadening the research applications of CRISPR base editors.
Editorial summary
Base editor that concurrently modifies both adenine and cytosine broadens the potential applications of base editing.
Adenine and cytosine base editors (ABEs and CBEs) enable programmable A-to-G or C-to-T transitions in DNA, respectively1–5. To create a dual-function base editor, we engineered a single protein harboring adenosine and cytidine deaminases from the previously described miniABEmax-V82G6 and Target-AID5 editors, respectively (Fig. 1a and Extended Data Fig. 1a). We chose to combine parts of these particular editors because the deaminase domains were located on opposite ends of the Streptococcus pyogenes Cas9 (SpCas9)-D10A nickase in these fusions (Fig. 1a) and were previously shown to exhibit substantially reduced or minimal off-target RNA editing in human cells6. In addition, the single monomer TadA variant in miniABEmax-V82G is smaller than the dimeric wild-type TadA-TadA variant present in other ABEs, thereby helping to minimize the overall size of the resulting SPACE fusion. To maximize the purity of on-target cytosine base edits induced by SPACE, we also included two uracil glycosylase inhibitors (UGIs) (Fig. 1a), a strategy first described with the previously published BE4 CBE3.
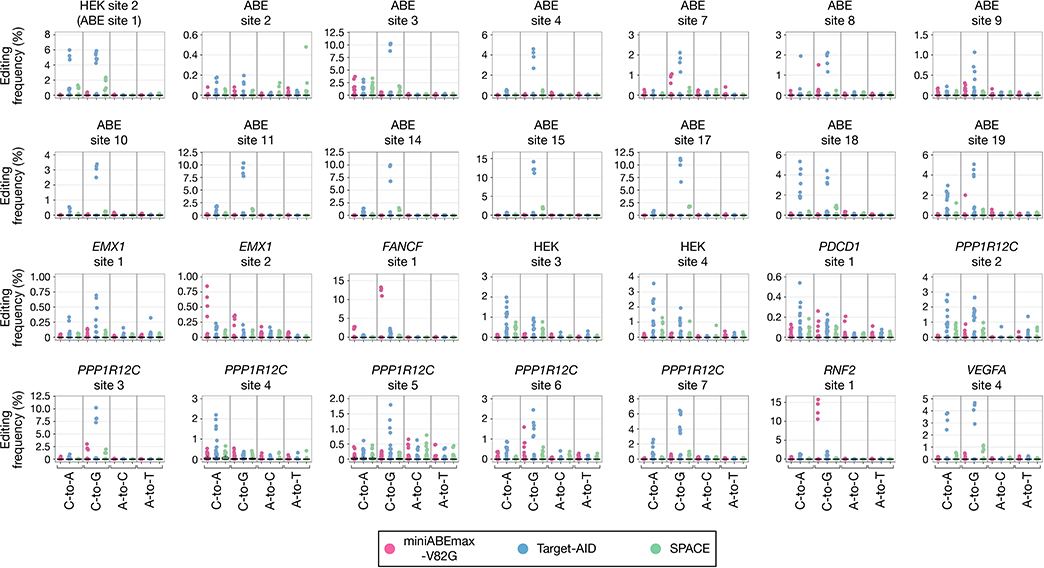
Figure 1. SPACE induces concurrent A-to-G and C-to-T base edits in human HEK293T cells.
a, Schematic illustrating adenine base editor miniABEmax-V82G, cytosine base editor Target-AID, and dual-deaminase base editor SPACE. A-to-G (pink halo) and C-to-T (blue halo) base edits are illustrated. Light blue shape indicates Steptococcus pyogenes Cas9 (D10A) nickase, purple structure in Cas9 represents the guide RNA, pink circle represents the TadA 7.10 (V82G) adenosine deaminase monomer, blue triangle indicates the cytidine deaminase pmCDA1, and yellow circles represent uracil glycosylase inhibitors (UGIs). N and C labels denote the N and C termini of the base editor. b, Heat maps showing on-target DNA A-to-G (pink) and C-to-T (blue) editing frequencies of nCas9 (Control), miniABEmax-V82G, Target-AID, and SPACE with 28 gRNAs (n= 4 independent replicates). Editing windows shown represent the most highly edited adenines and cytosines, not the entirety of the protospacer. Numbering at the bottom represents the position of the respective base in the protospacer sequence with 1 being the most PAM-distal. c, Box and dot plots indicating the aggregate distribution of A-to-G (pink) and C-to-T (blue) edits across the entire protospacer with SPACE using 28 gRNAs. In box plots, the box spans the interquartile range (IQR) (first to third quartiles), the horizontal line shows the median (second quartile) and whiskers extend to ± 1.5 × IQR. Single dots represent individual replicates. Graph shows the same data as shown in b (n=4). d, Composition of alleles with frequencies of 1% or higher that result from SPACE-induced on-target DNA editing with a gRNA targeting HEK site 2 (ABE site 1). Data is shown for one replicate from the HEK site 2 on-target experiment shown in b. Numbering indicates the position in the protospacer with 1 being the most PAM-distal.
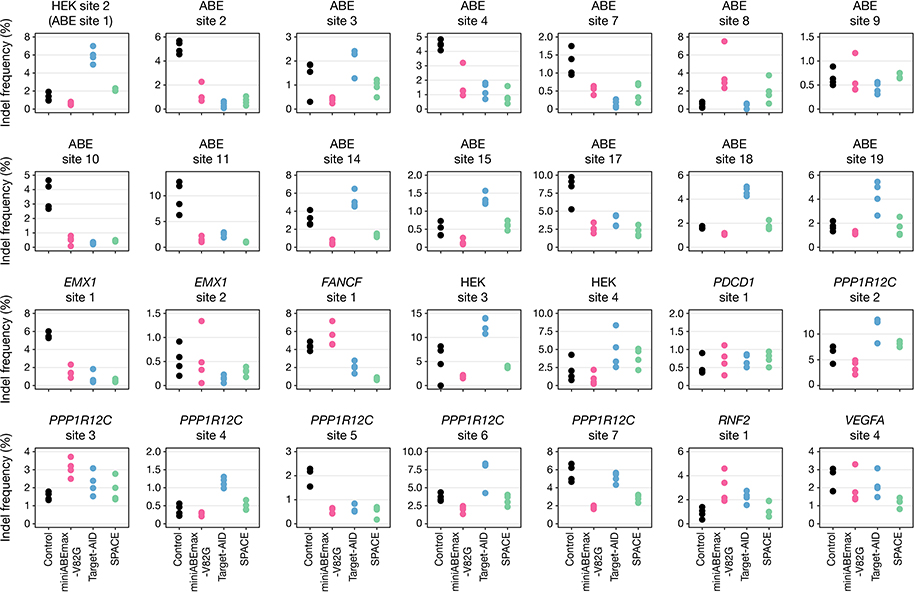
We directly compared the on-target DNA editing activities of SPACE with those of miniABEmax-V82G and Target-AID using 28 different gRNAs in human HEK293T cells (Fig. 1b and Supplementary Table 1). Overall, SPACE induced A-to-G editing at 25 out of 26 genomic sites that were edited by miniABEmax-V82G, and C-to-T editing at all 28 genomic sites that were edited by Target-AID (Fig. 1b). The efficiencies of A-to-G editing by SPACE (range: 0.03–71.33%, mean: 13.07%) were modestly reduced relative to those of miniABEmax-V82G (range: 0.06–70.82%, mean: 18.1%), whereas the efficiencies of C-to-T editing by SPACE (range: 0.3–69.98%, mean: 22.13%) were comparable to those observed with Target-AID (range: 0.28–70.32%, mean: 24.82%). We did not observe a strict requirement by SPACE for the identity of the base present just 5’ to an edited A or C (Fig. 1b). The editing windows of SPACE were slightly narrowed compared to those of miniABEmax-V82G and Target-AID (Fig. 1c, Extended Data Fig. 1b and c). Among the 28 target sites we examined, A-to-G and C-to-T edits by SPACE were mostly induced at positions 4–7 and 2–7, respectively, within the protospacer (with 1 being the most PAM-distal base) (Fig. 1c). Our analysis also showed that for all 28 gRNAs, the product purities and indel frequencies at on-target sites edited by SPACE were comparable to or better than those of miniABEmax-V82G or Target-AID individually (Extended Data Figs. 2 and 3). Although we do not know the precise causes of modest differences in editing efficiencies we observed between SPACE and the individual miniABEmax-V82G or Target-AID editors, we note that there are differences in protein expression (codon and promoter usage), nuclear localization signal (NLS) architecture, and the number of UGIs among these fusions (Extended Data Fig. 1a, Supplementary Table 2, Online Methods).
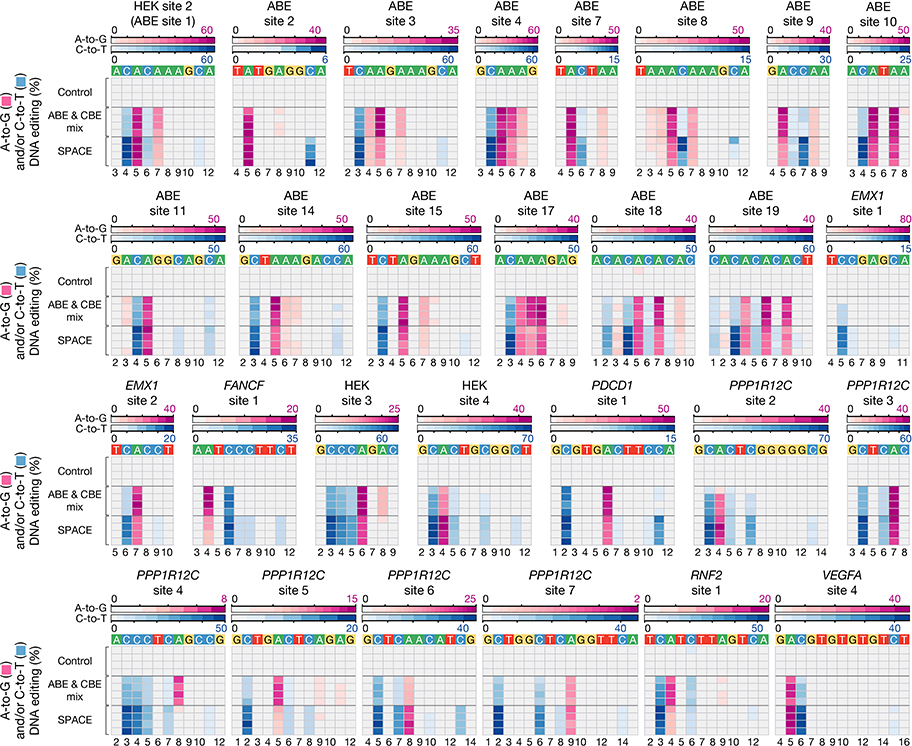
An important potential advantage of SPACE relative to single-deaminase editors is the capability to concurrently introduce more than one type of base edit. Among the 25 gRNAs that induced both A-to-G and C-to-T on-target edits with SPACE, we observed that for 18 of these gRNAs the summed frequencies of dual edited alleles were greater than 15% (Fig. 1d, Extended Data Fig. 4, and Supplementary Table 3). For 10 of these 25 gRNAs, the most efficiently edited on-target allele had both types of edits (for the replicate shown in Fig. 1d and Extended Data Fig. 4). To test whether SPACE is more efficient at inducing dual edits than the combined effects of separate adenine and cytosine base editors, we also performed experiments in HEK293T cells in which we directly compared SPACE with co-expressed miniABEmax-V82G and Target-AID (“ABE & CBE mix”) for each of the 28 gRNAs (Extended Data Fig. 5, and Supplementary Table 4, Online Methods). For 22 of the 28 gRNAs, the summed frequency of dual edited on-target alleles was higher with SPACE than with the “ABE & CBE mix” (Extended Data Fig. 6a, and Supplementary Fig. 1). Interestingly, the summed frequencies of on-target alleles harboring only A-to-G edits was higher with the “ABE & CBE mix” condition than with SPACE for 21 of these same 22 gRNAs (Extended Data Fig. 6a) whereas the summed frequencies of on-target alleles with only C-to-T edits was higher with SPACE than with the “ABE & CBE mix” for 16 of the 22 gRNAs (Extended Data Fig. 6a). Unwanted indels induced by SPACE (range: 0.02–7.1%, mean: 1.44%) were lower or comparable to those observed with the “ABE & CBE mix” (range: 0.13–11.92%, mean: 2.88%) at 27 out of 28 sites tested (Extended Data Fig. 6b). Although we cannot rule out that differences in the architecture of SPACE, miniABEmax-V82G, and Target-AID may affect their expression levels and/or activities (Supplementary Table 2), our results demonstrate that SPACE generally yields higher frequencies of dual-edited alleles and lower frequencies of indels at on-target sites compared to co-expression of standard editors harboring the same adenosine and cytidine deaminases individually.
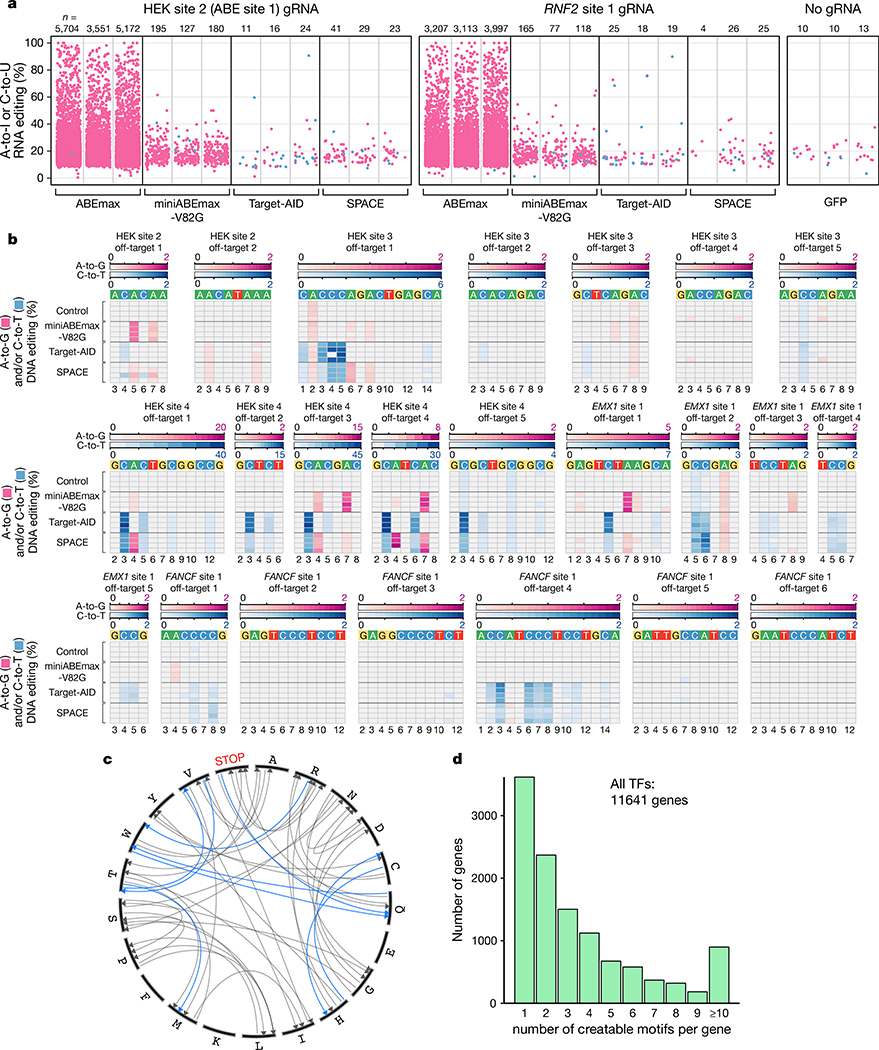
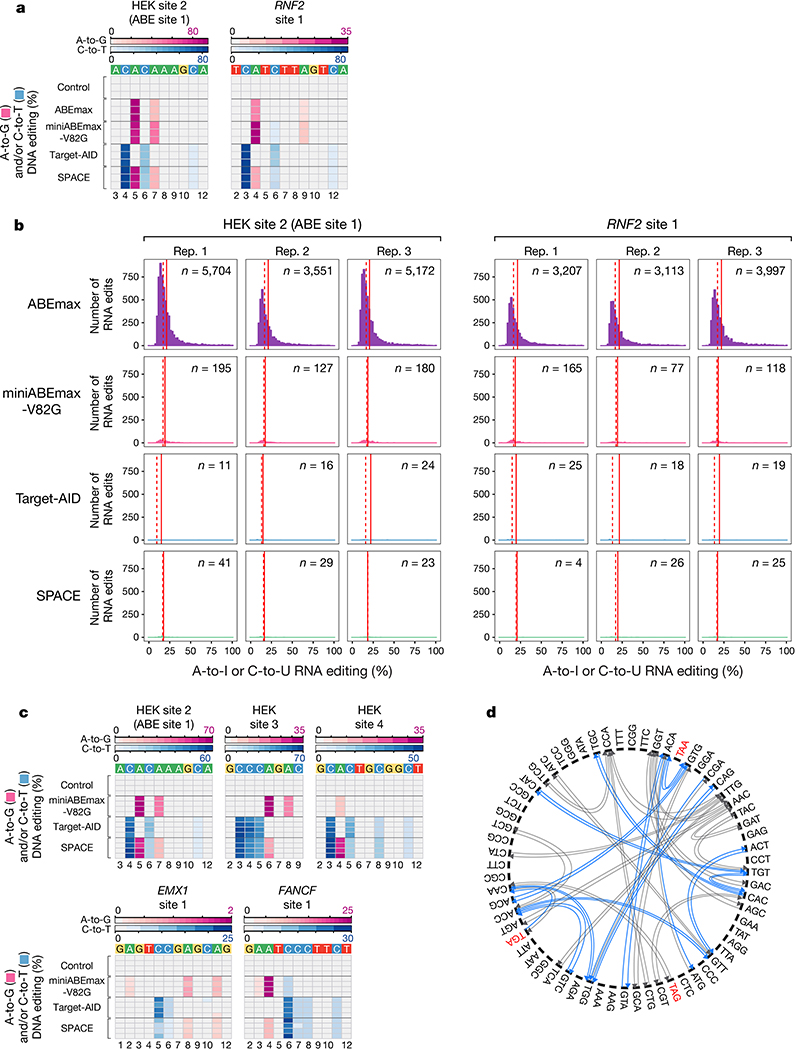
To characterize the transcriptome-wide RNA off-target activity of SPACE, we performed RNA-seq from HEK293T cells co-expressing SPACE with a gRNA targeting HEK site 2 or RNF2 site 1 (Online Methods). We also performed matched side-by-side RNA-seq experiments with HEK293T cells expressing miniABEmax-V82G, Target-AID, ABEmax (a positive control for RNA editing), and GFP (a negative control). Analysis of on-target DNA editing in the cells used for RNA-seq showed efficient editing with SPACE, miniABEmax-V82G, and Target-AID with both gRNAs (Extended Data Fig. 7a). As expected, GFP negative control experiments showed very few RNA C-to-U edits (range of 1–3) and A-to-I edits (range of 7–12) while ABEmax induced relatively high numbers of A-to-I edits (range of 3,105–5,696), miniABEmax-V82G induced low numbers of A-to-I edits (range of 73–194), and Target-AID induced even lower numbers of C-to-U edits (range of 6–11) (Fig. 2a, Extended Data Fig. 7b, and Supplementary Table 5). Cells expressing SPACE showed very few C-to-U edits (range of 0–4) and only small numbers of A-to-I edits (range of 4–37) edits (Fig. 2a, Extended Data Fig. 7b, and Supplementary Table 5). (The generally lower numbers of RNA edits we observed in our current experiments relative to previously published studies6,7 are due to reduced sequencing depth we used here (~14–18 million reads/sample) compared with our earlier work (˜80–120 million reads/sample) (Online Methods).) Based on these results, we conclude that SPACE retains the reduced RNA-editing activities observed with miniABEmax-V82G and Target-AID, inducing very low numbers of unwanted RNA edits throughout the transcriptome.
Figure 2. RNA and DNA off-target editing, potential amino acid modifications, and number of genes with potential transcription factor binding sites induced by SPACE.
a, Jitter plots show transcriptomic A-to-I (pink) and C-to-U (blue) mutations detected in RNA-seq experiments from HEK293T cells in which ABEmax, miniABEmax-V82G, Target-AID, or SPACE were co-expressed with either a HEK site 2 or RNF2 site 1 gRNA. GFP control cells express no gRNA. Data are shown from three independent replicates. n= number of combined adenines and cytosines modified. b, Heat maps showing A-to-G (pink) and C-to-T (blue) DNA off-target editing frequencies of nCas9 (Control), miniABEmax-V82G, Target-AID, or SPACE co-expressed in HEK293T cells with gRNAs targeted to HEK sites 2–4, EMX1 site 1, or FANCF site 1 (n= 4 independent replicates). The respective off-target editing windows for each site are shown. Location in the protospacer is indicated at the bottom with 1 being the most PAM-distal position. c, Circos plot showing amino acid changes that can be induced by SPACE (gray), including amino acid changes in specific codon context, that are uniquely enabled (blue). d, Bar plots showing computationally determined number of genes (y-axis) that could be targeted by SPACE to install 1–10 (or more) transcription factor (TF) binding sites (x-axis) in proximity of the transcription start site of coding genes in the human genome. Sites were filtered to contain a preferential SPACE editing window (C3-C4-A5 with 1 being the most PAM-distal position) and a canonical NGG PAM (Online Methods).
We also assessed Cas9/gRNA-dependent DNA off-target activities of SPACE by using targeted amplicon sequencing to quantify editing frequencies at 23 previously defined Cas9 off-target sites for five gRNAs (targeting HEK sites 2, 3, and 4, EMX1 site 1, and FANCF site 1)8. We directly compared SPACE editing frequencies at these 23 off-target sites to those observed with expression of miniABEmax-V82G, Target-AID, or a Cas9-D10A nickase (as a negative control). For 17 of these 23 off-target sites, editing efficiencies observed with SPACE were comparable or lower relative to those observed with miniABEmax-V82G or Target-AID (Fig. 2b and Supplementary Table 6). This was also true for the remaining six off-target sites with the exception of the adenines at positions 6 and 8 for HEK site 3 gRNA off-target site 1, the adenines at position 4 for HEK site 4 gRNA off-target sites 1 and 4, the cytosine at position 6 for EMX1 site 1 gRNA off-target site 2, the cytosine at position 8 for FANCF site 1 gRNA off-target site 1, and the adenine at position 4 for FANCF site 1 gRNA off-target site 4 (Fig. 2b and Supplementary Table 6). We also confirmed that on-target DNA editing was present with the five different gRNAs and SPACE, miniABEmax-V82G, or Target-AID co-expression (Extended Data Fig. 7c and Supplementary Table 6). This initial assessment suggests that SPACE does not induce dramatically different edit frequencies at known Cas9-dependent DNA off-target sites, but more comprehensive studies will be needed to fully define the genome-wide Cas9-dependent and gRNA-independent DNA off-target profiles of SPACE.
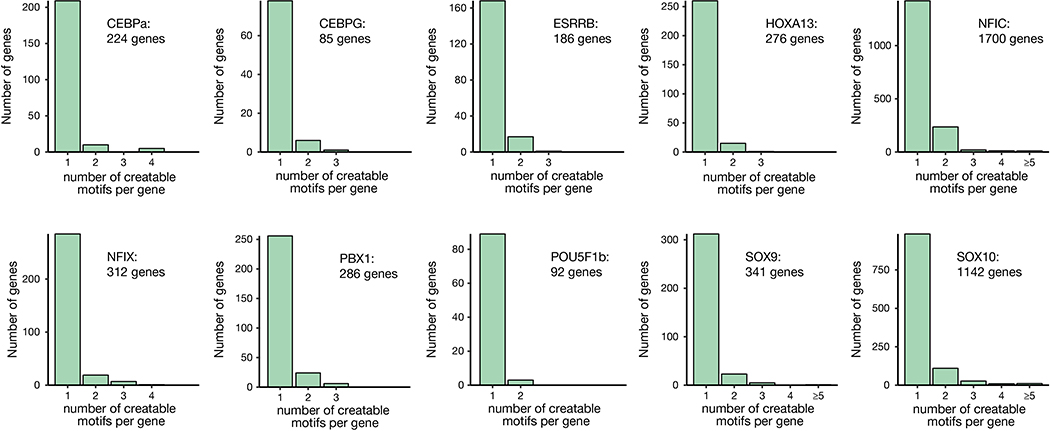
Our results demonstrate that SPACE enables the efficient and concurrent introduction of A-to-G and C-to-T edits, thereby expanding the range of targeted DNA edits that can be created with base editor technologies. This expanded editing capability will be broadly useful for a number of different research applications. For example, SPACE adds 60 additional codon changes (resulting in 18 amino acid substitutions) that cannot be created with existing single-action CBEs and ABEs (Extended Data Fig. 7d, Fig. 2c and Supplementary Table 7). In addition, SPACE could be useful for creating or reverting multi-nucleotide variants (MNVs), a newly emerging category of sequence variants associated with disease9,10 (also see: https://www.biorxiv.org/content/10.1101/573378v1) (Supplementary Table 8). Notably, among MNVs, TG-to-CA and CA-to-TG (both inducible by SPACE) are the most frequent consecutively arising adjacent dinucleotide MNVs9. Furthermore, the greater combinatorial diversity of mutations that result with SPACE as compared with single-deaminase base editors could make it attractive for molecular recording systems (e.g., lineage tracing)11,12 as well as for saturation mutagenesis screens, directed evolution, and protein engineering13,14. Finally, we envision that the concurrent editing of cytosines and adenines may enable the programmable installation of specific CA/TG-rich transcription factor (TF) binding sites. Our computational analysis indicates that thousands of genes harbor one or more SPACE-targetable CCA motifs in protospacer positions 3,4, and 5 with respect to an NGG-PAM, within −500 bp to 0 bp distance relative to the transcription start site, that can be targeted to create TF binding sites (Fig. 2d and Extended Data Fig. 8). Further protein engineering efforts may lead to expanded editing windows and higher editing efficiencies for future iterations of SPACE. While this work was being reviewed, another similar dual-deaminase architecture was reported by Gao and colleagues15. In summary, our development of SPACE should further expand the scope of research applications enabled by base editor technology.
Online Methods:
Plasmid cloning
All constructs (reported in Supplementary Table 2) were cloned into the CMV backbone (all ABE and SPACE constructs) from ABEmax-P2A-EGFP-NLS (AgeI/NotI digest; Addgene #112101) or into the CAG backbone (Target-AID only) from SQT817 (AgeI/NotI/EcoRV digest; Addgene #53373). All constructs with P2A-EGFP were cloned using either P2A-EGFP-NLS from ABEmax-P2A-EGFP-NLS or P2A-EGFP from BPK4335 (BE3-P2A-EGFP; Target-AID only) serving as the template. SPACE was cloned using Gibson assembly with bpNLS-TadA7.10(V82G)-SpCas9(D10A)-bpNLS from miniABEmax-V82G (Addgene #131313), pmCDA1 from Target-AID (Addgene #131300), and dual UGIs from BE4max (Addgene #112093). As reported in our previous study6, the pmCDA1 used in Target-AID contains an R187W single residue modification as compared to the reference sequence of pmCDA1 from NCBI (ABO15149.1). The codon usage for SpCas9-D10A in Target-AID differs from the codon usage of SpCas9-D10A in ABEmax, miniABEmax-V82G and SPACE (which share the same codon usage). The nCas9 (ABEmax without TadA domains; Addgene #123616) and GFP-only (enhanced GFP) constructs were used as negative controls. All guide RNA plasmids were cloned by ligation into the pUC19-based entry vector BPK1520 (BsmbI digest; Addgene #65777). All plasmids were midi or maxi prepped with the Qiagen Midi/Maxi Plus kits.
Cell culture
HEK293T cells (CRL-3216) were purchased from and STR-authenticated by ATCC. HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) with 10% (v/v) fetal bovine serum (FBS, Gibco) and 1% (v/v) penicillin-streptomycin (Gibco), passaged every 3–4 days, and tested for mycoplasma with MycoAlert PLUS (Lonza) every 4 weeks. HEK293T cells were used until passage 20 for all experiments.
Transfections
For DNA on-target experiments with 28 gRNAs (Fig. 1b), 1.25×104 HEK293T cells were seeded into 96-well Flat Bottom cell culture plates (Corning), transfected 24 h post-seeding with 30 ng base editor or control, 10 ng gRNA, and 0.3 μL TransIT-X2 (Mirus), and harvested 72 h after transfection to obtain genomic DNA (gDNA). For DNA off-target experiments (Fig. 2b, Extended Data Fig. 7c), 6.25×104 HEK293T cells were seeded into 24-well cell culture plates (Corning), transfected 24 h post seeding with 150 ng base editor or control, 50 ng gRNA, and 1.5 μL TransIT-X2, and harvested 72 h after transfection to obtain gDNA. For RNA off-target experiments (Fig. 2a, Extended Data Fig. 7a), 6.5×106 HEK293T cells were seeded into 150 mm cell culture dishes (Corning), transfected 24 h post-seeding with 37.5 μg base editor or control, 12.5 μg gRNA, and 150 μL TransIT-293 (Mirus), and sorted 36–40 h after transfection. For co-expression of miniABEmax-V82G and Target-AID (ABE & CBE mix) vs SPACE experiments (Extended Data Fig. 5), 1.25×104 HEK293T cells were seeded into 96-well cell culture plates, transfected 24 h post-seeding with 15 ng miniABEmax-V82G and 15 ng Target-AID for ABE & CBE mix, and 30 ng for both SPACE and the nCas9 control, 10 ng gRNA, and 0.3 μL TransIT-X2, and harvested 72 h after transfection to obtain gDNA. We have also conducted this experiment with twice the concentration of base editors and gRNAs, where cells were transfected with 30 ng miniABEmax-V82G and 30 ng Target-AID for ABE & CBE mix, and 60 ng for SPACE and the control, 20 ng gRNA, and 0.3 μL TransIT-X2; however, no increase of editing efficiencies was observed (data not shown).
Fluorescence-activated cell sorting (FACS)
As previously described7, after washing with Phosphate Buffer Saline (PBS, Corning), HEK293T cells were trypsinized and resuspended to 1×107 cells per ml in PBS supplemented with 10% (v/v) FBS and filtered through 35 μm cell strainer caps (Corning) to prepare for sorting 36–40 h post-transfection. After gating for single live cells (Supplementary Note), all GFP-positive cells were sorted into pre-chilled FBS with a FACSAria II (BD Biosciences) using FACSDiva version 6.1.3 (BD Biosciences).
DNA and RNA extraction
For DNA on-target experiments in 96-well plates, 72 h post-transfection, cells were washed with PBS, lysed with freshly prepared 43.5μL DNA lysis buffer (50 mM Tris HCl pH 8.0, 100 mM NaCl, 5 mM EDTA, 0.05% SDS), 5.25 μL Proteinase K (NEB), and 1.25 μL 1M DTT (Sigma). For DNA off-target experiments in 24-well plates, cells were lysed in 174 μL DNA lysis buffer, 21 μL Proteinase K, and 5 μL 1M DTT. For RNA off-target experiments, GFP sorted cells were split 20 % for DNA and 80 % for RNA extraction. Cells were centrifuged (200g, 8 min) and lysed as above for DNA or with 350 μL RNA lysis buffer LBP (Macherey-Nagel) for RNA. DNA lysates were incubated at 55°C on a plate shaker overnight, then gDNA was extracted with 2x paramagnetic beads (as previously described7), washed 3 times with 70% EtOH, and eluted in 30–80 μL 0.1X EB buffer (Qiagen). RNA lysates were extracted with the NucleoSpin RNA Plus kit (Macherey-Nagel) following the manufacturer’s instructions.
Library preparation for DNA targeted amplicon sequencing
Next-generation sequencing (NGS) of DNA targeted amplicons was performed as previously described7. In summary, the first PCR amplified the genomic site of interest with primers containing Illumina adapter sequences (Supplementary Table 9), and the second PCR added barcodes with primers containing unique pairs of p5/p7 Illumina barcodes that are analogous to TruSeq CD indexes. The libraries were pooled based on Pico Green (Promega) measurements on a BioTek Synergy HT plate reader, and the final pool was sequenced paired-end (PE) 2×150 on a MiSeq machine using 300-cycle MiSeq Reagent Kit v2 (Illumina). Demultiplexed FASTQs were downloaded from Basespace (illumina) and analyzed using the batch version of CRISPResso216.
Library preparation for RNA sequencing
Transcriptome sequencing of RNA (RNA-seq) was performed as previously described7. In summary, RNA libraries were prepared with the TruSeq Stranded Total RNA Library Prep Gold kit (Illumina), SuperScript III (Invitrogen) for first-strand synthesis, and IDT for Illumina TruSeq RNA unique dual indexes (96 indexes). The libraries were pooled based on Qubit measurements (Thermo Fisher), and the final pool was sequenced PE 2×76 with a NextSeq 500 machine using a NextSeq 500/550 High Output Kit v2 (150 cycles). In contrast to previous RNA-seq experiments with NovaSeq and HiSeq machines (80–120M reads/sample), we used a NextSeq machine for RNA-seq in this study resulting in reduced sequencing depth (14–18M reads/sample).
Targeted amplicon sequencing analysis
Amplicon sequencing data were analyzed with CRISPResso2 2.0.3016. Heat maps display an editing window that includes the edited As or Cs, and a gray background for editing efficiencies less than 2%. This background cut-off was relaxed for the heat maps showing DNA off-target editing (Fig. 2b). The following samples from the ABE & CBE mix vs SPACE on-target assays with 28 gRNAs (Extended Data Fig. 5) have fewer than 1,000 reads and are reported in Supplementary Table 4 (tab “read depth”): Control (nCas9) Rep3–4 for ABE site 14; Control Rep2–4, Mix (ABE & CBE mix) Rep2–4, and SPACE Rep4 for RNF2 site 1. The following samples from the DNA off-target assays (Fig. 2b) have fewer than 10,000 reads and are reported in Supplementary Table 6 (tab “read depth”): Target-AID Rep2–4 and SPACE Rep2–4 for EMX1 site 1 off-target 1; Target-AID Rep3–4 and SPACE Rep2–4 for EMX1 site 1 off-target 2; Target-AID Rep2–3 and SPACE Rep3 for FANCF site 1 off-target 1; Target-AID Rep2–4 and SPACE Rep2–4 for FANCF site 1 off-target 3. The percentage of alleles that contain an insertion or deletion was estimated with CRISPResso2, using the “CRISPRessoBatch_quantification_of_editing_frequency.txt” output table. For each experiment, indels were quantified across the protospacer by setting parameters -wc 10 and -w 10, and calculating Insertions + Deletions - Ìnsertions and Deletions`/Reads_aligned from the table columns.
RNA sequencing analysis
All computational analyses were performed as previously detailed7. Briefly, NextSeq reads were aligned to the human reference genome hg38 using STAR17, followed by removal of PCR duplicates and base recalibration. Mutations were then called using GATK HaplotypeCaller18,19. Mutations thus identified were then filtered to include only those for which coverage in control experiments was greater than the 90th percentile of read coverage across all mutations in the overexpression experiment and where 99% of control reads covering the edited base contained the reference allele. Furthermore, we filtered out those edits with fewer than 10 reads or 0% alternate allele frequencies. A-to-G/C-to-T edits include A-to-G/C-to-T edits identified on the positive strand as well as T-to-C/G-to-A edits on the negative strand.
Analysis of potential targets for the correction or generation of multi-nucleotide variants (MNVs) by SPACE
A list of multi-nucleotide variants (MNVs) was obtained from Wang et. al. “Landscape of multi-nucleotide variants in 125,748 human exomes and 15,708 genomes” (pre-print at http://dx.doi.org/10.1101/573378; data file at https://storage.googleapis.com/gnomad-public/release/2.1/mnv/gnomad_mnv_coding.tsv). These were filtered to detect disease correcting or disease generating modifications enabled by SPACE, which are defined as annotated MNVs with a nearby PAM. Disease correcting conversions are defined as having targetable Cs and As in the ALT position with matching Ts and Gs in the REF position; whereas disease generating conversions are defined as the reverse scenario, with targetable Cs and As in the REF position with matching Ts and Gs in the ALT position. Patterns for selected disease correcting MNV codons include “GNT>ANC”, “GTN>ACN”, “NGT>NAC”, “NTG>NCA”, “TGN>CAN”, and “TNG>CNA”; whereas patterns for disease generating include “ACN>GTN”, “ANC>GNT”, “CAN>TGN”, “CNA>TNG”, “NAC>NGT”, and “NCA>NTG”. PAMs considered include NGG, NGA, and NG.
Analysis of potential SPACE-encodable transcription factor binding sites
Annotated transcription start sites from hg38 refseq genes were obtained and filtered to exclude micro-RNAs, small NF90 associated RNAs (SNARs), long non-coding RNAs, small nucleolar RNAs, and anti-sense transcripts. These RNAs were filtered in part due to redundant annotations at the same transcription start sites (TSS) and to focus on protein-coding genes. Next, the remaining TSSes were padded to include the region −500 bp to 0 bp relative to the start site. From this 500 bp per-gene window, we found all matches of the “NNCCANNNNNNNNNNNNNNNNGG” motif on either strand that contain a preferential dual-editing window for SPACE and a canonical SpCas9-PAM (NGG). In total, this defined 53,750 protospacers (45,383 unique). To assess the potential for TF motif creation, we used the reference sequence to create “SPACE-edited” sequences for each of the protospacers by modifying the CCA to TTG. Using a set of 386 transcription motifs from the JASPAR2016 motif list20 we determined which motifs could be created with the SPACE modification for each transcription factor. Motif matching was performed using the motifmatchr package using default parameters as part of the chromVAR suite of tools21. Created motifs were those that did not occur in the reference sequence but were matches in the SPACE-edited sequence.
Circos plotting
Amino-acid and codon modification plots were constructed using Circos22.
Statistics & data reporting
Sample sizes were determined based on the published work of others in the field who perform similar experiments and achieve reproducible results. Investigators were not blinded to experimental conditions or analysis of experimental data. We did not use any specific statistical tests. In box plots, the box spans the interquartile range (IQR) (first to third quartiles) whereas the horizontal line shows the median (second quartile) and whiskers extend to ± 1.5 × IQR.
Data availability
Plasmids encoding SPACE will be made available on Addgene. All RNA-seq NGS data generated for this study have been deposited in the GEO data repository (series GSE137411), accessible via: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137411. All targeted amplicon sequencing data (DNA on- and off-target editing) have been deposited at the Sequence Read Archive (SRA), accessible via: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA609075.
Code availability
The authors will make all previously unreported custom computer code used in this work available upon request.
Life sciences reporting summary
Further information on statistics, methods, and study design can be found in the Nature Research Reporting Summary that is attached to this article.
Extended Data
Extended Data Fig. 1: Architectures of miniABEmax-V82G, Target-AID, and SPACE, and A-to-G or C-to-T editing distributions of miniABEmax-V82G or Target-AID.
a, Schematic illustration of miniABEmax-V82G, Target-AID, and SPACE architectures. Orange boxes = bipartite NLS for miniABEmax-V82G and SPACE or NLS for Target-AID; TadA* = mutant TadA 7.10 with V82G mutation, light grey box = SH3–3xFLAG for Target-AID, CDA1 = pmCDA1 with R187W mutation, and yellow boxes = UGIs. b, c, Box and dot plots indicating the distributions of A-to-G (pink, b) and C-to-T (blue, c) edits across 28 pooled genomic sites with miniABEmax-V82G (b) and Target-AID (c) including the entire protospacer. In box plots, the box spans the interquartile range (IQR) (first to third quartiles), the horizontal line shows the median (second quartile), and the whiskers extend to ± 1.5 × IQR. Single dots represent individual replicates. Graph was made using the same data shown in Fig. 1b (n=4).
Extended Data Fig. 2: On-target C-to-R (A/G) and A-to-Y (C/T) editing of miniABEmax-V82G, Target-AID, and SPACE.
Box and dot plots showing on-target DNA C-to-R (A/G) and A-to-Y (C/T) editing frequencies of miniABEmax-V82G (pink), Target-AID (blue), and SPACE (green) with 28 gRNAs (n=4). Box, horizontal line, and whisker are defined as in Fig. 1c. Single dots represent all Cs or As across the entire protospacer for all four replicates at each genomic site. Data from the same experiment as shown in Fig. 1b. Please note that box plots are mostly contained within the horizontal line (median, second quartile) close to or at the value “0“.
Extended Data Fig. 3: On-target indel frequencies induced by miniABEmax-V82G, Target-AID, and SPACE.
Dot plots showing on-target DNA indel frequencies induced by nCas9-D10A (Control, black), miniABEmax-V82G (pink), Target-AID (blue), and SPACE (green) with 28 gRNAs (n=4). Single dots represent individual replicates. Data from the same experiment as shown in Fig. 1b.
Extended Data Fig. 4: Allele frequency tables of DNA on-target editing by SPACE.
Composition of alleles with frequencies of 1% or higher that result from SPACE editing with 27 gRNAs (HEK site 2 data shown in Fig. 1d). Data are taken from the first replicate obtained for each gRNA on-target experiment shown in Fig. 1b. Numbering indicates the position in the protospacer with 1 being the most PAM-distal location.
Extended Data Fig. 5: On-target DNA editing of SPACE compared to co-expression of miniABEmax-V82G and Target-AID (ABE & CBE mix) at 28 genomic sites.
Heat maps showing on-target DNA A-to-G (pink) and C-to-T (blue) editing frequencies induced by nCas9 (Control), co-expression of miniABEmax-V82G and Target-AID (ABE & CBE mix), or SPACE with 28 gRNAs (n=4 independent replicates). Editing windows shown represent the edited adenines and cytosines, not the entirety of the protospacer. Numbering at the bottom represents the position of the respective base in the protospacer sequence with 1 being the most PAM-distal location.
Extended Data Fig. 6: On-target C-to-T, A-to-G and dual editing, and indel frequencies induced by co-expression of miniABEmax-V82G and Target-AID compared with SPACE.
a, Bar and dot plots showing mean sum of allele frequencies of all edited alleles with A-to-G only, C-to-T only, and concurrent A-to-G and C-to-T editing resulting from co-expression of miniABEmax-V82G and Target-AID (ABE & CBE mix, grey) or SPACE (green) with 28 gRNAs (n=4). Single dots represent individual replicates. Error bars represent the standard deviation (SD). Data are from the same experiment as shown in Extended Data Fig. 5. b, Dot plots showing on-target DNA indel frequencies of nCas9 (Control, black), co-expression of miniABEmax-V82G and Target-AID (ABE & CBE mix, grey) and SPACE (green) with 28 gRNAs (n=4). Single dots represent individual replicates. Data are from the same experiment as shown in Extended Data Fig. 5.
Extended Data Fig. 7: Additional data and analysis from DNA and RNA off-target experiments and SPACE-inducible amino acid modifications.
a, Heat maps showing the on-target DNA A-to-G (pink) and C-to-T (blue) editing frequencies of nCas9 (Control), ABEmax, miniABEmax-V82G, Target-AID, or SPACE with HEK site 2 and RNF2 site 1 gRNAs (n=3 independent replicates) for the RNA-seq experiments shown in Fig. 2a. Editing windows shown represent the most highly edited adenines and cytosines, not the entire protospacer. Numbering at the bottom represents the position of the respective base in the protospacer sequence with 1 being the most PAM-distal location. b, Histograms showing the total number of RNA A-to-I or C-to-U edits observed (y-axis) with different editing efficiencies (x-axis) for ABEmax, miniABEmax-V82G, Target-AID, or SPACE, each tested with the HEK site 2 and RNF2 site 1 gRNAs. n = number of modified adenines and cytosines. Experiments were performed in triplicate (data are from the same experiments shown in Fig. 2a). c, Heat maps showing on-target DNA A-to-G (pink) and C-to-T (blue) editing efficiencies of nCas9 (Control), miniABEmax-V82G, Target-AID, or SPACE with HEK sites 2–4, EMX1 site1, and FANCF site 1 gRNAs (n=4 independent replicates) for DNA off-target experiments shown in Fig. 2b. Editing windows shown represent the most highly edited adenines and cytosines, not the entire protospacer. Numbering at the bottom represents the position of the respective base in the protospacer sequence with 1 being the most PAM-distal location. d, Circos plot showing 60 unique codon changes (with respect to the start codon) that can be induced by dual editing of adenines and cytosines by SPACE (grey), 18 of which (blue) lead to unique SPACE-inducible amino acid changes with respect to the original codon (also see Fig. 2c and Supplementary Table 7).
Extended Data Fig. 8: Potential transcription factor binding sites that can be created with SPACE.
Bar plot showing computationally determined number of genes (y-axis) that could be targeted by SPACE to install 1–5 (or more) transcription factor binding sites (x-axis) in proximity of the transcription start site of coding genes in the human genome for 10 transcription factors. Sites were filtered to contain a preferential SPACE editing window (C3-C4-A5) and a canonical NGG-PAM (Online Methods).
Supplementary Material
Acknowledgements
Support for this work was provided by the National Institutes of Health (RM1 HG009490 to J.K.J. and R35 GM118158 to J.K.J. and M.J.A.). J.K.J. is additionally supported by the Desmond and Ann Heathwood MGH Research Scholar Award and the Robert B. Colvin, M.D. Endowed Chair in Pathology. J.G. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer 416375182. L.M.L. was supported by a Boehringer Ingelheim Fonds (BIF) MD fellowship. We thank M.K. Clement for technical advice, K. Petri, and P.K. Cabeceiras for discussions and technical advice, and L. Paul-Pottenplackel for assistance with editing the manuscript.
Footnotes
Competing Financial Interests Statement
J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Excelsior Genomics, Pairwise Plants, Poseida Therapeutics, Transposagen Biopharmaceuticals, and Verve Therapeutics (f/k/a Endcadia). M.J.A. has financial interests in Excelsior Genomics. J.K.J.’s and M.J.A.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. is a member of the Board of Directors of the American Society of Gene and Cell Therapy. J.G., R.Z., and J.K.J. are co-inventors on a patent application that has been filed by Partners Healthcare/Massachusetts General Hospital on engineered programmable multi-deaminase base editor architectures to enable concurrent editing of distinct bases on DNA.
References
- 1.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424, doi: 10.1038/nature17946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471, doi: 10.1038/nature24644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komor AC et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774, doi: 10.1126/sciadv.aao4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rees HA & Liu DR Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770–788, doi: 10.1038/s41576-018-0059-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, doi: 10.1126/science.aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Grunewald J et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat Biotechnol 37, 1041–1048, doi: 10.1038/s41587-019-0236-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grunewald J et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437, doi: 10.1038/s41586-019-1161-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197, doi: 10.1038/nbt.3117nbt.3117 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplanis J et al. Exome-wide assessment of the functional impact and pathogenicity of multinucleotide mutations. Genome Res 29, 1047–1056, doi: 10.1101/gr.239756.118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besenbacher S et al. Multi-nucleotide de novo Mutations in Humans. PLoS Genet 12, e1006315, doi: 10.1371/journal.pgen.1006315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907, doi: 10.1126/science.aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan MM et al. Molecular recording of mammalian embryogenesis. Nature 570, 77–82, doi: 10.1038/s41586-019-1184-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canver MC et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature, doi: 10.1038/nature15521 nature15521 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess GT et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 13, 1036–1042, doi: 10.1038/nmeth.4038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol, doi: 10.1038/s41587-019-0393-7 (2020). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 16.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226, doi: 10.1038/s41587-019-0032-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303, doi: 10.1101/gr.107524.110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498, doi: 10.1038/ng.806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathelier A et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44, D110–115, doi: 10.1093/nar/gkv1176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schep AN, Wu B, Buenrostro JD & Greenleaf WJ chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14, 975–978, doi: 10.1038/nmeth.4401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzywinski M et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645, doi: 10.1101/gr.092759.109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids encoding SPACE will be made available on Addgene. All RNA-seq NGS data generated for this study have been deposited in the GEO data repository (series GSE137411), accessible via: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137411. All targeted amplicon sequencing data (DNA on- and off-target editing) have been deposited at the Sequence Read Archive (SRA), accessible via: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA609075.