Abstract
Background
This work was aimed at exploring the regulatory network of non-coding RNA (ncRNA) especially circular RNA (circRNA) and microRNA (miRNA), in the sensitivity of non-small cell lung cancer (NSCLC) cells to low linear energy transfer (LET) X-ray and high-LET carbon ion irradiations.
Methods
The radioresistant NSCLC cell line A549-R11 was obtained from its parental cell line A549 through irradiation with X-rays of 2.0 Gy per fraction for 30 times. The sensitivities of A549, A549-R11 and H1299 cells exposed to X-rays and carbon ions were verified using the colony formation assay. A comprehensive circRNA-miRNA-mRNA network was constructed through the sequencing data in parental A549, acquired radioresistant A549-R11 and intrinsic radioresistant H1299 cells, and the network was further optimized according to the prognostic results from the TCGA and GEO databases.
Results
Based on high-throughput sequencing of circRNAs, we found that 40 circRNAs were up-regulated while 184 circRNAs were down-regulated in the intersection of the sets of A549-R11 and H1299 cells. Subsequently, a circRNA- miRNA-mRNA network, including 14 interactive pairs and 8 circRNAs, 4 overall survival-associated miRNAs, and 4 mRNAs, was constructed through the high-throughput data screening and bioinformatics methods.
Conclusions
Our results provide a complete understanding to the regulatory mechanism of the sensitivities to low-LET X-ray and high-LET carbon ion irradiations, and might be helpful to screen potential biomarkers for predicting the Carbon-ion radiotherapy (CIRT) and X-ray radiotherapy responses in NSCLC.
Keywords: Carbon-ion radiotherapy (CIRT), radiosensitivity, circRNA-miRNA-mRNA network, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is the most frequent primary malignance and the leading cause of cancer-related death worldwide. It has been reported that non-small cell lung cancer (NSCLC) accounts for approximately 80% of lung cancers (1). Radiotherapy as a universal treatment modality, however, produces a 5-year survival rate of merely 10–30% because of poor control of the primary tumor (2,3). Radioresistance has been considered as one of the most important reasons for treatment failure or local tumor recurrence (4). Carbon-ion radiotherapy (CIRT) is expected to be more effective even for photon-resistant tumors because of its physical and biological advantages (5,6). For early NSCLC patients, CIRT has demonstrated that local control and overall survival rates at 5 years are 80–90% and 40–50%, respectively (7). However, only a small proportion of the patients can benefit from CIRT because of high building and running costs of the accelerator facility. Therefore, it is absolutely necessary to screen potential biomarkers for predicting the CIRT and X-ray radiotherapy responses in locally-advanced NSCLC cases.
Over the past decade, with a great advance in the high-throughput sequencing, many non-coding RNAs (ncRNAs) such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) have been identified and found to play essentially regulatory role in the carcinogenesis (8-11). CircRNA, unlike the well-known linear RNA, forms a ring structure through the linkage of the 3′ and 5′ ends with a covalent bond (12). Recently, increasing studies have revealed that circRNAs play regulatory roles mainly at the transcriptional and post-transcriptional levels, including acting as miRNA sponges (13), interaction with RNA-binding proteins (RBPs) (14), and the modulation of gene transcription (15). The recently uncovered circRNA has protein-coding potential in eukaryotes (16). A few circRNAs, such as circRNA 100876 (17), hsa_circ_0043256 (18), circRNA HIPK3 (19), have been reported to be involved in the regulation of NSCLC progression.
In present study, we developed a lung carcinoma cell line (A549-R11) by irradiating human lung carcinoma A549 cells with fractionated irradiation (2 Gy × 30 fractions), which is resistant to low- linear energy transfer (LET) X-rays. Moreover, these two cell lines (A549 and A549-R11) showed a similar sensitivity to high-LET carbon ions. To discover the underlying molecular regulation mechanisms of circRNAs, miRNAs, and mRNAs in the radiosensitivity, the next-generation sequencing (NGS) technology was used to detect the differentially expressed circRNAs among the radioresistant NSCLC cell line A549-R11, its parental cell line A549 and an intrinsic radioresistant NSCLC cell line H1299. The differential expressions of circRNAs might be useful predictors for the differential responses of NSCLC to low-LET X-rays and high-LET carbon ions.
Methods
Establishment and identification of radioresistant cell line
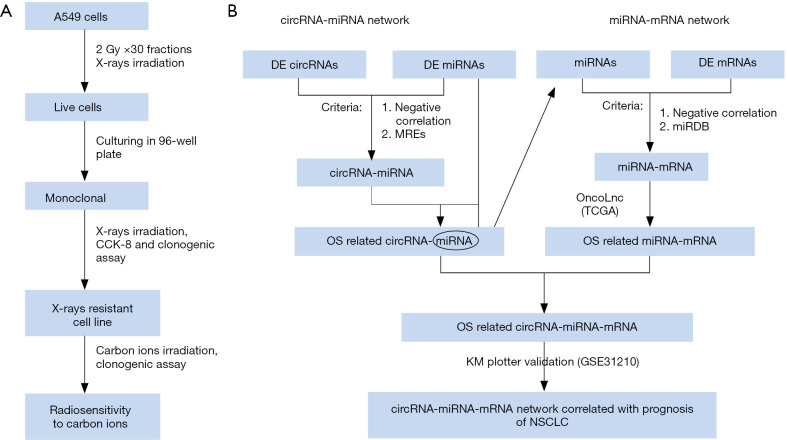
Human NSCLC cell lines A549 and H1299 were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 Medium (Thermo Fisher Scientific Inc.) containing 10% heat-inactivated fetal bovine serum (FBS; Bailing Bio, Lanzhou, China) at 37 °C in a humidified 5% CO2 atmosphere. The radioresistant NSCLC cell line A549-R11 was obtained from its parental cell line A549 through irradiation with X-rays of 2.0 Gy per fraction for 30 times (the process was depicted in Figure 1A).
Figure 1.
The flow charts of the establishment of the radioresistant NSCLC cell line and the construction of circRNA-miRNA-mRNA network. (A) The screening procedure of the radioresistant cell line A549-R11. (B) The flowchart for constructing the circRNA-miRNA-mRNA network. DE, differentially expressed; MREs, miRNA response elements; OS, overall survival.
The sensitivities of A549, A549-R11 and H1299 cells exposed to X-rays and carbon ions were verified using the colony formation assay (20).
RNA extraction and identification of radioresistance-associated circRNAs, miRNAs and mRNAs
Three samples were obtained from each of the three cell cultural groups (A549, A549-R11 and H1299 cells). Total RNA was isolated using the TRIzol (Invitrogen, USA) according to the manufacturer’s protocol. All the sequencing procedures and analyses were performed in Ribobio (Guangzhou, China). Differentially expressed circRNAs, miRNAs and mRNAs between two groups (A549 cells vs. A549-R11 cells and A549 cells vs. H1299 cells) with statistical significance were defined as |fold change| ≥2 and P<0.05 estimated with the Student t-test.
qRT-PCR validation
qRT-PCR was used to confirm the expression of differentially expressed circRNAs obtained from the sequencing data. Total RNA was extracted and assays were done in triplicate using the primers indicated in Table S1.
Kaplan-Meier analysis
To investigate the impact of the differentially expressed miRNAs and mRNAs on prognostic overall survival (OS) of patients with lung cancer, Kaplan-Meier survival analysis was performed using OncoLnc (http://www.oncolnc.org/), which is a database for interactively exploring survival correlations and contains survival data for more than 8,000 patients from 21 cancer studies by The Cancer Genome Atlas (TCGA) (21). The OSs of patients based on the expression level of the miRNAs and mRNAs in the A549-R11 set were compared. Briefly, patients were classified into the high or low expression group based on the expression level of each differentially expressed miRNA or mRNA, and then the OS was analyzed.
To further confirm the above-mentioned results, another tool, Kaplan-Meier plotter was employed (22). The Kaplan-Meier plotter (http://kmplot.com/analysis/) is capable to assess the effect of 54,000 genes on survival in 21 cancer types. In particular, we focused on a cohort of patients with NSCLC (GSE31210), which includes 246 patients, to evaluate the prognostic value of the differentially expressed mRNAs.
Prediction of circRNA-miRNA-mRNA networks
CircRNA, serving as a miRNA sponge to bind miRNA competitively, indirectly regulates target gene, which is known as competing endogenous RNA (ceRNA) (23). Based on this theory, circRNA-miRNA-mRNA networks were constructed. The interactions between miRNAs and circRNAs were predicted through miRNA response elements (MREs) in the miRanda, RNA hybrid, and TargetScan databases. Moreover, these circRNA-miRNA pairs were further screened using the OncoLnc toolkit according to the correlation between the differentially expressed miRNAs and OS of patients with lung adenocarcinoma (LUAD). Next, we used miRDB, an online database for miRNA target prediction and functional annotations (http://www.mirdb.org/), to predict target mRNAs. These interactions were similarly screened using the OncoLnc and Kaplan-Meier plotter toolkits. The flowchart of the circRNA-miRNA-mRNA interaction construction is depicted in Figure 1B and the network was visually displayed using the Cystoscope software V3.7.1 (San Diego, CA, USA).
Statistical analysis
Data are represented as the mean ± standard deviation (SD). Statistical analysis was conducted using the unpaired Student’s t-test. A difference was considered significant when P<0.05.
Results
Radiosensitivities of A549, A549-R11, and H1299 cells to low- and high-LET radiations
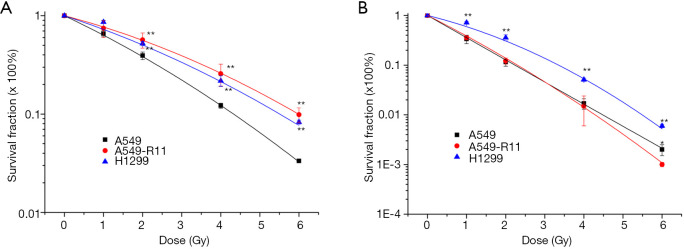
Radioresistance was identified by comparing the survival fraction of A549-R11 or H1299 with A549 cells. Shown in Figure 2A are the survival curves of the three cell lines, after exposure to X-rays. A549 cells represented the most significant sensitivity among the three cell lines, whereas A549-R11 and H1299 cells showed obvious radioresistance. For carbon ion irradiation (Figure 2B), H1299 exhibited the most significant radioresistance, while A549 and A549-R11 cells displayed a similar cell survival curve. The survival fractions at 2Gy of the three cell lines exposed to X-rays and carbon ions are summarized in Table 1. A549-R11 and H1299 cells were more resistant to low-LET X-rays than A549 cells; however, A549-R11 cells possessed a similar sensitivity to high-LET carbon ions like their paternal A549 cells.
Figure 2.
Survival fractions of A549, A549-R11 and H1299 cells exposed to low-LET X-rays (A) and high-LET carbon ions (B) measured with the clonogenic survival assay. *, P<0.05; **, P<0.01 vs. A549 cells.
Table 1. Summary of survival fraction and RR at 2 Gy.
| Treatment | X-rays | RR | Carbon ions | RR |
|---|---|---|---|---|
| SF2 (A549) | 0.393 | – | 0.117 | |
| SF2 (A549-R11) | 0.568 | 1.445 | 0.122 | 1.043 |
| SF2 (H1299) | 0.523 | 1.331 | 0.358 | 3.060 |
The data calculated from the survival curves. RR, radiosensitivity ratios.
Overview of circRNAs profiles
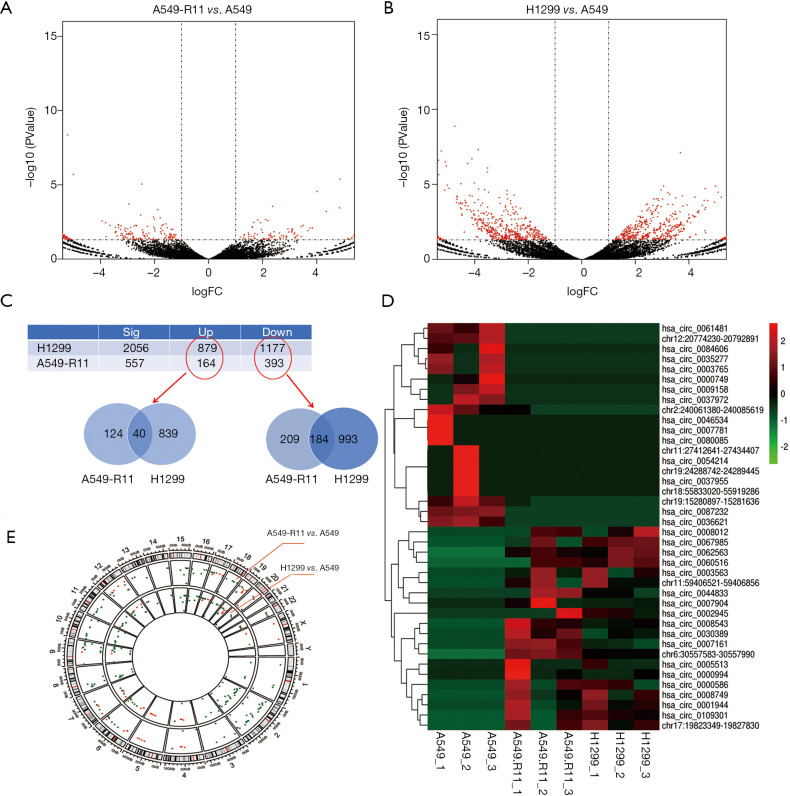
High-throughput next-generation RNA sequencing data were analyzed to explore the circRNA expression profile in radiosensitive A549, acquired radioresistant A549-R11 and intrinsic radioresistant H1299 cell lines. Using the CIRI2 and CIRCexplorer2 software, 28,082 distinct circRNA candidates containing at least one unique back- spliced read were found in the three cell lines. These circRNAs and their host genes are located in various genomic regions (Figure S1). The expression analysis of these transcripts revealed that a series of circRNAs were differentially expressed in the acquired radioresistant A549-R11 cells and intrinsic radioresistant H1299 cells compared with the radiosensitive A549 cells (Figure 3A,B). In the A549-R11 set, a total of 557 circRNAs were differentially expressed (|fold change| ≥2 and P<0.05), consisting of 164 up-regulated circRNAs and 393 down-regulated circRNAs in A549-R11 cells compared with A549 cells. In the H1299 set, the expression profiles of 2,056 circRNAs were different between H1299 and A549 cells, where 879 circRNAs were up-regulated while 1,177 circRNAs were down-regulated in H1299 cells. Then the different expressions were taken at the intersection of the sets of A549-R11 and H1299 (Figure 3C). We found that 40 were up-regulated and 184 were down-regulated in both sets, whereas 17 were inconsistent in two sets. The top twenty up- and down-regulated circRNAs ranked by fold change in an intersected set are shown in a hierarchical clustering map (Figure 3D). The distribution of the differentially expressed circRNAs on the chromosomes and the fold change are exhibited in Figure 3E.
Figure 3.
CircRNA expression profile. Volcano plots exhibit significantly dysregulated miRNAs in A549-R11 (A) and H1299 (B) sets, where the horizontal lines represent 2-fold up and down expressed circRNAs, and the vertical lines indicate P=0.05. (C) The intersection was taken in the different expressions of A549-R11 and H1299 sets. (D) Hierarchical clustering shows the top twenty up-regulated and down-regulated circRNAs. (E) The distribution of the differentially expressed circRNAs on the chromosomes and the fold change. The green and red dots represent the down- and up-regulated circRNAs, respectively. Center distance represents the fold change.
Validation of circRNA expressions
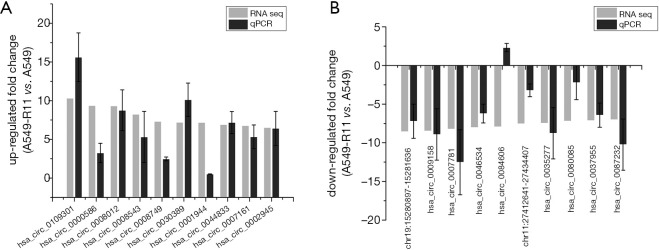
To verify the sequencing data, twenty differentially expressed circRNAs were selected, including the top 10 up-regulated circRNAs and the top 10 down-regulated circRNAs in the paired radioresistant A549-R11 and radiosensitive A549 cell lines. The results demonstrated that the expression levels of the circRNAs were consistent with the sequencing data, except hsa_circ_0084606, which was down-regulated in the sequencing data but up-regulated in the qPCR experiment (Figure 4A,B). These results suggest the high reliability of the sequencing data.
Figure 4.
Validation of the sequencing data. Twenty differentially expressed circRNAs including 10 up-regulated (A) and 10 down-regulated (B) were validated by qRT-PCR.
Expression profile of miRNAs and construction of OS-related circRNA-miRNA network
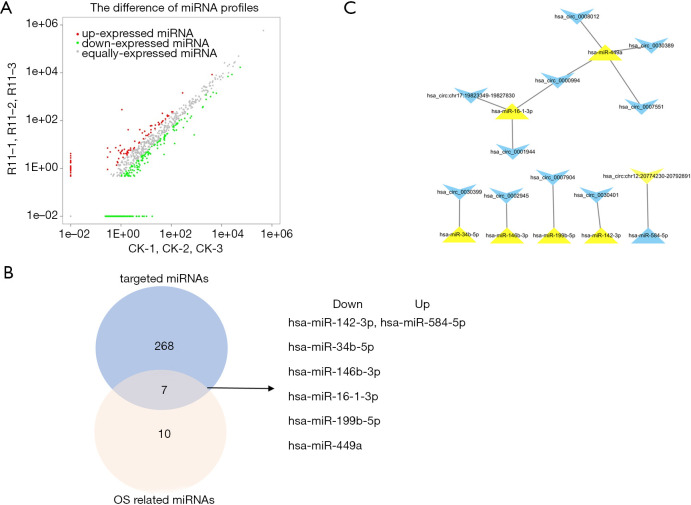
In our sequencing data, 399 miRNAs were identified to be differentially modulated. Among them, 314 miRNAs were down-regulated whereas 85 miRNAs were up-regulated (Figure 5A). The MiRanda, RNA hybrid, and TargetScan databases were used to predict the circRNA-miRNA pairs. We found that 81 of the above-mentioned total 224 intersected circRNAs had target relationships with 277 of the 399 differentially expressed miRNAs and formed 934 circRNA–miRNA pairs (Figure S2). However, these data were too large to further analysis. So, OS as a prognostic factor about differentially expressed miRNAs was introduced to screen these pairs. We found 17 miRNAs among the 399 differentially expressed miRNAs were significantly correlated with the survival of patients (Table S2). Figure 5B shows an intersection of the two miRNA groups (targeted miRNAs and OS related miRNAs). There were 7 miRNAs consisting of 6 down-regulated miRNAs and 1 up-regulated miRNA in this intersection. Their Kaplan-Meier survival analysis is shown in Figure S3. Finally, a circRNA-miRNA related OS network was constructed, including 11 circRNAs, 7 miRNAs, and 12 pairs (Figure 5C).
Figure 5.
Construction of the OS-related circRNA-miRNA networks. (A) A scatter plot shows the sequencing data distribution of miRNAs between A549-R11 and A549 cells. The red and green points show that the miRNAs were up-regulated and down-regulated, respectively. (B) The intersection was taken between the OS-related miRNAs and targeted-miRNAs in NSCLC. (C) The circRNA-miRNA interaction network associated with OS. Blue symbols represent up-regulated expressions, whereas yellow symbols represent down-regulated expression. Triangular and quadrilateral nodes represent miRNAs and circRNAs, respectively.
Differentially expressed mRNA profiles and construction of circRNA-miRNA- mRNA network correlated with the prognosis of NSCLC
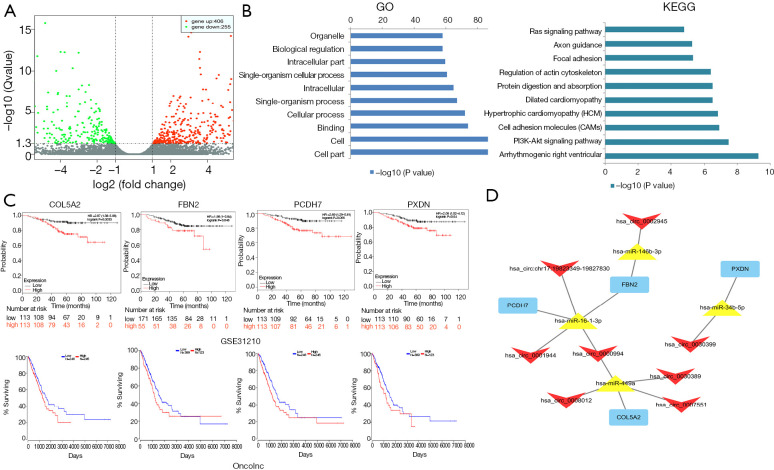
A total of 661 differentially expressed mRNAs were identified between A549 and A549-R11 cells, including 406 up-regulated and 255 down-regulated mRNAs (Figure 6A). The GO analysis revealed that, for these altered mRNAs, the top 3 enriched terms were cell part, cell and binging between A549 and A549-R11 cells. The KEGG analysis showed the top 10 pathways associated with the different mRNAs in the A549-R11 group (Figure 6B). Among the pathways, we identified the PI3K-Akt signaling pathway, cell adhesion molecules (CAMs) and the Ras signaling pathway, which have been reported to be related to tumorigenesis, progression, invasion and metastasis and drug resistance.
Figure 6.
A regulatory network of circRNA-miRNA-mRNA associated with prognosis. (A) A volcano plot exhibits significantly dysregulated mRNAs between A549-R11 and A549 groups. The red points show that the miRNAs were up-regulated more than two-fold. The green points show that the miRNAs were down-regulated more than two-fold. (B) Enrichment of the top 10 GO terms (left) and KEGG pathways (right) of differentially expressed mRNAs in A549-R11 and A549 groups. (C) The Kaplan-Meier analysis of NSCLC-specific overall survival of patients with tumors expressing different levels of mRNAs. (D) A circRNA-miRNA-mRNA network correlated with the prognosis of NSCLC. Quadrilateral, triangular and rectangle nodes represent circRNAs, miRNAs and mRNAs, respectively.
As the above-mentioned 7 prognostic signatures and circRNA-targeted miRNAs were employed to predict miRNA-mRNA interactions, we found that 32 of the 661 differentially expressed mRNAs could form 58 pairs with the miRNAs, 27 of which were up-regulated and 5 of which were down-regulated as shown in Table S3 and Figure S4. Likewise, prognostic survival analysis was conducted for the 32 mRNAs to examine whether the mRNAs expression significantly correlate to the survival of patients with NSCLC using the Oncolnc and Kaplan-Meier plotter datasets. However, only 4 mRNAs (COL5A2, FBN2, FCDH7 and PXDN) were found to be significantly associated with OS, both indicating worse prognosis in the high expression group (Figure 6C). Subsequently, a circRNA-miRNA-mRNA visualized regulatory network was constructed using the Cytoscape 3.7.1 (Figure 6D), containing 14 circRNA-miRNA-mRNA interactive pairs and 8 circRNAs, 4 OS-associated miRNAs, and 4 mRNAs.
Discussion
Radioresistance remains a significant clinical problem for radiotherapy with conventional radiation such as X-rays, because various tumors display widely varying radiosensitivities. To explore the radiation response in NSCLC, we made an isogenic model of radioresistance via chronic exposure to fractionated radiations, which has been previously reported (24-28). Compared with cell lines of the same origin but with obviously different radiosensitivities, the advantage of this type of model is that it avoids the influence of confusing factors such as genetic and inherent variation, allowing for the clarification of molecules that are involved in radioresistance. Our results also verified this point. The number of different circRNA expressions in the A549-R11 set accounted for merely 27% of those in the H1299 set. Additionally, this model mimics what occurs in the clinical setting, because standard radiotherapy consists of about 2 Gy once a day, 5 days a week, a period for 5–8 weeks (26). This produced a subclone cell line A549-R11, which exhibited enhanced clonogenic survival after irradiation compared to its parental cell line A549. Although A549-R11 cells were specifically resistant to low-LET X-rays, they possessed a similar sensitivity to high-LET carbon ions like their parental A549 cells. Besides, we also introduced another NSCLC cell line H1299, which has intrinsic resistance to X-rays and carbon ions compared with A549 cells, into our study to verify our sequencing data from the A549-R11 set.
It has been reported that radiation could induce a change of circRNA expressions in tumor and normal cells (29-31). Additionally, circRNAs could regulate cellular radiosensitivity. CircRNA_014511 over-expression has been linked to radioresistance in bone marrow mesenchymal stem cells (32). Shuai et al. (33) demonstrated that circRNA_0000285 located in the HIPK3 gene locus involved in the radiosensitivity of nasopharyngeal carcinoma. Different from the studies mentioned above, we believe that some circRNAs could be taken as the biomarkers for NSCLC diagnosis and prognosis in CIRT or radiotherapy with conventional radiation (X- or γ-rays). In this study, we detected the circRNA expressions among the different radiosensitive NSCLC cell lines (including acquired and intrinsic radioresistance) rather than compared the expression changes before and after irradiation. This may produce more realistic environment and improve diagnostic accuracy. We identified a large number of differentially expressed circRNAs among the cell lines from various genomic locations. The circRNA sequencing data revealed that 40 circRNAs were significantly up-regulated and 184 circRNAs were down-regulated in the radioresistant cells compared with their parental cells. Moreover, there was another characteristic for the radioresistant A549-R11 cell line, that is it had a similar sensitivity to carbon ions compared with its parental A549 cell line. This suggests that the circRNAs with different expressions between the two cell lines might be potential biomarkers to distinguish the radiation response after irradiation with X-rays or carbon ions. In fact, we have already obtained a circRNA hsa_circ_0103301, which is highly expressed in A549-R11 and H1299 cells. It could regulate the sensitivity to X-rays but has no effect on NSCLC cells after carbon ion irradiation (unpublished data).
Generally, interruption of a single ncRNA may only regulate a suppression effect, whereas joint destruction of multiple ncRNAs may be more effective. Based on this viewpoint, an interfering circRNA targeting multiple miRNAs was proposed and subjected to trials for NSCLC radiotherapy. Firstly, 934 circRNA-miRNA pairs and 58 miRNA-mRNA pairs were individually predicted using the bioinformatics method. Next, given the significant OS about differentially expressed miRNAs and mRNAs, only 12 circRNA-miRNA pairs and 5 miRNA- mRNA pairs, were selected to generate a global regulatory network. Finally, a circRNA-miRNA-mRNA network including 8 circRNAs, 4 miRNAs, and 4 mRNAs was constructed based on negative correlations between miRNA and mRNA as well as miRNA and circRNA.
In the network we constructed, some ncRNAs and mRNAs have been reported to probably be involved in the initiation and progression of cancer. For example, Zheng et al. found that PXDH was highly expressed in ovarian cancer, which promoted proliferation, invasion and migration and was associated with poor prognosis (34). The similar results were observed in metastatic melanoma tumor (35). PCDH7 upregulation was observed in castrate-resistant prostate cancer (CRPC) and CRPC cells. The knockdown of PCDH7 decreased the activity of the PI3K/Akt pathway and inhibited cell proliferation (36). Recently, several studies showed that there was clinical significance of COL5A2 in patients with adenomas (37), breast cancer (38), colorectal cancer (39), and osteosarcoma (40), especial bladder cancer (41-43). Furthermore, some studies reported that the miRNAs in the constructed network might be differentially expressed in some cancers. Ma et al. uncovered 15 miRNAs, including hsa-miR-16-1-3p, were down-regulated in the plasma of EGFR-TKI resistant patients with NSCLC compared with that of EGFR-TKIs sensitive patients (44). Hsa-miR-16-1 was also mostly down-regulated in lung adenocarcinoma cell lines, and induced cell cycle arrest (45). It has been reported that over-expression of miR-34b could decrease proliferation, migration, and invasion and promote apoptosis in lung cancer (46) and colon cancer cells (47) by targeting different pathways. Lower miR-34b was significantly associated with an aggressive phenotype in urothelial carcinoma of the bladder patients compared with the nonaggressive subject (48). Moreover, down-regulated hsa-miR-146b-3p was significantly correlated with recurrence in osteosarcoma (49), and OS in hepatocellular carcinoma (22). MiR-449 mimics strongly inhibited proliferation, promotes apoptosis, and leads to cell cycle arrest in prostate cancer (50,51) and hepatocellular carcinoma cells (52). Also, we searched only one paper about 4 circRNAs in our network, namely up-regulated has_circ_0001944 may be involved in breast cancer brain metastasis (53). Other circRNAs detected in our ceRNA network have not been reported previously.
In addition, another cohort, CAARRY including 68 patients who received radiotherapy in the Kaplan-Meier plotter, was used to verify the 4 mRNAs mentioned above. CLO5A2 and PXDN were observed to have a significant correlation with the first progression of the lung cancer patients after radiotherapy (Figure S5), suggesting the accuracy of our prediction clearly.
Conclusions
We united high-throughput data screening and bioinformatics to construct a ncRNA network describing the possible regulatory mechanisms of NSCLC sensitivity to low-LET X-rays and high-LET carbon ions. Importantly, our results provide promising potential molecular markers for distinguishing the patients with NSCLC who will gain from radiotherapy with conventional X-rays or CIRT.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work is jointly supported by the National Key Research and Development Program of China (Grant No. 2018YFC0115702), the Key Deployment Project of Chinese Academy of Sciences (Grant No. KFZD-SW-222) and the National Natural Science Foundation of China (Grant No. 11705245, 11875299, U1532264 and U1730133).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2002
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2002
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2002). The authors have no conflicts of interest to declare.
References
- 1.Grutters JP, Pijls-Johannesma M, Ruysscher DD, et al. The cost-effectiveness of particle therapy in non-small cell lung cancer: exploring decision uncertainty and areas for future research. Cancer Treat Rev 2010;36:468-76. 10.1016/j.ctrv.2010.02.018 [DOI] [PubMed] [Google Scholar]
- 2.Arriagada R, Le Chevalier T, Quoix E, et al. ASTRO (American Society for Therapeutic Radiology and Oncology) plenary: Effect of chemotherapy on locally advanced non-small cell lung carcinoma: a randomized study of 353 patients. GETCB (Groupe d'Etude et Traitement des Cancers Bronchiques), FNCLCC (Federation Nationale des Centres de Lutte contre le Cancer) and the CEBI trialists. Int J Radiat Oncol Biol Phys 1991;20:1183-90. 10.1016/0360-3016(91)90226-T [DOI] [PubMed] [Google Scholar]
- 3.Dosoretz DE, Katin MJ, Blitzer PH, et al. Radiation therapy in the management of medically inoperable carcinoma of the lung: results and implications for future treatment strategies. Int J Radiat Oncol Biol Phys 1992;24:3-9. 10.1016/0360-3016(92)91013-D [DOI] [PubMed] [Google Scholar]
- 4.Ding M, Zhang E, He R, et al. Newly developed strategies for improving sensitivity to radiation by targeting signal pathways in cancer therapy. Cancer Sci 2013;104:1401-10. 10.1111/cas.12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz-Ertner D, Tsujii H. Particle radiation therapy using proton and heavier ion beams. J Clin Oncol 2007;25:953-64. 10.1200/JCO.2006.09.7816 [DOI] [PubMed] [Google Scholar]
- 6.Lopez Perez R, Nicolay NH, Wolf JC, et al. DNA damage response of clinical carbon ion vs. photon radiation in human glioblastoma cells. Radiother Oncol 2019;133:77-86. 10.1016/j.radonc.2018.12.028 [DOI] [PubMed] [Google Scholar]
- 7.Demizu Y, Fujii O, Iwata H, et al. Carbon ion therapy for early-stage non-small-cell lung cancer. Biomed Res Int 2014;2014:727962. 10.1155/2014/727962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev 2015;36:25-64. 10.1210/er.2014-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol 2017;10:2. 10.1186/s13045-016-0370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou LD, Zhang J. Circular RNAs: An emerging type of RNA in cancer. Int J Immunopathol Pharmacol 2017;30:1-6. 10.1177/0394632016686985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 14.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 16.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract 2017;213:453-56. 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Zhuang Y, Jiang S, et al. Cinnamaldehyde induces apoptosis and reverses epithelial-mesenchymal transition through inhibition of Wnt/beta-catenin pathway in non-small cell lung cancer. Int J Biochem Cell Biol 2017;84:58-74. 10.1016/j.biocel.2017.01.005 [DOI] [PubMed] [Google Scholar]
- 19.Tian F, Wang Y, Xiao Z, et al. Circular RNA CircHIPK3 Promotes NCI-H1299 and NCI-H2170 Cell Proliferation through miR-379 and its Target IGF1. Zhongguo Fei Ai Za Zhi 2017;20:459-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin X, Liu Y, Ye F, et al. Role of autophagy in high linear energy transfer radiation-induced cytotoxicity to tumor cells. Cancer Sci 2014;105:770-8. 10.1111/cas.12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci 2016;2:e67. 10.7717/peerj-cs.67 [DOI] [Google Scholar]
- 22.Nagy Á, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 24.Shimura T, Kakuda S, Ochiai Y, et al. Acquired radioresistance of human tumor cells by DNA-PK/AKT/GSK3beta-mediated cyclin D1 overexpression. Oncogene 2010;29:4826-37. 10.1038/onc.2010.238 [DOI] [PubMed] [Google Scholar]
- 25.Lynam-Lennon N, Reynolds JV, Pidgeon GP, et al. Alterations in DNA repair efficiency are involved in the radioresistance of esophageal adenocarcinoma. Radiat Res 2010;174:703-11. 10.1667/RR2295.1 [DOI] [PubMed] [Google Scholar]
- 26.Kuwahara Y, Li L, Baba T, et al. Clinically relevant radioresistant cells efficiently repair DNA double-strand breaks induced by X-rays. Cancer Sci 2009;100:747-52. 10.1111/j.1349-7006.2009.01082.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Luo H, Jiang Z, et al. Fractionated irradiation-induced EMT-like phenotype conferred radioresistance in esophageal squamous cell carcinoma. J Radiat Res 2016;57:370-80. 10.1093/jrr/rrw030 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Sato K, Imai T, Okayasu R, et al. Heterochromatin domain number correlates with X-ray and carbon-ion radiation resistance in cancer cells. Radiat Res 2014;182:408-19. 10.1667/RR13492.1 [DOI] [PubMed] [Google Scholar]
- 29.Yu D, Li Y, Ming Z, et al. Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. PeerJ 2018;6:e5011. 10.7717/peerj.5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He N, Sun Y, Yang M, et al. Analysis of Circular RNA Expression Profile in HEK 293T Cells Exposed to Ionizing Radiation. Dose Response 2019;17:1559325819837795. 10.1177/1559325819837795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Yuan B, Wu Z, et al. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene 2017;629:35-42. 10.1016/j.gene.2017.07.078 [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang J, Li J, et al. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn J Basic Med Sci 2019;19:155-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuai M, Hong J, Huang D, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett 2018;16:6495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng YZ, Liang L. High expression of PXDN is associated with poor prognosis and promotes proliferation, invasion as well as migration in ovarian cancer. Ann Diagn Pathol 2018;34:161-5. 10.1016/j.anndiagpath.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 35.Jayachandran A, Prithviraj P, Lo PH, et al. Identifying and targeting determinants of melanoma cellular invasion. Oncotarget 2016;7:41186-202. 10.18632/oncotarget.9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shishodia G, Koul S, Koul HK. Protocadherin 7 is overexpressed in castration resistant prostate cancer and promotes aberrant MEK and AKT signaling. Prostate 2019;79:1739-51. 10.1002/pros.23898 [DOI] [PubMed] [Google Scholar]
- 37.Guillen-Ahlers H, Buechler SA, Suckow MA, et al. Sulindac treatment alters collagen and matrilysin expression in adenomas of ApcMin/+ mice. Carcinogenesis 2008;29:1421-7. 10.1093/carcin/bgn123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargas AC, McCart Reed AE, Waddell N, et al. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res Treat 2012;135:153-65. 10.1007/s10549-012-2123-4 [DOI] [PubMed] [Google Scholar]
- 39.Fischer H, Stenling R, Rubio C, et al. Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis 2001;22:875-8. 10.1093/carcin/22.6.875 [DOI] [PubMed] [Google Scholar]
- 40.Wu D, Chen K, Bai Y, et al. Screening of diagnostic markers for osteosarcoma. Mol Med Rep 2014;10:2415-20. 10.3892/mmr.2014.2546 [DOI] [PubMed] [Google Scholar]
- 41.Zeng XT, Liu XP, Liu TZ, et al. The clinical significance of COL5A2 in patients with bladder cancer: A retrospective analysis of bladder cancer gene expression data. Medicine (Baltimore) 2018;97:e0091. 10.1097/MD.0000000000010091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng XY, Shi MJ, Zeng ZH, et al. The Role of COL5A2 in Patients With Muscle-Invasive Bladder Cancer: A Bioinformatics Analysis of Public Datasets Involving 787 Subjects and 29 Cell Lines. Front Oncol 2019;8:659. 10.3389/fonc.2018.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Liu X, Liu T, et al. Identification of Biomarkers Correlated with the TNM Staging and Overall Survival of Patients with Bladder Cancer. Front Physiol 2017;8:947. 10.3389/fphys.2017.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Y, Pan X, Xu P, et al. Plasma microRNA alterations between EGFR-activating mutational NSCLC patients with and without primary resistance to TKI. Oncotarget 2017;8:88529-36. 10.18632/oncotarget.19874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bandi N, Zbinden S, Gugger M, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res 2009;69:5553-9. 10.1158/0008-5472.CAN-08-4277 [DOI] [PubMed] [Google Scholar]
- 46.Zhuang XF, Zhao LX, Guo SP, et al. miR-34b inhibits the migration/invasion and promotes apoptosis of non-small-cell lung cancer cells by YAF2. Eur Rev Med Pharmacol Sci 2019;23:2038-46. [DOI] [PubMed] [Google Scholar]
- 47.Ye K, Xu C, Hui T. MiR-34b inhibits the proliferation and promotes apoptosis in colon cancer cells by targeting Wnt/beta-catenin signaling pathway. Biosci Rep 2019;39:BSR20191799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Inamoto T, Uehara H, Akao Y, et al. A Panel of MicroRNA Signature as a Tool for Predicting Survival of Patients with Urothelial Carcinoma of the Bladder. Dis Markers 2018;2018:5468672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, Ma J, Wang A, et al. A support vector machine and a random forest classifier indicates a 15-miRNA set related to osteosarcoma recurrence. Onco Targets Ther 2018;11:253-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009;28:1714-24. 10.1038/onc.2009.19 [DOI] [PubMed] [Google Scholar]
- 51.Noonan EJ, Place RF, Basak S, et al. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget 2010;1:349-58. 10.18632/oncotarget.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buurman R, Gurlevik E, Schaffer V, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 2012;143:811-20.e15. 10.1053/j.gastro.2012.05.033 [DOI] [PubMed] [Google Scholar]
- 53.Fu B, Zhang A, Li M, et al. Circular RNA profile of breast cancer brain metastasis: identification of potential biomarkers and therapeutic targets. Epigenomics 2018;10:1619-30. 10.2217/epi-2018-0090 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as