Abstract
Background
Previous data have confirmed that disordered long non-coding ribonucleic acid (lncRNA) expression is evident in many cancers and is correlated with tumor progression. The present study aimed to investigate the function of long non-coding RNA00844 (LINC00844) in hepatocellular carcinoma (HCC).
Methods
The expression levels of target genes were detected with real-time polymerase chain reaction (PCR) and western blotting. The biologic function of HCC cells was determined with cell viability assay, colony formation assay, cell cycle analysis, apoptosis detection, and Transwell migration assay in vitro. Tumorigenesis was performed with cell injection in vivo. The relationship between LINC00844 and survival outcomes was determined with the Cox proportional hazards model. A RNA precipitation assay was conducted to reveal the types of LINC00844 that potentially bind with proteins.
Results
LINC00844 was found to be significantly decreased in HCC tissue and was correlated with poor tumor characteristics, such as portal vein invasion, high α-fetoprotein (AFP), and a high rate of tumor recurrence. Exotic LINC00844 expression in HCC cell lines significantly suppressed proliferation and migration, as well as invasiveness, whereas LINC00844 deletion had the opposite effect. LINC00844 overexpression significantly inhibited HCC tumorigenesis in vivo. Mechanistic analyses indicated that the mitogen-activated protein kinase (MAPK) signaling pathway was remarkably inactivated by LINC00844. Furthermore, the immunoprecipitation assay verified that LINC00844 can bind to zinc-alpha-2-glycoprotein (AZGP1) and interfere with its translocation. LINC00844 can also promote AZGP1 expression, leading to the suppression of the transforming growth factor-β1 (TGF-β1)-extracellular signal-regulated kinase (ERK) pathway.
Conclusions
LINC00844 is a novel anti-oncogene in the development of HCC and a potentially promising therapeutic target in HCC.
Keywords: Hepatocellular carcinoma (HCC), long non-coding RNA00844 (LINC00844), mitogen-activated protein kinase signaling pathway (MAPK signaling pathway), transforming growth factor-β1 (TGF-β1), zinc-alpha-2-glycoprotein (AZGP1)
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies globally and has a high mortality rate (1). Approximately half of all new HCC cases in China are caused by chronic hepatitis B virus infection and liver cirrhosis, which are highly prevalent. Liver transplantation (LT) is considered the gold standard in the treatment of HCC. LT can allow the tumor tissue and cirrhotic liver to be totally removed, and achieve satisfactory liver function, leading to improved quality of life and survival outcomes for patients. Unfortunately, despite advances in surgical techniques and immunosuppressive therapies, the rate of tumor recurrence after LT is still >20%, which impacts patients’ long-term survival (2). Moreover, the molecular mechanisms underlying tumorigenesis and the recurrence of HCC have yet to be fully understood. Therefore, exploration of the essential regulators of HCC would contribute to a deeper understanding of the pathogenesis of the disease and the identification of new therapeutic targets.
Among the numerous ribonucleic acid (RNA) molecules transcribed from mammalian genomes, long non-coding RNAs (lncRNAs; >200 nucleotides) have no or low protein-coding potential but are important regulators in multiple biological processes. Recent advances have further suggested that the role of lncRNAs in basic, translational, and clinical oncology is equal to, or even surpasses, that of protein-coding messenger RNAs (mRNAs). The US Food and Drug Administration has approved antisense oligonucleotides (ASOs) for the treatment of familial hypercholesterolemia and transthyretin amyloidosis (3). Therefore, lncRNAs are considered potential therapeutic targets because of their correlation with the initiation and development of tumors. Arun et al. found that Malat knockdown using ASOs resulted in slower tumor growth and reduced metastasis in a preclinical breast cancer model (4). Wang et al. also targeted the Malat1/AR-v7 axis in their preclinical study and found that Malat1 siRNA decreased anti-androgen enzalutamide resistance in a mouse model of metastatic castration-resistant prostate cancer (5). These studies provide a basis for the use of lncRNAs in oncology.
The role of lncRNAs in modulating drug resistance, metastasis, and immune surveillance in HCC has also been investigated. For instance, Wu et al. reported that lnc histone deacetylase 2 is highly expressed in HCC tissue and is recruited via the nucleosome remodeling and deacetylase complex onto the promoter of PTCH1 to suppress PTCH1 transcription, leading to Hedgehog signaling activation and CD13+CD133+ cell expansion (6). Meanwhile, lncRNA HANR was found to promote HCC progression and enhance chemosensitivity to doxorubicin by binding to GSKIP to increase GSK3β phosphorylation (7). Ye et al. showed that the expression of lncRNA cyclooxygenase-2 (COX2) was higher in M1 macrophages than in M2 macrophages, and that lncRNA COX2 siRNA transfection inhibited M1 macrophage function and strengthened M2 macrophage function, leading to increased immune evasion of HCC (8). There has been a lot of research on the underlying mechanisms and clinical implications of lncRNAs in HCC; however, most of this research has focused on tumor-promoting lncRNAs. Therefore, the present study aimed to investigate the expression of long non-coding RNA00844 (LINC00844) in HCC.
We found that LINC00844 was decreased in HCC tissue and was correlated with higher tumor recurrence after LT. Ectopic LINC00844 expression inhibited proliferation, invasiveness, migration, and tumorigenesis in HCC. The RNA precipitation assay revealed that LINC00844 bound to zinc-alpha-2-glycoprotein (AZGP1), leading to the retention of AZGP1 in the cytoplasm. Moreover, LINC00844 expression was found to be closely related to AZGP1 expression in HCC tissue. In vitro, LINC00844 knockdown or overexpression resulted in changes in AZGP1 expression. Combined with the results of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, LINC00844 was shown to regulate the biological function of HCC cells, leading to the suppression of the transforming growth factor-β1 (TGF-β1)-extracellular signal-regulated kinase (ERK) pathway by targeting AZGP1.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3848).
Methods
Antibodies and cells
HCCLM3 (RRID:CVCL_6832, catalog no. GDC214) was obtained from China Center for Type Culture Collection, and Huh7 (RRID:CVCL_0336, catalog no. TCHu182) cell lines were purchased from the Council Cell Bank of Typical Culture Preservation. Rabbit anti-p-MEK polyclonal antibody (RRID:AB_331648, catalog no. 9121), mouse anti-ERK monoclonal antibody (RRID:AB_390780, catalog no. 4696), rabbit anti-p-ERK polyclonal antibody (RRID:AB_2315112, catalog no. 4370), and rabbit anti-GAPDH monoclonal antibody (RRID:AB_10622025, catalog no. 5174) were purchased from Cell Signal Technology. Rabbit anti-tubulin polyclonal antibody (RRID:AB_10575456, catalog no. MBS530753) was obtained from MyBiosource. Rabbit anti-AZGP1 polyclonal antibody (RRID:AB_11159784, catalog no. ab117275); rabbit anti-cyclin D1 monoclonal antibody (RRID:AB_2750906, catalog no. ab134175); rabbit anti-CDK4 monoclonal antibody (RRID:AB_10867218, catalog no. ab108357); rabbit anti-histone polyclonal antibody, nuclear loading control, and ChIP grade (RRID:AB_302613, catalog no. ab1791); rabbit anti-Ki-67 polyclonal antibody (RRID:AB_443209, catalog no. ab15580); goat anti-mouse immunoglobulin G (IgG) polyclonal antibody (RRID:AB_954556; catalog no. ab205718); and goat anti-rabbit IgG polyclonal antibody (RRID:AB_955447, catalog no. ab6721) were purchased from Abcam (USA).
Clinical materials and sample preparation
The present study was performed in accordance with the principles of the Declaration of Helsinki (as revised in 2013) and approved by the Ethical Committee at the First Affiliated Hospital, School of Medicine, Zhejiang University (No. 2018658). Individual consent for this retrospective analysis was waived. All clinical samples used in the study were obtained from HCC patients at the First Affiliated Hospital, School of Medicine, Zhejiang University, between 2014 and 2016. Patient data were collected from the LT database on the hospital information system.
Cell lines and cell transfection
Two human HCC cell lines, HCC-LM3 and Huh7, were cultured with Dulbecco’s Modified Eagle Medium (DMEM) in 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a 37 °C, 5% CO2 humidified incubator; the medium was replaced every 2 days.
Lentivirus encoding LINC00844 and green fluorescent protein (GFP) was constructed by GeneChem (Shanghai), along with a GFP lentiviral vector with a scramble sequence (Scr-siRNA/GFP) as the negative control. These vectors were respectively transfected into cells according to the manufacturer’s instructions, and stable cell lines were selected with puromycin. The siRNA was transfected with Lipofectamine 3000 reagent (Thermo Fisher Scientific, USA), and the siRNA target sequences for each gene were as follows: (I) si-LINC00844: si-LINC00844-1: 5'-3'GGGUUCAAGCAAGUAAAUUTTAAUUUACUUGTUUGAACCCTT; si-LINC00844-2: 5'-3'GCGCAGAGAACAUUGAAAUTTAUUUCAAUGUUCUCUGCGCTT; and (II) si-AZGP1: si-AZGP1-1: 5'-3'GGUCGUUACUCUCUGACCUTTAGGUCAGAGAGUAACGACCTT; si-AZGP1-2: 5'-3'GGAGCAUUCUGGAAAUAUUTTAAUAUUUCCAGAAUGCUCCTT.
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from clinical samples and HCC cells with TRIzol reagent (Invitrogen), and cDNA was synthesized using Takara PrimeScript RT Reagent Kit (Takara Bio, Japan) according to the manufacturer’s instructions. mRNA expression was determined with the Bio-Rad PCR (Thermo Fisher Scientific, USA) instrument using SYBR Premix Ex Taq (Takara Bio, Japan).
The specific primer pairs used for the target genes were as follows: (I) LINC00844: forward 5'-3': CAGTTTTAGCAAGGAGGTTTCT; reverse 5'-3': ATGCTTCTATGTGACCTGTGA; (II) AZGP1: forward 5'-3': GACATCTTTATGGAGACCCTGAAAGACAT; reverse 5'-3': CTGTTATTCTCGATCTCACAACCAAACCT; and (III) GAPDH: forward 5'-3': CCTGGTATGACAACGAATTTG; reverse 5'-3': CAGTGAGGGTCTCTCTCTTCC.
The expression level was calculated using the 2–ΔΔCt method, and the results were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which was used as an internal control.
Western blot analysis
Total protein was extracted using RIPA buffer (Thermo Scientific, USA) containing protease and phosphatase inhibitors (Thermo Scientific). The protein concentration was detected with the BCA protein assay kit (Thermo Scientific); 10–50 µg protein was loaded on 4–20% ExpressPlus PAGE gels (GenScript, USA) for separation and then transferred to 0.45-µm polyvinylidene difluoride membranes (Millipore, USA). After washing three times, the membrane was incubated with primary antibodies at 4 °C overnight. The membrane was then incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Immunoblots were detected by EZ-ECL (Biological Industries, Israel), and GAPDH was used as the loading control.
Cell viability assay
Cell viability was measured using a Cell Counting Kit-8 assay (Dojindo Laboratories), according to manufacturer’s instructions. Briefly, HCC cells were seeded onto a 96-well plate at a density of 1×103 cells/well. After culture for 24, 48, 72, or 96 h, the absorbance was detected at 450 nm.
Colony formation assay
HCC cells were seeded onto a 6-well plate at a density of 1×103. After 14 days of incubation, the cells were stained with 0.5% crystal violet solution (Thermo Scientific), and the colonies were counted and photographed.
Cell migration and invasion assays
For the migration and invasion assays, Transwell membrane filters (24 wells and 8-µm pore size; Corning) were coated with Matrigel matrix (Becton Dickinson, USA) or left uncoated. Then, 5×104 cells with 100 µL serum-free medium were added to the upper chamber, and 10% FBS medium (500 µL) was placed in the lower chamber. After 48 h of incubation at 37 °C, the cells remaining on the upper surface were removed with cotton swabs, and the cells on the lower surface of the membrane were stained with a 0.5% crystal violet solution for counting and photographing.
Cell cycle analysis
HCC cells were digested and fixed in 75% precooled ethanol at 4 °C overnight. After washing three times with cold phosphate-buffered saline, the cells were stained with a cell cycle staining kit (Multi Sciences, USA) in the dark at room temperature for 30 min. Cell cycle distribution was detected by flow cytometry (Beckman, USA). The percentage of the cell population in each phase was calculated with ModFit LT software (Verity Software House, USA).
Cell apoptosis analysis
HCC cells were collected and stained with the Annexin V-APC/7-AAD Apoptosis kit (Multi Sciences) in the dark at 4 °C. After incubation for 15 min, the cells were quantified with a flow cytometer (FACSCanto II; BD Biosciences, USA).
RNA pull-down assay
Six biotinylated probes were designed for LINC00844 absorbance; 1×107 cells infected with Lv-LINC00844 were lysed in RIPA buffer, and 1 U/µL RNAase inhibitor (Thermo Fisher Scientific) was added. The assay was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Scientific) following the manufacturer’s instructions. The probe sequences (5' to 3') used were as follows: probe 1, CTATCAGACTGCATCAAGCA; probe 2, TGGTTCAGATTTTCTTCCTA; probe 3, TTCTCATGCTTCTATGTGAC; probe 4, TACACCCTGTTATTTAGTCT; probe 5, TCCTTGCTAAAACTGAACGA; and probe 6, ACTGGACATTGCAAACACTT.
Chromatin isolation by RNA purification (ChiRP): MS/MS
Cells (1×107) infected with Lv-LINC00844 were digested on plates, and reversibly cross-linked in situ with formaldehyde, followed by hybridization with biotin-labeled probes specifically designed for LINC00844 (prefixed with non-specific antisense or pretreated with RNase in the control group). After one night, non-specific binding proteins were washed under strong denaturing conditions, and RBP was obtained, which could be identified and quantified by LC-MS/MS. After data searching and quantifying, the proteins specifically interacting with LINC00844 had a significant difference between the two groups, and non-specific interacting proteins appeared in both groups. The ratio of non-specific proteins between the groups was close to 1:1. The interacting protein profile of LINC00844 was then constructed.
Tumor xenograft experiment
Experiments were performed under a project license (No. 2018705) granted by institutional ethics board of Zhejiang University, in compliance with Chinese guidelines for the care and use of animals. All of the animal experiments were performed following the National Institutes of Health Guide for the Care and Use of Laboratory Animals. In total, 1×106 cells with LINC00844 knockdown or scramble shRNA transcription were suspended in 100 µL medium and respectively injected into the lateral flank of 6-week-old immunodeficient female mice. Ten mice were divided randomly into two groups. The tumor volume was measured weekly with a caliper and calculated using the following equation: tumor size = (width2 × length)/2. After 6 weeks, all of the mice were sacrificed, and the tumors were harvested for immunohistochemistry.
Statistical analysis
All of experiments were independently repeated three times. The data were presented as mean ± standard deviation, and differences between the two groups were determined by two-tailed Student’s t-tests. The potential predictive value of LINC00844 on survival outcomes was analyzed by Kaplan-Meier curves and the log-rank test. LINC00844, α-fetoprotein (AFP), portal vein tumor thrombus (PVTT), largest tumor diameter ≥3 cm, and tumor number >3 were incorporated into the Cox proportional hazards model (backward selection likelihood function) to determine their independent effects. Data analysis was performed using SPSS 19.0 software (IBM, USA). P<0.05 was considered to be statistically significant.
Results
Pattern and significance of LINC00844 expression in HCC
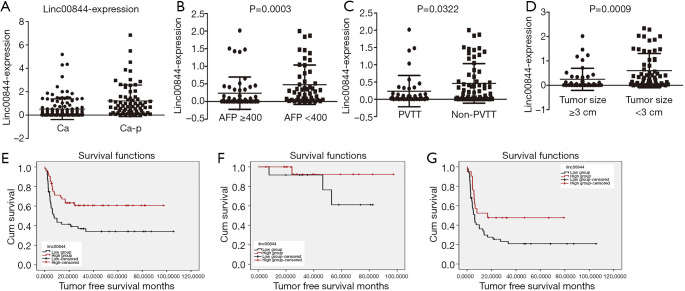
In a previous study, we profiled mRNA and lncRNA in 10 paired samples of HCC and adjacent liver tissue (9); LINC00844 was found to be one of the top 20 significantly downregulated lncRNAs in HCC, relative to adjacent liver tissue. In a cohort of LT patients, we further confirmed the decrease of LINC00844 expression in HCC tissue compared with corresponding adjacent non-tumor tissue (Figure 1A). Moreover, the suppressed expression of LINC00844 was observed in tumors with portal vein invasion and a high AFP value; the greater the tumor number, the greater the suppression (Figure 1B,C,D). To determine the relationship between LINC00844 and tumor-free survival (TFS), HCC patients with LT were divided into high and low LINC00844-expression groups. After adjusting for differences in clinicopathological features and demographic characteristics, Kaplan-Meier analysis was performed and revealed that patients with lower LINC00844 expression developed more frequent recurrence and poorer long-term TFS after LT (Figure 1E). The cohort was further stratified into within and beyond Milan criteria; the related high LINC00844 expression significantly reduced the tumor recurrence rate and increased TFS (Figure 1F,G). Tumor features correlated with LINC00844 were further analyzed using Cox’s proportional hazards regression analysis model; suppressed LINC00844 expression was found to be an independent predictor of HCC prognosis [relapse-free survival (RFS): hazard ratio (HR): 2.157, 95% confidence interval (CI): 1.129–4.121, P=0.020] (Table 1). The results indicated that LINC00844 might be a critical antitumor gene that inhibits the recurrence and progression of HCC.
Figure 1.
LINC00844 expression was significantly decreased in HCC tissue compared with corresponding adjacent non-tumor tissue (A). Suppressed expression of LINC00844 was correlated with portal vein invasion (B), number of tumor sites ≥3 (C), and AFP ≥400 ng/mL (D). Kaplan-Meier analysis showed that patients with lower LINC00844 expression had a higher rate of HCC recurrence than those with higher LINC00844 expression (E). This trend was maintained in patients regardless of whether they fell within (F) or beyond the Milan criteria (G). LINC00844, long non-coding RNA00844; HCC, hepatocellular carcinoma; AFP, α-fetoprotein.
Table 1. Cox proportional hazards regression for Predictors of HCC recurrence following LT.
| Variables | TFS | |
|---|---|---|
| P value | HR (95% CI) | |
| AFP >400 | 0.144 | 1.609 (0.850–3.044) |
| PVTT | 0.004 | 2.252 (1.291–3.927) |
| Diameter ≥3 cm | <0.001 | 3.262 (1.744–6.101) |
| Tumor number >3 | 0.001 | 3.314 (1.647–6.670) |
| LINC00844 | 0.020 | 2.157 (1.129–4.121) |
HCC, hepatocellular carcinoma; LT, liver transplantation; TFS, tumor-free survival; HR, hazard ratio; CI, confidence interval; AFP, α-fetoprotein; PVTT, portal vein tumor thrombus; LINC00844, long non-coding RNA00844.
Modulation of LINC00844 expression altered HCC proliferation
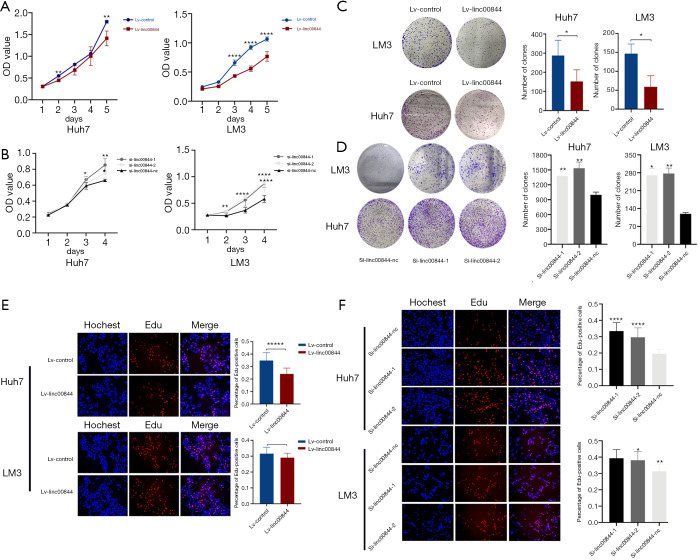
To investigate the potential role of LINC00844 in HCC progression, we first evaluated the altered LINC00844 expression on proliferation and cell cycle. A specific LINC00844 lentivirus and siRNA targeting LINC00844 were respectively used to modulate LINC00844 expression (Figure S1). When LM3 and Huh7 cells were upregulated in LINC00844 expression, cell viability decreased after 48 h in Huh7 cells and 72 h in LM3 cells (Figure 2A); however, HCC cells with LINC00844 knockdown had increased cell viability (Figure 2B). For the colony assay, we observed larger and more colonies in cells with LINC00844 suppression (Figure 2C), but smaller and fewer colonies in cells with LINC00844 overexpression (Figure 2D), compared with the corresponding control cells. Moreover, after increasing LINC00844 expression, the cells showed lower concentrations of EdU staining (Figure 2E). The opposite effect was observed in cells with LINC00844 knockdown (Figure 2F). These data confirmed that LINC00844 plays an essential role in suppressing HCC cell proliferation.
Figure 2.
Cell viability was significantly decreased in HCC cells with upregulated LINC00844 expression (A). HCC cells with LINC00844 knockdown had increased cell viability (B). LINC00844 knockdown promoted colony formation of HCC cells (C), but this effect was reversed by LINC00844 overexpression (D); the cells were stained with crystal violet. Compared with control cells, the cells with LINC00844 overexpression had a lower concentration of EdU stain (E, 100×), but LINC00844 knockdown increased the positive EdU staining rate (F, 100×). *, P<0.05; **, P<0.01; ****, P<0.001; *****, P<0.0001. HCC, hepatocellular carcinoma; LINC00844, long non-coding RNA00844.
LINC00844 promoted apoptosis and suppresses HCC cell metastasis
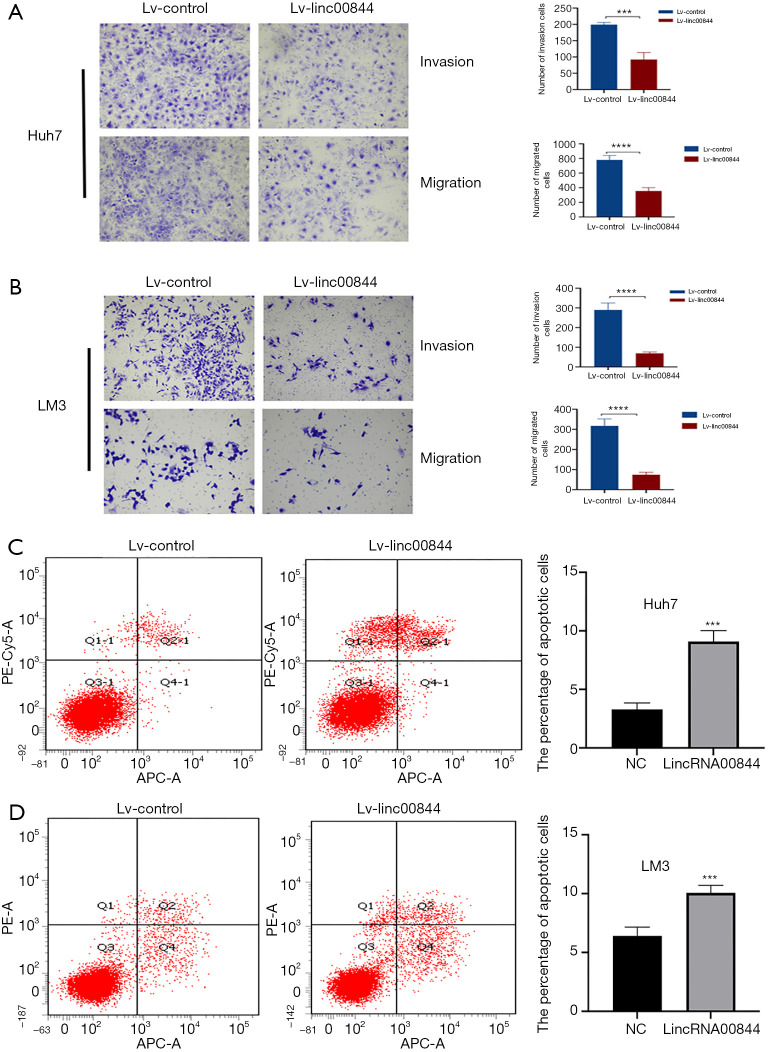
HCC cell migration and invasion is necessary for HCC metastasis, which was detected with a Transwell assay in the present study. Exotic LINC00844 was transfected into Huh7 and LM3 cells, resulting in significantly decreased invasion ability, as evidenced by an increase in the average number of invading cells in the Matrigel matrix, compared with the negative controls (Figure 3A,B). As expected, the migration ability of both the Huh7 and LM3 cells was also suppressed.
Figure 3.
Exotic LINC00844 expression significantly decreased the invasion and migration abilities of Huh7 (A) and LM3 cells (B). Huh7 (C) and LM3 cells (D) with LINC00844 overexpression had a higher rate of apoptosis. The cells were stained with crystal violet. The magnification was 100×. ***, P<0.005; ****, P<0.001. LINC00844, long non-coding RNA00844.
Flow cytometry was used to determine the effect of LINC00844 overexpression on HCC cell apoptosis. We found that LINC00844 upregulation could induce apoptosis of LM3 and Huh7 cells, as quantified with APC/7AAD staining. LM3 cells with LINC00844 overexpression had greater apoptosis rate (10.07±0.72) than the negative control cells (6.8±0.65, P<0.05) or the Huh7 cell line (9.1±0.84 vs. 3.3±0.43, P<0.05) (Figure 3C,D).
LINC00844 inhibited HCC cell tumorigenesis in vivo
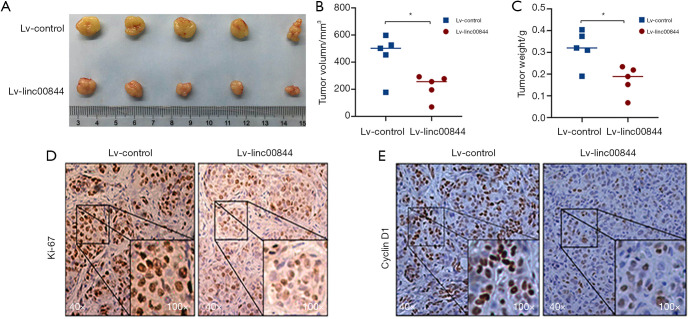
A xenograft mouse model was established to explore the antitumor role of LINC00844 in HCC further. The findings showed that LINC00844 overexpression in Huh7 cells inhibited subcutaneous tumor growth (Figure 4A), which was indicated by the decreased tumor volume and tumor weight compared to the negative control group (Figure 4B,C). After the tumors were harvested, Ki-67 and cyclin D1 immunohistochemistry was performed. In the LINC00844 overexpression group, the staining intensities of Ki-67 and cyclin D1 were much lighter compared to control group (Figure 4D,E). Our results demonstrated a positive role of LINC00844 in suppressing HCC cell proliferation, limiting tumor progression.
Figure 4.
The tumorigenic capability of Huh7 cells was inhibited by LINC00844 overexpression (A), which was indicated by the tumor volume (B) and tumor weight (C). Positive rates of Ki-67 (D) and cyclin D1 (E) immunohistochemistry stain were higher in the LINC00844 knockdown group than in the LINC00844 upregulation group. *, P<0.05. LINC00844, long non-coding RNA00844.
LINC00844 inactivated the mitogen-activated protein kinase (MAPK) signaling pathway
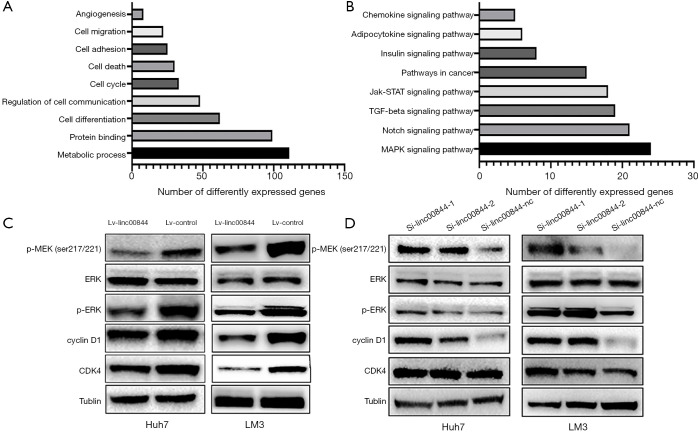
To study the molecular mechanisms underlying the function of LINC00844, we profiled the gene expression pattern by RNA sequencing. Overall, LINC00844 overexpression induced the differential expression of 870 genes (≥1.5-fold change, P<0.05). The data were further analyzed using an online database (http://david.abcc.ncifcrf.gov/). In Gene Ontology analysis, these differentially expressed genes are involved in biological processes relevant to cancer pathogenesis, such as metabolic processes, protein binding, and cell cycle, growth, and death (Figure 5A). Moreover, KEGG pathway analysis indicated that the MAPK signaling pathway was the top canonical pathway identified in this process (Figure 5B). Protein detection assay was then performed, and revealed that the levels of MEK and ERK1/2 phosphorylation were significantly lower in cells with LINC00844 overexpression (Figure 5C), but higher in cells transfected with LINC00844 siRNA (Figure 5D).
Figure 5.
Gene Ontology analysis indicated LINC00844 was involved in biological processes related to cancer pathogenesis, such as metabolic processes, protein binding, and cell cycle, growth, and death (A). KEGG pathway analysis indicated that LINC00844 inactivated the MAPK signaling pathway (B). Western blotting showed that MEK and ERK 1/2 phosphorylation were inhibited in LINC00844 overexpression cells (C), but were promoted in LINC00844 siRNA transfection cells (D). LINC00844, long non-coding RNA00844; KEGG, Kyoto Encyclopedia of Genes and Genomes pathway; MAPK, mitogen-activated protein kinase; MEK, mitogen activated kinase; ERK, extracellular signal-regulated kinase.
LINC00844 blocked the AZGP1 mediated TGF-β1-ERK signaling pathway
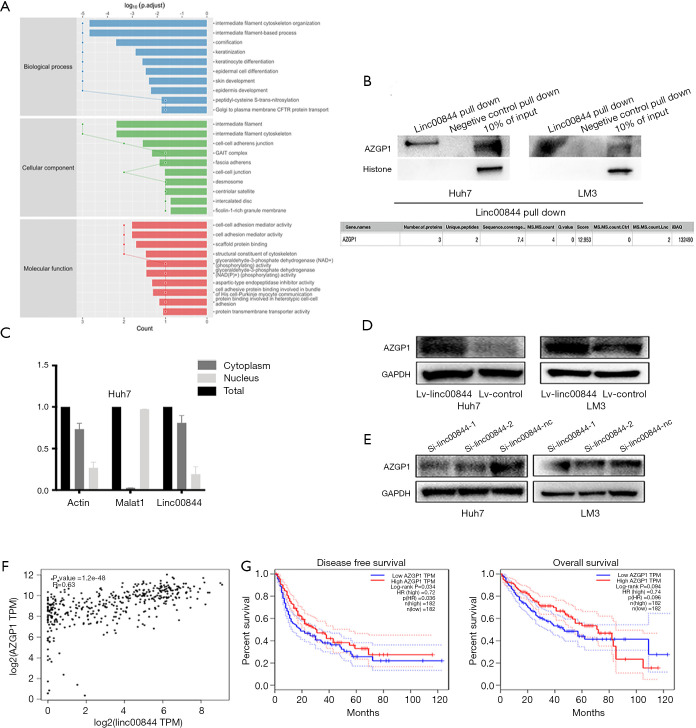
To further investigate the underlying molecular mechanism of the LINC00844-inactivating MAPK signaling pathway, the ChiRP assay was performed. The ChiRP mixture underwent mass spectrometry analysis, and the results revealed 42 proteins that could potentially combine with LINC00844. These proteins were analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) online database (http://gepia.cancer-pku.cn/) and were involved in the regulation of several cell biological functions, such as adhesion and differentiation (Figure 6A). Previous studies have verified AZGP1 to be an important upstream mediator in the inhibition of TGF-β1-mediated ERK phosphorylation. Therefore, we identified AZGP1 as a candidate for LINC00844-regulating MAPK signaling. The binding of AZGP1 and LINC00844 was further verified using RNA pull-down and western blot assays (Figure 6B). Liu et al. stated that AZGP1 could directly bind to and inhibit TGF-β1 in temporal lobe epilepsy (10). Based on this, we detected the LINC00844 expression pattern in the cytoplasm and nucleus, and found LINC00844 to be mainly located in the cytoplasm (Figure 6C). Moreover, in HCC cells with LINC00844 overexpression, we found decreased expression of AZGP1; however, the opposite effect was observed in LINC00844 knockdown cells (Figure 6D,E). The relationship between AZGP1 and LINC00844 expression was also confirmed using the GEPIA online database (Figure 6F). GEPIA also demonstrated that the expression level of AZGP1 was significantly correlated with shorter disease-free survival and overall survival in HCC patients (Figure 6G). These data confirmed that LIMC00844 could inhibit ERK phosphorylation by binding to AZGP1, as well as regulating AZGP1 expression.
Figure 6.
Chromatin isolation by RNA purification and MS indicated that LINC00844 potentially combines with 42 proteins involved in the regulation of several cell biological functions, such as adhesion and differentiation (A). RNA pull-down assay verified the binding of AZGP1 and LINC00844 (B). RT-PCR showed that LINC00844 is mainly located in the cytoplasm (C). LINC00844 inhibition increased AZGP1 expression (D). LINC00844 overexpression decreased AZGP1 expression (E). Data from the GEPIA online database confirmed the close relationship between LINC00844 and AZGP1 (F), and verified that AZGP1 was correlated with poorer survival outcomes (G). MS, mass spectrum; LINC00844, long non-coding RNA00844; AZGP1, zinc-alpha-2-glycoprotein; RT-PCR, reverse transcription polymerase chain reaction; GEPIA, Gene Expression Profiling Interactive Analysis.
AZGP1 knockdown eliminated the inhibitory effect of LINC00844 on cell cycle
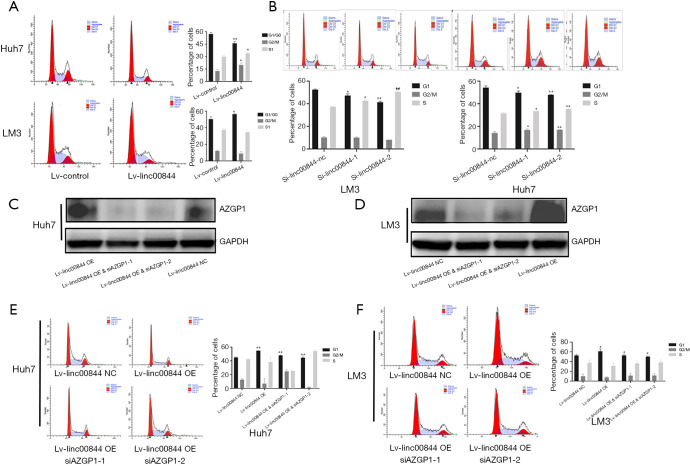
Cell cycle analysis was also performed to evaluate the effect of AZGP1 on LINC00844-mediated cell proliferation inhibition. Following LINC00844 overexpression, HCC cells showed cell cycle arrest and lower levels of CK1 and CDK expression (Figure 7A). However, in cells with LINC00844 knockdown, advanced cell cycle and up-regulated expression of CK1 and CDK were observed (Figure 7B). To verify whether LINC00844 induces HCC cell suppression and is dependent on AZGP1, we transfected AZGP1 siRNA into HCC cells with high LINC00844 overexpression (Figure 7C,D). The cell cycle analysis revealed that downregulated AZGP1 eliminated the inhibitory effect of LINC00844 on cell cycle arrest (Figure 7E,F).
Figure 7.
LINC00844 overexpression promotes the cell cycle arrest of HCC cells (A), and this effect is reversed by LINC00844 knockdown (B). Huh7 (C) and LM3 cells (D) with high LINC00844 expression were transfected with siRNA AZGP1. SiAZGP1 eliminated the inhibitory effect of LINC00844 on cell cycle in Huh7 (E) and LM3 cells (F). *, P<0.05; **, P<0.01. LINC00844, long non-coding RNA00844; AZGP1, zinc-alpha-2-glycoprotein; HCC, hepatocellular carcinoma.
Discussion
HCC is a highly aggressive and intractable malignancy (11). LT is considered the gold standard treatment for HCC, but it still has a high incidence of tumor recurrence and metastasis, which limits the long-term survival outcomes of patients (12). Multiple genetic and epigenetic alterations, as well as extrinsic microenvironment factors, contribute to the recurrence of HCC. To date, no single effective strategy against tumor recurrence following LT has been devised (13). Therefore, a better understanding of the underlying molecular mechanisms regulating HCC recurrence is required.
Recent evidence suggests that many lncRNAs are involved in tumor metastasis; however, the mechanisms are not well understood (14-16). It is well known that HCC recurrence is likely the result of microscopic extrahepatic metastatic foci (17), even many clinical parameters for stratifying patients with high risk was identified and the LT process totally removed the primary liver. Metastasis is a complex process involving the activation of several signaling pathways and epithelial–mesenchymal transition (18,19). Cancer stem cells are believed to be able to resist antitumor immunity and recover months or years after surgery; they are also responsible for disease recurrence. Wang et al. identified several lncRNAs from cancer stem cells of HCC cell lines and investigated their roles in the regulation of HCC stemness. They found that lncTCF7 is highly expressed in HCC and cancer stem cells, and that it could promote the self-renewal of HCC cells and tumor propagation via the activation of Wnt signaling (20). Furthermore, a lncRNA named lncRNA-ATB, which is activated by TGF-β1, was not only found to be upregulated in HCC metastases, but was also observed to increase the expression of ZEB1 and ZEB2 via competitively binding with miR-200 family members to promote EMT and invasion; lncRNA-ATB therefore appears to be essential for TGF-β signaling (21). This evidence supports the essential role of lncRNAs in regulating extrahepatic metastasis of HCC.
To further identify the lncRNAs physiologically involved in HCC recurrence, we identified LINC00844, as a novel tumor suppressor from previous transcriptome sequence data, and suggested that its inhibition might be a new characteristic of HCC development. In our large cohort of LT patients, qRT-PCR revealed that the expression of LINC00844 was sometimes decreased but was heterogeneous in primary tumors when compared with adjacent non-tumor tissue. HCC patients with LINC00844 inhibition had an increased risk of recurrence after surgery, which directly correlated with patients’ long-term survival outcomes. Furthermore, in patients who exceeded the Milan criteria, those with low LINC00844 expression were still prone to earlier HCC recurrence after LT. Some new clinic pathological characteristics, including tumor grade and AFP, are considered to be important predictors of HCC recurrence and have been utilized to expand the Milan criteria (2,22); however, due to the high rate of post-LT tumor recurrence, the utilization of these characteristics alone is insufficient. Therefore, molecular-based tumor staging is crucial for the further improvement of LT criteria, individualized diagnoses, and therapy. Our data indicated that LINC00844 is a precise predictor that can be used in the molecular classification of HCC patients to improve Milan criteria restrictions (23,24) and therapies following surgery.
In vivo and in vitro methods were also applied to determine the involvement of LINC00844 in a regulatory network of metastatic properties. After LINC00844 up-regulation, HCC cells showed significantly reduced invasive and migratory abilities, leading to a less malignant phenotype. We found that the overexpression of LINC00844 induced inhibitory cell viability, promoted cell cycle arrest, and increased apoptosis, while silencing of LINC00844 had the opposite effect, which suggested LINC00844 plays a crucial role in regulating proliferation. Previous studies have found that cell proliferation and resistance to apoptosis are closely correlated with tumor progression and metastasis. These results confirm that LINC00844 can inhibit multiple processes that are essential for the extrahepatic metastasis of HCC.
To explore the molecular mechanism underlying LINC00844 regulating HCC cell function, a gene expression microarray assay was carried out. The pathway analysis suggested that LINC00844 overexpression inhibits the MAPK signaling pathway. Previously published studies have confirmed that this pathway is associated with the metastatic phenotypes associated with malignancy. Walter et al. suggested that MAPK signaling activation could promote the phosphorylation of retinoblastoma, resulting in the inactivation of retinoblastoma and accelerating the acquisition of metastatic competency (25). Chen et al. suggested that the MAPK pathway is frequently aberrantly activated in metastatic prostate cancer, potentially due to genomic amplification of the PPP1CA gene, which acts as a B-Raf activating phosphatase (26). We conducted a CO-IP and MS analysis to investigate the mechanism of LINC00844 regulating the MAPK pathway and found that LINC00844 tightly binds with AZGP1.
AZGP1 is a 42-kDa, soluble, secretory protein with a homologous structure and similar amino acid sequence to proteins in the major histocompatibility complex class I family (27). Previous studies have identified AZGP1 as an anti-inflammatory adipocytokine that is able to suppress tumor development (28). Huang et al. found that AZGP1 expression was decreased in HCC and was correlated with several clinicopathological parameters, including high AFP, poorer tumor differentiation, and reduced RFS (29). Moreover, AZGP1 was reported to be actively involved in the suppressive regulation of tumor growth and proliferation (30). Interestingly, some studies found that AZGP1 could block TGF-β1-mediated, ERK phosphorylation (31). Well-known induction of metastatic phenotype by TGF-β in tumor cells needed alteration in additional signaling cascades including MAPK, and TGF-β1 was also an activator of ERK phosphorylation, these two signaling pathways had inextricable links (32,33). In their study, Kong et al. suggested that AZGP1 blocked TGF-β1-mediated MAPK activation, which was dependent on Smad4, rather than Ras/ERK signaling (34). In a CO-IP assay, direct binding between p-ERK, TGF-β1, and AZGP1 was observed, which resulted in an anti-inflammatory effect (10). Moreover, our results revealed that AZGP1 knockdown also inhibits TGF-β1 expression. These data provide evidence for the crucial role of AZGP1 in regulating the TGF-β1-ERK2 pathway.
Conclusions
The suppression of LINC00844 expression plays a significant role in the pathogenesis and pathophysiology of HCC by promoting the AZGP1-mediated TGF-β1-ERK pathways, which results in HCC recurrence and unsatisfactory survival outcomes. Epigenetically regulated AZGP1 might be a promising therapy against HCC for maintaining epithelium.
Supplementary
The article’s supplementary files as
Acknowledgments
The author would like to thank Dr. Danjing Guo and Dr. Rong Su for their assistance with the study.
Funding: The present study was supported by the Key Research Development Program of Zhejiang Province (No. 2018C03018), Science and Technology Projects of Medicine and Health in Zhejiang Province (No. WKJ-ZJ-1923 and 2020383364), National Key R&D Program of China (No. 2017YFCO114102), and Science and Technology Project of Jiaxing (No. 2018AD32058 and 2019AD32208).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was performed in accordance with the principles of the Declaration of Helsinki (as revised in 2013) and approved by the Ethical Committee at the First Affiliated Hospital, School of Medicine, Zhejiang University (No. 2018658). Individual consent for this retrospective analysis was waived. Experiments were performed under a project license (No. 2018705) granted by institutional ethics board of Zhejiang University, in compliance with Chinese guidelines for the care and use of animals.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3848
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3848
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3848). The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Xu X, Lu D, Ling Q, et al. Liver transplantation for hepatocellular carcinoma beyond the Milan criteria. Gut 2016;65:1035-41. 10.1136/gutjnl-2014-308513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coelho T, Adams D, Silva A, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med 2013;369:819-29. 10.1056/NEJMoa1208760 [DOI] [PubMed] [Google Scholar]
- 4.Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 2016;30:34-51. 10.1101/gad.270959.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R, Sun Y, Li L, et al. Preclinical Study using Malat1 Small Interfering RNA or Androgen Receptor Splicing Variant 7 Degradation Enhancer ASC-J9((R)) to Suppress Enzalutamide-resistant Prostate Cancer Progression. Eur Urol 2017;72:835-44. 10.1016/j.eururo.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Zhu P, Lu T, et al. The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J Hepatol 2019;70:918-29. 10.1016/j.jhep.2018.12.015 [DOI] [PubMed] [Google Scholar]
- 7.Xiao J, Lv Y, Jin F, et al. LncRNA HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem 2017;43:1926-38. 10.1159/000484116 [DOI] [PubMed] [Google Scholar]
- 8.Ye Y, Xu Y, Lai Y, et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem 2018;119:2951-63. 10.1002/jcb.26509 [DOI] [PubMed] [Google Scholar]
- 9.Lin B, Chen T, Zhang Q, et al. FAM83D associates with high tumor recurrence after liver transplantation involving expansion of CD44+ carcinoma stem cells. Oncotarget 2016;7:77495-507. 10.18632/oncotarget.12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Wang T, Liu X, et al. Neuronal zinc-alpha2-glycoprotein is decreased in temporal lobe epilepsy in patients and rats. Neuroscience 2017;357:56-66. 10.1016/j.neuroscience.2017.05.043 [DOI] [PubMed] [Google Scholar]
- 11.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 2014;63:844-55. 10.1136/gutjnl-2013-306627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verna EC, Patel YA, Aggarwal A, et al. Liver transplantation for hepatocellular carcinoma: management after the transplant. Am J Transplant 2020;20:333-47. 10.1111/ajt.15697 [DOI] [PubMed] [Google Scholar]
- 13.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin 2012;62:394-9. 10.3322/caac.21161 [DOI] [PubMed] [Google Scholar]
- 14.Mitra R, Chen X, Greenawalt EJ, et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun 2017;8:1604. 10.1038/s41467-017-01781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol 2017;19:1105-15. 10.1038/ncb3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigoutsos I, Lee SK, Nam SY, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol 2017;18:98. 10.1186/s13059-017-1224-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmerman MA, Ghobrial RM, Tong MJ, et al. Recurrence of hepatocellular carcinoma following liver transplantation: a review of preoperative and postoperative prognostic indicators. Arch Surg 2008;143:182-8; discussion 188. 10.1001/archsurg.2007.39 [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Rogoff HA, Keates S, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci U S A 2015;112:1839-44. 10.1073/pnas.1424171112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med 2019;216:1016-26. 10.1084/jem.20181827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015;16:413-25. 10.1016/j.stem.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 21.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666-81. 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 22.Nemes B, Gaman G, Polak WG, et al. Extended-criteria donors in liver transplantation Part II: reviewing the impact of extended-criteria donors on the complications and outcomes of liver transplantation. Expert Rev Gastroenterol Hepatol 2016;10:841-59. 10.1586/17474124.2016.1149062 [DOI] [PubMed] [Google Scholar]
- 23.Yao FY, Ferrell L, Bass NM, et al. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology 2001;33:1394-403. 10.1053/jhep.2001.24563 [DOI] [PubMed] [Google Scholar]
- 24.Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol 2009;10:35-43. 10.1016/S1470-2045(08)70284-5 [DOI] [PubMed] [Google Scholar]
- 25.Walter DM, Yates TJ, Ruiz-Torres M, et al. RB constrains lineage fidelity and multiple stages of tumour progression and metastasis. Nature 2019;569:423-7. 10.1038/s41586-019-1172-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Wan L, Zhang J, et al. Deregulated PP1alpha phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat Commun 2018;9:159. 10.1038/s41467-017-02272-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgi W, Schmid K. Preparation and properties of Zn-alpha 2-glycoprotein of normal human plasma. J Biol Chem 1961;236:1066-74. [PubMed] [Google Scholar]
- 28.Leal VO, Mafra D. Adipokines in obesity. Clin Chim Acta 2013;419:87-94. 10.1016/j.cca.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 29.Huang Y, Li LZ, Zhang CZ, et al. Decreased expression of zinc-alpha2-glycoprotein in hepatocellular carcinoma associates with poor prognosis. J Transl Med 2012;10:106. 10.1186/1479-5876-10-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hale LP. Zinc alpha-2-glycoprotein regulates melanin production by normal and malignant melanocytes. J Invest Dermatol 2002;119:464-70. 10.1046/j.1523-1747.2002.01813.x [DOI] [PubMed] [Google Scholar]
- 31.Xu MY, Chen R, Yu JX, et al. AZGP1 suppresses epithelial-to-mesenchymal transition and hepatic carcinogenesis by blocking TGFbeta1-ERK2 pathways. Cancer Lett 2016;374:241-9. 10.1016/j.canlet.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 32.Reichl P, Haider C, Grubinger M, et al. TGF-beta in epithelial to mesenchymal transition and metastasis of liver carcinoma. Curr Pharm Des 2012;18:4135-47. 10.2174/138161212802430477 [DOI] [PubMed] [Google Scholar]
- 33.Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-beta receptor function. Trends Cell Biol 2009;19:385-94. 10.1016/j.tcb.2009.05.008 [DOI] [PubMed] [Google Scholar]
- 34.Kong B, Michalski CW, Hong X, et al. AZGP1 is a tumor suppressor in pancreatic cancer inducing mesenchymal-to-epithelial transdifferentiation by inhibiting TGF-beta-mediated ERK signaling. Oncogene 2010;29:5146-58. 10.1038/onc.2010.258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as