Abstract
Background
Ferroptosis is a novel form of regulated cell death that can inhibit the progression of chemotherapy-resistant tumors. However, the types of cancer most susceptible to ferroptosis induction and the role of ferroptosis regulators in cancers, especially pancreatic cancer, remain unclear.
Methods
RNA sequencing data of 31 cancers were collected from The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx). A nomogram integrating patients’ clinical information and risk scores based on the expression levels of ferroptosis regulators was depicted. Correlations among the activity levels of 29 immunity-associated gene sets, immune scores, infiltrating immune cells and key ferroptosis regulators were assessed.
Results
We performed a pan-cancer analysis and identified 14 distinct cancers that may show a robust response to ferroptosis inducers. Interestingly, the Xc-complex, which is the major target of ferroptosis induction, was upregulated in gemcitabine-resistant pancreatic cancer cells (P<0.05). Furthermore, we focused on the role of ferroptosis regulators in mediating the survival of patients with pancreatic cancer and constructed a prognostic model with good accuracy (AUC =0.713). We also correlated elevated sensitivity to ferroptosis with higher scores for CD8+ T cells (P<0.001), the type two interferon response (P<0.001) and immune checkpoints (P<0.05).
Conclusions
We hypothesized that the ferroptosis pathway plays an important role in the prognosis of pancreatic cancer. Immuno- and chemotherapy combined with a ferroptosis inducer is a feasible therapeutic approach for pancreatic cancer.
Keywords: Ferroptosis, pancreatic cancer, immunity, gemcitabine resistance, prognosis
Introduction
Ferroptosis is a newly identified form of regulated cell death (RCD) characterized by iron-dependent accumulation of lipid peroxidation products (1-3). Since the concept was first proposed by Dixon et al. in 2012, ferroptosis has been found to participate in many physiopathological processes, including the development of many kinds of cancers (1). Some studies have indicated that ferroptosis induction inhibits the progression of chemotherapy-resistant tumors (4-6). Several underlying mechanisms, including lncRNA regulation, immune evasion, immune surveillance and TP53-related pathway modulation, may explain the anticancer benefits observed after ferroptosis induction (7-9).
Pancreatic cancer is one of the most lethal malignancies with annual mortality rates almost equal to incidence rates (10). In recent years, the incidence of pancreatic cancer has increased annually with an annual percent change of 0.56–1.23% (11), which has imposed heavy health and economic burdens on most countries. Although immunotherapy has gradually become a popular and effective treatment method for some malignancies, pancreatic cancer is insensitive to programmed cell death-1/programmed cell death-ligand 1 (PD1/PD-L1) blockade-mediated targeted therapy, which is considered to be immunotherapy “cold” (12,13). Moreover, gemcitabine, the key drug in first-line chemotherapeutic regimens for pancreatic cancer, shows effective and anticancer ability at first but fails to control the disease (14,15). Hence, the development of a new strategy to enhance the sensitivity of pancreatic cancer to immunotherapy and reduce its resistance to gemcitabine is an urgent task.
Several regimens have been reported to induce ferroptosis via inhibition of specific targets in the ferroptosis pathway (16). For example, erastin, a typical ferroptosis inducer, directly inhibits SLC7A11, which depletes glutathione, a major antioxidant in cells (1). RSL3 induces ferroptosis through depletion of GPX4, an important reductase in reactive oxygen species (ROS) clearance (17). In addition, sorafenib, a multitargeted anticancer agent, induces ferroptosis via blockade of the Xc-complex, which consists of SLC7A11 and SLC3A2, hindering intracellular cysteine influx and thus glutathione synthesis (17). Ferroptosis can be manipulated by many anticancer regimens, and some of which have been approved by the U.S. Food and Drug Administration (17,18). We believe that safe and effective ferroptosis inducers have potential roles in adjuvant chemotherapy for cancers, but the types of cancers that are sensitive to ferroptosis induction remain unknown. ACSL4 dictates the sensitivity to ferroptosis in tumor cells (19). Thus, tumors overexpressing ASCL4 may be sensitive to ferroptosis induction. In addition, only a few studies to date have focused on the role of ferroptosis in pancreatic cancer (18,20-22), and most of these studies were only based on cell experiments.
Here, we conducted a human sample-based study to investigate the expression levels of ferroptosis-related genes in 31 distinct cancers and to determine the potential relationships between ferroptosis and the prognosis, immune microenvironment and gemcitabine resistance of pancreatic cancer.
Methods
Data sources and online tools
Forty-three genes involved in the ferroptosis process (hereafter, ferroptosis regulators) according to previous reports were identified and analyzed in this study. Data from The Cancer Genome Atlas pancreatic adenocarcinoma (TCGA-PAAD) dataset combined with data from the Genotype-Tissue Expression (GTEx) project were collected to investigate the differential expression of these 43 ferroptosis regulators in 31 cancers. In detail, transcriptome data for 176 pancreatic cancer samples and four normal pancreas tissues were collected from the TCGA-PAAD dataset, and data for 167 normal samples were collected from the GTEx project to compensate for the insufficiency of normal tissues in the TCGA-PAAD dataset. Fold change (FC) refers to the ratio of gene expression levels between two groups. The threshold for significant differential gene expression was established as P<0.05 and |logFC|>1. The abovementioned analysis was performed in Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn), an online tool universally recognized by researchers. We acquired the list of Gene Expression Omnibus (GEO) datasets pertaining to pancreatic cancer from the study conducted by Xie et al. (23). We then removed the datasets containing nonpancreatic samples, such as saliva and peripheral blood from patients with pancreatic cancer. Finally, seven GEO datasets (GSE15471, GSE62452, GSE11838, GSE32676, GSE41368, GSE43795 and GSE43288) were used as validation cohorts (Table S1). The GSE80617 GEO dataset was used to assess differential gene expression in gemcitabine-resistant pancreatic cancer cells. Mutation analysis of the 43 ferroptosis regulators in pancreatic cancer was performed in cBioPortal (www.cbioportal.org/). Immunohistochemical data were collected from the Human Protein Atlas (https://www.proteinatlas.org/) for validation at the protein level. For construction of the prognostic model, we selected only tumor samples with a tumor purity greater than 0.6 to ensure the reliability of our study. One GEO dataset (GSE57495) containing 63 tumor samples was used as the validation cohort.
The information of 29 immune gene sets is summarized in Table S2. Data pertaining to immune cell infiltration were downloaded from CIBERSORT (https://cibersort.stanford.edu/), a commonly used online tool for analysis of the immune microenvironment. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
Linear data in two groups were compared using an unpaired t test. If a variable was being compared across more than two groups, one-way analysis of variance (ANOVA) was applied. Cox regression analysis was conducted to calculate the hazard ratio of each ferroptosis regulator for the overall survival time. Lasso regression was conducted to calculate the risk coefficient of each gene after removal of genes with an overfitting trend according to the partial likelihood deviance and lambda value. The lambda value is determined as the value that minimizes the likelihood deviance, and the coefficient-lambda curve shows the genes that are eligible when the lambda value is determined. The lasso risk was calculated using the following formula: . Finally, the remaining genes were utilized to construct a predictive model for the prognosis of patients in the PAAD dataset. When we evaluated the accuracy of our prognostic model, we adjusted the expression level of each ferroptosis regulator in the separate GEO datasets due to the different sequencing platforms used, which ensured optimized comparability between the validation and TCGA cohorts. First, we used the following formula to standardize the expression level of each gene: , , . Then, we used the following formula to adjust each Xstd to match the TCGA training data: . All correlation analyses were conducted with GraphPad Prism 7.04, and the Pearson coefficient (r) reflected the strength of the correlation. Other statistical analyses were performed with R software (version 3.6.1). Survival analysis was implemented using the R package “survival”. A receiver operating characteristic (ROC) curve was depicted using the R package “survivalROC”. The “glmnet” package was used to perform Cox proportional hazards regression analysis with least absolute shrinkage (glmnet, version 2.0-18). Nomograms and calibration curves were constructed with the package “rms 5.1-4”. The package “Estimate” was used to calculate the immune score and tumor purity of each tumor sample. The co-expression mean value of SLC3A2 and SLC7A11 was defined as the sum of their adjusted expression levels, which was calculated according to the following formula: . Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted with the package “org.Hs.eg.db 3.10”. P<0.05 was considered statistically significant.
Role of funding sources
All funding sources are listed in the acknowledgement section. Sponsors are supposed to pay the fee for publication.
Results
Ferroptosis regulators are differentially expressed in tumor samples compared to adjacent normal tissues
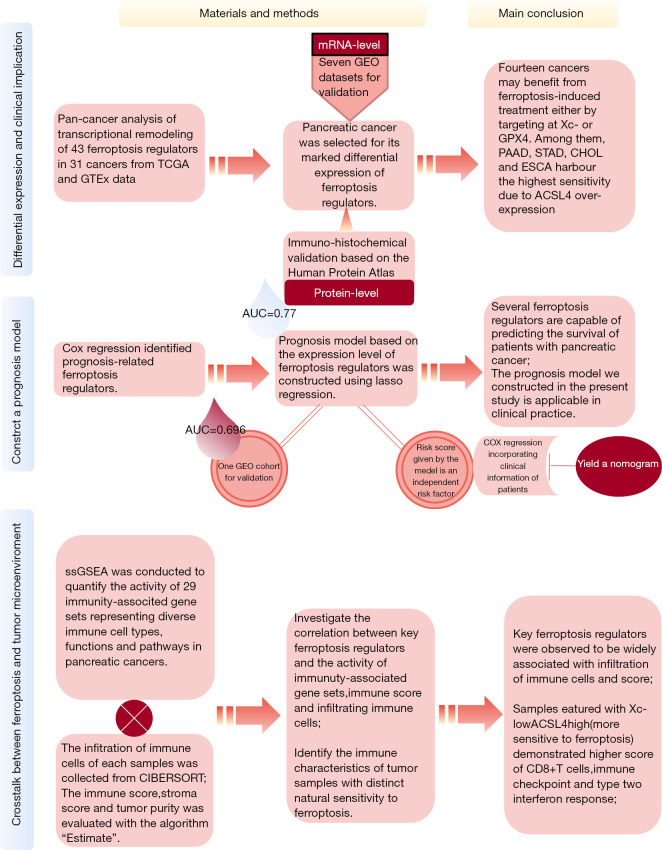
A flow chart is provided in Figure 1 to summarize the entire study.
Figure 1.
Study design and flow chart.
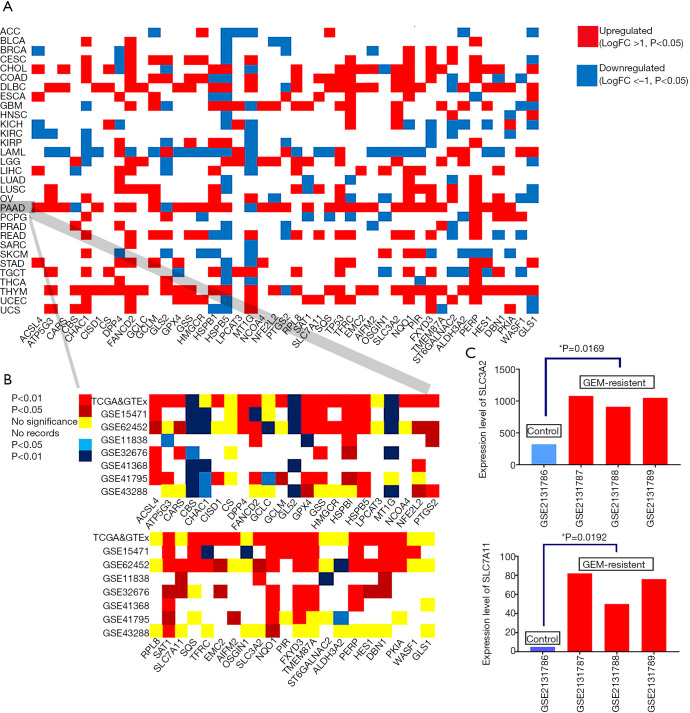
Overall, each of the 31 cancers had several differentially expressed ferroptosis regulators in the pan-cancer analysis (Figure 2A). Ferroptosis regulators were broadly upregulated in most tumor samples, except for acute myeloid leukemia samples in which only 8 genes were upregulated. Notably, the Xc-complex and GPX4, the key targets for ferroptosis regulation, were upregulated in 14 cancers, indicating that specific antagonists of the Xc-complex and GPX4 may benefit patients with these cancers v[cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophore (KICH), brain lower grade glioma (LGG), lung squamous cell carcinoma (LUSC), PAAD, rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thymoma (THYM), and uterine corpus endometrial carcinoma (UCEC)]. This analysis revealed that pancreatic cancer exhibited marked variation in the expression profile of ferroptosis regulators. Hence, we further investigated the role of this change in the prognosis of patients with pancreatic cancer and the potential crosstalk between the immune microenvironment and ferroptosis.
Figure 2.
Differential expression profiles of 43 ferroptosis regulators in cancer and normal samples. (A) The differential expression profiles of 43 ferroptosis regulators in 31 distinct cancer and normal samples. (B) The expression profiles of 43 ferroptosis regulators in seven Gene Expression Omnibus (GEO) cohorts were similar to the results in the The Cancer Genome Atlas (TCGA)/The Genotype-Tissue Expression (GTEx) cohort. (C) The expression levels of SLC3A2 and SLC7A11 were upregulated in gemcitabine-resistant pancreatic cancer cells.
Further validation of the expression profile of ferroptosis regulators in pancreatic cancer
The validation cohort consisted of seven GEO datasets with a distribution similar to that of TCGA cohort regarding the differential expression of ferroptosis regulators (Figure 2B). Overall, most ferroptosis regulators, including SLC3A2, the major component of the Xc-complex that regulates the ferroptosis process, were highly expressed in pancreatic cancer. Unexpectedly, the other component of the Xc-complex, SLC7A11, did not exhibit significant upregulation in the TCGA cohort, but it was upregulated in three of the GEO datasets (GSE62452, GSE11838, and GSE32676). Another key regulator of ferroptosis, GPX4, was also upregulated in tumor samples, supporting the biological plausibility of agents targeting GPX4. NCOA4 and CISD1 were upregulated in pancreatic cancer and in a few other cancer types, suggesting that they play roles specifically in the pathophysiology of pancreatic cancer and these few other cancers through ferroptosis regulation. In addition, we analyzed the proteomic data of the ferroptosis regulators in the Human Protein Atlas and found that the protein expression levels of these ferroptosis regulators were similar to their mRNA levels (Figure S1).
Mutations in ferroptosis regulators in pancreatic cancer samples are associated with shorter overall and disease-free survival times
Mutation data for 43 ferroptosis regulators in 850 samples were extracted from 4 databases [International Cancer Genome Consortium (ICGC), Queensland Centre for Medical Genomics (QCMG), TCGA and University of Texas Southwestern Medical Center (UTSW)]. Mutation of ferroptosis regulators was found in 171 samples (20%). Survival analysis showed that mutations in ferroptosis regulators in tumor samples were associated with shorter overall and disease-free survival times (Figure S2A,B). Compared to samples with copy number alterations in ferroptosis regulator genes, their unaltered counterparts had fewer copy number alterations in other genes as shown in the volcano plot (Figure S2C,D). CDKN2A, TP53, KRAS and SMAD4 are the four driver genes in pancreatic cancer initiation and development. In samples harboring mutations in ferroptosis regulators, the copy number alteration frequency of the four driver genes was increased (Figure S2E).
The Xc-complex and five other ferroptosis regulators are upregulated in gemcitabine-resistant pancreatic ductal adenocarcinoma cells
We analyzed the mRNA expression profiles of parental Panc1 cell lines and gemcitabine-resistant Panc1 cell lines in a GEO dataset (GSE80617). Interestingly, seven ferroptosis regulators were upregulated after continuous exposure to gemcitabine for 2 months (Table S3). Among these upregulated ferroptosis regulators, two components of the Xc-complex were dramatically upregulated, suggesting that ferroptosis-inducing agents targeting the Xc-complex may be promising treatments for gemcitabine-resistant pancreatic cancer (Figure 2C).
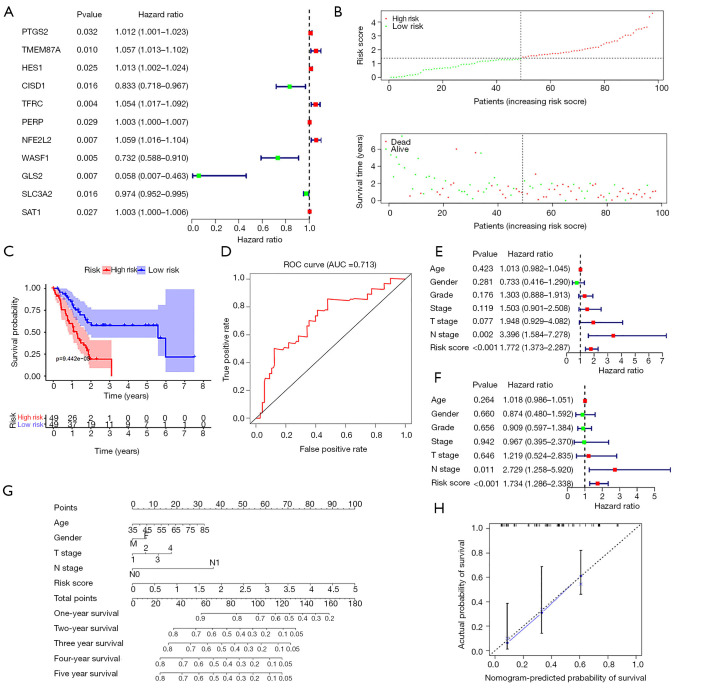
A predictive model based on ferroptosis regulators accurately evaluates pancreatic cancer patient survival time
To ensure the reliability of this study, we first selected 98 tumor samples with a tumor purity greater than 0.6 for the subsequent analysis. From the set of 43 ferroptosis regulators, univariate Cox regression analysis identified 11 genes associated with the prognosis of pancreatic cancer (Figure 3A). Among these genes, CISD1, WASF1, GLS2 and SLC3A2 were negatively associated with the overall survival time, while overexpression of the other genes dictated a prolonged survival time. Lasso regression was conducted to calculate the risk value of each sample in terms of survival prognosis based on the expression level of ferroptosis regulators. To avoid the overfitting phenomenon, only five genes (CISD1, WASF1, SAT1, TMEM87A and SLC3A2) were retained in the predictive model after lasso regression (Figure S3). The risk score curve demonstrated the distribution of tumor samples based on the lasso risk values calculated by our model (Figure 3B). As the survival curve shows (Figure 3C), samples with low lasso risk values were associated with a significant increase in survival time compared to those with high lasso risk values (P<0.01). Next, we generated a ROC curve to evaluate the accuracy of our predictive model. The area under the curve (AUC) value of this model was 0.713 (Figure 3D), indicating intermediate statistical accuracy. One GEO cohort (GSE57495) was used as a validation cohort for our predictive model. Survival analysis also confirmed a significant survival advantage for tumor samples with low lasso risk values (Figure S4A) despite a slight decrease in the AUC value (which decreased to 0.696) (Figure S4B). Furthermore, univariate and multivariate Cox analyses were combined to determine whether the lasso risk is an independent predictive factor for pancreatic cancer prognosis in our model. Both the N stage and lasso risk value independently predicted survival expectancy (Figure 3E,F). Finally, we constructed a nomogram to help clinicians conveniently use our model in combination with some clinical characteristics, such as age, T stage and N stage, to predict the survival expectancy of a specific patient with pancreatic cancer (Figure 3G). The calibration curve indicated the high accuracy of our nomogram (Figure 3H).
Figure 3.
Prognostic model for pancreatic cancer based on ferroptosis regulators. (A) Forest plot showing the ferroptosis regulators associated with the survival of patients with pancreatic cancer; (B) risk score curve showing the distribution of patients with distinct lasso risk coefficients calculated by our model; (C) survival curve presenting the significant difference between the high- and low-risk groups in terms of survival outcomes; (D) receiver operating characteristic (ROC) curve showing the good accuracy of our model for the prediction of pancreatic cancer survival; (E,F) univariate and multivariate regression analyses identified that the risk value calculated by our model is an independent indicator of pancreatic cancer survival; (G) the nomogram combining ferroptosis-based risk values with several clinical factors accurately predicted the survival of patients with pancreatic cancer; (H) the calibration curve confirmed the accuracy of the nomogram.
Elevated sensitivity to ferroptosis is correlated with enhanced immune activity and potential for antitumor immune therapy
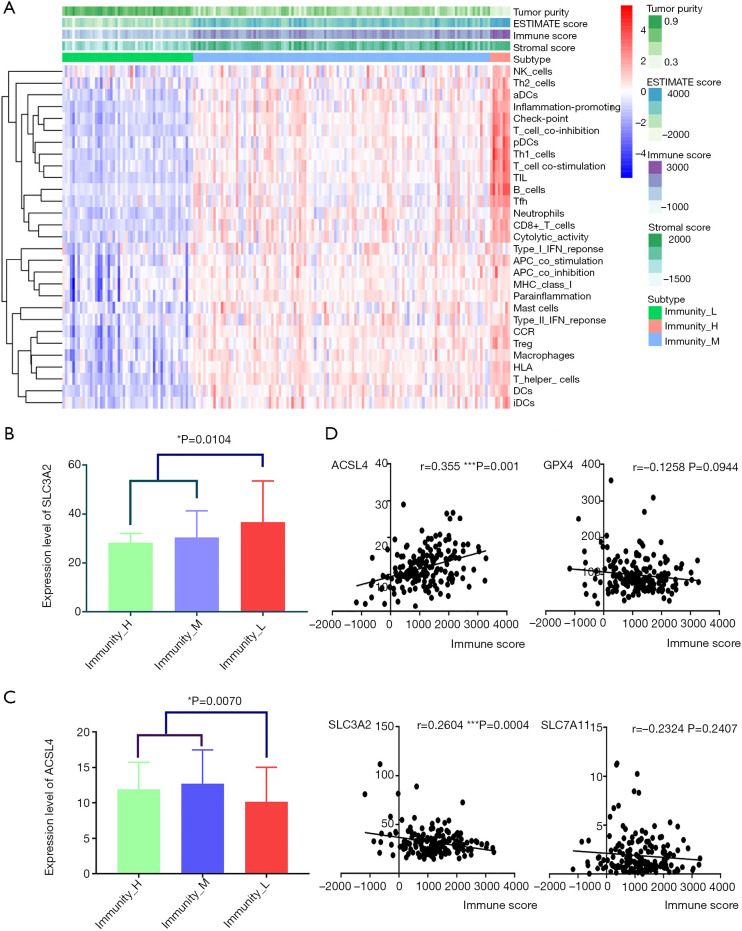
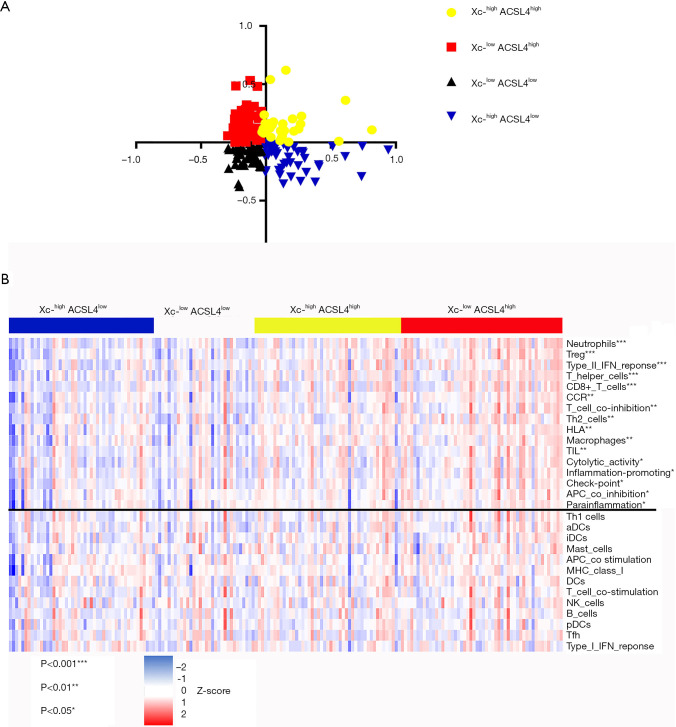
Single-sample gene set enrichment analysis (ssGSEA) was conducted to quantify the activity or enrichment levels of immune cells, functions or pathways in pancreatic cancer samples by analyzing 29 immune-associated gene sets representing diverse immune cell types, functions and pathways. Then, 178 tumor samples were clustered into three groups based on immune activity (Figure 4A). Interestingly, when we compared the expression levels of four essential ferroptosis regulators (ACSL4, GPX4, SLC3A2 and SLC7A11) in each immunity-based group, we found that SLC3A2 was significantly upregulated in the Immunity_low group (Figure 4B), while ACSL4 showed the opposite pattern (Figure 4C). Hence, we further explored potential associations among immune scores, immune cell infiltration and the expression levels of three key ferroptosis regulators.
Figure 4.
Single-sample gene set enrichment analysis (ssGSEA) classified pancreatic cancer samples into three immunity-based groups. (A) Samples were classified based on the activity of 29 immunity-associated pathways; (B) SLC3A2 was highly expressed in the groups with lower immune activity; (C) ACSL4 was downregulated in the groups with lower immune activity; (D) the correlation between immune scores and key ferroptosis regulators.
Linear regression analysis showed that SLC3A2 was negatively associated with the immune score (Figure 4D) but that ACSL4 was positively correlated with the immune score (Figure 4D). Next, we investigated correlations between six types of common infiltrating immune cells and the expression of ACSL4, GPX4, SLC3A2 and SLC7A11. As shown in Figure S5, ACSL4 was positively correlated with the numbers of B cells, CD8+ T cells, dendritic cells, macrophages and neutrophils. GPX was negatively associated with the numbers of CD8+ T cells, dendritic cells, macrophages and neutrophils. SLC3A2 was negatively associated only with the number of neutrophils, and SLC7A11 was negatively correlated with the numbers of CD4+ T cells and macrophages.
We further divided 178 tumor samples into 4 groups based on the adjusted expression levels of ACSL4 and the Xc-complex (Figure 5A). The expression level of the Xc-complex is defined as the co-expression mean value of SLC3A2 and SLC7A11. Samples with Xc-lowACSL4high exhibited enhanced scores for CD8+ T cells, the type two interferon response and immune checkpoints, which are indicators of better efficacy of checkpoint inhibitor-based antitumor therapy (Figure 5B).
Figure 5.
Samples with distinct sensitivity to ferroptosis exhibited different immune characteristics. (A) Samples were divided into four groups with distinct sensitivity to ferroptosis; (B) groups with distinct sensitivity to ferroptosis showed varying activity in 16 immunity-associated pathways.
GO and KEGG analyses of the 43 ferroptosis regulators reveal alteration of some ferroptosis-related pathways in pancreatic cancer
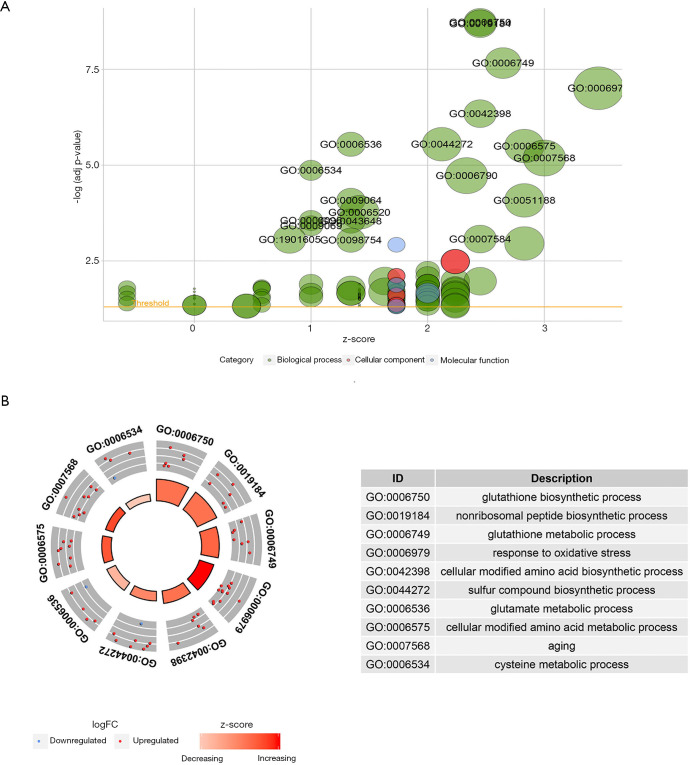
GO analysis showed that the 43 analyzed ferroptosis regulators are enriched mostly in the following biological processes: “response to oxidative stress”, “glutathione synthesis and metabolism” and “sulfur compound and amino acid synthesis and metabolism”. Most translation products of these genes were distributed at the outer membrane of organelles. Regulation of ligase and antioxidant activity was the main molecular function of these ferroptosis regulators (Figure 6). KEGG analysis revealed that the 43 genes are involved in the following signaling pathways: “ferroptosis”, “glutathione metabolism” and “cysteine and methionine metabolism” (Figure S6). Thus, both the GO and KEGG analysis results confirmed that the 43 selected genes are ferroptosis-related and emphasized the role of glutathione biosynthesis and metabolism in the development of pancreatic cancer.
Figure 6.
Gene Ontology (GO) analyses of the 43 ferroptosis regulators.
Discussion
While an increasing number of studies have explored the role of ferroptosis in the initiation and progression of cancers in the past decade, many questions remain unanswered, and more data remain to be mined (2,24). For example, it is important to understand which types of cancer are most sensitive to ferroptosis induction. A primary consideration is that susceptibility to ferroptosis induction differs dramatically among cancer cells. Xie et al. identified DLBC cells as the most susceptible to erastin-induced ferroptosis among several types of cancer cells (breast cancer cells, lung cancer cells, colon cancer cells, melanocytes, central nervous system cancer cells and ovarian cancer cells) (25). We sought to determine the molecular features that a certain type of cancer should exhibit for susceptibility to ferroptosis induction. We investigated the expression levels of 43 ferroptosis regulators in resected human cancer samples via bioinformatic analysis and identified 14 types of cancer with upregulation of either the Xc-complex or GPX4 (CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, LGG, LUSC, PAAD, READ, STAD, THYM and UCEC). High expression of the Xc-complex and GPX4 endows these cancers with natural robust resistance to ferroptosis. Given that almost all ferroptosis inducers target either the Xc-complex or GPX4, however, we speculated that high expression of SLC7A11, SLC3A2 and GPX4 may be the molecular signature of promising efficacy of ferroptosis-based treatment. In addition, among the 14 abovementioned cancers, 4 types (PAAD, STAD, CHOL and ESCA) exhibit ACSL-4 upregulation, which may enhance ferroptosis sensitivity by enriching cellular membranes with long polyunsaturated ω6 fatty acids, indicating that these cancers may be susceptible to ferroptosis-inducing treatment (19). Our findings have important clinical implications. For example, STAD, one type of tumor, overexpresses only GPX4 and not the Xc-complex. Thus, when designing the plan for treating STAD with ferroptosis inducers, regimens including agents specifically targeting GPX4, such as RSL-3, ML162 and some kinds of diverse pharmacological inhibitor (DPI) compounds, should be prioritized. Recently, many drugs have been reported to induce ferroptosis via other targets. For instance, statins block CoQ10 biosynthesis by targeting HMGCR, which leads to accumulation of lipid peroxidation products and cell death (26). Therefore, our results also provide guidance for the use of these regimens in cancer treatment.
Given the associations between some ferroptosis regulators and pancreatic cancer, we constructed a prognostic model with good predictive accuracy. In addition, our model showed intermediate accuracy in a GEO validation cohort, suggesting its capability to predict pancreatic cancer survival in other data sources. However, we constructed a nomogram integrating patients’ age, sex, T stage, N stage and risk score based on the expression levels of ferroptosis regulators, providing a convenient approach for clinicians to predict the survival of patients with pancreatic cancer.
The mechanism by which ferroptosis inducers cause cancer cell death is clear. For example, binding of erastin to the Xc-complex blocks the import of cysteine. Glutathione biosynthesis then decreases, leading to insufficient levels of glutathione, an essential cofactor for GPX4 in lipid peroxide elimination. Finally, excessive accumulation of lipid peroxides rapidly causes cell death. In fact, other ferroptosis inducers exert their effects through a similar mechanism. However, because ferroptosis is a form of RCD occurring in physiological cells, the mechanism underlying whether and how its dysregulation participates in tumor initiation and progression is unclear. A recent study has demonstrated that a single-nucleotide polymorphism at codon 47 in TP53 (referred to as S47) inhibits ferroptosis in hepatocellular carcinoma (HCC) (27). This study concluded that this variant of p53 causes a defect in ferroptosis induction, resulting in an increased risk of cancer in a mouse model. In patients with breast and lung cancers harboring wild-type p53, low expression of the lncRNA P53RRA is significantly correlated with poor survival. Furthermore, Mao et al. found that the cytosolic P53RRA-G3BP1 interaction displaces p53 from the G3BP1 complex, resulting in increased nuclear accumulation of p53, which leads to ferroptosis (8). Here, we also discovered that the copy number alteration frequency was higher in samples harboring mutations in ferroptosis regulators than in control samples (Figure 3E). Therefore, TP53 mutation may be a biologically plausible origin of ferroptosis inhibition in pretumor cells, which initiates tumorigenesis. Some researchers have referred to ferroptosis as autophagic cell death (28). NCOA4-mediated autophagic degradation of iron storage proteins (called ferritinophagy) controls the accumulation of cellular labile iron and further induces ferroptosis (28,29). In this context, the disturbance of cellular autophagic homeostasis also mitigates ferroptotic activity, which probably promotes tumorigenesis. Other mechanisms of ferroptosis modulation in cancers, such as lipid storage (30), regulation of the OTUB1 deubiquitylase (31) and inhibition of Nrf2 (5), are also interesting.
The tumor microenvironment has become a central factor focused on by almost all oncologists. Until recently, however, the crosstalk between the immune system and ferroptosis has not been studied. Wang et al. found that CD8+ T cells induce ferroptosis during cancer immunotherapy by releasing interferon gamma, which downregulates the expression of the Xc-complex (32). Pancreatic cancer is thought to be a “cold” tumor in terms of immunotherapy. Hence, developing a new strategy to stimulate the response of these tumors to immunotherapy is an urgent task. Here, we found that ACSL4, a biomarker for ferroptosis sensitivity, is positively associated with the immune score and the number of CD8+ T cells. SLC7A11, SLC3A2 and GPX4 are three biomarkers whose overexpression indicates resistance to ferroptosis induction. Among these biomarkers, SLC3A2 was negatively associated with the immune score, and each biomarker was negatively correlated with the numbers of certain immune cells. Based on the ssGSEA results, we found that tumor samples with XclowACSL4high exhibited higher scores for CD8+ T cells, the type two interferon response and immune checkpoints. All these findings suggested that samples with high sensitivity to ferroptosis may also be sensitive to checkpoint inhibitor-based immunotherapy and vice versa. Under these circumstances, immunotherapy combined with a ferroptosis inducer is a feasible therapeutic approach for pancreatic cancer, but more studies are needed to confirm this conclusion.
Gemcitabine is the first-line adjuvant chemotherapeutic drug for pancreatic cancer. Here, we found that both SLC7A11 and SLC3A2 are upregulated in gemcitabine-resistant cancer cells. Previous studies have proposed many explanations for chemotherapeutic resistance. For example, scientists have shown that cytotoxic, chemotherapy-associated release of prostaglandin E2 (PGE2) promotes the repopulation of neighboring cancer stem cells (33). Here, we hypothesized the following potential mechanism by which tumor cells acquire drug resistance: a decline in ferroptosis sensitivity mediated by upregulated expression of the Xc-complex. However, more studies are needed to confirm our hypothesis.
Our study has some strengths. First, in this human tissue-based study, we systematically explored the expression of 43 ferroptosis regulators in 31 distinct cancers. Thus, this study is more convincing and valuable than other cell-based and single-cancer analyses in the literature. Second, our prognostic model and nomogram have considerable clinical implications, especially for clinicians asked to provide a potential patient survival time. Third, we are the first to correlate enhanced immune activity with enhanced ferroptosis sensitivity and robust gemcitabine resistance with diminished ferroptosis sensitivity in pancreatic cancer. Certainly, our study also has some limitations. First, we used only the sample information recorded in TCGA, GEO and other existing online databases, and we did not analyze fresh samples. In addition, due to the severe lack of data for the M stage in pancreatic cancer samples in TCGA database, we analyzed only the T and N stages for nomogram construction.
In conclusion, our study outlined the expression profiles of 43 ferroptosis regulators in 31 cancers and constructed a ferroptosis regulator-based prognostic model for pancreatic cancer. We also discussed the potential relationships among the immune microenvironment, gemcitabine resistance and ferroptosis sensitivity. Future studies are expected to elucidate the underlying mechanism by which immune-related factors mediate the regulation of ferroptosis sensitivity.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported in part by the National Natural Science Foundation of China (No. 81802352), the National Science Foundation for Distinguished Young Scholars of China (No. 81625016), the Shanghai Sailing Program (No. 17YF1402500), the Scientific Innovation Project of Shanghai Education Committee (2019-01-07-00-07-E00057) and the Clinical and Scientific Innovation Project of Shanghai Hospital Development Center (SHDC12018109).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2554a
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2554a). The authors have no conflicts of interest to declare.
References
- 1.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Z, Song J, Yung BC, et al. Emerging Strategies of Cancer Therapy Based on Ferroptosis. Adv Mater 2018;30:e1704007. 10.1002/adma.201704007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol 2016;26:165-76. 10.1016/j.tcb.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao M, Deng J, Liu F, et al. Triggered ferroptotic polymer micelles for reversing multidrug resistance to chemotherapy. Biomaterials 2019;223:119486. 10.1016/j.biomaterials.2019.119486 [DOI] [PubMed] [Google Scholar]
- 5.Roh JL, Kim EH, Jang H, et al. Nrf2 inhibition reverses the resistance of cisplatin-resistant head and neck cancer cells to artesunate-induced ferroptosis. Redox Biol 2017;11:254-62. 10.1016/j.redox.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin D, Kim EH, Lee J, et al. Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer. Free Radic Biol Med 2018;129:454-62. 10.1016/j.freeradbiomed.2018.10.426 [DOI] [PubMed] [Google Scholar]
- 7.Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520:57-62. 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao C, Wang X, Liu Y, et al. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res 2018;78:3484-96. 10.1158/0008-5472.CAN-17-3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang X, Green MD. Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer Discov 2019;9:1673-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore A, Donahue T. Pancreatic Cancer. JAMA 2019;322:1426. 10.1001/jama.2019.14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon-Dseagu VL, Devesa SS, Goggins M, et al. Pancreatic cancer incidence trends: evidence from the Surveillance, Epidemiology and End Results (SEER) population-based data. Int J Epidemiol 2018;47:427-39. 10.1093/ije/dyx232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Wang Y, Ding J, et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature 2020;579:421-6. 10.1038/s41586-020-2079-1 [DOI] [PubMed] [Google Scholar]
- 14.Binenbaum Y, Na'ara S, Gil Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist Updat 2015;23:55-68. 10.1016/j.drup.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 15.Wei L, Ye H, Li G, et al. Cancer-associated fibroblasts promote progression and gemcitabine resistance via the SDF-1/SATB-1 pathway in pancreatic cancer. Cell Death Dis 2018;9:1065. 10.1038/s41419-018-1104-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ 2016;23:369-79. 10.1038/cdd.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sui X, Zhang R, Liu S, et al. RSL3 Drives Ferroptosis Through GPX4 Inactivation and ROS Production in Colorectal Cancer. Front Pharmacol 2018;9:1371. 10.3389/fphar.2018.01371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eling N, Reuter L, Hazin J, et al. Identification of artesunate as a specific activator of ferroptosis in pancreatic cancer cells. Oncoscience 2015;2:517-32. 10.18632/oncoscience.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 2017;13:91-8. 10.1038/nchembio.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Guo G, Zhou X, et al. Potential mechanism of ferroptosis in pancreatic cancer. Oncol Lett 2020;19:579-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Zhang Z, Wang M, et al. Role of GRP78 inhibiting artesunate-induced ferroptosis in KRAS mutant pancreatic cancer cells. Drug Des Devel Ther 2019;13:2135-44. 10.2147/DDDT.S199459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi Y, Kasukabe T, Kumakura S. Piperlongumine rapidly induces the death of human pancreatic cancer cells mainly through the induction of ferroptosis. Int J Oncol 2018;52:1011-22. 10.3892/ijo.2018.4259 [DOI] [PubMed] [Google Scholar]
- 23.Xie ZC, Dang YW, Wei DM, et al. Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: a comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases. Onco Targets Ther 2017;10:3991-4005. 10.2147/OTT.S136878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu B, Chen XB, Ying MD, et al. The Role of Ferroptosis in Cancer Development and Treatment Response. Front Pharmacol 2018;8:992. 10.3389/fphar.2017.00992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Y, Song X, Sun X, et al. Identification of baicalein as a ferroptosis inhibitor by natural product library screening. Biochem Biophys Res Commun 2016;473:775-80. 10.1016/j.bbrc.2016.03.052 [DOI] [PubMed] [Google Scholar]
- 26.Chen JJ, Galluzzi L. Fighting Resilient Cancers with Iron. Trends Cell Biol 2018;28:77-8. 10.1016/j.tcb.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 27.Jennis M, Kung CP, Basu S, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev 2016;30:918-30. 10.1101/gad.275891.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M, Monian P, Pan Q, et al. Ferroptosis is an autophagic cell death process. Cell Res 2016;26:1021-32. 10.1038/cr.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou W, Xie Y, Song X, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 2016;12:1425-8. 10.1080/15548627.2016.1187366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, Meng L, Han L, et al. Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun 2019;508:997-1003. 10.1016/j.bbrc.2018.12.039 [DOI] [PubMed] [Google Scholar]
- 31.Liu T, Jiang L, Tavana O, et al. The Deubiquitylase OTUB1 Mediates Ferroptosis via Stabilization of SLC7A11. Cancer Res 2019;79:1913-24. 10.1158/0008-5472.CAN-18-3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. 10.1038/s41586-019-1170-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurtova AV, Xiao J, Mo Q, et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 2015;517:209-13. 10.1038/nature14034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as