Abstract
BACKGROUND
Gut tryptophan (Trp) metabolites are produced by microbiota and/or host metabolism. Some of them have been proven to promote or inhibit colorectal cancer (CRC) in vitro and animal models. We hypothesized that there is an alteration of gut Trp metabolism mediated by microbiota and that it might be involved in the pathogenesis of cancer in patients with CRC.
AIM
To investigate the features of Trp metabolism in CRC and the correlation between fecal Trp metabolites and gut microbiota.
METHODS
Seventy-nine patients with colorectal neoplastic lesions (33 with colon adenoma and 46 with sporadic CRC) and 38 healthy controls (HCs) meeting the inclusion and exclusion criteria were included in the study. Their demographic and clinical features were collected. Fecal Trp, kynurenine (KYN), and indoles (metabolites of Trp metabolized by gut microbiota) were examined by ultraperformance liquid chromatography coupled to tandem mass spectrometry. Gut barrier marker and indoleamine 2,3-dioxygenase 1 (IDO1) mRNA were analyzed by quantitative real-time polymerase chain reaction. Zonula occludens-1 (ZO-1) protein expression was analyzed by immunohistochemistry. The gut microbiota was detected by 16S ribosomal RNA gene sequencing. Correlations between fecal metabolites and other parameters were examined in all patients.
RESULTS
The absolute concentration of KYN [1.51 (0.70, 3.46) nmol/g vs 0.81 (0.64, 1.57) nmol/g, P = 0.036] and the ratio of KYN to Trp [7.39 (4.12, 11.72) × 10-3 vs 5.23 (1.86, 7.99) × 10-3, P = 0.032] were increased in the feces of patients with CRC compared to HCs, while the indoles to Trp ratio was decreased [1.34 (0.70, 2.63) vs 2.46 (1.25, 4.10), P = 0.029]. The relative ZO-1 mRNA levels in patients with CRC (0.27 ± 0.24) were significantly lower than those in HCs (1.00 ± 0.31) (P < 0.001), and the relative IDO1 mRNA levels in patients with CRC [1.65 (0.47-2.46)] were increased (P = 0.035). IDO1 mRNA levels were positively associated with the KYN/Trp ratio (r = 0.327, P = 0.003). ZO-1 mRNA and protein levels were positively correlated with the indoles/Trp ratio (P = 0.035 and P = 0.009, respectively). In addition, the genera Asaccharobacter (Actinobacteria) and Parabacteroides (Bacteroidetes), and members of the phylum Firmicutes (Clostridium XlVb, Fusicatenibacter, Anaerofilum, and Anaerostipes) decreased in CRC and exhibited a positive correlation with indoles in all subjects.
CONCLUSION
Alteration of fecal Trp metabolism mediated by microbiota is associated with intestinal barrier function and tissue Trp metabolism, and may be involved in the pathogenesis of CRC.
Keywords: Tryptophan metabolism, Colorectal cancer, Kynurenine, Indoles, Microbiota, Colorectal adenoma
Core Tip: This study comprehensively assessed the profiles of fecal tryptophan (Trp) metabolism in patients with colorectal cancer (CRC) and to explore the potential correlations between the gut microbiome, intestinal barrier function, tissue kynurenine pathway (KP), and alterations in fecal Trp metabolism. We found that CRC gut Trp metabolism was characterized by a decreased Trp indole pathway, which was positively correlated with bowel gut barrier function, and an increased KP in colon tissue. In addition, the decreased indoles-producing bacteria may lead to downregulation of the Trp indole metabolic pathway, allowing more Trp to be metabolized along the KP.
INTRODUCTION
Population aging has resulted in a substantial increase in the number of new colorectal cancer (CRC) cases globally. It is estimated that in 2030, approximately 2.2 million new cases will be diagnosed with CRC and that 1.1 million will die from the disease worldwide[1]. Despite a slight decrease in the death rate[2], CRC is still ranking third in morbidity and second among all cancer-related death cases[3,4]. The development of CRC results from the progressive accumulation of genetic and environmental alterations, which drive the malignant evolution of the colon from normal mucosa through early adenoma/polyp to cancer[5]. The complicated process is considered to be associated with the following risk factors: Diet (high intake of fat/protein and low consumption of dietary fibers), sociodemographic and medical factors, lifestyle (exercise, smoking, and drinking history), and so on[6].
A growing number of prior studies have indicated that diet, gut microbiota, and metabolite interactions are involved in the pathogenesis of CRC via damage to the epithelial barrier and mucus barrier, gut inflammation, immune escape, genetic/epigenetic alteration[7-9], etc. Most of the topics were focused on short-chain fatty acids[10], amino acids[11], and bile acids (BAs)[12]. Although tryptophan (Trp) metabolites have been proven to promote or inhibit CRC in vitro and animal models[13,14], few studies on gut Trp metabolism have been found, especially its interaction with gut microbiota. Therefore, it is meaningful to study the characteristics of gut Trp metabolism in patients with CRC.
In addition to being excreted in the stool and used for protein synthesis, gut Trp is catabolized mainly along the following three pathways[15]: (1) Trp indole pathway: Trp in feces can be metabolized into indole and indole derivatives by gut microorganisms, such as Clostridium sporogenes and E. coli[16], Achromobacter liquefaciens, Bacteroides spp[17], and Bifidobacterium spp[18]. Indoles, including indole, indole-3-acid-acetic (IAA), skatole (3-methylindole), indole-3-propionic acid (IPA), indole-3-aldehyde (IALD), indole-3-acetaldehyde (IAALD), and so on[11], play an important role in modulating the expression of inflammation-related genes[19], strengthening the epithelial cell barrier[20], and inhibiting the growth of CRC cells[21] in an aryl hydrocarbon receptor (AHR)-dependent way; (2) Kynurenine (KYN) pathway (KP): Most Trp in tissue is metabolized to KYN along the KP, which is regulated by the main rate-limited enzymes, including indoleamine 2,3-dioxygenase 1 (IDO1) in almost all tissue and Trp 2,3-dioxygenase in the liver[22]. Intestinal bacteria can indirectly participate in the KP by affecting the activity of IDO or the concentration of other metabolites[23]; and (3) Serotonin pathway (SP): Trp can be converted to 5-hydroxytryptophan (5-HTP) by Trp hydroxylase, followed by the decarboxylation of 5-HTP through the activation of aromatic L-amino acid decarboxylase[15]. Spore-forming bacteria promote serotonin synthesis from enterochromaffin cells (ECs) in the colon[24]. Since the first two signaling pathways have been shown to be associated with an impaired gut barrier, overactivation of WNT/β-catenin, and immune escape in CRC mice[13,25,26], we hypothesized that they may also be involved in the development of CRC in humans.
The primary aim of this study was to investigate the alteration of gut Trp metabolism and microbiota in patients with CRC. The secondary purpose was to explore the potential correlations between the gut microbiome, intestinal barrier function, tissue KP, and alterations in fecal Trp metabolism, respectively.
MATERIALS AND METHODS
Subjects and sample collection
In this study, we used the biological samples of 79 patients with colorectal neoplastic lesions [33 with colon adenoma (ADA) and 46 with sporadic CRC] and 38 age-, sex-, and body mass index (BMI)-matched healthy controls (HCs). The patients were recruited between March 2019 and December 2019 at The China-Japan Friendship Hospital. The HCs were recruited through public posters. We estimated the sample size with PASS 08.0.3 (NCSS LLC, Kaysville, UT, United Satets) based on the KYN content in the pretest.
All the patients included in our study were newly diagnosed and did not receive chemotherapy or any other therapy. All patients were hospitalized in the gastroenterology department and underwent neoplastic lesion resection.
The inclusion criteria were as follows: (1) Patients whose tumors were removed and pathologically diagnosed with adenomatous polyp or colorectal adenocarcinoma; (2) HCs were recruited from asymptomatic individuals who had undergone complete colonoscopy and had negative results; and (3) Age of 18-80 years old, regardless of sex. Subjects with the following conditions were excluded: (1) Subjects who had a history of psychiatric disorders or were pregnant or lactating; (2) Subjects with other primary digestive disorders or patients in whom neoplastic lesions developed in the context of hereditary disease; (3) Patients with current evidence of inflammatory or infective diseases; and (4) Those unable to come off any of the following medications: Antibiotics, nonsteroidal anti-inflammatory drugs, immune modulators, probiotics, corticosteroids, prokinetics, or antispasmodics within 4 wk. All patients and HCs volunteered from similar geographic areas and had similar eating habits.
The demographic and clinical data, such as age, sex, BMI, history of smoking, drinking, and detailed family history, were collected. Pathological data were also included for analysis. The tumors were staged according to the American Joint Commission on Cancer (AJCC) TNM staging system[27].
After signing an informed consent form, fecal samples and colon tissues of each subjects were collected. Fecal samples were collected from asymptomatic volunteers before colonoscopy or patients before resection. Each qualified fecal sample was collected in 7:00-9:00 in the morning and divided into two parts with sterile stool collection tubes. Then, samples were frozen in liquid nitrogen immediately and stored at -80 °C for further testing. Two pieces of colon tissues were taken from the adenoma or cancer of patients and the rectosigmoid junction of HCs. One specimen was immediately fixed in formalin for 3 d, embedded in paraffin, and sectioned (4 μm) for immunohistochemistry. Other tissues were then immediately immersed in RNAstorage reagent (RNAwait; Solarbio, Beijing, China) and stored at -80 °C for real-time polymerase chain reaction (RT-PCR).
Our study was approved by the Ethics Committee of China-Japan Friendship Hospital (No. 2018-116-K85), and written informed consent was obtained from all subjects.
Targeted metabolomics profiling
Chemicals and reagents: We used targeted metabolomics methods to quantitate the Trp and Trp metabolites in this study. The metabolites detected are as follows: L-Trp, L-KYN, indole, skatole, indole-3-carboxylic acid (I3CA), IALD, IAA, IPA, indoxyl-3-sulfate (I3S), and IAALD. All standards used were obtained from Sigma-Aldrich (St. Louis, MO, United States) and Steraloids Inc. (Newport, RI, United States). Formic acid (Optima LC-MS) was obtained from Sigma-Aldrich (St. Louis, MO, United States). Methanol, isopropanol, and acetonitrile (Optima LC-MS) were purchased from Thermo-Fisher Scientific (Fair Lawn, NJ, United States).
Sample preparation: Standards were precisely weighed and dissolved in 50% methanol, mixed into a concentration of 5.0 mg/mL, and then diluted into a series of standard samples to obtain standard curves of concentration. Fecal samples were thawed in an ice bath to diminish degradation. For indoles, 10-mg fecal samples were accurately weighed and transferred into a grinding tube, 15 μL of water was added and homogenized for 3 min, and then 150 μL of methanol containing internal standards was added. After 3 min of homogenizing, the supernatant was transferred to a 96-well plate by high-speed centrifugation (18000 g, 20 min). Next, 50 μL of ultra-pure water was added, and the samples were oscillated at a speed of 650 rpm for 10 min at 10 °C. For Trp and KYN, 5 mg of samples were weighed and transferred to a new tube. After homogenization and centrifugation, 20 μL of ultrapure water was added, and then the plate was sealed. The derivatized samples were oscillated at a speed of 650 rpm for 10 min at 10 °C. After centrifugation, 135 μL of supernatant was transferred to a new plate for LC-MS analysis.
Ultra-performance liquid chromatography coupled to tandem mass spectrometry data acquisition and processing: In addition to indole and skatole, other metabolites were detected with an ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, United States). The concentrations of indole and skatole were detected by UPLC-Photo-Diode Array at a wavelength of 218 nm.
The optimized instrument settings of indoles are briefly described below: A 10-uL sample at a temperature of 10 °C was injected in the splitless mode into an ACQUITY UPLC Cortecs C18 1.7 um VanGuard precolumn (2.1 mm × 5 mm) and an ACQUITY UPLC Cortecs C18 1.7 um analytical column (2.1 mm × 100 mm). The mobile phase consisted of 10 mmol/L ammonium acetate with water with 0.1% formic acid (mobile phase A) and acetonitrile/IPA (70:30) or acetonitrile with 0.1% formic acid (mobile phase B) run at a flow rate of 0.3 mL/min. The elution gradients were 0-3 min (10%-30% B), 3-4.5 min (30%-45% B), 4.5-6.5 min (45%-100% B), 6.5-7 min (100% B), 7-8 min (100%-10% B), and 8-9 min (10% B). The column was maintained at 40 °C. The mass spectrometer was operated with source and desolvation temperatures set at 150 °C and 550 °C, respectively. The capillary voltage was 2.0 (ESI-/+) kV mode. The settings for Trp and KYN are similar to those of indoles. However, they have a different mobile phase gradient: 0-1 min (5% B), 1-11 min (5%-78% B), 11-13.5 min (78%-95% B), 13.5-14 min (95%-100% B), 14-16 min (100% B), 16-16.1 min (100%-5% B), and 16.1-18 min (5% B).
The sample control procedure and data control procedure refer to criterion (ISO9001, QAIC/CN/170149). The raw data files generated by UPLC-MS/MS were processed using MassLynx software (v4.1, Waters, Milford, MA, United States) to perform peak integration, calibration, and quantitation for each metabolite.
Zonula occludens-1 and IDO1 mRNA analysis by quantitative RT-PCR
Gut barrier function was assessed by measuring Zonula occludens-1 (ZO-1). The levels of ZO-1 and IDO1 mRNA in tissues were analyzed by RT-PCR. Total RNA in colon tissues was isolated with TRIzol Reagent (Servicebio, Wuhan, China). A RevertAid First Strand cDNA Synthesis Kit was used to perform reverse transcription (Thermo Scientific, Waltham, MA, United States). Then, RT-PCR was performed using the StepOnePlus Real-Time PCR System. The primers (GAPDH: forward, 5’-GGAAGCTTGTCATCAATGGAAATC-3’and reverse, 5’-TGATGACCCT TTTGGCTCCC-3’. IDO1: forward, 5’-GGATTCTTCCTG GTCTCTCTATT GG-3’ and reverse, 5’-GGTCTTCCC AGAACCCTTCATAC-3’. ZO-1: forward, 5’-TTCCAG CCAGCCTGCTAAAC-3’ and reverse, 5’-CAATA GCGTAGCCCGTTCATCT-3’) for PCR were from Biotechnology (Shanghai, China). All samples were amplified with the following thermal cycling conditions: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and then 60 °C for 60 s. All results were normalized to the reference gene as a control using the 2-∆∆Ct method[28]. The levels of ZO-1 and IDO1 mRNA are expressed as fold changes relative to the mean level of HCs.
ZO-1 protein analysis by immunohistochemistry
Paraffin sections were processed for immunohistochemistry. They were incubated with the primary antibody at a dilution of 1:200 (ZO-1 mouse monoclonal antibody, Proteintech, Wuhan, China). More details were described as previous studies[29,30].
16S ribosomal RNA gene sequencing
DNA extraction and sequencing: Genomic DNA was extracted from 200 mg of fecal samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Valencia, United States) following the instructions. The amplification selection region of 16S rDNA was V3-V4, and the general primers used were 341F and 806R. The sequences are 341F (5’-CCTACGGGRSGCAGCAG-3’) and 806R (5’-GGACTACVVGGGTATCTAATC-3’). Individual amplification products were purified, barcoded, and pooled to construct the sequencing library (KAPA HiFi Hotstart ReadyMix PCR kit). Then, Illumina NovaSeq PE250 was used for sequencing (Illumina, CA, United States). The sequenced raw data were then spliced and filtered to obtain clean data. Clustering and species classification were performed. After quality control was performed, reads were first sorted according to their abundance from large to small and then clustered through 97% similarity to obtain operational taxonomic units (OTUs).
Statistical analysis
Chemometric analyses of clinicopathologic and metabolite data were performed using SPSS (IBM-SPSS Statistics, United States) version 22. 0. Quantitative data analyses were carried out by the Kolmogorov-Smirnov to test whether the data were in accord with the normal distribution and are presented as the mean ± SD or the median (Q1, Q3). When comparing between groups, ANOVA were used to analyze data that obeyed the normal distribution. The Mann-Whitney U test or Kruskal-Wallis test was used to analyze abnormally distributed data. Qualitative data were analyzed using the chi-squared test or nonparametric test. False discovery rate (FDR) correction was applied when comparing the relative abundances of gut microbiota. The microbiota data were analyzed with R statistical software. Correlations between parameters were explored using Spearman’s correlation analysis. A two-tailed P < 0.05 was considered statistically significant.
RESULTS
Characteristics of participants
A total of 117 participants were included in the study. There were 46 patients with CRC (32 males and 14 females) and 38 HCs (24 males and 14 females) who participated in the study. In order to gain more insight into the adenoma-carcinoma sequence, 33 patients with ADA (23 males and 10 females) were also included. No significant differences were found between every two groups with regard to age, sex, or BMI (P > 0.05). Their demographic and clinical information is presented in Table 1.
Table 1.
Demographic and clinical information of participants included in the study
|
Feature
|
HCs (n = 38)
|
ADA patients (n = 33)
|
CRC patients (n = 46)
|
P
value
|
| Age (yr) | 56.85 ± 10.99 | 61.18 ± 8.53 | 63.63 ± 11.39 | 0.095 |
| Gender (male:female) | 24:14 | 23:10 | 32:14 | 0.799 |
| BMI (kg/m2) | 23.86 ± 3.84 | 24.817 ± 3.31 | 23.66 ± 2.88 | 0.676 |
| Smoking history (%) | 10 (26.3) | 15 (45.5) | 17 (37.0) | 0.241 |
| Drinking history (%) | 14 (36.8) | 9 (27.3) | 12 (26.1) | 0.522 |
| CRC of FDRs (%) | 6 (21.4) | 8 (24.2) | 8 (17.4) | 0.753 |
The data are presented as the mean ± SD. HC: Healthy control; ADA: Colorectal adenoma; CRC: Colorectal cancer; BMI: Body mass index; FDR: False discovery rate.
Fecal Trp and Trp metabolites
Stool samples with insufficient quantity and unqualified quality were excluded. A total of 91 samples were tested for Trp and Trp metabolites. Metabolites of the 91 subjects were used for correlation analysis with corresponding parameters of colon tissue and shifted microbiota of feces. I3S and IAALD were excluded for analysis due to a detection rate of lower than 85%. The indoles here are the sum of indole, skatole, IAA, IPA, I3CA, and IALD. The absolute concentrations of metabolites are shown in Table 2. The concentration of fecal KYN was higher in patients with ADA and CRC than in HCs (P = 0.036 and P = 0.627, respectively) but only reached a statistically significant difference in CRC. In detail, the concentrations of IALD and I3CA were significantly elevated in patients with CRC (P = 0.044 and P = 0.084, respectively), while IPA was decreased in both patients with ADA and CRC (P < 0.005). There was no significant difference in the levels of total indoles between patients with ADA and HCs or patients with CRC and HCs (P > 0.05).
Table 2.
Concentrations of tryptophan and tryptophan metabolites in patients and healthy controls
| Metabolite (nmol/L) | Controls (n = 28) | ADA patients (n = 24) | CRC patients (n = 39) |
Intergroup comparisons (P value)
|
||
|
HC/ADA
|
HC/CRC
|
ADA/CRC
|
||||
| Trp | 196.67 (125.10, 353.30) | 289.46 (206.2, 393.72) | 241.88 (165.98, 429.16) | 0.081 | 0.118 | 0.611 |
| KYN | 0.81 (0.64, 1.57) | 0.94 (0.66, 2.25) | 1.51 (0.70, 3.46) | 0.627 | 0.036 | 0.140 |
| Indole | 252.00 (170.00, 545.00) | 251.00 (66.50, 366.75) | 356.00 (128.50, 729.25) | 0.213 | 0.878 | 0.170 |
| Skatole | 210.00 (172.00, 223.00) | 137.00 (115.00, 206.00) | 231.00 (147.50, 265.50) | 0.111 | 0.495 | 0.019 |
| IAA | 6.56 (1.25, 20.59) | 6.63 (3.35, 18.82) | 7.61 (1.85, 16.1) | 0.401 | 0.943 | 0.876 |
| IPA | 10.67 (6.36, 15.48) | 7.75 (4.55, 10.38) | 7.65 (4.19, 13.11) | 0.025 | 0.025 | 0.348 |
| IALD | 8.00 (4.25 15.50) | 19.50 (8.25, 31.75) | 37.00 (13.00, 63.00) | 0.044 | 0 | 0.024 |
| I3CA | 1.13 (0.81, 1.71) | 1.10 (0.74, 2.11) | 3.64 (1.79, 8.15) | 0.971 | 0.011 | 0.028 |
| Indoles | 428.24 (276.45, 963.95) | 325.50 (179.82, 504.33) | 547.87 (247.97, 823.64) | 0.100 | 0.955 | 0.044 |
| Indoles/Trp ratio | 2.46 (1.25, 4.10) | 1.48 (0.58, 1.97) | 1.34 (0.70, 2.63) | 0.003 | 0.029 | 0.413 |
| KYN/Trp ratio (× 10-3) | 5.23 (1.86, 7.99) | 3.73 (1.99, 11.39) | 7.39 (4.12, 11.72) | 0.845 | 0.032 | 0.105 |
The data are presented as the median (Q1, Q3). HC: Healthy control; ADA: Colorectal adenoma; CRC: Colorectal cancer; BMI: Body mass index; Trp: Tryptophan; KYN: Kynurenine; IAA: Indole-3-acid-acetic; IPA: Indole-3-propionic acid; IALD: Indole-3-aldehyde; I3CA: Indole-3-carboxylic acid.
Through further analysis, we found that the ratio of KYN to Trp (KYN/Trp ratio) was significantly higher in patients with CRC than in HCs (P < 0.005). In addition, the ratio of indoles to Trp (indoles/Trp ratio) in feces decreased in both patients with ADA and CRC when compared with HCs (P = 0.003 and P = 0.029, respectively). Regarding the concentrations of Trp, indole, IAA, and skatole, no statistically significant difference was found between HCs and patients (P > 0.005). The details are shown in Table 2.
IDO1 mRNA and tight junction protein ZO-1 mRNA
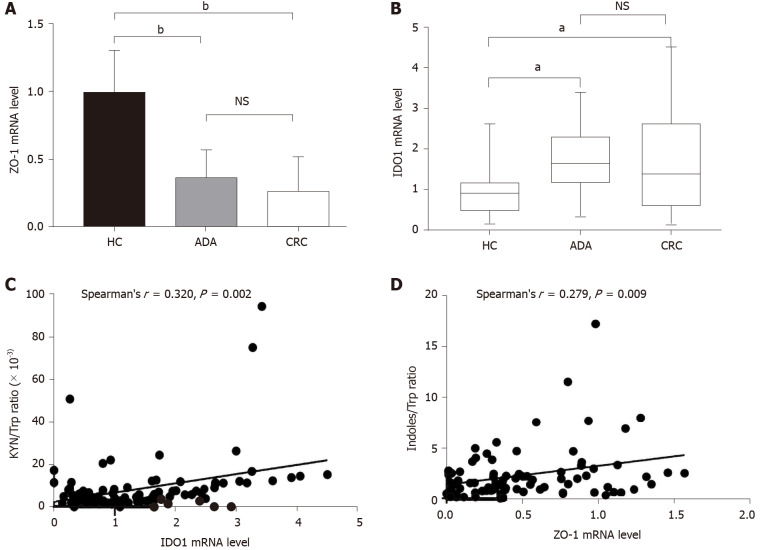
IDO1 is the main rate-limiting enzyme for Trp metabolism in colon tissues. ZO-1 is a well-accepted marker of gut barrier function. IDO1 mRNA and ZO-1 mRNA were detected in colon tissues. The relative ZO-1 mRNA levels in patients with ADA (0.36 ± 0.21) and CRC (0.27 ± 0.24) were significantly lower than those in HCs (1.00 ± 0.31) (P < 0.001) (Figure 1A). In comparison to HCs [0.93 (0.46-1.28)], the relative IDO1 mRNA levels in patients with ADA [1.68 (1.28-2.49)] and CRC [1.65 (0.47-2.46)] were increased (P = 0.004 and P = 0.035, respectively) (Figure 1B).
Figure 1.
Levels of Zonula occludens-1 and indoleamine 2,3-dioxygenase 1 mRNA in colon tissue and their correlations with fecal tryptophan metabolites. A: The relative Zonula occludens-1 (ZO-1) mRNA levels in colorectal adenoma and colorectal cancer (CRC) were significantly lower than those in controls (P < 0.001); B: The relative indoleamine 2,3-dioxygenase 1 (IDO1) mRNA levels in ARA and CRC were higher than those in controls (P =0.04 and P = 0.035, respectively); C: Fecal kynurenine/tryptophan (Trp) ratio was positively correlated with the level of IDO1 mRNA in all subjects; D: There was a positive correlation between fecal indoles/Trp ratio and ZO-1 mRNA level in all subjects. KYN: Kynurenine; Trp: Tryptophan; HC: Healthy control; ADA: Colorectal adenoma; CRC: Colorectal cancer; ZO-1: Zonula occludens-1; IDO1: Indoleamine 2,3-dioxygenase 1.
Correlations between fecal Trp metabolism and ZO-1 and IDO1 mRNA
We analyzed the correlations between fecal Trp metabolism and other parameters (IDO1 and ZO-1 mRNA levels) in all subjects. IDO1 mRNA levels were positively associated with the KYN/Trp ratio (r = 0.327, P = 0.003), and ZO-1 mRNA levels were positively correlated with the indoles/Trp ratio (r = 0.279, P = 0.009) (Figure 1C and D).
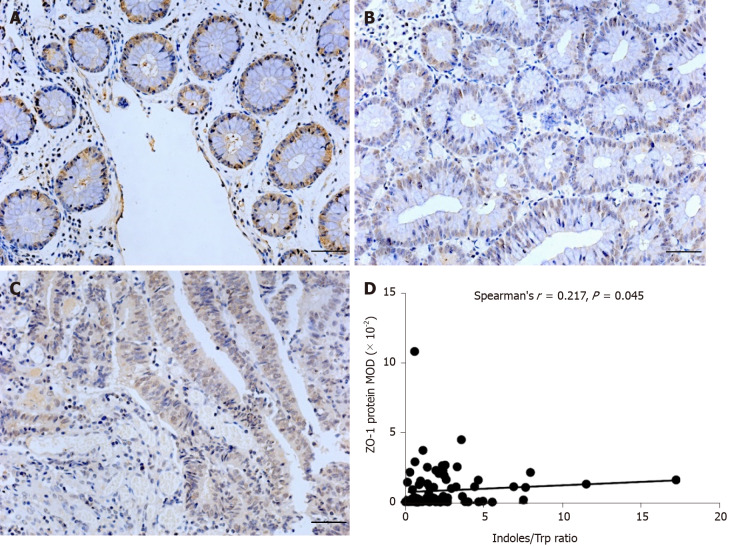
ZO-1 protein and correlation between indole/Trp ratio and ZO-1 protein
Representative photos of the immunoreactivity of ZO-1 are showed in Figure 2A-C. Mean optical density of ZO-1 tended to be decreased in ADA [0.34E-2 (0.02E-2 to 0.47E-2)] and CRC [0.08E-2 (0.06E-2 to 0.13E-2)] compared to controls [1.84E-2 (1.22E-2 to 2.56E-2)] (P < 0.001). There was a positive correlation between fecal indoles/Trp ratio and ZO-1 protein in all subjects (r = 0.217, P = 0.045) (Figure 2D).
Figure 2.
Immunohistochemical staining for Zonula occludens-1 and its correlation with fecal indoles/tryptophan ratio (× 200 magnification). Immunoreactivity of Zonula occludens-1 (ZO-1) tended to be decreased in colorectal adenoma (B) and colorectal cancer (C) compared to controls (A) as shown in the figure (scale bar = 100 μm); D: There was a positive correlation between fecal indoles/tryptophan ratio and ZO-1 protein in all subjects (P = 0.045). MOD: Mean optical density; Trp: Tryptophan; ZO-1: Zonula occludens-1.
Features of gut microbiome of patients with colorectal neoplastic lesions
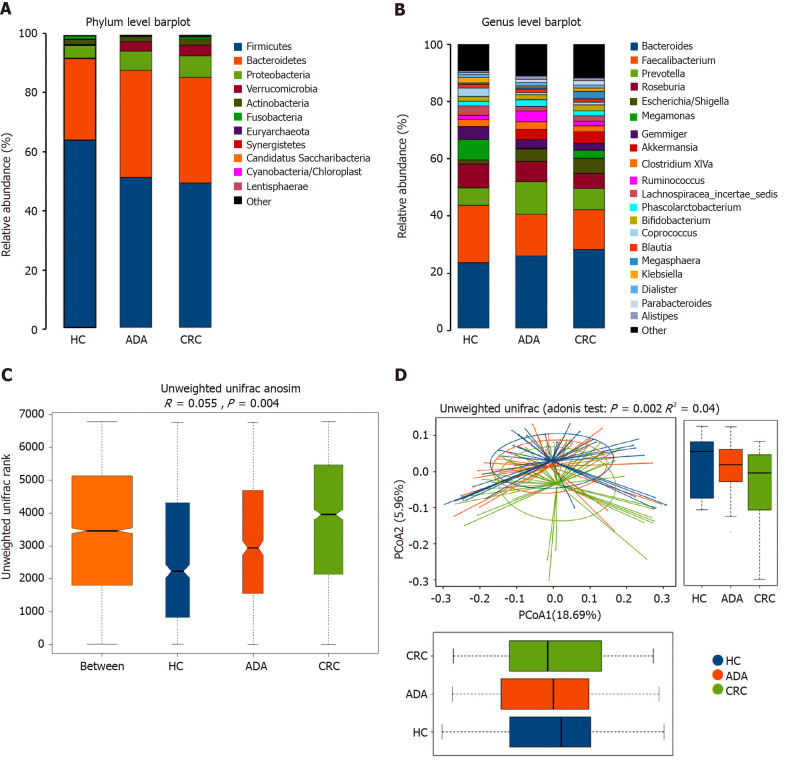
We performed 16S ribosomal RNA gene sequencing on the fecal samples of the three groups of subjects. Microbiome signatures along the adenoma-carcinoma sequence were detected. A total of 883 OTUs were generated from 117 samples. The relative abundances of the microbiota in the three groups at the phylum and genus levels are shown in Figure 3A and B. Firmicutes and Bacteroidetes were the most predominant phyla. The Firmicutes/Bacteroidetes ratio in patients with CRC (49.48%:36.20%) was lower than that in HCs (64.25%:28.06%). Although the richness of flora decreased in the disease groups, it did not reach statistical significance (chao1, P > 0.05). Both the Adonis and Anosim analyses indicated that the global microbiota structure of the samples of patients with ADA and CRC differed significantly from that of HCs (P < 0.05). There was a trend along the HC-adenoma-carcinoma sequence (Figure 3C and D).
Figure 3.
Fecal bacterial structure of patients with colorectal adenoma, colorectal cancer, and healthy controls. A and B: Relative bacterial abundance of fecal bacterial at the phylum and genus levels in the three groups; C and D: Unweighted Unifrac (Anosim and Adonis) analysis of fecal bacterial in the patients and healthy control group (P < 0.05). Different colors represent different groups, and the distance between points indicates the degree of difference; boxes indicate the interquartile range; lines inside the boxes indicate the medians; the two whiskers indicate the maximum and minimum of the data; the points outside the box indicate outliers. HC: Healthy control; ADA: Colorectal adenoma; CRC: Colorectal cancer.
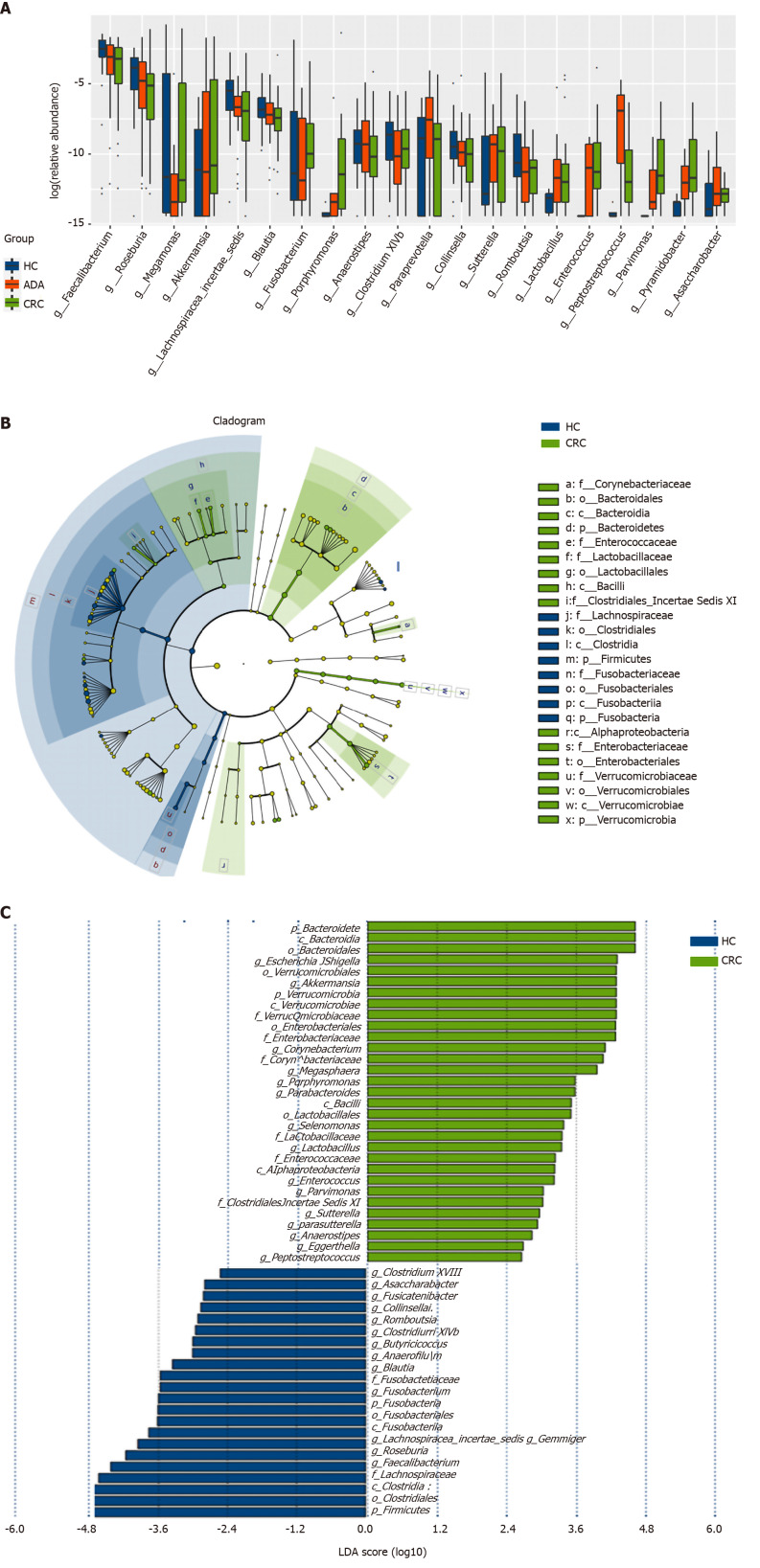
The top 20 different genera among the three groups are shown in Figure 4A (P < 0.05). The abundance of six genera decreased gradually, and three genera increased gradually in the HC-adenoma-carcinoma sequence. Then, we focused on differences in bacteria between CRC and HCs. At the genus level, a total of 29 genera were found to have different abundances between the two groups using the Mann-Whitney U analysis. After FDR correction, only ten genera reached a significant difference (PAdjueted < 0.05).
Figure 4.
Different genus among the three groups and linear discriminant analysis effect size analysis of fecal bacterial of colorectal cancer patients and healthy controls. A: Top 20 different genus among the three groups with a Kruskal-Wallis’s P value < 0.05; B: Cladogram showing enriched taxa in the colorectal cancer (CRC) patient and healthy control (HC) groups; the taxa represented by the English letters in the cladogram are shown in the legend on the right; C: Taxa enriched in HCs are indicated with a negative score and the taxa enriched in the CRC group are indicated with a positive linear discriminant analysis (LDA) score; only taxa with an LDA effect size’s P value < 0.05 and an LDA score ≥ 2.0 are showed. LDA: Linear discriminant analysis; HC: Healthy control; CRC: Colorectal cancer.
Linear discriminant analysis (LDA) integrated with effect size (LEfSe) was used to identify the specific bacteria associated with CRC (Figure 4B and C). On the one hand, several bacteria, including the Bacteroidetes phylum, class Bacteroidia, order Bacteroidales, genus Escherichia/Shigella (phylum Proteobacteria), and order Verrucomicrobiales (phylum Verrucomicrobia), were all significantly overrepresented [all LDA scores (log10) > 3.6] in the feces of patients with CRC. On the other hand, the phylum Firmicutes, order Clostridiales (phylum Firmicutes), and family Lachnospiraceae (phylum Firmicutes) were the most abundant microbiota in HCs [LDA scores (log10) > 4.8].
Correlations between bacterial genera and fecal metabolites
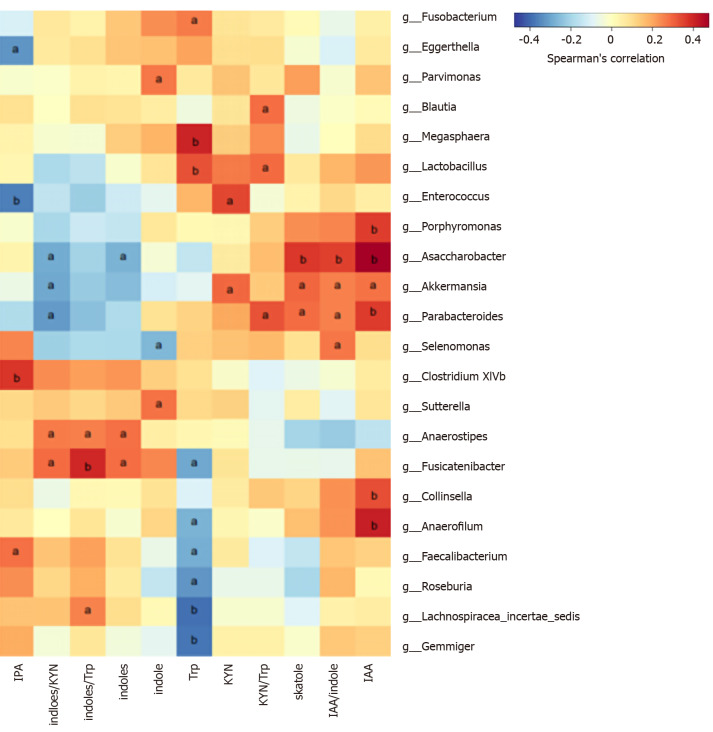
We analyzed the correlation between differentially abundant bacterial genera (LEfSe analysis) and Trp metabolites in all subjects. Remarkably, the genera Asaccharobacter (Actinobacteria) and Parabacteroides (Bacteroidetes) and members of the phylum Firmicutes (Clostridium XlVb, Fusicatenibacter, Anaerofilum, and Anaerostipes) exhibited a positive correlation with indoles, and the genus Lactobacillus was positively correlated with fecal Trp. Other details are shown in Figure 5.
Figure 5.
Correlations between differential bacteria at the genus level using linear discriminant analysis effect size analysis and fecal metabolites. The heatmap presents the Spearman correlation coefficients between fecal metabolites and genera with significantly different relative abundance in the colorectal cancer (CRC) patients and healthy controls (HCs). Indoles were positively related to the genera Asaccharobacter (Actinobacteria) and Parabacteroides (Bacteroidetes), and members of the phylum Firmicutes (such as Clostridium XlVb, Fusicatenibacter, Anaerofilum, and Anaerostipes), whose relative abundances were lower in CRC patients than in HCs in the linear discriminant analysis effect size analysis. aP < 0.05, bP < 0.01. HC: Healthy control; CRC: Colorectal cancer. IPA: Indole-3-propionic acid; IAA: Indole-3-acid-acetic; KYN: Kynurenine; Trp: Tryptophan.
DISCUSSION
This is the first study to investigate the profiles of fecal Trp metabolism in patients with CRC by LC-MS and to explore the potential correlations between the gut microbiome, intestinal barrier function, tissue Trp KP, and alterations in fecal Trp metabolism. We found that CRC fecal Trp metabolism was characterized by a decreased Trp indole pathway, which is positively correlated with bowel gut barrier function, and an increased fecal KYN/Trp ratio, which can partially reflect KP activity in colon tissue. In addition, correlations between differentially abundant bacterial genera and imbalanced fecal Trp metabolism were also found in this study. This study provides a preliminary exploration for possible involvement of Trp metabolites in the tumorigenesis of CRC in humans.
In recent years, accumulating evidence suggests that gut indoles, mostly Trp metabolites, are decreased in the feces of patients with metabolic syndrome[31] and play a protective role in DSS-induced colitis or gut pathogen infection[32,33]. There are few clinical studies on indoles in CRC, and one early study detected an increase in indoles in CRC feces and attributed this high concentration to differences in diet[34]. In this study, the fecal indoles/Trp ratio in patients with CRC was much lower than that in healthy subjects, although no differences were found in the absolute indole concentration. This result is supported by a number of in vitro and in vivo studies. I3C can induce apoptosis and decrease cell viability in several human CRC cell lines[35]. The impaired Trp indole pathway plays a catalytic role in the early stages of CRC[13]. Interruption of Trp indole AHR signaling results in markedly increased TNF-α, IL-1β, and IL-6 mRNA levels in the inflammation-associated colorectal tumorigenesis model. Once activated by indoles, AHR acts directly on intestinal stem cells to strengthen intestinal barrier function and maintain mucin production[25]. Exposure to physiological concentrations of indole results in increased expression of the anti-inflammatory cytokines IL-10[36] and IL-22[37]. Moreover, the positive correlation between the indoles/Trp ratio and ZO-1 in our study is further evidence that indoles are involved in intestinal barrier function. In addition, indole derivatives have anticancer effects via the PI3K/Akt/mTOR signaling pathway[38]. Collectively, we concluded that there is an inhibitory effect of fecal indoles on CRC tumorigenesis.
KYN is a key metabolite of Trp along the KP[22] and has been shown to be elevated in colon cancer tissue and serum in previous studies[39,40]. KYN metabolized in host epithelial cells or mucus can enter the feces and fecal KYN/Trp ratio can partially reflect KP activity in colon tissue. The KYN/Trp ratio is regarded as a promising surveillance biomarker for cancer[41]. Given its rate-limiting function, the activity of IDO1 can be roughly assessed by the KYN/Trp ratio in serum[42]. The high expression of IDO1 in CRC is consistent with our results and has a positive correlation with poor overall survival outcomes[43]. IDO (mainly IDO1)/KP promotes tumorigenesis through two independent mechanisms. On the one hand, KP metabolites result in the activation of Wnt/β-catenin signaling independent of the effect on adaptive immunity[44]. New discoveries have shown that KP metabolites have the direct function of promoting cancer cell proliferation and inhibiting apoptosis by activating PI3K/AKT signaling in the neoplastic colon epithelium[14]. It has been validated that Wnt/β-catenin and PI3K/AKT are canonical pathways in CRC. On the other hand, activating the KP is thought to facilitate tumor progression by promoting immune suppression or tolerance. IDO1-expressing cells (mainly APCs) help create a tolerogenic microenvironment in the tumor and the tumor-draining lymph nodes that contributes to tumor immune escape and metastasis[26]. We observed not only an increase in KYN and the KYN/Trp ratio in CRC feces but also a positive correlation between the fecal KYN/Trp ratio and tissue IDO1. Therefore, changes in fecal KYN and the KYN/Trp ratio, which can be detected noninvasively and partially reflect tissue KP, may be involved in the development of CRC.
Since Trp in feces is mainly metabolized by bacteria, we analyzed the microflora profiles of healthy people and patients with CRC as well as the relationship between metabolites and differentially abundant bacteria. We observed a decrease in Firmicutes (Clostridiales and Clostridium) and an increase in Proteobacteria (Escherichia/Shigella genus and Enterobacteriaceae genus), which was consistent with the results of previous studies[45,46]. The former (mainly Clostridium) are known to play an anti-inflammatory role in the gut, while a member of the phylum Proteobacteria Escherichia coli [with expression of the genomic island polyketide synthase (pks+)] can alkylate the DNA of epithelial cells and thus lead to CRC[47,48]. Consistent with a recent study[45], we observed a decline in the Firmicutes/Bacteroidetes ratio in CRC, although an upregulated ratio has been suggested as an indicator of several disorders[49]. In addition, Adonis and Anosim analyses showed a difference between patients with CRC and HCs in bacterial composition. Therefore, the tumor-promoting effects of the gut microbiome may be caused not only by a specific pathogen but also by holistic dysbiosis[50].
In the Trp indole pathway, most gut Trp is converted to indole by tryptophan decarboxylase, to indoleacetamide via tryptophan monooxygenase, and to trytamine. In further steps, these metabolites can be metabolized to skatole, IPA, or I3S, serving as the end products of Trp metabolism in the gut[11]. All these enzymes involved in these reactions are produced by the expression of functional genes in gut microbes. We found that indoles were positively related to the genera Asaccharobacter (Actinobacteria) and Parabacteroides (Bacteroidetes) and members of the phylum Firmicutes (such as Clostridium XlVb, Fusicatenibacter, Anaerofilum, and Anaerostipes), whose relative abundances were lower in CRC patients than in HCs in the LEfSe analysis. Actinobacteria[51] and Clostridium of Firmicutes[16] have been proven to be involved in the production of indoles. This suggests that decreased indoles-producing or indoles-related bacteria may lead to downregulation of the Trp indole metabolic pathway, allowing more Trp to be metabolized along the KP. In addition, the significant positive correlation between Trp and Lactobacillus can be explained by Trp’s ability to promote the growth of Lactobacillus[33]. The diversity and complexity of intestinal bacteria and metabolites limit our ability to explain all the results and require further study.
To learn more about the adenoma-carcinoma sequence, we also studied the characteristics of gut microbiota and Trp metabolism in patients with colon ADA, which is different from other studies. As expected, some increasing and decreasing changes along the HC-adenoma-carcinoma sequence were found. Changes in the intestinal microenvironment caused by microbiota dysbiosis and shifts in Trp metabolism occurred in the early events of CRC pathogenesis. A previous study suggested that adding indoles to the diet has a certain antitumor effect in ApcMin/+ mice[52]. Therefore, supplementation with probiotics or indoles may also have an inhibitory effect on the HC-adenoma-carcinoma sequence in humans.
The limitations of the study are as follows: First, the fecal microbiota and metabolites were not equivalent to the concept of the “tumor microenvironment”, and they tend to be influenced by diet and lifestyle[7,53]. Nevertheless, the noninvasiveness makes the results easily verifiable in a larger sample in the future. Second, the conclusions based on this observational study prevent us from determining the causal relationships. Our results showed that fecal Trp metabolism was correlated with shifted microbiota, intestinal barrier function, and tissue Trp KP. There is no evidence that they have participated in the pathogenesis of CRC. It is possible that CRC induced the changes of microbiota and fecal Trp metabolis. Despite some support from preclinical studies[13,52], the results need to be further verified by more evidence. Third, although the levels of mRNA and protein have a similar trend in tissue[54,55], the expression of IDO1 represented by the mRNA level was not accurate. Since this is a preliminary exploratory study, we will detect IDO1 protein expression and other indexes of intestinal barrier function in subsequent validation experiments. Finally, subjects were not given a standardized diet during the study period as in some other studies[45], but were required to maintain their dietary habits before fecal samples were collected. The standardized diet minimizes diet-induced deviations in microbiota and Trp metabolites but does not reflect the long-term status of the patient’s gut microenvironment. Although a standardized diet in this study is important, the need for scheduled surgery for patients with colon cancer makes this difficult to implement.
CONCLUSION
In conclusion, this study provides new evidence that gut Trp metabolism and microbiome compositions are different in subjects with CRC compared to HCs. Reduced fecal Trp indole metabolism and increased tissue KP may be involved in the pathogenesis of CRC by reducing the intestinal barrier and increasing immune escape, respectively. Collectively, these data suggest that restoring the homeostasis of Trp metabolism and the microbiome may have preventive or therapeutic effects on CRC.
ARTICLE HIGHLIGHTS
Research background
Population aging has resulted in a substantial increase in the number of new colorectal cancer (CRC) cases globally. Gut microbiota and metabolite interactions are involved in the pathogenesis of CRC via various genetic and epigenetic alterations. Gut tryptophan (Trp) metabolites are produced by microbiota and/or host metabolism. Some of them were thought to play a role in CRC in animal and in vitro studies.
Research motivation
Few studies on fecal Trp metabolism have been found, especially its interaction with gut microbiota. Therefore, it is meaningful to study the characteristics of gut Trp metabolism in patients with CRC.
Research objectives
To investigate the features of Trp metabolism in CRC patients and the correlation between fecal Trp metabolites and gut microbiota.
Research methods
Subjects meeting the inclusion and exclusion criteria were included in the study. Their demographic and clinical features were collected. Fecal Trp, kynurenine (KYN), and indoles (metabolites of Trp metabolized by gut microbiota) were examined by ultraperformance liquid chromatography coupled to tandem mass spectrometry. Gut barrier marker and indoleamine 2,3-dioxygenase 1 (IDO1) mRNA were analyzed by quantitative real-time polymerase chain reaction. Zonula occludens-1 (ZO-1) protein expression was analyzed by immunohistochemistry. The gut microbiota was detected by 16S ribosomal RNA gene sequencing. Correlations between fecal metabolites and other parameters were examined in all patients.
Research results
The absolute concentration of KYN and the ratio of KYN to Trp were increased in the feces of patients with CRC compared to HCs, while the indoles to Trp ratio was decreased. Colon IDO1 mRNA levels were positively associated with fecal KYN/Trp ratio and ZO-1 mRNA levels were positively correlated with the indoles/Trp ratio. The genera Asaccharobacter (Actinobacteria) and Parabacteroides (Bacteroidetes), and several members of the phylum Firmicutes (Clostridium XlVb, Fusicatenibacter, Anaerofilum, and Anaerostipes) decreased in CRC and exhibited a positive correlation with indoles in all subjects.
Research conclusions
CRC gut Trp metabolism was characterized by a decreased Trp indole pathway in feces, which is positively correlated with bowel gut barrier function, and an increased kynurenine pathway activity in colon tissue. In addition, correlations between differentially abundant bacterial genera and imbalanced fecal Trp metabolism were also found in this study.
Research perspectives
This preliminary study investigated the alteration of gut Trp metabolism and the possible mechanism of Trp metabolites in CRC pathophysiology. In the future, we will focus on the following aspects. First, we will detect Trp and Trp metabolites in both feces and colon tissues to further study the “tumor microenvironment”. Second, it is necessary to study the dietary habits of CRC patients and explore the relationship between the diet and gut Trp metabolism. Supplementation with indoles in diet may also have an inhibitory effect on the HC-adenoma-carcinoma sequence in humans.
ACKNOWLEDGEMENTS
We thank Dr. Du SY, Dr. Chen S, and Dr. Wang HF for enrollment of participants.
Footnotes
Institutional review board statement: This study was approved by the Ethics Committee of China-Japan Friendship Hospital (No. 2018-116-K85).
Informed consent statement: All patients signed an informed consent form before study enrollment.
Conflict-of-interest statement: All authors report no conflicts of interest.
STROBE statement: The authors have read the STROBE Statement—checklist of items, and the manuscript was prepared and revised according to the STROBE Statement—checklist of items.
Manuscript source: Unsolicited manuscript
Peer-review started: September 20, 2020
First decision: September 29, 2020
Article in press: November 2, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gazouli M, Wu M S-Editor: Huang P L-Editor: Wang TQ P-Editor: Ma YJ
Contributor Information
Xi-Zhen Sun, Graduate School, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing 100730, China; Department of Gastroenterology, China-Japan Friendship Hospital, Beijing 100029, China.
Dong-Yan Zhao, Graduate School, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing 100730, China; Department of Gastroenterology, China-Japan Friendship Hospital, Beijing 100029, China.
Yuan-Chen Zhou, Department of Gastroenterology, China-Japan Friendship Hospital, Beijing 100029, China; Peking University China-Japan Friendship School of Clinical Medicine, Peking University, Beijing 100029, China.
Qian-Qian Wang, Peking University China-Japan Friendship School of Clinical Medicine, Peking University, Beijing 100029, China.
Geng Qin, Department of Gastroenterology, China-Japan Friendship Hospital, Beijing 100029, China.
Shu-Kun Yao, Department of Gastroenterology, China-Japan Friendship Hospital, Beijing 100029, China. shukunyao@126.com.
Data sharing statement
No additional data are available.
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Henley SJ, Ward EM, Scott S, Ma J, Anderson RN, Firth AU, Thomas CC, Islami F, Weir HK, Lewis DR, Sherman RL, Wu M, Benard VB, Richardson LC, Jemal A, Cronin K, Kohler BA. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer. 2020;126:2225–2249. doi: 10.1002/cncr.32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 5.Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015; 149: 1177-1190. :e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Yu J. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell. 2018;9:474–487. doi: 10.1007/s13238-018-0543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Vogtmann E, Ahlquist DA, Devens ME, Kisiel JB, Taylor WR, White BA, Hale VL, Sung J, Chia N, Sinha R, Chen J. Fecal Metabolomic Signatures in Colorectal Adenoma Patients Are Associated with Gut Microbiota and Early Events of Colorectal Cancer Pathogenesis. mBio. 2020;11 doi: 10.1128/mBio.03186-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coker OO, Wu WKK, Wong SH, Sung JJY, Yu J. Altered Gut Archaea Composition and Interaction With Bacteria Are Associated With Colorectal Cancer. Gastroenterology 2020; 159: 1459-1470. :e5. doi: 10.1053/j.gastro.2020.06.042. [DOI] [PubMed] [Google Scholar]
- 10.Niccolai E, Baldi S, Ricci F, Russo E, Nannini G, Menicatti M, Poli G, Taddei A, Bartolucci G, Calabrò AS, Stingo FC, Amedei A. Evaluation and comparison of short chain fatty acids composition in gut diseases. World J Gastroenterol. 2019;25:5543–5558. doi: 10.3748/wjg.v25.i36.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Hou Y, Wang G, Zheng X, Hao H. Gut Microbial Metabolites of Aromatic Amino Acids as Signals in Host-Microbe Interplay. Trends Endocrinol Metab. 2020;31:818–834. doi: 10.1016/j.tem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Song X, Khan S, Li Y, Guo Z, Li C, Wang S, Dong W, Liu W, Wang B, Cao H. The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: An old story, yet mesmerizing. Int J Cancer. 2020;146:1780–1790. doi: 10.1002/ijc.32563. [DOI] [PubMed] [Google Scholar]
- 13.Díaz-Díaz CJ, Ronnekleiv-Kelly SM, Nukaya M, Geiger PG, Balbo S, Dator R, Megna BW, Carney PR, Bradfield CA, Kennedy GD. The Aryl Hydrocarbon Receptor is a Repressor of Inflammation-associated Colorectal Tumorigenesis in Mouse. Ann Surg. 2016;264:429–436. doi: 10.1097/SLA.0000000000001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishnupuri KS, Alvarado DM, Khouri AN, Shabsovich M, Chen B, Dieckgraefe BK, Ciorba MA. IDO1 and Kynurenine Pathway Metabolites Activate PI3K-Akt Signaling in the Neoplastic Colon Epithelium to Promote Cancer Cell Proliferation and Inhibit Apoptosis. Cancer Res. 2019;79:1138–1150. doi: 10.1158/0008-5472.CAN-18-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 17.Keszthelyi D, Troost FJ, Masclee AA. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil. 2009;21:1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 18.Aragozzini F, Ferrari A, Pacini N, Gualandris R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol. 1979;38:544–546. doi: 10.1128/aem.38.3.544-546.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, Garner AL, Mohammadi S, O'Connell DJ, Abubucker S, Arthur TD, Franzosa EA, Huttenhower C, Murphy LO, Haiser HJ, Vlamakis H, Porter JA, Xavier RJ. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017; 22: 25-37. :e6. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott SA, Fu J, Chang PV. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2020;117:19376–19387. doi: 10.1073/pnas.2000047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Megna BW, Carney PR, Depke MG, Nukaya M, McNally J, Larsen L, Rosengren RJ, Kennedy GD. The aryl hydrocarbon receptor as an antitumor target of synthetic curcuminoids in colorectal cancer. J Surg Res. 2017;213:16–24. doi: 10.1016/j.jss.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comai S, Bertazzo A, Brughera M, Crotti S. Tryptophan in health and disease. Adv Clin Chem. 2020;95:165–218. doi: 10.1016/bs.acc.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Martin-Gallausiaux C, Larraufie P, Jarry A, Béguet-Crespel F, Marinelli L, Ledue F, Reimann F, Blottière HM, Lapaque N. Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells. Front Immunol. 2018;9:2838. doi: 10.3389/fimmu.2018.02838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, Li V, Maradana MR, Schiering C, Stockinger B. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity 2018; 49: 353-362. :e5. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117:1147–1154. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Qin G, Liu DR, Wang Y, Yao SK. Increased expression of brain-derived neurotrophic factor is correlated with visceral hypersensitivity in patients with diarrhea-predominant irritable bowel syndrome. World J Gastroenterol. 2019;25:269–281. doi: 10.3748/wjg.v25.i2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu XJ, Zhang YL, Liu L, Pan L, Yao SK. Increased expression of nerve growth factor correlates with visceral hypersensitivity and impaired gut barrier function in diarrhoea-predominant irritable bowel syndrome: a preliminary explorative study. Aliment Pharmacol Ther. 2017;45:100–114. doi: 10.1111/apt.13848. [DOI] [PubMed] [Google Scholar]
- 31.Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, Michel ML, Chong-Nguyen C, Roussel R, Straube M, Jegou S, McQuitty C, Le Gall M, da Costa G, Lecornet E, Michaudel C, Modoux M, Glodt J, Bridonneau C, Sovran B, Dupraz L, Bado A, Richard ML, Langella P, Hansel B, Launay JM, Xavier RJ, Duboc H, Sokol H. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab 2018; 28: 737-749. :e4. doi: 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, Wang Q, Ma Y, Li L, Yu K, Zhang Z, Chen G, Li X, Xiao W, Xu P, Yang H. Aryl Hydrocarbon Receptor Activation Modulates Intestinal Epithelial Barrier Function by Maintaining Tight Junction Integrity. Int J Biol Sci. 2018;14:69–77. doi: 10.7150/ijbs.22259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol. 1985;109:135–141. doi: 10.1007/BF00391888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Megna BW, Carney PR, Nukaya M, Geiger P, Kennedy GD. Indole-3-carbinol induces tumor cell death: function follows form. J Surg Res. 2016;204:47–54. doi: 10.1016/j.jss.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011; 141: 237-248, 248. :e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Popolo A, Pinto A, Daglia M, Nabavi SF, Farooqi AA, Rastrelli L. Two likely targets for the anti-cancer effect of indole derivatives from cruciferous vegetables: PI3K/Akt/mTOR signalling pathway and the aryl hydrocarbon receptor. Semin Cancer Biol. 2017;46:132–137. doi: 10.1016/j.semcancer.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Walczak K, Dąbrowski W, Langner E, Zgrajka W, Piłat J, Kocki T, Rzeski W, Turski WA. Kynurenic acid synthesis and kynurenine aminotransferases expression in colon derived normal and cancer cells. Scand J Gastroenterol. 2011;46:903–912. doi: 10.3109/00365521.2011.579159. [DOI] [PubMed] [Google Scholar]
- 40.Engin AB, Karahalil B, Karakaya AE, Engin A. Helicobacter pylori and serum kynurenine-tryptophan ratio in patients with colorectal cancer. World J Gastroenterol. 2015;21:3636–3643. doi: 10.3748/wjg.v21.i12.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SH, Mahendran R, Tham SM, Thamboo TP, Chionh BJ, Lim YX, Tsang WC, Wu QH, Chia JY, Tay MHW, Goh BYS, Chen KW, Mallari JZ, Periaswami R, Raman L, Choo SN, Kioh DYQ, Chiong E, Esuvaranathan K, Chan ECY. Tryptophan-kynurenine ratio as a biomarker of bladder cancer. BJU Int. 2020 doi: 10.1111/bju.15205. [DOI] [PubMed] [Google Scholar]
- 42.Botticelli A, Cerbelli B, Lionetto L, Zizzari I, Salati M, Pisano A, Federica M, Simmaco M, Nuti M, Marchetti P. Can IDO activity predict primary resistance to anti-PD-1 treatment in NSCLC? J Transl Med. 2018;16:219. doi: 10.1186/s12967-018-1595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma WJ, Wang X, Yan WT, Zhou ZG, Pan ZZ, Chen G, Zhang RX. Indoleamine-2,3-dioxygenase 1/cyclooxygenase 2 expression prediction for adverse prognosis in colorectal cancer. World J Gastroenterol. 2018;24:2181–2190. doi: 10.3748/wjg.v24.i20.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thaker AI, Rao MS, Bishnupuri KS, Kerr TA, Foster L, Marinshaw JM, Newberry RD, Stenson WF, Ciorba MA. IDO1 metabolites activate β-catenin signaling to promote cancer cell proliferation and colon tumorigenesis in mice. Gastroenterology 2013; 145: 416-25. :e1–4. doi: 10.1053/j.gastro.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, Cai S, Qin H, Goel A, Li X, Ma Y. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9:4101–4114. doi: 10.7150/thno.35186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Cheng Y, Shao L, Ling Z. Alterations of the Predominant Fecal Microbiota and Disruption of the Gut Mucosal Barrier in Patients with Early-Stage Colorectal Cancer. Biomed Res Int. 2020;2020:2948282. doi: 10.1155/2020/2948282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wick EC, Wang H, Wu S, Pardoll DM, Housseau F, Sears CL. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masumoto S, Terao A, Yamamoto Y, Mukai T, Miura T, Shoji T. Non-absorbable apple procyanidins prevent obesity associated with gut microbial and metabolomic changes. Sci Rep. 2016;6:31208. doi: 10.1038/srep31208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin L, Xu X. Indole-3-acetic acid production by endophytic Streptomyces sp. En-1 isolated from medicinal plants. Curr Microbiol. 2013;67:209–217. doi: 10.1007/s00284-013-0348-z. [DOI] [PubMed] [Google Scholar]
- 52.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, Akiyama T, Kurosumi M, Poellinger L, Kato S, Fujii-Kuriyama Y. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci USA. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 54.Xiang Z, Li J, Song S, Wang J, Cai W, Hu W, Ji J, Zhu Z, Zang L, Yan R, Yu Y. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res. 2019;38:314. doi: 10.1186/s13046-019-1318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim Y, West GA, Ray G, Kessler SP, Petrey AC, Fiocchi C, McDonald C, Longworth MS, Nagy LE, de la Motte CA. Layilin is critical for mediating hyaluronan 35kDa-induced intestinal epithelial tight junction protein ZO-1 in vitro and in vivo. Matrix Biol. 2018;66:93–109. doi: 10.1016/j.matbio.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.