Abstract
PURPOSE
CALGB 40601 assessed whether dual versus single human epidermal growth factor receptor 2 (HER2) –targeting drugs added to neoadjuvant chemotherapy increased pathologic complete response (pCR). Here, we report relapse-free survival (RFS), overall survival (OS), and gene expression signatures that predict pCR and survival.
PATIENTS AND METHODS
Three hundred five women with untreated stage II and III HER2-positive breast cancer were randomly assigned to receive weekly paclitaxel combined with trastuzumab plus lapatinib (THL), trastuzumab (TH), or lapatinib (TL). The primary end point was pCR, and secondary end points included RFS, OS, and gene expression analyses. mRNA sequencing was performed on 264 pretreatment samples.
RESULTS
One hundred eighteen patients were randomly allocated to THL, 120 to TH, and 67 to TL. At more than 7 years of follow-up, THL had significantly better RFS and OS than did TH (RFS hazard ratio, 0.32; 95% CI, 0.14 to 0.71; P = .005; OS hazard ratio, 0.34; 95% CI, 0.12 to 0.94; P = .037), with no difference between TH and TL. Of 688 previously described gene expression signatures, significant associations were found in 215 with pCR, 45 with RFS, and only 22 with both pCR and RFS (3.2%). Specifically, eight immune signatures were significantly correlated with a higher pCR rate and better RFS. Among patients with residual disease, the immunoglobulin G signature was an independent, good prognostic factor, whereas the HER2-enriched signature, which was associated with a higher pCR rate, showed a significantly shorter RFS.
CONCLUSION
In CALGB 40601, dual HER2-targeting resulted in significant RFS and OS benefits. Integration of intrinsic subtype and immune signatures allowed for the prediction of pCR and RFS, both overall and within the residual disease group. These approaches may provide means for rational escalation and de-escalation treatment strategies in HER2-positive breast cancer.
INTRODUCTION
Human epidermal growth factor receptor 2 (HER2) is amplified and overexpressed in 15% to 20% of all breast cancers and, when untreated, is the most aggressive breast cancer phenotype. Over the past 20 years, the prognosis of HER2-positive breast cancer has been markedly improved with the implementation of anti-HER2 targeted therapies. In neoadjuvant randomized trials, dual HER2 blockade is associated with a higher pathologic complete response (pCR) rate compared with that of single agents.1-5 In the absence of chemotherapy, dual HER2 blockade alone is associated with pCR rates of approximately 25%6,7; however, large adjuvant trials have demonstrated only marginal survival benefit of dual HER2 blockade compared with standard trastuzumab added to chemotherapy.8,9
CONTEXT
Key Objective
Here, we report CALGB 40601 prespecified secondary end points of relapse-free survival (RFS) and overall survival with a median follow-up of 7 years and a comprehensive exploratory analysis testing the ability of hundreds of genomic biomarkers to predict not only pathologic complete response (pCR), but also RFS.
Knowledge Generated
In CALGB 40601, there was a significant improvement in RFS and overall survival at 7 years with dual (lapatinib and trastuzumab) versus single anti-HER2 therapy. Biomarker analysis showed that the HER2-enriched subtype, a key independent predictor of pCR, was also an inverse predictor of RFS, whereas immune gene expression signatures were significantly correlated with higher pCR rates and better RFS.
Relevance
For future escalation and de-escalation strategies in HER2-positive breast cancer, we may need to integrate the information provided by clinical parameters, intrinsic subtype, and immune signatures to best predict response and survival.
Both the intrinsic complexity of HER2-positive tumors and the properties of its tumor microenvironment are responsible for the differences in response to anti-HER2 targeted therapies. HER2-positive disease is highly heterogeneous. All the intrinsic subtypes—Luminal A, Luminal B, HER2 Enriched, Basal like, and Normal like—can be identified within HER2-positive tumors by gene expression. Among them, the HER2-Enriched subtype has been systematically associated with higher pCR rates to anti-HER2 targeted therapies across multiple neoadjuvant clinical trials.3,6,10-17 At a DNA level, PIK3CA mutations have also been associated with lower treatment response.18,19 Apart from intrinsic tumor characteristics, different immune system features have been correlated with a higher pCR rate in HER2-positive breast cancer, including the presence of tumor-infiltrating lymphocytes (TILs),14,20-24 programmed death ligand-1 protein expression,23 T-cell receptor diversity metrics,25 and immune gene-expression signatures.3,12,26 TILs in the NeoALTTO trial22 showed a significant association with survival.
Patients with clinical stage II and III HER2-positive breast cancer are typically treated with neoadjuvant chemotherapy plus HER2 targeting because of the effect of neoadjuvant treatment on surgical end points (smaller surgeries and less need for axillary lymph node dissection) and the tailoring of therapy by pCR status now that patients with residual disease (RD) receive ado-trastuzumab emtansine.27 Thus, pCR is no longer merely an intermediate biomarker of outcome, rather both pCR and survival are clinically meaningful end points.
We have previously reported the pCR end points for CALGB 40601 (now part of the Alliance for Clinical Trials in Oncology), including finding a significant contribution to pCR of intrinsic subtype and immune gene signatures.3 Here, we report CALGB 40601 prespecified secondary end points of relapse-free survival (RFS) and overall survival (OS), with a median follow-up of 7 years, and the association between pCR and RFS/OS. Using the transcriptome analyses from pretreatment biopsies, we performed an exploratory analysis to test the ability of clinical and genomic biomarkers to predict pCR and RFS at 7 years. Understanding which biomarkers are going to have an influence on both pCR and survival outcomes will help build prognostic tools to design future escalation and de-escalation trials in HER2-positive breast cancer.
PATIENTS AND METHODS
CALGB 40601 Study Design and Patients
The CALGB 40601 study design and pCR results have been previously published.3 A total of 305 women with stage II and III HER2-positive breast cancer were randomly assigned to receive paclitaxel (T) at 80 mg/m2 once per week, with the addition of trastuzumab (H; 4 mg/kg loading dose followed by 2 mg/kg), lapatinib (L; 1,500 mg/d), or both (L at 1,000 mg/d plus the same dose of H) for 16 weeks. On the basis of reports of inferiority and higher toxicity, the TL arm was closed early; patients who were randomly assigned to that arm completed treatment. Protocol-defined therapy ended at surgery. It was recommended that all patients receive dose-dense doxorubicin and cyclophosphamide (AC) and complete 1 year of trastuzumab adjuvantly.
The primary end point was pCR, defined as no invasive tumor in the breast at surgery. Secondary end points included RFS and OS. RFS was defined as the interval from surgery to ipsilateral invasive breast tumor recurrence, regional recurrence, distant recurrence, or death of any cause, whichever occurred first. Patients without an event were censored at the date of the last clinical assessment. OS was defined as the interval from random assignment to death or last follow-up. Clinical data collection and statistical analyses were conducted by the Alliance Statistics and Data Center.
Sample Acquisition and Biospecimen Processing
Participants underwent four pretreatment 16-gauge core biopsies for research. The CONSORT diagram (Data Supplement) shows the flow of participants from the intention-to-treat (ITT) population to the gene expression cohort. Patients with inadequate RNA quality were excluded from the final gene expression cohort, which consisted of 264 patients. All 264 patients signed an institutional review board–approved, protocol-specific informed consent document following federal and institutional guidelines. This document also included consent for biomarker research.
Tumor Gene Expression Analyses
Gene expression profiles from pretreatment core biopsies were generated by RNA sequencing using an Illumina HiSEquation 2000 (Data Supplement).28 Intrinsic subtype biomarkers were obtained by the PAM50 predictor29 after applying a new HER2/estrogen receptor subgroup-specific gene normalization method (Data Supplement).30 Gene expression signatures and single genes representing multiple biologic pathways and cell types (Data Supplement) were also assessed following different specifications (Data Supplement).
Data Analysis and Interpretation
ANOVA and Fisher exact test were used to compare clinicopathologic variables between different treatment and biomarker groups. Proportions and 95% CIs were provided. We estimated 7-year RFS and OS rates for each treatment arm and pCR group using the Kaplan-Meier method. The relationship between clinical and genomic biomarkers with pCR and/or RFS was assessed using univariable and multivariable logistic and Cox proportional hazards regression models, respectively. Odds ratios, hazard ratios (HRs) and 95% CIs were calculated for each response variable. The significance level was set to a two-sided α of .05, and P values were unadjusted for multiplicity. The Akaike Information Criterion and Bayesian Information Criterion were used to analyze the goodness of fit of two competing statistical models. Collinearity between gene expression continuous variables was evaluated using Pearson correlation. High-dimensional modeling for pCR and RFS was done by Elastic Net (R package glmnet31). A detailed description can be found in the Data Supplement. Analyses were based on the clinical study database frozen on April 3, 2019, and all statistical analyses were performed using R 3.5.2 and Python 3.6.
RESULTS
Clinicopathologic Characteristics
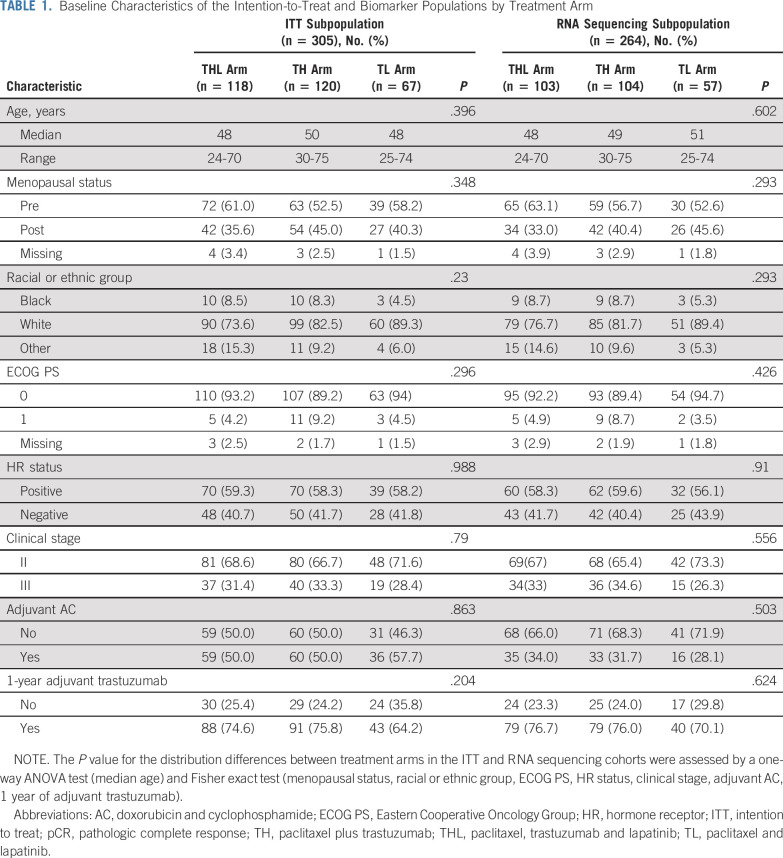
From December 2008 to February 2012, 305 patients were randomly assigned to one of three treatment groups: 118 to trastuzumab plus lapatinib (THL), 120 to TH, and 67 to TL. Of the 305 patients, 299 began protocol treatment; pCR was evaluable in 295 patients. RNA sequencing was performed on 264 pretreatment tumor samples (Data Supplement). Patient characteristics were balanced by treatment arm in both the RNA sequencing and the ITT populations (Table 1). pCR rates in the ITT subpopulation were 57% (95% CI, 47% to 66%) in the THL arm, 45% (95% CI, 36% to 54%) in the TH arm, and 30% (95% CI, 19% to 42%) in the TL arm, slightly different from the original pCR rates3 that were calculated using the pCR-evaluable cohort (n = 295). After surgery, as recommended by the protocol, 51% received AC and 73% completed 1 year of trastuzumab. There was no imbalance by treatment arm in either the RNA sequencing and the ITT cohorts (Table 1).
TABLE 1.
Baseline Characteristics of the Intention-to-Treat and Biomarker Populations by Treatment Arm
RFS and OS Analyses
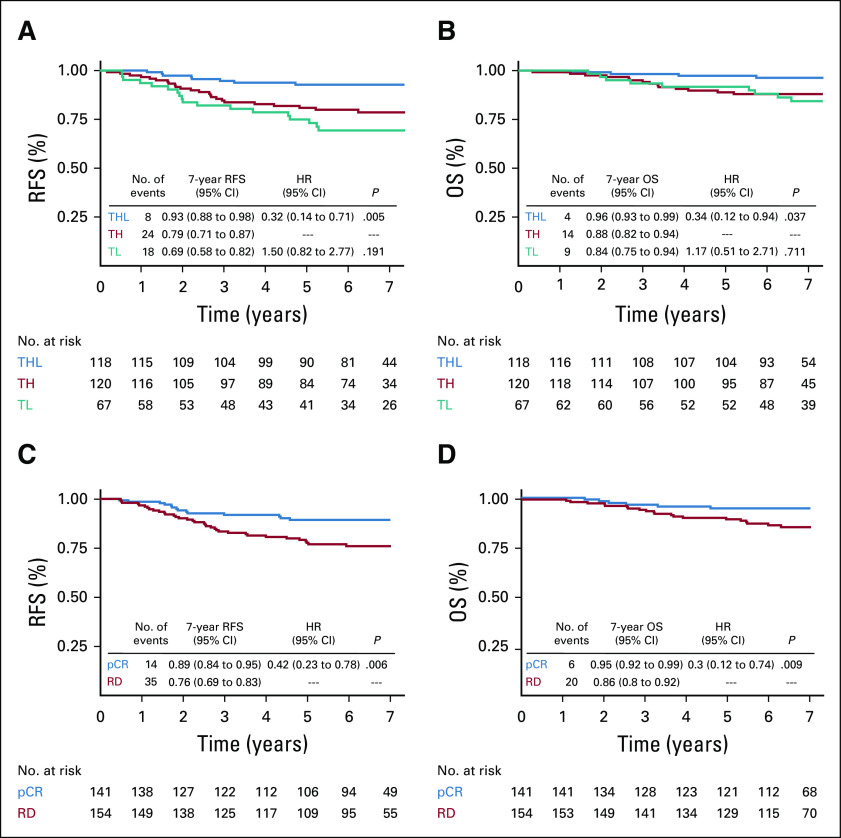
With a median follow-up of 83 months from random assignment (interquartile range, 71 to 90 months), RFS events were recorded in 16% of participants: 18 (26.9%) in the TL arm, 24 (20%) in the TH arm, and eight (6.8%) in the THL arm, with corresponding 7-year RFS rates of 69% (95% CI, 58% to 82%; TL), 79% (95% CI, 71% to 87%; TH), and 93% (95% CI, 88% to 98%; THL). The RFS difference between the THL and control TH arms was highly statistically significant (HR, 0.32; 95% CI, 0.14 to 0.71; P = .005; Fig 1A). Nine deaths (13.4%) occurred in the TL arm, 14 (11.7%) in the TH group, and four (3.4%) in the THL group, with corresponding 7-year OS rates of 84% (TL), 88% (TH), and 96% (THL). OS was significantly higher in the THL compared with the TH arm (HR, 0.34; 95% CI, 0.12 to 0.94; P = .037; Fig 1B). Neither receipt of adjuvant AC, nor whether the full year of adjuvant trastuzumab was completed, altered these relationships (Data Supplement).
FIG 1.
Kaplan-Meier curves for relapse-free survival (RFS) and overall survival (OS) in the intention-to-treat (ITT) population. (A) Kaplan-Meier estimates of RFS at 7 years by treatment arm in the ITT population, showing a significant benefit of THL (paclitaxel, trastuzumab, and lapatinib) versus TH (paclitaxel plus trastuzumab) treatment arms. (B) Kaplan-Meier estimates of OS at 7 years by treatment arm in the ITT population, showing a significant benefit of THL versus TH treatment arms. (C) Kaplan-Meier estimates of RFS at 7 years by pathologic complete response (pCR) status in the ITT population, showing a significant improvement in outcome in pCR versus residual disease (RD). (D) Kaplan-Meier estimates of OS at 7 years by pCR status in the ITT population, showing a significant improvement in outcome in pCR versus RD. HR, hazard ratio; TL, paclitaxel and lapatinib.
When all treatment groups were combined, a significant association between pCR and RFS was found. Of the 141 patients who achieved pCR, 14 (9.9%) had an RFS event compared with 35 (23%) of 154 patients with RD (HR, 0.42; 95% CI, 0.23 to 0.78; P = .006; Fig 1C). Patients who achieved pCR also had improved OS benefit compared with patients with RD (HR, 0.3; 95% CI, 0.12 to 0.74; P = .009; Fig 1D). pCR and treatment effect in RFS and OS were preserved also in the RNA sequencing subpopulation (Data Supplement).
Intrinsic Subtype Association With Response and RFS
Using a new and improved PAM50 normalization method (Data Supplement), the majority of tumors were HER2 Enriched (146 of 264; 55%), followed by Luminal B (13%), Normal like (12%), Luminal A (11%), and Basal like (8%; Data Supplement). Although subtype distribution significantly differed by hormone receptor status (P < .001), the HER2-Enriched subtype was the most frequent subtype identified in both hormone receptor–positive and –negative groups (44% and 72%, respectively; Data Supplement).
pCR in the breast was achieved in 89 (61%) of 146 HER2-Enriched patients compared with 29 (25%) of 118 non–HER2-Enriched patients (odds ratio, 3.8; 95% CI, 2.23 to 6.72; P < .001; Data Supplement). Significant RFS differences were found among the different subtypes. Luminal A tumors, with the lowest pCR rate (14.3%), carried the best RFS outcome, with no events recorded after 7 years of follow-up. In contrast, HER2-Enriched patients, with the highest pCR rate, carried a significantly worse RFS outcome, with 30 (20.5%) of 146 RFS events recorded (Data Supplement). Of interest, only 10 (11%) of 89 HER2-Enriched pCR patients had an RFS event compared with 20 (35%) of 57 HER2-Enriched RD patients.
Genomic Modeling for pCR and RFS
In our previous analyses,3,26 prespecified gene expression signatures—intrinsic subtype, immune activation—significantly and independently contributed to pCR. Building on this, we sought to create a unified predictive model for both pCR and RFS. OS data, with a low number of events (n = 28), were not mature enough for high-quality predictive modeling. We first built 688 logistic regression models to test the ability of each genomic variable to predict pCR in the presence of clinical information, including treatment arm, clinical stage, and hormone receptor status. A total of 215 genomic variables (31.25%) were significantly associated with pCR (Data Supplement). In concordance with our previous results,3,26 subtype-related biomarkers, like the HER2-Enriched signature, the PAM50 risk of recurrence, ERBB2 gene expression, and B cell/immunoglobulin G (IgG) immune signatures were found to powerfully and positively predict pCR. In contrast, luminal parameters, like ESR1 gene expression, the Luminal A signature, the Luminal Tumor Score,32 the chemo-endocrine score (CES),33 and the LumA-HER2-E scores were negative predictors of response.
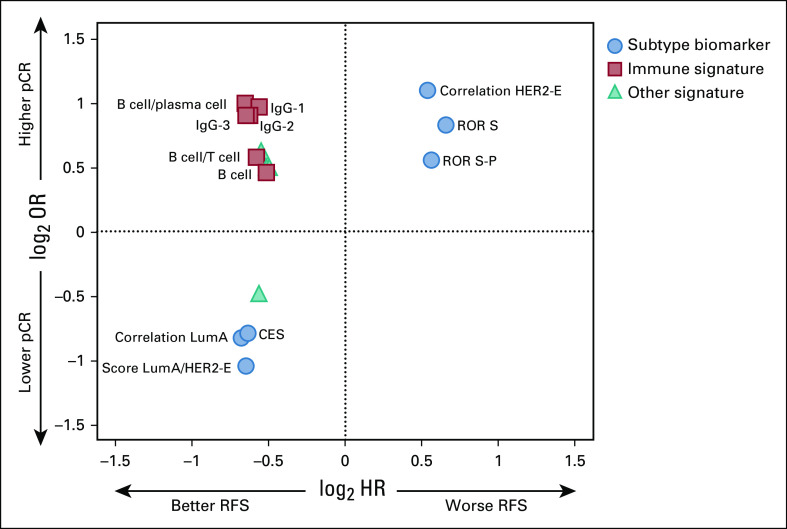
Similarly, we created Cox proportional hazards regression models to predict RFS at 7 years. Only 45 (6.54%) of the 688 biomarkers tested were significantly associated with RFS, and neither receipt of adjuvant AC, nor completion of adjuvant trastuzumab altered this association (Data Supplement). Twenty-two (3.2%) were significantly associated with both pCR and RFS (Fig 2 and Data Supplement), including seven intrinsic subtype-related biomarkers and eight immune signatures. Of interest, all the immune signatures, which were strongly correlated with higher pCR rate, were also associated with longer RFS. In contrast, all the tumor-related biomarkers worked in opposite directions for the prediction of pCR and RFS. Akaike Information Criterion and Bayesian Information Criterion tests showed a better fit of the univariable LumA-HER2-E score pCR and RFS models compared with the HER2-Enriched and Luminal A models (Data Supplement).
FIG 2.
Association between odds ratio (OR) and hazard ratio (HR) in 16 significant biomarkers of response and survival. OR and HR values have been log transformed. Whereas all the immune-related biomarkers predicted a higher response and a longer relapse-free survival (RFS), subtype-related biomarkers showed the opposite direction for the prediction of response and survival. For subtype-related biomarkers, the correlation to each PAM50 centroid was used. CES, chemoendocrine score; HER2-E, HER2-enriched; IgG, immunoglobulin G; LumA, Luminal A; P, proliferation; pCR, pathologic complete response; ROR, risk of recurrence; S, subtype.
A high Pearson correlation among the different genomic biomarkers was observed (Data Supplement). To handle this collinearity, we used Elastic Net to build two high-dimension models for predicting pCR and RFS at 7 years using as outcome time to an event. The best pCR model included 55 (7.9%) of 696 (eight clinical and 688 genomic) variables and an area under the curve value of 0.82 in the train set and 0.75 in the test set, in the line of the expected values from cross-validation (Data Supplement). This model included clinical features, such as treatment arm, as well as several subtype and immune B-cell and T-follicular helper signatures, ESR1 and ERBB2 gene expression, and some supervised signatures, such as the CES33 and LumA-HER2-E scores. The optimal RFS model included 46 (6.6%) of 697 (nine clinical and 688 genomic) variables, with a C-index of 0.88 in the train set and 0.63 when applied to the test set, in line with the expected values from cross-validation (Data Supplement). In this model, clinical parameters, such as pCR status and treatment arm, carried the greatest weights. Subtype-related biomarkers and multiple B-cell and T-cell signatures were also present. From all 688 genomic variables tested, only nine (1.3%) were present in both models (Data Supplement). In particular, five immune signatures were included within the significant positive features for response and RFS prediction: two IgG signatures, one B-cell/plasma cell signature, one T-helper signature, and a T-cell/B-cell cooperation signature.34 In contrast, all subtype-related biomarkers worked in opposite directions for the prediction of pCR and RFS. Of interest, the LumA-HER2-E score was selected together with the HER2-Enriched and the Luminal A signatures for both models. The CES33 score and the ESR1 and ERBB2 gene expression, present in the pCR model, were not selected by Elastic Net as RFS predictors.
The predictive value of HER2-Enriched and the IgG signatures, or whether these two biomarkers can predict the benefit of dual over single HER2 blockade was also explored (Data Supplement). HER2-Enriched and IgG-high patients treated with THL had a significant RFS benefit compared with TH (HR, 0.28; 95% CI, 0.1 to 0.77; P = .014; and HR, 0.09; 95% CI, 0.01 to 0.72; P = .023) with no significant differences in non–HER2-Enriched and IgG-low subgroups; however, both interaction tests were nonsignificant. Consequently, we could not establish the predictive value of these two biomarkers.
Genomic Signatures for RFS Within Patients With RD
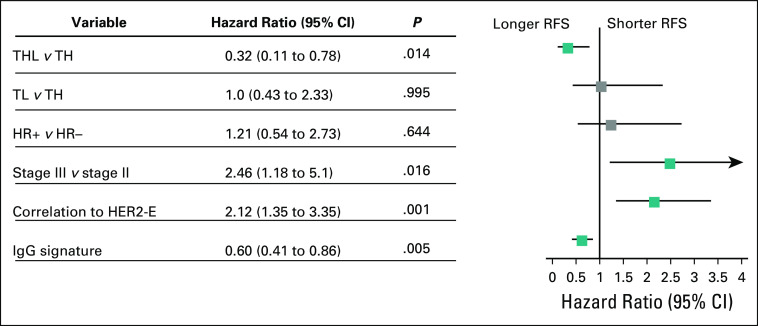
The RD subset was of particular interest given the therapeutic and prognostic implications, as well as the discordance of the pCR and RFS relationship in key signatures. In two univariable Cox proportional hazards regression models in patients with RD, the HER2-Enriched signature was significantly correlated with shorter RFS (HR, 1.77; 95% CI, 1.19 to 2.62; P = .005), whereas the IgG signature was correlated with longer RFS (HR, 0.65; 95% CI, 0.46 to 0.93; P = .019). Moreover, HER2-Enriched or IgG-low RD patients had significantly shorter RFS interval (Data Supplement). In multivariable Cox analysis, the IgG signature remained an independent good prognostic factor (HR, 0.60; 95% CI, 0.41 to 0.86; P = .005), whereas patients with RD with higher HER2-Enriched signature had significantly worse RFS (HR, 2.12; 95% CI, 1.35 to 3.35; P = .001; Fig 3). Of interest, hormone receptor status was not significantly associated with RFS in this group.
FIG 3.
Forest plot representing a multivariable Cox proportional hazards regression analysis within the residual disease group of patients. HER2-enriched (HER2-E) subtype was correlated with shorter relapse-free survival (RFS), whereas immunoglobulin G (IgG) signature was an independent good prognosis factor. The correlation to the HER2-E centroid and the IgG gene expression signature, both as continuous variables, were used for the multivariable Cox proportional hazards regression analysis. HR, hormone receptor; TH, paclitaxel plus trastuzumab; THL, paclitaxel, trastuzumab, and lapatinib; TL, paclitaxel and lapatinib.
DISCUSSION
In CALGB 40601, women whose tumors had pCR to neoadjuvant chemotherapy plus HER2 targeting had significantly better RFS and OS than did women with RD, a finding consistent with many other neoadjuvant trials. Of interest, there was a significant improvement in RFS and OS at 7 years with dual therapy in this trial, a surprising finding given that a large adjuvant trial, ALTTO, which included a lower clinical risk but otherwise similar patient population, demonstrated only a modest and statistically nonsignificant effect (disease-free survival HR, 0.84) of adding lapatinib administered for a longer duration.8 A trial similar in population and intervention to CALGB 40601, NeoALTTO, found numerically higher but nonsignificant EFS differences with dual therapy (84% v 76%)35; as in CALGB 40601, survival outcome was a secondary end point. For these reasons, our statistically significant effect of dual therapy on relapse and survival should be considered in the context of its secondary analytic nature and the results of other trials suggesting a far more limited impact of dual therapy.
Accumulating evidence supports the clinical validity of two prognostic biomarkers in HER2-positive breast cancer: intrinsic subtype and immune cell features. The HER2-Enriched subtype, a key and independent predictor of pCR, was also found to be an inverse predictor of RFS. Specifically, HER2-Enriched patients, with the highest pCR rate of the tumor intrinsic subtypes, had significantly worse RFS than did patients with Luminal A tumors, even in the presence of HER2-targeted drugs. This finding, which fails to conform to the known association of pCR with outcome, is consistent with emerging data regarding the complexity of the relationship of pCR with RFS and the effect of confounding but unmeasured variables when the focus is entirely on pCR. This is an example of Simpson’s paradox,36 in which the confounding variable is the substantially worse RFS outcome among HER2-Enriched tumors with RD. Genomically, the HER2-Enriched subtype is the most HER2 oncogene addicted of the subtypes, which likely explains the high sensitivity to anti-HER2 therapies and may also explain why an HER2-Enriched tumor resistant to anti-HER2 therapies administered neoadjuvantly carries a particularly poor prognosis. Luminal tumors, which comprise the majority of non–HER2-Enriched among clinically HER2-positive tumors, are genomically driven by hormone receptor–related pathways and are usually hormone receptor positive, likely benefiting from 5 to 10 years of adjuvant endocrine therapy.
Conversely, evidence of immune activation using multiple RNA-based signatures was significantly and independently directly associated with a higher likelihood of pCR and better RFS. Whether a pathology-based approach, such as TILs, will perform similarly as well as multigene expression profiling is unknown. In NeoALTTO, TILs and immune signatures seemed to predict higher pCR,12,22 whereas only TILs portended statistically significant better event-free survival.22
The combination of HER2-Enriched and IgG signatures provided more prognostic information than either alone; to build a useful prognostic tool for patients with HER2-positive breast cancer, both biomarkers should be taken into account. We also found that these signatures may provide augmented clinical value in patients with RD.
Our study has several strengths. First, the clinical trial has mature RFS and OS estimates. Second, we were able to perform a comprehensive genomic analysis on a significant proportion (86.6%) of the pretreatment specimens. Third, we performed an integrated analysis of clinical parameters and 688 genomic biomarkers, testing their ability to predict not only response but also survival. Although the gene expression correlative analysis of the NOAH and the NeoALTTO trials previously identified an association between a small number of signatures with pCR and event-free survival,10,12 the current study is the most comprehensive effort to date to correlate pCR and RFS predictors in HER2-positive breast cancer. Efforts of this type are key to identifying mechanisms to more precisely select patients who are appropriate for de-escalation strategies, as was done in the APT trial using only low-risk clinical features37 or escalation strategies, such as the use of adjuvant ado-trastuzumab emtansine.
In contrast, our study also has several limitations. First, CALGB 40601 was not powered to detect a survival benefit, and it was not designed to be prescriptive regarding adjuvant therapy other than the recommendation of completion of AC chemotherapy and of 1 year of adjuvant trastuzumab. For this reason, RFS biomarker analysis should be also interpreted with caution. Second, the anti-HER2 approaches used in the trastuzumab-containing arms are consistent with the current standard of care, but lapatinib is not used in the early breast cancer setting because of the negative results of the ALTTO trial.8 Finally, TILs assessment in the pretreatment specimens has not been performed yet and will be reported in a future analysis.
To conclude, in CALGB 40601, dual HER2 blockade with lapatinib added to trastuzumab and chemotherapy demonstrated a significant effect on RFS compared with trastuzumab plus chemotherapy alone, and patients who achieved pCR had significantly better outcomes than patients with RD. However, most patients with RD did not experience relapse, and some pCR patients did experience relapse. Our genomic data suggest that future escalation and de-escalation strategies may benefit from integrating the information provided by clinical parameters, intrinsic subtype, and immune signatures to predict not only response, but also survival.
PRIOR PRESENTATION
Presented in part at the 40th San Antonio Breast Cancer Symposium, San Antonio, CA, December 5-9, 2017; and at the ESMO Congress, Barcelona, Spain, September 27-October 1, 2019.
SUPPORT
Supported by National Cancer Institute, National Institutes of Health, Grants No. U10CA180821, U10CA180882, and U24CA196171 (to the Alliance for Clinical Trials in Oncology); R01CA229409, UG1CA233160, UG1CA233180, UG1CA233290, UG1CA233329, UG1CA233333, UG1CA233373, and P50CA58223 (L.A.C. and C.M.P.); U10CA180888 and UG1CA233160 (SWOG); and by The Breast Cancer Research Foundation (Alliance, L.A.C., and C.M.P.), Susan G. Komen (L.A.C. and C.M.P.), GlaxoSmithKline and SEOM (Becas FSEOM para Formación en Investigación en Centros de Referencia en el Extranjero 2016 to AF-M). For additional support acknowledgment: https://acknowledgments.alliancefound.org.
CLINICAL TRIAL INFORMATION
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
Conception and design: Mei-Yin Polley, Lyndsay Harris, Donald Berry, Olwen Hahn, Clifford Hudis, Eric Winer, Charles M. Perou, Lisa A. Carey
Financial support: Clifford Hudis, Charles M. Perou, Lisa A. Carey
Administrative support: Clifford Hudis, Lisa A. Carey
Provision of study materials or patients: Ian E. Krop, Chau Dang, Eric Winer, Ann Partridge, Charles M. Perou, Lisa A. Carey
Collection and assembly of data: Aranzazu Fernandez-Martinez, David W. Hillman, Mei-Yin Polley, Katherine A. Hoadley, Sara Tolaney, Chau Dang, Donald Berry, Olwen Hahn, Clifford Hudis, Charles M. Perou, Lisa A. Carey
Data analysis and interpretation: Aranzazu Fernandez-Martinez, Ian E. Krop, David W. Hillman, Mei-Yin Polley, Joel S. Parker, Lucas Huebner, Katherine A. Hoadley, Jonathan Shepherd, N. Lynn Henry, Chau Dang, Donald Berry, Clifford Hudis, Eric Winer, Ann Partridge, Charles M. Perou, Lisa A. Carey
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Survival, Pathologic Response, and Genomics in CALGB 40601 (Alliance), a Neoadjuvant Phase III Trial of Paclitaxel-Trastuzumab With or Without Lapatinib in HER2-Positive Breast Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Ian E. Krop
Employment: AMAG Pharmaceuticals (I), Freeline Therapeutics (I)
Leadership: AMAG Pharmaceuticals (I), Freeline Therapeutics (I)
Stock and Other Ownership Interests: AMAG Pharmaceuticals (I), Freeline Therapeutics (I), Vertex (I)
Honoraria: Genentech, AstraZeneca, Celltrion
Consulting or Advisory Role: Genentech, Seattle Genetics, Daiichi Sankyo, Macrogenics, Taiho Pharmaceutical, Context Therapeutics, Novartis, Merck, ION Pharma, Bristol Myers Squibb
Research Funding: Genentech (Inst), Pfizer (Inst)
Joel S. Parker
Stock and Other Ownership Interests: GeneCentric
Consulting or Advisory Role: Medivation
Patents, Royalties, Other Intellectual Property: Authored patents related to the PAM50 algorithm which are licensed to NanoString Technologies.
Sara Tolaney
Consulting or Advisory Role: Novartis, Pfizer, Merck, Eli Lilly, Nektar, NanoString Technologies, AstraZeneca, Puma Biotechnology, Genentech, Eisai, Immunomedics, Sanofi, Celldex, Bristol Myers Squibb, Paxman, Seattle Genetics, Odonate Therapeutics, AbbVie, Silverback Therapeutics, G1 Therapeutics, OncoPep, Kyowa Hakko Kirin, Samsung Bioepis, CytomX Therapeutics, Daiichi Sankyo, Athenex
Research Funding: Genentech (Inst), Merck (Inst), Exelixis (Inst), Pfizer (Inst), Eli Lilly (Inst), Novartis (Inst), Bristol Myers Squibb (Inst), Eisai (Inst), AstraZeneca (Inst), NanoString Technologies (Inst), Cyclacel (Inst), Nektar (Inst), Immunomedics (Inst), Odonate Therapeutics (Inst), Sanofi (Inst), Seattle Genetics (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Eli Lilly, Merck, Nektar, Novartis, Pfizer, Genentech, Immunomedics, Eisai, NanoString Technologies, Puma Biotechnology, Celldex
N. Lynn Henry
Research Funding: Innocrin Pharma (Inst), Pfizer (Inst), AbbVie (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/27894/summary
Chau Dang
Honoraria: Eli Lilly, Daiichi Sankyo, Puma Biotechnology, Genentech
Consulting or Advisory Role: Genentech, Puma Biotechnology, Eli Lilly, Daiichi Sankyo
Research Funding: Genentech (Inst), Puma Biotechnology (Inst)
Travel, Accommodations, Expenses: Genentech, Puma Biotechnology, Eli Lilly, Daiichi Sankyo
Lyndsay Harris
Patents, Royalties, Other Intellectual Property: Philips Healthcare
Donald Berry
Employment: Berry Consultants
Leadership: Berry Consultants
Stock and Other Ownership Interests: Berry Consultants
Consulting or Advisory Role: Berry Consultants
Research Funding: Daiichi Sankyo
Travel, Accommodations, Expenses: Berry Consultants
Olwen Hahn
Leadership: Via Oncology
Stock and Other Ownership Interests: Teleflex Medical
Honoraria: Cardinal Health (I)
Consulting or Advisory Role: Pfizer
Travel, Accommodations, Expenses: Cardinal Health (I)
Clifford Hudis
Uncompensated Relationships: Alliance Foundation, Columbia University External Scientific Advisory Board, Memorial Sloan Kettering Cancer Center
Open Payments Link: https://openpaymentsdata.cms.gov/physician/471974/summary
Eric Winer
Honoraria: Genentech, Genomic Health
Consulting or Advisory Role: Leap Therapeutics, Seattle Genetics, Jounce Therapeutics, GlaxoSmithKline, Carrick Therapeutics, Eli Lilly, G1 Therapeutics, Syros Pharmaceuticals, Genentech
Research Funding: Genentech (Inst)
Other Relationship: InfiniteMD
Ann Partridge
Patents, Royalties, Other Intellectual Property: Royalty payments for coauthoring the breast cancer survivorship section of UpToDate
Travel, Accommodations, Expenses: Novartis
Charles M. Perou
Leadership: GeneCentric
Stock and Other Ownership Interests: Bioclassifier, GeneCentric
Consulting or Advisory Role: Bioclassifier, GeneCentric, NanoString Technologies, Veracyte
Patents, Royalties, Other Intellectual Property: Royalties from PAM50 breast cancer gene patent application, and from lung gene signature patent
Travel, Accommodations, Expenses: Takeda, Chugai Pharma
Lisa A. Carey
Research Funding: Innocrin Pharma (Inst), Syndax (Inst), Immunomedics (Inst), Novartis (Inst), NanoString Technologies (Inst), AbbVie (Inst), Seattle Genetics (Inst)
Patents, Royalties, Other Intellectual Property: Royalty-sharing agreement, investorship interest in licensed intellectual property to startup company, Falcon Therapeutics, that is designing neural stem cell–based therapy for glioblastoma multiforme (I)
Uncompensated Relationships: Sanofi (Inst), Novartis (Inst), G1 Therapeutics (Inst), Genentech (Inst), GlaxoSmithKline (Inst), Exact Sciences (Inst), AstraZeneca (Inst), Daiichi Sanyo (Inst), Aptitude Health (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/179671
No other potential conflicts of interest were reported.
REFERENCES
- 1.Robidoux A, Tang G, Rastogi P, et al. Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): An open-label, randomised phase 3 trial. Lancet Oncol. 2013;14:1183–1192. doi: 10.1016/S1470-2045(13)70411-X. [DOI] [PubMed] [Google Scholar]
- 2.Baselga J, Bradbury I, Eidtmann H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–640. doi: 10.1016/S0140-6736(11)61847-3. [Erratum: Lancet 379:616, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542–549. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarneri V, Frassoldati A, Bottini A, et al. Preoperative chemotherapy plus trastuzumab, lapatinib, or both in human epidermal growth factor receptor 2-positive operable breast cancer: Results of the randomized phase II CHER-LOB study. J Clin Oncol. 2012;30:1989–1995. doi: 10.1200/JCO.2011.39.0823. [DOI] [PubMed] [Google Scholar]
- 5.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 6.Llombart-Cussac A, Cortés J, Paré L, et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): An open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18:545–554. doi: 10.1016/S1470-2045(17)30021-9. [DOI] [PubMed] [Google Scholar]
- 7.Rimawi MF, Mayer IA, Forero A, et al. Multicenter phase II study of neoadjuvant lapatinib and trastuzumab with hormonal therapy and without chemotherapy in patients with human epidermal growth factor receptor 2-overexpressing breast cancer: TBCRC 006. J Clin Oncol. 2013;31:1726–1731. doi: 10.1200/JCO.2012.44.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piccart-Gebhart M, Holmes E, Baselga J, et al. Adjuvant lapatinib and trastuzumab for early human epidermal growth factor receptor 2-positive breast cancer: Results from the randomized phase III adjuvant lapatinib and/or trastuzumab treatment optimization trial. J Clin Oncol. 2016;34:1034–1042. doi: 10.1200/JCO.2015.62.1797. [Erratum: J Clin Oncol 37:356, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Minckwitz G, Procter M, de Azambuja E, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N Engl J Med. 2017;377:122–131. doi: 10.1056/NEJMoa1703643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Bianchini G, Thomas M, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin Cancer Res. 2014;20:511–521. doi: 10.1158/1078-0432.CCR-13-0239. [DOI] [PubMed] [Google Scholar]
- 11.Swain SM, Tang G, Lucas PC, et al. Pathologic complete response and outcomes by intrinsic subtypes in NSABP B-41, a randomized neoadjuvant trial of chemotherapy with trastuzumab, lapatinib, or the combination. Breast Cancer Res Treat. 2019;178:389–399. doi: 10.1007/s10549-019-05398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fumagalli D, Venet D, Ignatiadis M, et al. RNA sequencing to predict response to neoadjuvant anti-HER2 therapy: A secondary analysis of the NeoALTTO randomized clinical trial. JAMA Oncol. 2017;3:227–234. doi: 10.1001/jamaoncol.2016.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prat A, Slamon D, Hurvitz S, et al. Association of intrinsic subtypes with pathological complete response (pCR) in the KRISTINE neoadjuvant phase 3 clinical trial in HER2-positive early breast cancer (EBC) Cancer Res. 2018;78(abstr PD3-06) [Google Scholar]
- 14.Dieci MV, Prat A, Tagliafico E, et al. Integrated evaluation of PAM50 subtypes and immune modulation of pCR in HER2-positive breast cancer patients treated with chemotherapy and HER2-targeted agents in the CherLOB trial. Ann Oncol. 2016;27:1867–1873. doi: 10.1093/annonc/mdw262. [DOI] [PubMed] [Google Scholar]
- 15.Bianchini G, Parker J, Carey L, et al. Research-based PAM50 predicts risk of relapse in residual disease after anti-HER2 therapies. Ann Oncol. 2018;29(suppl 8):VIII61. [Google Scholar]
- 16.Prat A, Pascual T, De Angelis C, et al. HER2-Enriched subtype and ERBB2 expression in HER2-positive breast cancer treated with dual HER2 blockade. J Natl Cancer Inst. 2020;112:46–54. doi: 10.1093/jnci/djz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schettini F, Pascual T, Conte B, et al. HER2-enriched subtype and pathological complete response in HER2-positive breast cancer: A systematic review and meta-analysis. Cancer Treat Rev. 2020;84:101965. doi: 10.1016/j.ctrv.2020.101965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogue-Geile KL, Song N, Jeong JH, et al. Intrinsic subtypes, PIK3CA mutation, and the degree of benefit from adjuvant trastuzumab in the NSABP B-31 trial. J Clin Oncol. 2015;33:1340–1347. doi: 10.1200/JCO.2014.56.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majewski IJ, Nuciforo P, Mittempergher L, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015;33:1334–1339. doi: 10.1200/JCO.2014.55.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 21.Loi S, Michiels S, Salgado R, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 22.Salgado R, Denkert C, Campbell C, et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: A secondary analysis of the NeoALTTO trial JAMA Oncol 1448–454.2015 [Errata: JAMA Oncol 1:544, 2015; JAMA Oncol 1:1172, 2015; JAMA Oncol 5:122, 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barroso-Sousa R, Barry WT, Guo H, et al. The immune profile of small HER2-positive breast cancers: A secondary analysis from the APT trial. Ann Oncol. 2019;30:575–581. doi: 10.1093/annonc/mdz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuciforo P, Pascual T, Cortés J, et al. A predictive model of pathologic response based on tumor cellularity and tumor-infiltrating lymphocytes (CelTIL) in HER2-positive breast cancer treated with chemo-free dual HER2 blockade. Ann Oncol. 2018;29:170–177. doi: 10.1093/annonc/mdx647. [DOI] [PubMed] [Google Scholar]
- 25.Powles RL, Redmond D, Sotiriou C, et al. Association of T-cell receptor repertoire use with response to combined trastuzumab-lapatinib treatment of HER2-positive breast cancer: Secondary analysis of the NeoALTTO randomized clinical trial. JAMA Oncol. 2018;4:e181564. doi: 10.1001/jamaoncol.2018.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanioka M, Fan C, Parker JS, et al. Integrated analysis of RNA and DNA from the phase III trial CALGB 40601 identifies predictors of response to trastuzumab-based neoadjuvant chemotherapy in HER2-positive breast cancer. Clin Cancer Res. 2018;24:5292–5304. doi: 10.1158/1078-0432.CCR-17-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Minckwitz G, Huang CS, Mano MS, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 28.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Rødland EA, Tibshirani R, et al. Molecular subtyping for clinically defined breast cancer subgroups. Breast Cancer Res. 2015;17:29. doi: 10.1186/s13058-015-0520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. 2005;67:301–320. [Google Scholar]
- 32.Garcia-Recio S, Thennavan A, East MP, et al. FGFR4 regulates tumor subtype differentiation in luminal breast cancer and metastatic disease. J Clin Invest. 2020;130:4871–4887. doi: 10.1172/JCI130323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prat A, Lluch A, Turnbull AK, et al. A PAM50-based chemoendocrine score for hormone receptor-positive breast cancer with an intermediate risk of relapse. Clin Cancer Res. 2017;23:3035–3044. doi: 10.1158/1078-0432.CCR-16-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollern DP, Xu N, Thennavan A, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179:1191–1206.e21. doi: 10.1016/j.cell.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Azambuja E, Holmes AP, Piccart-Gebhart M, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): Survival outcomes of a randomised, open-label, multicentre, phase 3 trial and their association with pathological complete response. Lancet Oncol. 2014;15:1137–1146. doi: 10.1016/S1470-2045(14)70320-1. [DOI] [PubMed] [Google Scholar]
- 36.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 37.Tolaney SM, Barry WT, Dang CT, et al. Adjuvant paclitaxel and trastuzumab for node-negative, HER2-positive breast cancer. N Engl J Med. 2015;372:134–141. doi: 10.1056/NEJMoa1406281. [DOI] [PMC free article] [PubMed] [Google Scholar]