Abstract
Although 80% of individuals infected with the severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) recover without antiviral treatments, the other 20% progress to severe forms of pulmonary disease, suggesting that the host’s immune response to the virus could influence the outcome of coronavirus disease 2019 (COVID-19). SARS-CoV-2 infects alveolar epithelial type 2 cells expressing angiotensin-converting enzyme 2, and these infected epithelial cells recruit dendritic cells, neutrophils and monocytes /macrophages, leading to the activation of CD4+ and CD8+ T cells. These cells launch an antiviral immune response, but are able to completely suppress viral replication or completely eradicate virus in a limited proportion of infected patients. In other patients, viral suppression is incomplete and the numbers of circulating B and T cells are subsequently reduced by as yet unknown mechanisms. Some patients with sustained viral replication progress to a severe condition called cytokine storm. Although antiviral drug(s) should be considered early in infection to prevent progression, there have been no antiviral therapies proven to be effective for significantly inhibiting the viral replication in vivo and suppressing the progression to cytokine storm. Blocking the action of cytokines with dexamethasone or anti-interleukin-6 could have a pivotal role in treatment of those patients. Therapeutic strategy should therefore be based on viral kinetics and the immunopathology of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Immune response, Antibody, Viral kinetics, Treatment
Core Tip: Since the coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), the core therapeutic strategy should be aimed at the eradication of the virus. Unfortunately, there have been no antiviral drugs proven to be effective for this viral infection. The pathogenesis of SARS-CoV-2 is formed by the interaction between the viral infection and the immune response to the virus, and the various clinical features observed in patients with COVID-19 could be due to differences in host immune responses. Moreover, therapeutic strategies should be based on both viral kinetics and the immune response. This editorial summarizes current understanding about immune responses in patients infected with SARS-CoV-2 and provides clues to therapeutic strategies based on this information.
INTRODUCTION
A novel coronavirus, severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), which emerged in 2019 in Wuhan, China, has been found to cause acute respiratory syndrome called coronavirus disease 2019 (COVID-19), with some patients developing a severe form of this disease leading to death. This virus is the third highly pathogenic coronavirus reported to date and has caused a worldwide outbreak of infection called a pandemic. Around 80% of individuals infected with SARS-CoV-2 appear to recover without antiviral treatments[1], indicating that an adequate host immune response against the virus may be sufficient to cure the viral infection. In contrast, the other 20% of patients develop severe forms of this disease, suggesting that an inadequate immune response may predispose to poorer outcomes. Understanding host immune responses against SARS-CoV-2 may be important in designing therapeutic strategies to fight viral infection.
UNDERSTANDING THE IMMUNOPATHOGENESIS OF COVID-19
Virology of SARS-CoV-2
Coronaviruses are enveloped viruses with a single-stranded positive sense RNA. For the virus to enter host cells, the spike (S) protein on the surface of SARS-CoV-2, after priming by the host serine protease TMPRSS2, binds to the host cell angiotensin-converting enzyme 2 (ACE2) receptor[2]. The structure of the receptor-binding domain (RBD) of S protein that binds to the ACE2 receptor has been determined[3], and antibody (Ab) against this RBD is thought to have the ability to neutralize the virus. Cells abundantly expressing ACE2 include alveolar type 2 cells of the lungs and gastrointestinal epithelial cells[4], but any cells expressing ACE2 could be targeted by the virus. Whether SARS-CoV-2 has cytopathic effects (CPE) is unknown, but Middle East respiratory syndrome coronavirus has been reported to induce apoptosis of lung cells through upregulation of Smad7 and fibroblast growth factor 2 expression[5]. Therefore, high replication of SARS-CoV-2 may also induce CPE, which could be involved in the pathogenesis of viral infection. In contrast, alveolar type 2 cells infected with SARS-CoV-2 or alveolar macrophages have been reported to produce cytokines and chemokines. Although these molecules may activate immune responses against the virus, they could also contribute to the pathogenesis of lung diseases and thrombosis[6]. The cellular mechanism underlying the pathogenesis induced by SARS-CoV-2 infection is thus thought to be complex, involving viral replication and immune responses against infected cells.
Immune response to SARS-CoV-2
Innate immunity: Following viral infection, the innate immune system is immediately activated to eliminate pathogens without harming host cells. This system is responsible for host defenses until the development of acquired immunity, with T and B cell responses being fully activated 7 or more days after infection[7]. Cells involved in innate immunity include dendritic cells, macrophages, neutrophils and natural killer (NK) cells. Although little is known about innate immune responses to SARS-CoV-2 infection, the expression of type I interferon (IFN), which is usually produced by virus-infected cells and can suppress viral replication and limit the dissemination of infected cells, is thought to be suppressed in COVID-19[8]. However, an analysis of temporal dynamics in viral shedding showed that the peak of viral shedding from the throat occurs prior to the onset of symptoms and gradually decreases thereafter[9], suggesting that an antiviral innate immune response suppresses viral replication during early phases of COVID-19. The innate immune response, coupled with subsequent acquired immune responses, may be sufficient to eradicate the virus in 80% of infected patients, who recover without antiviral treatments, but may not be vigorous enough to eradicate the virus in the remaining infected patients. Sustained replication of the virus in the latter patients may trigger subsequent inflammatory responses and recruit excess numbers of dendritic cells, T cells, B cells, NK cells, neutrophils and monocytes/macrophages[10], leading to the development of moderate and severe pathology of the lungs. Cells are thought to be recruited by cytokines, such as tumor necrosis factor (TNF)-α and interleukin (IL)-6, and chemokines, such as CCL2/MCP-1, CCL3/MIP-1α, and CXCL10/IP-10, which are produced by infected airway epithelial cells and alveolar macrophages, as reported in a mouse model of SARS-CoV infection[6].
SARS-CoV-2 specific B cell responses: B cell populations were shown to be lower in patients with severe COVID-19 than in control individuals[11]. The role of B cells in the pathogenesis of COVID-19 remains unclear, but a report of SARS-CoV-2 infection in patients with B cell defects may provide important information on their role. SARS-CoV-2 infection of patients with agammaglobulinemia lacking B cells resulted in a short course of infection with mild severity[12]. Although evidence in additional patients is needed to determine the role of B cells in the pathogenesis of COVID-19, these findings suggest that B cells may not be required to control infection.
Abs against the RBD of the S protein of SARS-CoV-2 have been observed frequently in infected patients, with seroconversion rates of putative neutralizing total, IgM, and IgG antibodies being 93.1%, 87.2% and 64.7%, respectively, and their median seroconversion times being 11 d, 12 d and 14 d, respectively[13]. These findings suggest that B cell function is preserved, despite reductions in the numbers of circulating B cells. An analysis of the temporal profile of serum Abs found that rates of seropositivity for anti-nucleocapsid protein IgG and IgM Abs were 94% and 88%, respectively, while the rates of seropositivity for anti-RBD IgG and IgM Abs were 100% and 94%, respectively[14]. Ab responses were found to be faster and higher in severe than in mild cases. Although IgM responses generally precede IgG responses, both were found to occur almost simultaneously in subjects infected with SARS-CoV-2[14]. Therefore, measuring both IgM and IgG Abs would have high sensitivity for the diagnosis of acute infection. However, the absence of Abs does not preclude recent infection, as not all patients develop Ab responses and the levels of neutralizing Abs detected after infection were found to be decreased or disappear in the early convalescent phase[15].
Because neutralizing IgG Abs are present in the blood of many patients who recover from acute SARS-CoV-2 infection[16], the plasma from convalescing patients has been utilized to treat those actively infected with SARS-CoV-2[17-19]. Infusion of convalescent plasma was found to be effective, even in critically ill patients with acute respiratory distress syndrome (ARDS)[19]. Because cytokine storm is the main pathogenic mechanism involved in the development of ARDS, the effect observed by infusion of convalescent plasma may be due not only to neutralizing Abs but to putative immunoregulatory molecules that can inhibit cytokine overproduction.
It is unclear, however, whether patients who recover from COVID-19 are protected from the effects of re-exposure to the virus. The World Health Organization stated that there is currently no evidence showing that recovered patients are protected from a second infection[20]. However, a model of SARS-CoV-2 infection in rhesus macaques demonstrated that primary infection with SARS-CoV-2 induced successful protection from re-infection[21]. Additional studies are required to determine whether protective immunity against SARS-CoV-2 develops after primary infection, because these data are important in determining whether vaccines against SARS-CoV-2 would be effective in preventing re-infection.
SARS-CoV-2 and T cell responses: Both CD4+ and CD8+ T cells are actively involved in the pathogenesis of COVID-19[22], and the percentages, but not the absolute numbers, of circulating CD4+ and CD8+ T cells producing IFN-γ were found to be higher in severely infected than in mildly infected patients[11]. Moreover, S protein-specific CD4+ T cells have been detected in infected patients[23]. These CD4+ T cells may be cross-reactive clones against SARS-CoV-2, which may have developed in response to endemic coronavirus infection, because these T cells have been detected in some uninfected persons[23].
Many studies have indicated that both CD4+ and CD8+ T cell populations are decreased in response to SARS-CoV-2 infection, especially in severe cases, indicating that overall T cell response becomes impaired during the progression of the disease[22,24]. The mechanism responsible for T cell decrease in COVID-19 is unclear, but T cell apoptosis may result from the defective activation of T cells by virus-infected dendritic cells[25].
Virus-specific memory CD8+ T cells were shown to protect infected individuals from the lethality of COVID-19[26]. Therefore, a decreased CD8+ T cell response in COVID-19 patients may lead to inadequate control of viral replication. Moreover, the increased expression of NKG2A in CD8+ T cells and NK cells of patients with COVID-19 indicated that these cells may have been exhausted, possibly from the high viral load in inflamed tissues[27]. These findings suggest an inadequate antiviral CD8+ T cell response to SARS-CoV-2 infection is associated with progression to the severe form of the disease.
CD4+ T cells also play an important role in controlling viral replication[16]. Both the numbers and functions of CD4+ T cells were found to be reduced in patients with COVID-19[22]. However, the role of CD4+ T cells in the control of SARS-CoV-2 infection is unclear, because the course of disease was mild in patients co-infected with SARS-CoV-2 and human immunodeficiency virus (HIV) and having low CD4+ T cell counts, similar to findings in immunocompetent persons[28-30]. The immunosuppressive state with low CD4+ T cell counts observed in HIV-infected patients may enable their escape from harmful immune over-responses. Similarly, a mild course of infection was observed in SARS-CoV-2-infected patients with chronic arthritis who were being treated with immunosuppressive agents[31]. Moreover, a mouse model of SARS-CoV infection showed that viral clearance could occur even after the depletion of both CD4+ and CD8+ T cells, with viral clearance resulting from the accumulation of dendritic cells and neutrophils, recruited by MCP-1/CCL2 and MIP-2β/CXCL2[32]. Collectively, although the numbers of functions of both CD4+ T cells and CD8+ T cells are decreased in COVID-19 patients, additional investigations are required to determine the precise mechanisms underlying these reductions and the roles of both T cell subsets in the control and pathogenesis of COVID-19.
Cytokine storm: Of 138 patients hospitalized for SARS-CoV-2-infected pneumonia, 36 (26.1%) were transferred to the intensive care unit during the clinical course of the disease because of complications, including ARDS, arrhythmia, and shock[33]. The disease rapidly progressed in these patients, with a median time from first symptom to ARDS being 8.0 d[33]. Because most deaths of SARS-CoV-2-infected patients are caused by its complications, especially ARDS[34], understanding the mechanism by which these infected patients develop ARDS is important for their management and can reduce mortality rates. Although relatively little is known about the factors associated with progression to severe disease, the main pathophysiology of ARDS was shown to be cytokine release syndrome (CRS)[25], also called cytokine storm. Cytokines and chemokines overproduced in a cytokine storm include IL-1β, IL-2, IL-6, IL-10, TNF-α, IFN-γ, IP-10, MIP-1, and MIP-1α[35]. IL-6 is thought to be a key factor, because serum IL-6 concentration was associated with disease severity[36] and mortality[37]. IL-6 is synthesized by macrophages and dendritic cells upon recognition of pathogens through toll-like receptors at the site of infection[38]. Circulating IL-6 binds to the soluble form of IL-6 receptor, forming a complex with a gp130 dimer on potentially all cell surfaces. The complex activates the JAK-STAT3 signaling pathway in various cell types, including endothelial cells, resulting in CRS and finally leading to ARDS[38]. Blocking this pathway could have therapeutic effects against CRS and ARDS in patients with a severe form of COVID-19.
FROM UNDERSTANDING OF IMMUNE RESPONSE TO THERAPEUTIC STRATEGY
The pathogenesis of SARS-CoV-2 is formed by the interaction between the viral infection and the immune response to the virus as mentioned so far. Therefore, understanding the immune response in the viral infection is important to manage the viral infection.
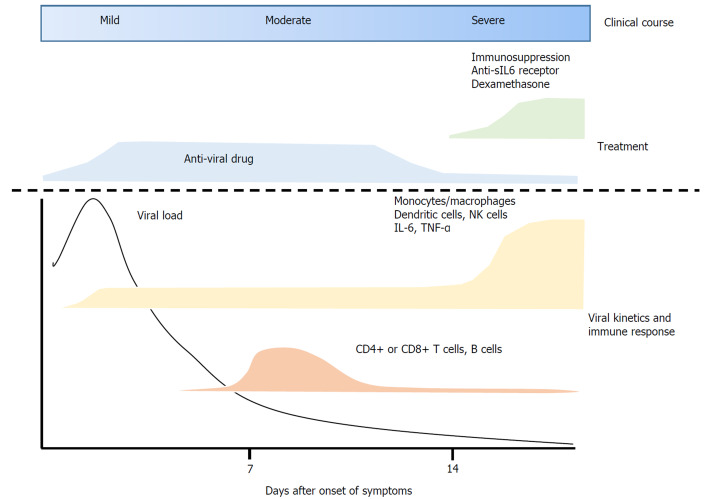
Immune responses to SARS-CoV-2 infection can be summarized as follows: After infection with SARS-CoV-2, the virus enters ACE2-expressing alveolar epithelial type 2 cells. These infected epithelial cells recruit dendritic cells, neutrophils and monocytes/macrophages, leading to the activation of CD4+ and CD8+ T cells. These cells exert anti-viral immune responses, completely suppressing viral replication or eradicating the virus in almost 80% of infected patients. In the remaining 20%, however, T cell populations are reduced due to unclear mechanisms. Rather, a cytokine storm occurs, resulting in various complications such as ARDS. Various cytokines including IL-6, produced by activated dendritic cells and monocytes/macrophages, are thought to be key factors causing the severe pulmonary disease (Figure 1).
Figure 1.
Therapeutic strategy based on viral kinetics and immunopathogenesis during the course of coronavirus disease 2019. sIL-6 receptor: Soluble form of interleukin-6 receptor; NK cells: Natural killer cells; TNF-α: Tumor necrosis factor-alpha.
Because COVID-19 is caused by SARS-CoV-2, the core therapeutic strategy should be aimed at the eradication of the virus[39]. Antiviral drugs clinically used so far include remdesivir[40], favipiravir[41], chloroquine[42], hydroxychloroquine[43], and lopinavir/ ritonavir[44]. Remdesivir and favipiravir are RNA-dependent RNA polymerase inhibitors shown to inhibit viral replication in vitro. The United States Food and Drug Administration has allowed the “Emergency Use” of remdesivir as an experimental drug in the United States. However, there have been no antiviral drugs proven to be virally effective for this viral infection.
Theoretically, effective antiviral drugs should be administered during an early phase of the disease, after the onset of symptoms, when viral replication seems still active based on temporal changes in viral load[14] and immune response[22]. However, immediate use of antivirals after the onset of symptoms is questionable, because antiviral drugs administered under conditions of high viral replication can predispose to the emergence of drug-resistant mutants. Moreover, not all patients require antiviral drugs as 80% of patients have shown spontaneous eradication of the virus in the absence of antiviral drugs[1]. Therefore, candidates for treatment with antiviral drugs include patients in the acute phase of the disease with mild to moderate severity, and those with persistent symptoms lasting for several days without improvements or showing indications of progression to the severe form of the disease.
The mortality rate in patients who progress to the severe form of the disease is 50%[34]. Antiviral drugs do not play a pivotal role in those patients because viral replication is not the main pathogenesis at the time of cytokine storm. Rather, these patients may benefit from immunomodulatory treatment. Drugs that block IL-6 activity may be effective. For example, treatment with tocilizumab, a monoclonal antibody that binds the soluble form of IL-6 receptor, markedly improved the respiratory condition and resolved computed tomography findings of the lungs in 80% of severely ill COVID-19 patients[45]. Recently, dexamethasone has been shown to reduce the progression of severe lung injury and mortality of the patients[46,47]. Although those data are promising, immunosuppression by those agents may delay viral clearance[23], suggesting that co-administration of an effective antiviral drug may be beneficial.
Other therapeutic options for cytokine storm include convalescent plasma[8-10] and the plasma has shown promise in the treatment of the severe form of COVID-19[16-18]. However, because plasma may contain unknown viruses transmittable to the recipient, this treatment should be considered only if no other treatments result in patient improvement.
CONCLUSION
In conclusion, the treatment strategy for patients with COVID-19 should be carefully designed based on both viral kinetics and immunopathogenesis. However, pandemic infection with SARS-CoV-2 may persist for several months, even if treatments are effective. The development of an effective vaccine will be necessary for termination of this pandemic infection.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interest.
Manuscript source: Invited manuscript
Peer-review started: May 15, 2020
First decision: November 3, 2020
Article in press: November 21, 2020
Specialty type: Medicine, research and experimental
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhuo ZQ S-Editor: Chen XF L-Editor: A P-Editor: Ma YJ
References
- 1.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 4.Coma A, Jonte F, Zanabili Y, Soto I, Fanjul F, Carrera D, Rayón C, Luño E, Jalón S, Pinto V. [Low-grade non-Hodgkin's lymphomas. Study of 73 cases] Sangre (Barc) 1992;37:249–254. [PubMed] [Google Scholar]
- 5.Yeung ML, Yao Y, Jia L, Chan JF, Chan KH, Cheung KF, Chen H, Poon VK, Tsang AK, To KK, Yiu MK, Teng JL, Chu H, Zhou J, Zhang Q, Deng W, Lau SK, Lau JY, Woo PC, Chan TM, Yung S, Zheng BJ, Jin DY, Mathieson PW, Qin C, Yuen KY. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris G, Bortolasci CC, Puri BK, Olive L, Marx W, O'Neil A, Athan E, Carvalho AF, Maes M, Walder K, Berk M. The pathophysiology of SARS-CoV-2: A suggested model and therapeutic approach. Life Sci. 2020;258:118166. doi: 10.1016/j.lfs.2020.118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Rokni M, Ghasemi V, Tavakoli Z. Immune responses and pathogenesis of SARS-CoV-2 during an outbreak in Iran: Comparison with SARS and MERS. Rev Med Virol. 2020;30:e2107. doi: 10.1002/rmv.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang S, Tanaka T, Narazaki M, Kishimoto T. Targeting Interleukin-6 Signaling in Clinic. Immunity. 2019;50:1007–1023. doi: 10.1016/j.immuni.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M, Liu W, Zhu Y, Lin Q, Mao L, Fang M, Zhang H, Sun Z. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5:e137799. doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinti I, Lougaris V, Milito C, Cinetto F, Pecoraro A, Mezzaroma I, Mastroianni CM, Turriziani O, Bondioni MP, Filippini M, Soresina A, Spadaro G, Agostini C, Carsetti R, Plebani A. A possible role for B cells in COVID-19? J Allergy Clin Immunol. 2020;146:211–213. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, Hu JL, Xu W, Zhang Y, Lv FJ, Su K, Zhang F, Gong J, Wu B, Liu XM, Li JJ, Qiu JF, Chen J, Huang AL. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Pang R, Xue X, Bao J, Ye S, Dai Y, Zheng Y, Fu Q, Hu Z, Yi Y. Anti-SARS-CoV-2 virus antibody levels in convalescent plasma of six donors who have recovered from COVID-19. Aging. 12:6536–6542. doi: 10.18632/aging.103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye M, Fu D, Ren Y, Wang F, Wang D, Zhang F, Xia X, Lv T. Treatment with convalescent plasma for COVID-19 patients in Wuhan, China. J Med Virol. 2020;92:1890–1901. doi: 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, van Buskirk C, Grossman BJ, Joyner M, Henderson JP, Pekosz A, Lau B, Wesolowski A, Katz L, Shan H, Auwaerter PG, Thomas D, Sullivan DJ, Paneth N, Gehrie E, Spitalnik S, Hod EA, Pollack L, Nicholson WT, Pirofski LA, Bailey JA, Tobian AA. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J Clin Invest. 2020;130:2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, Wei J, Xiao H, Yang Y, Qu J, Qing L, Chen L, Xu Z, Peng L, Li Y, Zheng H, Chen F, Huang K, Jiang Y, Liu D, Zhang Z, Liu Y, Liu L. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO "Immunity passports" in the context of COVID-19. 2020. Available from: https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19 .
- 21.Deng W, Bao L, Liu J, Xiao C, Liu J, Xue J, Lv Q, Qi F, Gao H, Yu P, Xu Y, Qu Y, Li F, Xiang Z, Yu H, Gong S, Liu M, Wang G, Wang S, Song Z, Liu Y, Zhao W, Han Y, Zhao L, Liu X, Wei Q, Qin C. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pia L. SARS-CoV-2-reactive T cells in patients and healthy donors. Nat Rev Immunol. 2020;20:353. doi: 10.1038/s41577-020-0333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender AE. Food additives. Sci Prog. 1988;72:549–562. [PubMed] [Google Scholar]
- 25.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 26.Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco JL, Ambrosioni J, Garcia F, Martínez E, Soriano A, Mallolas J, Miro JM COVID-19 in HIV Investigators. COVID-19 in patients with HIV: clinical case series. Lancet HIV. 2020;7:e314–e316. doi: 10.1016/S2352-3018(20)30111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascolo S, Romanelli A, Carleo MA, Esposito V. Could HIV infection alter the clinical course of SARS-CoV-2 infection? J Med Virol. 2020;92:1777–1778. doi: 10.1002/jmv.25881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Luo L, Bu H, Xia H. One case of coronavirus disease 2019 (COVID-19) in a patient co-infected by HIV with a low CD4+ T-cell count. Int J Infect Dis. 2020;96:148–150. doi: 10.1016/j.ijid.2020.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monti S, Balduzzi S, Delvino P, Bellis E, Quadrelli VS, Montecucco C. Clinical course of COVID-19 in a series of patients with chronic arthritis treated with immunosuppressive targeted therapies. Ann Rheum Dis. 2020;79:667–668. doi: 10.1136/annrheumdis-2020-217424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velavan TP, Meyer CG. Mild versus severe COVID-19: Laboratory markers. Int J Infect Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 39.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, Li H, Wang S, Ruan S, Yang C, Mei C, Wang Y, Ding D, Wu F, Tang X, Ye X, Ye Y, Liu B, Yang J, Yin W, Wang A, Fan G, Zhou F, Liu Z, Gu X, Xu J, Shang L, Zhang Y, Cao L, Guo T, Wan Y, Qin H, Jiang Y, Jaki T, Hayden FG, Horby PW, Cao B, Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, Liao X, Gu Y, Cai Q, Yang Y, Shen C, Li X, Peng L, Huang D, Zhang J, Zhang S, Wang F, Liu J, Chen L, Chen S, Wang Z, Zhang Z, Cao R, Zhong W, Liu Y, Liu L. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering. :(Beijing) 2020. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury MS, Rathod J, Gernsheimer J. A Rapid Systematic Review of Clinical Trials Utilizing Chloroquine and Hydroxychloroquine as a Treatment for COVID-19. Acad Emerg Med. 2020;27:493–504. doi: 10.1111/acem.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, Wang J, Liu X, Wang S, Wu X, Ge Q, He J, Zhan H, Qiu F, Guo L, Huang C, Jaki T, Hayden FG, Horby PW, Zhang D, Wang C. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Han M, Li T, Sun W, Wang D, Fu B, Zhou Y, Zheng X, Yang Y, Li X, Zhang X, Pan A, Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.RECOVERY Collaborative Group. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ. Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N Engl J Med. :2020. [Google Scholar]
- 47.Tomazini BM, Maia IS, Cavalcanti AB, Berwanger O, Rosa RG, Veiga VC, Avezum A, Lopes RD, Bueno FR, Silva MVAO, Baldassare FP, Costa ELV, Moura RAB, Honorato MO, Costa AN, Damiani LP, Lisboa T, Kawano-Dourado L, Zampieri FG, Olivato GB, Righy C, Amendola CP, Roepke RML, Freitas DHM, Forte DN, Freitas FGR, Fernandes CCF, Melro LMG, Junior GFS, Morais DC, Zung S, Machado FR, Azevedo LCP COALITION COVID-19 Brazil III Investigators. Effect of Dexamethasone on Days Alive and Ventilator-Free in Patients With Moderate or Severe Acute Respiratory Distress Syndrome and COVID-19: The CoDEX Randomized Clinical Trial. JAMA. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]