Abstract
Reprogramming scar fibroblasts into new heart muscle cells has the potential to restore function to the injured heart. However, the effectiveness of reprogramming is notably low. We have recently demonstrated that the effectiveness of reprogramming fibroblasts into heart muscle cells (cardiomyocytes) is increased by the addition of RNA-sensing receptor ligands. Clinical use of these ligands is problematic due to their ability to induce adverse inflammatory events. To overcome this issue, we sought to determine whether synthetic analogs of natural RNA-sensing receptor ligands, which avoid generating inflammatory insults and are nuclease resistant, would similarly enhance fibroblast reprogramming into cardiomyocytes. Indeed, one such stabilized RNA, ICR2, increased the expression of cardiomyocyte-specific mRNAs in reprogrammed fibroblasts. Moreover, ICR2 enhanced the ability of reprogramming factors to produce cardiomyocytes with mature sarcomeres. Knockdown assays indicated that the effects of ICR2 were mediated by the RNA-sensing receptors Rig-I and TLR3. In addition, ICR2 reduced the effective dose and number of reprogramming factors needed for efficient reprogramming. In summary, the synthetic RNA oligonucleotide ICR2 is a potential therapeutic agent to enhance cardiac reprogramming efficiency.
Keywords: reprogramming, cardiac, microRNAs, synthetic RNAs, innate immunity
Graphical Abstract

Fibroblast reprogramming to cardiomyocytes has the potential to restore function in the injured heart. Dzau and colleagues have discovered that the efficiency of fibroblast reprogramming to cardiomyocytes is enhanced by the synthetic RNA, ICR2. ICR2 was found to exert its effects via the receptors TLR3 and Rig-1 acting through NF-κB.
Introduction
The human heart has limited regenerative capacity. Following cardiac injury, cardiac muscle cells (cardiomyocytes) are lost and not replaced. In their place, fibroblasts invade the injury zone where they rapidly divide and secrete extracellular matrix proteins.1 These extracellular matrix proteins stiffen the wall of the heart and impair normal cardiac function. Over time, impairments in cardiac function often leads to heart failure.2 Considering that fibroblasts are a major component of scar tissue, these cells are an attractive target for therapeutic strategies that seek to regenerate heart tissue. Indeed, we and others have demonstrated that combinations of transcription factors or microRNAs (miRNAs) regenerate cardiac tissue by converting fibroblasts within the scar into functional cardiomyocytes.3, 4, 5, 6, 7, 8, 9 This approach offers important therapeutic advantages over stem cell transplantation, including ethical objections and potential tumorigenicity.10, 11, 12 However, converting fibroblasts into cardiomyocytes is relatively inefficient. We recently demonstrated that the effectiveness of conversion is enhanced by poly(I:C); a ligand of the RNA-sensing receptor TLR3.6 Poly(I:C) is structurally similar to double-stranded viral RNA and is considered a “natural” TLR3 agonist. Unfortunately for clinical applications, poly(I:C) is prone to inducing adverse inflammatory events and is very sensitive to nuclease degradation.13
Activation of RNA-sensing receptors is emerging as important modality for the treatment of cancer.14, 15, 16, 17 As mentioned above, poly(I:C) can induce adverse inflammatory events. Consequently, significant efforts have been devoted to developing synthetic analogs of the natural RNA-sensing receptor ligands, which do not induce adverse inflammatory events while retaining the ability of the natural ligands to induce tumor regression. One group of synthetic RNA-receptor ligands, termed ICRs, are particularly potent for tumor regression.18 Importantly, injection of these ICRs into mice did not lead to any adverse effects and moreover many chemically modified therapeutic oligonucleotides are making their way through clinical studies with a few recently approved.18, 19, 20
In this study, we wanted to determine whether synthetic, chemically stabilized analogs of natural RNA-sensing receptor ligands are capable of enhancing the efficiency of fibroblast reprogramming into cardiomyocytes. Indeed, we found that the synthetic RNA-sensing receptor ligand ICR2 enhanced cardiomyocyte-specific gene expression, as well as the maturation of the reprogrammed cells. Moreover, we identified that ICR2 exerts its beneficial effects via Rig-I, TLR3, and nuclear factor κB (NF-κB).
Results
The objective of this study was to determine whether synthetic RNA-sensing receptor ligands are able to enhance the effectiveness of fibroblast reprogramming into cardiomyocytes. In this study, fibroblasts were directly reprogrammed into cardiomyocytes via introduction of a combination of miRNAs called miR combo.3, 4, 5, 6, 7, 8, 9 Two synthetic RNA-sensing receptor ligands were used in this study: ICR2 and ICR420. ICR2 is a blunt-ended hairpin RNA 23 nucleotides in length, whereas ICR4 is predicted to form a double stem-loop structure 55 nucleotides in length.18 ICR2 strongly activates IRF3 and NF-κB. In contrast, ICR4 only activates IRF3.18
In the first instance, we incubated miR combo transfected fibroblasts with increasing doses of ICR2 or ICR4 and determined the effects on cardiomyocyte-specific gene expression. The ED100 dose for both ICR2 and ICR4 was 5 μg/μL. To identify the optimal dose for ICR2 and ICR4, we first carried out a set of pilot experiments increasing the dose of ICR (0, 0.1, 0.5, 1, 2, 5, and 10 μg/mL) in the culture media of miR combo transfected cells. The optimal ICR dose was defined as the minimum concentration of ICR necessary for the maximum stimulatory effect on miR combo with respect to cardiomyocyte-specific gene expression. This ICR dose, defined as the ED100 dose, was 5 μg/mL for both ICR2 and ICR4 (data not shown). As the fibroblasts used in this study were primary (i.e., non-tumor) cells, neither ICR2 nor ICR4 induced cell death. In control cells transfected with miR combo, there was significant upregulation of cardiomyocyte-specific gene expression (Figure 1). Interestingly, the addition of 5 μg/μL ICR2 to the culture media of miR combo transfected fibroblasts significantly enhanced cardiomyocyte-specific gene expression (Figure 1). In contrast, 5 μg/μL of ICR4 had no significant effect on cardiomyocyte-specific gene expression (Figure 1).
Figure 1.
The synthetic RNA oligonucleotide ICR2 improves the effectiveness of fibroblast reprogramming to cardiomyocytes
Fibroblasts were transfected with either the reprogramming cocktail miR combo or a non-targeting control miR (negmiR). 1 day later, transfection complexes were removed and the cells cultured for 4 days with the ED100 dose of ICR2 or ICR4 (5 μg/mL). 14 days after transfection, RNA was analyzed for the expression of the indicated cardiomyocyte-specific genes. Expression values were normalized to the housekeeping gene GAPDH and are represented as a fold change compared to the non-targeting control miR negmiR. N = 15. ∗Comparisons made to negmiR, ∗∗∗p < 0.001. †Comparisons made between miR combo + ICR and miR combo + vehicle, †† p < 0.01.
We then wanted to determine whether the effects of ICR2 were also seen with protein expression and cellular maturation. In accordance with our previous reports, miR combo increased the number of cells expressing the cardiomyocyte-specific protein Actn2 by 4-fold. As expected, incubating the miR combo transfected fibroblasts with poly(I:C) increased the number of Actn2+ cells by 15-fold (Figure 2A). Interestingly, equivalent numbers of Actn2+ cells were found with addition of ICR2 treatment following miR combo administration (Figure 2A). During cardiomyocyte maturation, proteins such as Actn2 self-assemble into striated structures known as sarcomeres. In miR combo transfected fibroblasts, sarcomeres were evident. However, as we have reported previously, the sarcomeres were relatively immature with limited striated patterning being evident (Figure 2B).3,6 In line with our previous report, poly(I:C) significantly enhanced cardiomyocyte maturation.6 Sarcomeres were more evident and even more importantly, they displayed the striated pattern observed in a mature cardiomyocyte (Figure 2B, with quantification provided in Figure 2C). This same striated sarcomere pattern characteristic of a mature cardiomyocyte was also observed in miR combo transfected fibroblasts that were incubated with the ICR2 oligonucleotide (Figure 2B, with quantification provided in Figure 2C).
Figure 2.
ICR2 enhances cardiomyocyte maturity
Fibroblasts were transfected with either the reprogramming cocktail miR combo or a non-targeting control miR (negmiR). 1 day later, transfection complexes were removed and the cells cultured for 4 days with ICR2 (5 μg/mL) or poly(I:C) (5 μg/mL). After 14 days, cells were incubated with antibodies that recognize the cardiomyocyte-specific protein Actn2. (A) The number of Actn2+ cells expressed as a percentage of the total number of cells. N = 3. ∗Comparisons made to negmiR, ∗p < 0.05, ∗∗p < 0.01. †Comparisons made between miR combo + ICR and miR combo + vehicle, †p < 0.05, †† p < 0.01. (B) Micrographs demonstrating striated sarcomere structure. Scale bar, 50 μm. N = 3. Representative images are shown. Nuclei stained with DAPI. (C) Quantification of (B). The area of the cell occupied by sarcomeres was expressed as a percentage of the total area of the cell. N = 3. ∗Comparisons made to negmiR, ∗∗∗p < 0.001. †Comparisons made between miR combo + ICR and miR combo + vehicle, ††† p < 0.001.
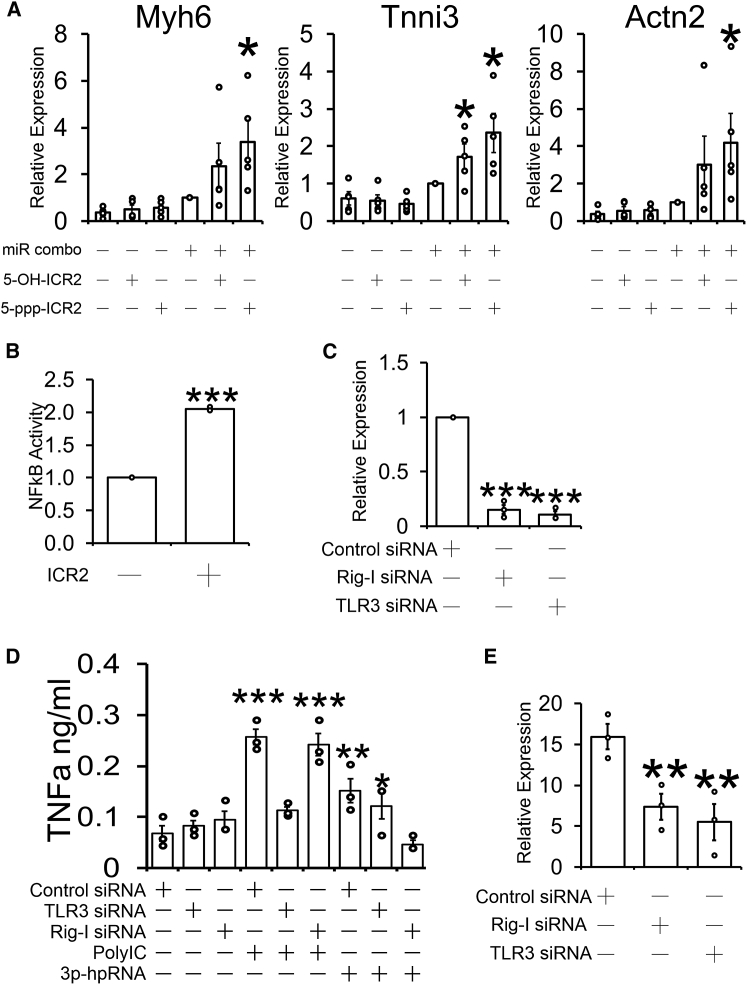
We then sought to determine how ICR2 was enhancing reprogramming effectiveness. ICR2 contains a 5′ triphosphate; a moiety that is recognized by several RNA-sensing receptors. Consequently, we asked what effect the removal of the 5′ triphosphate group would have on the ability of ICR2 to enhance miR combo mediated fibroblast reprogramming. In control cells, miR combo induced reprogramming in fibroblasts as evidenced by increased expression of cardiomyocyte-specific genes (Figure 3A). In accordance with data presented in Figure 1, the ability of miR combo to reprogram fibroblasts was enhanced by the addition of ICR2 containing a 5′ triphosphate group (Figure 3A). Interestingly, the ability of ICR2 to enhance fibroblast reprogramming was reduced but not totally eliminated when the 5′ triphosphate group was replaced with a hydroxyl group (Figure 3A).
Figure 3.
ICR2 enhances fibroblast reprogramming via Rig-I and TLR3
(A) Fibroblasts were transfected with either the reprogramming cocktail miR combo or a non-targeting control miR (negmiR). 1 day later, transfection complexes were removed and the cells cultured for 4 days with either 5 μg/mL 5′ triphosphate ICR2 (5-ppp-ICR2) or 5 μg/mL 5′ hydroxyl ICR2 (5-OH-ICR2). 14 days after transfection, RNA was analyzed for the expression of the indicated cardiomyocyte-specific genes. Expression values were normalized to the housekeeping gene GAPDH and are represented as a fold change compared to negmiR. N = 4–5. ∗Comparisons made to negmiR ∗p < 0.05; ∗∗p < 0.01. (B) Fibroblasts were transfected with a NF-κB-luciferase reporter plasmid. After 3 days, the cells were incubated with 5 μg/mL ICR2 for 6 h or an equivalent volume of vehicle as a control. Luciferase activity was determined spectrophotometry and normalized for protein content. Luciferase activity is represented as a fold change compared to control cells. N = 3. (C) Fibroblasts were transfected with siRNAs targeting TLR3 or Rig-I. Non-targeting siRNAs were used as a control. 3 days later, RNA levels of TLR3 and Rig-I were determined by qPCR. Expression values were normalized to the housekeeping gene GAPDH and are represented as a fold change compared to the control siRNA. N = 3. Comparisons made to control siRNA, ∗∗∗p < 0.05; ∗∗p < 0.01. (D) Fibroblasts were transfected with siRNAs targeting TLR3 or Rig-I. Non-targeting siRNAs were used as a control. 3 days later, the media were removed and the cells stimulated with the TLR3 ligand poly(I:C) (5 μg/mL), the Rig-I agonist 3p-hpRNA (5 μg/mL), or the equivalent volume of vehicle. The doses of poly(I:C) and Rig-I agonists used have been demonstrated to have the maximal effect on enhancing miR combo reprogramming. 1 day after stimulation, media was removed and TNF-α levels determined by ELISA. N = 3. Comparisons made to control siRNA plus vehicle, ∗∗∗p < 0.05; ∗∗p < 0.01. (E) Fibroblasts were transfected with siRNAs targeting TLR3 or Rig-I. Non-targeting siRNAs were used as a control. 3 days later, the cells were further transfected with miR combo. Following the removal of miR combo one day after transfection, the cells were incubated for a further 4 days with 5 μg/mL ICR2. 14 days after transfection, RNA was analyzed for the expression of the cardiomyocyte-specific gene Myh6. Expression values were normalized to the housekeeping gene GAPDH and are represented as a fold change compared to negmiR. N = 3. ∗p < 0.05; ∗∗p < 0.01.
To further investigate the mechanism by which ICR2 enhances fibroblast reprogramming into cardiomyocytes, we conducted assays to elucidate the signaling pathway. We have previously shown that poly(I:C) enhances cardiomyocyte maturation via NF-κB activation. Not surprisingly, ICR2 treatment also induces NF-κB expression (Figure 3B). The level of NF-κB induction achieved was similar to poly(I:C). To determine which NF-κB upstream RNA-sensing receptors potentially recognize ICR2, we employed specific small interfering RNAs (siRNAs) to knock down their expression. Knockdown of TLR3 and Rig-I was robust (Figure 3C). Moreover, knockdown was associated with a loss of functional activity. In TLR3 knockdown cells, the TLR3 ligand poly(I:C) was unable to induce tumor necrosis factor alpha (TNF-α) secretion (Figure 3D). Similarly, there was no TNF-α secretion in Rig-I knockdown cells stimulated with the Rig-I ligand 3p-hpRNA (Figure 3D). We determined that the loss of either Rig-I or TLR3 limited ICR2 from enhancing the expression of the cardiomyocyte-specific gene Myh6 (Figure 3E). Similar effects were observed with other cardiomyocyte-specific genes Actn2 and Tnni3; however, the effects did not reach significance (data not shown).
Finally, we wanted to determine whether ICR2 reduced the dose of miR combo needed for reprogramming and also reduced the number of miRNAs necessary for reprogramming. To address the first question, we transfected fibroblasts with 1/2 and 1/4 dose of miR combo and stimulated the cells with ICR2. As shown in Figure 4A, transfection of fibroblasts with a 1/2 and 1/4 dose of miR combo showed significantly lower reprogramming efficiency when compared to the standard miR combo transfection. However, the addition of ICR2 to fibroblasts transfected with 1/2 dose of miR combo gave rise to similar reprogramming efficiency as the standard miR combo transfection (Figure 4A). To address the second question, we transfected the fibroblasts with miR combo or the individual constituents of miR combo (miR-1, miR-133, miR-208, miR-499). When compared to miR combo, the individual constituents of miR combo were ineffective at inducing cardiomyocyte-specific gene expression (Figure 4B). The addition of ICR2 had no effect on the ability of miR-1, miR-208, or miR-499 to induce cardiomyocyte-specific gene expression (Figure 4B). In contrast, the combination of ICR2 and miR-133 was equivalent to standard miR combo transfection (Figure 4B).
Figure 4.
ICR2 reduces the reprogramming factor dose and number
(A) Fibroblasts were transfected with the standard concentration of miR combo, 1/2 the standard concentration and 1/4 the standard concentration. Control cells were transfected with the non-targeting miR, negmiR. 1 day later, transfection complexes were removed and the cells cultured for 4 days with ICR2 (5 μg/mL). After 7 days, cardiomyocyte-specific gene expression was measured by qPCR. Expression values were normalized to the housekeeping gene GAPDH. N = 3. Comparisons made to fibroblasts transfected with the standard concentration of miR combo, ∗p < 0.05; ∗∗p < 0.01. (B) Fibroblasts were transfected with miR combo or the individual components of miR combo (miR-1, miR-133, miR-208, miR-499). Control cells were transfected with the non-targeting miR, negmiR. 1 day later, transfection complexes were removed and the cells cultured for 4 days with ICR2 (5 μg/mL). After 7 days, cardiomyocyte-specific gene expression was measured by qPCR. Expression values were normalized to the housekeeping gene GAPDH and are expressed as a fold change compared to negmiR. N = 3. Comparisons made to fibroblasts transfected with negmiR, ∗p < 0.05; ∗∗∗p < 0.001.
Discussion
In this study we demonstrate that the synthetic RNA-sensing receptor ligand ICR2 enhances fibroblast reprogramming into cardiomyocytes.
When compared to ICR4, cancer cells exposed to ICR2 produce significantly higher levels of interferon-β (IFN-β) and pro-inflammatory cytokines.18 This effect is due to differences in the signaling pathways that are activated by the two synthetic RNA oligonucleotides. In cancer cells exposed to ICR2, NF-κB is strongly activated. In contrast, ICR4 only weakly stimulates NF-κB18. Considering that we, and others, have demonstrated the importance of NF-κB for cardiomyocyte gene expression, it is interesting that the degree of NF-κB activation by synthetic RNAs determines their effectiveness with respect to enhancing the efficiency of fibroblast reprogramming into cardiomyocytes.6,21,22 Beyond pro-inflammatory cytokines, ICR2 also has a greater effect on IFN-β levels when compared to ICR4. IFN-β induces JAK activation;23 however, we have previously demonstrated that reprogramming efficiency is increased by JAK inhibition.3 If NF-κB activation and JAK inhibition were equally potent with regards to reprogramming efficiency then ICR2 would be expected to have no effect on fibroblast reprogramming to cardiomyocytes. Thus, our data suggests that reprogramming efficiency is more sensitive to NF-κB activation rather than to JAK inhibition. Moreover, our data also suggests that ICR2 potency could be further enhanced by blocking IFN-β or JAK.
In this study, we demonstrated that ICR2 enhanced fibroblast reprogramming to cardiomyocytes via the RNA-sensing receptors RIG-I and TLR3. The ability of ICR2 to influence biological events via RIG-I may be cell-dependent. For example, ICR2 demonstrates similar effectiveness in Huh7.0 and RIG-I-deficient Huh7.5 cell lines.18 Furthermore, RIG-I knockdown in Huh7.0 cells did not diminish the ability of ICR2 to induce IFN-β expression.18 Considering that knockdown of either RIG-I or TLR3 impaired ICR2 activity in fibroblasts, it is likely that ICR2 binds to multiple receptors. In this regard, it is interesting that the activity of ICR2 was substantially reduced by removal of the 5′ triphosphate group. While it is possible that the effects of 5′ triphosphate removal were due to changes in the 3D conformation of the ICR2 molecule, it is known that 5′ triphosphorylation is necessary for RNA binding to Rig-I and IFIT1.24,25 Future studies will be focused on both 3D conformation changes and receptor binding. Similarly, another area of future study is the role of PKR. Like Rig-I, PKR also binds to RNA molecules with a 5′ triphosphate group.26 However the structural determinants for recognition are different between the two receptors. While Rig-I binds preferentially to 5′-triphosphorylated short blunt-end dsRNA, PKR binds predominantly to 5′-triphosphorylated single-stranded RNA (ssRNA) with short stem-loops.27,28 Considering the range of possible configurations ICR2 could adopt in an aqueous solution, investigating the role of PKR in mediating the effects of ICR2 is warranted.
Reprogramming fibroblasts to iPSCs and endothelial cells has also been shown to be dependent upon RNA-sensing receptor pathways.29,30 Conversion of fibroblasts into induced pluripotent stem cells (iPSCs) is efficient when the reprogramming factors Oct4, Sox2, Klf4, and c-Myc are delivered into fibroblasts via viruses. In contrast, expression of the reprogramming factors via cell permeable peptides is very inefficient. The Cooke laboratory found that these cell-permeable peptides demonstrated comparable efficiency to virus delivery methods when the fibroblasts were cultured in poly(I:C).29 Similar results were then obtained with reprogramming to endothelial cells.30 It was subsequently demonstrated that Rig-I, IRF3, and NF-κB were necessary for the effects that the authors had observed.31 Considering the role of IRF3 and NF-κB, it is interesting to speculate that ICR2 may also enhance reprogramming based methods of iPSC and endothelial cell generation.
The ability of ICR2 to enhance reprogramming efficiency was found to also have further benefits such as reducing the dosage of miR combo needed for reprogramming. Intriguingly, ICR2 combined with miR-133 displayed equivalent reprogramming efficiency as miR combo. This may suggest that ICR2 affects the same targets as miR-1, miR-208, and miR-499. It is also possible that ICR2 stimulation bypasses the need for these miRNAs; potentially by stimulating transcription factors that induce cardiomyocyte-specific genes. It will also be interesting to determine whether these effects are also true in vivo.
Materials and methods
Chemicals
Poly(I:C) LMW (low molecular weight) was purchased from Invivogen (Waltham, MA, USA). ICR2 and ICR4 were generated as described previously.18
miRNA transfection
Mouse (C57BL/6) neonatal cardiac fibroblasts were isolated from 2-day-old mouse neonates according to the method outlined in Jayawardena et al.32 Following isolation fibroblasts were cultured in growth media containing DMEM (ATCC, catalog number 30-2002) supplemented with 15% v/v fetal bovine serum (FBS; Thermo Scientific, Waltham, MA; Hyclone Fetal bovine serum, catalog number SH30071.03, lot number AXK49952) and 1% v/v penicillin/streptomycin (GIBCO, Waltham, MA, USA; catalog number 15140-122, 100 units penicillin, 100 μg/mL streptomycin). Fibroblasts were passaged once the cells had reached 70%–80% confluence using 0.05% w/v trypsin (GIBCO, Waltham, MA; catalog number 25300-054). Freshly isolated fibroblasts were labeled as passage 0. Experiments were conducted with cells at passage 2. For all experiments, cells were seeded at 5,000 cells/cm2 in growth media (DMEM, ATCC, Manassas, VA, USA; 15% FBS, 1% P/S). After 24 h, the cells were transfected with transfection reagent alone (Dharmafect-I, Thermo Scientific, Waltham, MA, USA), with transfection reagent plus non-targeting microRNAs (negmiR), or with transfection reagent plus our previously reported combination of cardiac reprogramming miRNAs (miR combo, miR-1, miR-133, miR-208, miR-499).32 In these studies, the final concentration of the non-targeting miRNA negmiR and miR combo was 50 nM. In brief, 5 nmol of miRNA was diluted into 150 μL of DMEM serum-free media. In a separate tube, 1.5 μL of Dharmafect-I (GE Healthcare, Chicago, IL, USA; catalog number T-2001-02) was diluted into 150 μL of DMEM serum free media. After 5 min at room temperature, the two tubes were combined. After a 20 min incubation, the complexes were added to one well of a 12-well plate. Growth media was added (450 μL) and after 24 h the complexes were removed. Cells were then cultured in growth media, media changes every 2 days, for the duration of the experiment. All studies were approved by the Duke University Division of Laboratory Animals (DLAR) and the Duke Institutional Animal Care and Use Committee (IACUC). Protocol number is A056-19-03.
ICR treatment
Sequences
ICR2: 5′-GGAUGCGGUACCUGACAGCAUCC-3′
ICR4: 5′-GGAUGCGGUACCUGACAGCAUCCUAAACUCAUGGUCCAUGUUUGUCCAUGGACCA-3′. 1 day after cells were transfected with miR combo, transfection complexes were removed and replaced with fibroblast growth media. ICR compounds were added directly to the culture media. Fresh media, vehicle, poly(I:C) or the ICR compounds were added daily for a total of 4 days after which cells were cultured in fibroblast growth media for the remainder of the experiment.
qPCR
500 ng of total RNA was extracted using Quick-RNA MiniPrep Kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA, USA). Total RNA was converted to cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems, Waltham, MA, USA) in a 20 μL reaction. cDNA (2 μL) was used in a standard qPCR reaction involving FAM conjugated gene specific primers and TaqMan Gene Expression Master Mix (Applied Biosystems, Waltham, MA, USA). The following qPCR primers were acquired from Thermo Fisher (Waltham, MA, USA): Gapdh (Mm99999915_m1), Tnni3 (Mm00437164_m1), Actn2 (Mm00473657_m1), and Myh6 (Mm00440359_m1). Sequence information is proprietary. Expression values were calculated by normalizing to the housekeeping gene GAPDH. Ct values for GAPDH were not altered by any experimental condition (data not shown).
Immunofluorescence
Cells were fixed with 2% v/v paraformaldehyde (EMS, Durham, NC, USA) as described previously.9 Fixed cells were blocked in antibody buffer (5% w/v BSA, 0.1% v/v Tween-20, in PBS) for 1 h at room temperature. Following blocking, cells were incubated overnight at 4°C with an Actn2 antibody (Sigma, A7811, 1:100) in antibody buffer. After the overnight incubation, cells were washed three times in antibody buffer. Following washing, cells were incubated with Alexa Fluor conjugated secondary antibodies (Invitrogen, Waltham, MA, USA, goat anti-mouse 594 nm) at a 1:500 dilution in antibody buffer for 1 h at room temperature. Nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI) at 1 μg/mL for 30 min at room temperature in antibody buffer. Following washing in PBS to remove unbound complexes, immunofluorescence was measured using a Zeiss Axiovert 200 inverted microscope. Images (six per well) were taken at random by a blinded investigator.
To calculate the area occupied by sarcomeres, we used CellProfiler to calculate the total area of the cell and ImageJ was used to measure the striated area.
NF-κB luciferase reporter transfection and determining NF-κB activity
An NF-κB-luciferase reporter plasmid pGreenFire1-NFkB was acquired from System Biosciences with Cat# TR012PA-1 and packaged into lentivirus by co-transfection of HEK293 cells with helper plasmids pSPAx2 (Addgene, Watertown, MA, USA; #12260) and pMD2G (Addgene #12259). After 3 days, lentivirus was collected by centrifugation (3,000 × g, 60 min) of the media through a 100 kDa cut-off filter. Viral titer was determined by qPCR.
For analysis of NF-κB activity, P1 cardiac fibroblasts (500,000 cells in a T150 flask) were infected with 100,000 viral particles per cell for 24 h. After removal of the viral particles, the cells were cultured for another 24 h before being seeded onto 24-well plate (12,500 cells per well) for further treatment. Luciferase activity was measured using Promega (Madison, WI, USA) Luciferase Assay System (Cat# E1500) and read with a luminometer. For quantification, the raw values were normalized to the protein content of the well. Protein content was determined by first extracting the cells in 100 μL lysis buffer (50 mM Tris-HCl pH7.5, 1% v/v SDS). Protein concentration was then determined by Lowry using BSA as a standard.
siRNA knockdown
siRNA pools (four siRNAs targeting each gene) and a negative control siRNA were purchased from Dharmacon (Chicago, IL, USA). siRNAs were made to 20 μM in nuclease free water, aliquoted, and stored −80°C until use. Fibroblasts were seeded into 12 well plates at 20,000 cells per well 1 day prior to transfection. On the day of transfection, siRNAs were diluted to 5 μM in nuclease free water. For each well, 5 μL of the working siRNA solution was diluted with 95 μL Opti-MEM-Serum Free media. In a separate tube 5 μL of Dharmafect-I (Dharmacon, Chicago, IL, USA) was diluted with 95 μL Opti-MEM-Serum Free media. After 5 min incubation, the two solutions were combined. After 20 min, complete media lacking antibiotics was added (800 μL) and the transfection complexes added to the cells.
TNF-α ELISA
One day before ELISA, media was replaced with fresh cytokine free growth media (1 mL per well of a 12-well plate). On the day of the ELISA, media was removed and the amount of TNF-α determined according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA, catalog number DY410). Raw values were adjusted for cell number by measuring the protein concentration as described for the analysis of NF-κB activity.
Statistics
Independent t tests were used for experiments with two groups. For experiments with more than two groups, ANOVA was used with Bonferroni correction to determine significances between groups.
Groups lower than N = 5 are plotted the individual data points, as well as the mean and SEM. Groups greater than N = 5 show only the mean and SEM.
Acknowledgments
Research conducted in these studies was supported by National Heart, Lung, and Blood Institute grant R01 HL131814-01A1 and the Edna and Fred L. Mandel, Jr. Foundation.
Author contributions
J.H., conceptualization, formal analysis, investigation, writing-review, and editing; C.P.H., conceptualization, formal analysis, investigation, supervision, writing-original draft, writing-review, and editing; R.E.P. conceptualization, writing-review, and editing; J.L. conceptualization, resources, writing-review, and editing; B.A.S. conceptualization, resources, writing-review, and editing; V.J.D. conceptualization, supervision, writing-review, and editing.
Declaration of interests
V.J.D., JL., B.A.S., and C.P.H. are co-founders of Recardia Therapeutics.
References
- 1.Fan D., Takawale A., Lee J., Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heidenreich P.A., Trogdon J.G., Khavjou O.A., Butler J., Dracup K., Ezekowitz M.D., Finkelstein E.A., Hong Y., Johnston S.C., Khera A., American Heart Association Advocacy Coordinating Committee. Stroke Council. Council on Cardiovascular Radiology and Intervention. Council on Clinical Cardiology. Council on Epidemiology and Prevention. Council on Arteriosclerosis. Thrombosis and Vascular Biology. Council on Cardiopulmonary. Critical Care. Perioperative and Resuscitation. Council on Cardiovascular Nursing. Council on the Kidney in Cardiovascular Disease. Council on Cardiovascular Surgery and Anesthesia, and Interdisciplinary Council on Quality of Care and Outcomes Research Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 3.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayawardena T.M., Finch E.A., Zhang L., Zhang H., Hodgkinson C.P., Pratt R.E., Rosenberg P.B., Mirotsou M., Dzau V.J. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dal-Pra S., Hodgkinson C.P., Dzau V.J. Induced cardiomyocyte maturation: Cardiac transcription factors are necessary but not sufficient. PLoS ONE. 2019;14:e0223842. doi: 10.1371/journal.pone.0223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgkinson C.P., Pratt R.E., Kirste I., Dal-Pra S., Cooke J.P., Dzau V.J. Cardiomyocyte Maturation Requires TLR3 Activated Nuclear Factor Kappa B. Stem Cells. 2018;36:1198–1209. doi: 10.1002/stem.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dal-Pra S., Hodgkinson C.P., Mirotsou M., Kirste I., Dzau V.J. Demethylation of H3K27 Is Essential for the Induction of Direct Cardiac Reprogramming by miR Combo. Circ. Res. 2017;120:1403–1413. doi: 10.1161/CIRCRESAHA.116.308741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Dal-Pra S., Mirotsou M., Jayawardena T.M., Hodgkinson C.P., Bursac N., Dzau V.J. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci. Rep. 2016;6:38815. doi: 10.1038/srep38815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Hodgkinson C.P., Lu K., Payne A.J., Pratt R.E., Dzau V.J. Selenium Augments microRNA Directed Reprogramming of Fibroblasts to Cardiomyocytes via Nanog. Sci. Rep. 2016;6:23017. doi: 10.1038/srep23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macia E., Boyden P.A. Stem cell therapy is proarrhythmic. Circulation. 2009;119:1814–1823. doi: 10.1161/CIRCULATIONAHA.108.779900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mummery C. Induced pluripotent stem cells--a cautionary note. N. Engl. J. Med. 2011;364:2160–2162. doi: 10.1056/NEJMcibr1103052. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita T., Kawai H., Tian F., Ohta Y., Abe K. Tumorigenic development of induced pluripotent stem cells in ischemic mouse brain. Cell Transplant. 2011;20:883–891. doi: 10.3727/096368910X539092. [DOI] [PubMed] [Google Scholar]
- 13.Naumann K., Wehner R., Schwarze A., Petzold C., Schmitz M., Rohayem J. Activation of dendritic cells by the novel Toll-like receptor 3 agonist RGC100. Clin. Dev. Immunol. 2013;2013:283649. doi: 10.1155/2013/283649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besch R., Poeck H., Hohenauer T., Senft D., Häcker G., Berking C., Hornung V., Endres S., Ruzicka T., Rothenfusser S., Hartmann G. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palchetti S., Starace D., De Cesaris P., Filippini A., Ziparo E., Riccioli A. Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells. J. Biol. Chem. 2015;290:5470–5483. doi: 10.1074/jbc.M114.601625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salaun B., Coste I., Rissoan M.C., Lebecque S.J., Renno T. TLR3 can directly trigger apoptosis in human cancer cells. J. Immunol. 2006;176:4894–4901. doi: 10.4049/jimmunol.176.8.4894. [DOI] [PubMed] [Google Scholar]
- 17.Duewell P., Steger A., Lohr H., Bourhis H., Hoelz H., Kirchleitner S.V., Stieg M.R., Grassmann S., Kobold S., Siveke J.T. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ. 2014;21:1825–1837. doi: 10.1038/cdd.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J., Lee Y., Xu L., White R., Sullenger B.A. Differential Induction of Immunogenic Cell Death and Interferon Expression in Cancer Cells by Structured ssRNAs. Mol. Ther. 2017;25:1295–1305. doi: 10.1016/j.ymthe.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nimjee S.M., Sullenger B.A. Therapeutic Aptamers: Evolving to Find their Clinical Niche. Curr. Med. Chem. 2020;27:4181–4193. doi: 10.2174/0929867326666191001125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodruff R.S., Sullenger B.A. Modulation of the Coagulation Cascade Using Aptamers. Arterioscler. Thromb. Vasc. Biol. 2015;35:2083–2091. doi: 10.1161/ATVBAHA.115.300131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Liu Z., Welch J.D., Gao X., Wang L., Garbutt T., Keepers B., Ma H., Prins J.F., Shen W. Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell. 2019;25:149–164. doi: 10.1016/j.stem.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biermann M., Cai W., Lang D., Hermsen J., Profio L., Zhou Y., Czirok A., Isai D.G., Napiwocki B.N., Rodriguez A.M. Epigenetic Priming of Human Pluripotent Stem Cell-Derived Cardiac Progenitor Cells Accelerates Cardiomyocyte Maturation. Stem Cells. 2019;37:910–923. doi: 10.1002/stem.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haji Abdolvahab M., Mofrad M.R., Schellekens H. Interferon Beta: From Molecular Level to Therapeutic Effects. Int. Rev. Cell Mol. Biol. 2016;326:343–372. doi: 10.1016/bs.ircmb.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbas Y.M., Pichlmair A., Górna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toroney R., Hull C.M., Sokoloski J.E., Bevilacqua P.C. Mechanistic characterization of the 5′-triphosphate-dependent activation of PKR: lack of 5′-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD. RNA. 2012;18:1862–1874. doi: 10.1261/rna.034520.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 29.Lee J., Sayed N., Hunter A., Au K.F., Wong W.H., Mocarski E.S., Pera R.R., Yakubov E., Cooke J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng S., Chanda P., Thandavarayan R.A., Cooke J.P. Transflammation: Innate immune signaling in nuclear reprogramming. Adv. Drug Deliv. Rev. 2017;120:133–141. doi: 10.1016/j.addr.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayed N., Ospino F., Himmati F., Lee J., Chanda P., Mocarski E.S., Cooke J.P. Retinoic Acid Inducible Gene 1 Protein (RIG1)-Like Receptor Pathway Is Required for Efficient Nuclear Reprogramming. Stem Cells. 2017;35:1197–1207. doi: 10.1002/stem.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayawardena T., Mirotsou M., Dzau V.J. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol. Biol. 2014;1150:263–272. doi: 10.1007/978-1-4939-0512-6_18. [DOI] [PubMed] [Google Scholar]