Abstract
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are genetically, pathologically and clinically-related progressive neurodegenerative diseases. Thus far, several SQSTM1 variations have been identified in patients with ALS and FTD. However, it remains unclear how SQSTM1 variations lead to neurodegeneration. To address this issue, we investigated the effects of ectopic expression of SQSTM1 variants, which were originally identified in Japanese and Chinese sporadic ALS patients, on the cellular viability, their intracellular distributions and the autophagic activity in cultured cells. Expression of SQSTM1 variants in PC12 cells exerted no observable effects on viabilities under both normal and oxidative-stressed conditions. Further, although expression of SQSTM1 variants in PC12 cells and Sqstm1-deficient mouse embryonic fibroblasts resulted in the formation of numerous granular SQSTM1-positive structures, called SQSTM1-bodies, their intracellular distributions were indistinguishable from those of wild-type SQSTM1. Nonetheless, quantitative colocalization analysis of SQSTM1-bodies with MAP1LC3 demonstrated that among ALS-linked SQSTM1 variants, L341V variant showed the significantly lower level of colocalization. However, there were no consistent effects on the autophagic activities among the variants examined. These results suggest that although some ALS-linked SQSTM1 variations have a discernible effect on the intracellular distribution of SQSTM1-bodies, the impacts of other variations on the cellular homeostasis are rather limited at least under transiently-expressed conditions.
Keywords: Amyotrophic lateral sclerosis (ALS), Frontotemporal dementia (FTD), SQSTM1, SQSTM1/p62-body, MAP1LC3/LC3, Autophagy
Abbreviations: ALS, amyotrophic lateral sclerosis; CCCP, carbonyl cyanide 3-chlorophenylhydrazone; CI, complete protease inhibitor; CQ, chloroquine; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; DMEM, Dulbecco's Modified Eagle's medium; DTT, dithiothreitol; EBSS, Earle's Balanced Salt Solution; GST, glutathione S-transferase; HA, hemagglutinin; HRP, horseradish peroxidase; IPTG, isopropyl thio-beta-D-galactoside; MEF, mouse embryonic fibroblast; MND, motor neuron disease; NGS, normal goat serum; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; PFA, paraformaldehyde; PVDF, polyvinylidene difluoride; RT, room temperature; SBMA, spinal and bulbar muscular atrophy; SDS, sodium dodecyl sulfate; WT, wild-type
Highlights
-
•
Ectopic expression of ALS-linked SQSTM1 variants does not affect cell viability.
-
•
Ectopic expression of SQSTM1 in cells results in formation of SQSTM1-body.
-
•
Ectopic expression of SQSTM1 in cells has marginal impacts on the autophagic activity.
-
•
SQSTM1L341V variant exhibits impaired association with LC3 in cultured cells.
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by progressive motor disturbance due to a selective loss of upper and lower motor neurons [1,2]. On the other hand, frontotemporal dementia (FTD) is a group of disorders that cause changes in social behavior, personality and loss of language skills due to progressive damage to the frontal and/or temporal lobes [3]. These disorders were previously regarded to be distinct from one another. However, several recent studies have uncovered that they are genetically, pathologically and clinically related [3,4], and might be on a continuum where ALS is at one end and FTD is at the other end. Indeed, several genes, such as C9orf72, TBK1, SQSTM1, TARDBP and OPTN, have been identified as common associated genes with both ALS and FTD [3,4].
Recently, we and others have identified several missense variations in SQSTM1 in patients with sporadic as well as familial ALS [[5], [6], [7], [8], [9], [10], [11], [12], [13]]. The SQSTM1 coded protein, SQSTM1 (also known as p62 or sequestosome-1), comprises multiple domains and motifs that mediate the interaction with various factors [14,15]. SQSTM1 interacts with TRAF6 E3 ligase, which in turn ubiquitinates mTORC1 on lysosomes in response to increased concentration of amino acids [16]. SQSTM1 also acts as an adaptor protein for ubiquitinated cargo molecules on selective autophagy [17]. It can interact not only with ubiquitinated cargo molecules via its C-terminal ubiquitin-associated (UBA) domain but also with MAP1LC3 (LC3) via its LC3-interacting region (LIR), thereby ubiquitinated cargo molecules are efficiently engulfed into autophagosomes and degraded [18]. Further, the N-terminal Phox and Bem 1 (PB1) domain, which is known to be involved in homo-oligomer formation of SQSTM1, is also required for proper localization of SQSTM1 on phagophore [19] and selection of ubiquitinated cargo molecules [20]. Indeed, we have previously shown that both loss of SQSTM1 and excessive SQSTM1 expression deregulate the autophagy-endolysosomal degradation system and accelerate the onset of disease in ALS mouse models [21,22]. On the other hand, under oxidative stressed and/or dysregulated proteostatic conditions, SQSTM1 expression is known to be upregulated, thereby activating the NFE2L2-dependent anti-oxidative stress signaling pathway through interaction between SQSTM1 and KEAP1, a negative regulator for NFE2L2 [23,24]. Notably, the LIR (residues 335–341) is located very close to the KEAP1-interaction region (KIR; residues 347–352) [25], suggestive of a possible competition in the binding by LC3 and KEAP1 in this region. These findings suggest that SQSTM1 has protective roles against various cellular stresses, such as autophagy-mediated proteostatic and oxidative stresses.
Thus far, several biochemical as well as pathological studies on disease-linked SQSTM1 variants have been published. The ALS-linked L341V LIR variation reduces the affinity of SQSTM1 to LC3 in vitro and retards the degradation of SQSTM1 in cultured cells [26]. The KIR variants, P348L and G351A, show reduced ability to activate NFE2L2 signaling [25]. Further, ALS/FTD-linked C-terminal variant, G427R, not only inhibits selective autophagy and but also disrupts NFE2L2 anti-oxidative stress response [27]. Under pathological conditions, it has been demonstrated that enlarged granular SQSTM1-positive structures, called SQSTM1-bodies, are frequently detected in motor neurons of ALS patients [2]. Interestingly, when artificial SQSTM1 mutants are expressed in cells, the morphology of SQSTM1-bodies is altered [28]. However, despite of increased number of studies on SQSTM1 variants, it still remains unclear whether disease-linked variations in SQSTM1 are associated with such abnormalities.
In this study, we focused on four SQSTM1 missense variants that were originally identified and as yet exclusively found in Japanese or Chinese patients with sporadic ALS [[7], [8], [9]]. Two variations, A53T and V90M, reside in the PB1 domain, and other two variations, D337E and L341V, are located in the LIR of SQSTM1. In order to clarify the effects of these SQSTM1 variants in cells, we performed cell viability assay under normal and stressed conditions, and also conducted a series of immunocytochemical and biochemical analyses on the intracellular distribution of SQSTM1-bodies as well as the autophagic activities.
2. Materials and methods
2.1. Plasmids
Human SQSTM1_wild-type (WT) cDNA were obtained by subcloning the reverse transcriptase-PCR-amplified full-length SQSTM1 cDNA. SQSTM1 mutants were generated by PCR-based site-directed mutagenesis using SQSTM1_WT as a template. Full length SQSTM1_WT and its mutant/variant cDNAs were subcloned into pCIneo and pGEX6P vectors. Plasmids generated were as follows; pCIneoFLAG-SQSTM1_WT, pCIneoFLAG-SQSTM1_A53T, pCIneoFLAG-SQSTM1_V90M, pCIneoFLAG-SQSTM1_D337E, pCIneoFLAG-SQSTM1_L341V, pCIneoFLAG-SQSTM1_K7A/D69A, pCIneoFLAF-SQSTM1_D335A/D336A/D337A, pCIneoHA-SQSTM1_WT, pCIneoHA-SQSTM1_A53T, pCIneoHA-SQSTM1_V90M, pCIneoHA-SQSTM1_D337E, pCIneoHA-SQSTM1_L341V, pCIneoHA-SQSTM1_K7A/D69A, pCIneoHA-SQSTM1_D335A/D336A/D337A, pGEX6P-SQSTM1_WT, pGEX6P-SQSTM1_A53T, pGEX6P-SQSTM1_V90M and pGEX6P-SQSTM1_K7A/D69A. DNA sequence of the insert as well as the flanking regions in each plasmid was verified by sequencing.
2.2. Reagents and antibodies
We used the following reagents for cell viability assay; menadione (or vitamin K3) (SIGMA), carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (SIGMA) and Alamar Blue Reagent (Thermo Fisher). All other regents were from commercial sources and of analytical grade.
Primary antibodies for Western blotting included rabbit anti-human SQSTM1 (1:3000, MBL, PM045), rabbit anti-human LC3 (1:3000, SIGMA, L8918), mouse monoclonal anti-FLAG M2 (1:5000, SIGMA, F1804), rabbit polyclonal anti-HA (1:2000, CST, 3724), rabbit anti-GAPDH (1:5000, SIGMA, G9545) and mouse monoclonal anti-β-actin (1:3000, SANTA CRUZ, sc-47778) antibodies. Secondary antibodies included horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:5000, GE Healthcare bioscience NA934) and HRP-conjugated anti-mouse IgG (1:5000, Jackson) antibodies.
Primary antibodies for immunocytochemistry included rabbit anti-human LC3 (1:2000, MBL PM036), guinea-pig anti-human SQSTM1 (1:2000, PROGEN, GP62-C) and mouse monoclonal anti-FLAG M2 (1:20000, SIGMA, F1804) antibodies. Secondary antibodies included Alexa 488 conjugated anti-guinea-pig IgG (1:500, Molecular Probes), Alexa 594 conjugated anti-rabbit IgG (1:500, Molecular Probes) and Alexa 488 conjugated anti-mouse IgG (1:500, Molecular Probes) antibodies.
2.3. Animals
We used Sqstm1-deficient (KO) mice, which were generously gifted from Drs. Tetsuro Ishii and Toru Yanagawa (University of Tsukuba) [29]. Genotypes of the mice were determined by PCR using primers as follows; A170#1F: 5′-CTGCATGTCTTCTCCCATGAC-3′, A170#1R: 5′-TAGATACCTAGGTGAGCTCTG-3′, A170#2F:5′-CTTACGGGTCCTTTTCCCAAC-3′ and A170#2R: 5′-TCCTCCTTGCCCAGAAGATAG-3′. Mice were housed at an ambient temperature of 22 °C with a 12 h light-dark cycle. Food and water were fed ad libitum. All animal experimental procedures were carried out in accord with the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and reviewed and approved by The Institutional Animal Care and Use Committee at Tokai University.
2.4. Cell culture
PC12 cells were cultured in Dulbecco's Modified Eagle's medium with High glucose (DMEM-HG) (Wako) supplemented with 7.5% (v/v) heat-inactivated fetal bovine serum (FBS) (PAA Laboratories), 7.5% (v/v) heat-inactivated horse serum (HS) (GIBCO), 100 U/ml penicillin G, 100 μg/ml streptomycin and 100 μg/ml sodium pyruvate. Mouse embryonic fibroblasts (MEFs) and COS-7 cells were cultured in the same condition as PC12 cells, except that 10% (v/v) FBS was used as a serum supplement.
2.5. Transfection
Transfection were performed as previously described [30,31]. In brief, PC12 cells and MEFs were seeded onto an 8 well-chamber slide at a density of 1 × 104 cells for immunocytochemistry and onto a 12 well-plate at a density of 1 × 105 cells for western blot analysis. PC12 cells, MEFs and COS-7 cells were transfected with plasmids using Lipofectamine 3000 Reagent (Invitrogen), Lipofectamine LTX Reagent (Invitrogen) and Effectene Transfection Reagent (Qiagen), respectively, according to the manufacturer's instructions.
2.6. Cell viability assay
PC12 cells transfected with FLAG-tagged plasmids were left untreated, or treated with 10 μM menadione (to assess cell viability in response to oxidative stress) for 4 h, or 750 μM CCCP (to assess cell viability in response to mitochondrial stress) for 10 h. Cells transfected with pCIneo-vector in the untreated condition were used as control in these assays. After treatment of these reagents, Alamar Blue Reagent [10% (v/v) of medium] was added. Resazurin, a principal component of Alamar Blue reagent is reduced to highly red fluorescent resorufin only in viable cells. We detected the fluorescence intensity of resorufin by Spectra Max i3 (Perkin Elmer). We determined the appropriate concentrations of menadione and CCCP, at which an approximately 75–80% of cells was survived, and used in these assays. Values for the cell viability were calculated after subtracting by those obtained from the cells treated with 0.1% (w/v) Triton X-100 (complete cell death), and represented as the mean percentage relative to control cells transfected with pCIneo-vector under normal conditions.
2.7. Protein purification
GST-fused SQSTM1_WT, _A53T, _V90M and _K7A/D69A were induced in E. coli BL21, which were transformed with pGEX6P-SQSTM1_WT, _A53T, _V90M and _K7A/D69A, by culturing in LB medium containing 50 μg/ml ampicillin and 100 μM isopropyl thio-beta-D-galactoside (IPTG), respectively. E. coli pellet was lysed in buffer A [20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 2 mM EDTA, 0.2% Tween-20, Complete Protease Inhibitor (CI) (Rhoche)], and sonicated. After centrifugation at 10,000 ×g for 30 min at 4 °C, supernatant was filtered with 0.2 μm syringe filter and then mixed with glutathione-Sepharose beads. Beads were washed three times with buffer A without CI at 4 °C. GST-fused proteins bound to the beads were treated with Pre-Scission protease in buffer B [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.2% Tween-20, 1 mM dithiothreitol (DTT)]. After centrifugation at 5000 ×g for 10 min at 4 °C, the supernatant was recovered. Concentration of proteins in the supernatant was determined by Micro BCA (Pierce).
2.8. Western blotting
Equal amount of protein samples was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (Wako Supersep Ace, 5–20% 17 well) in SDS-PAGE buffer [25 mM Tris-HCl, 192 mM glycine, 0.1% (w/v) SDS], and electrophoretically blotted onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad Laboratories) in transfer buffer [25 mM Tris-HCl, 192 mM glycine, 20% (w/v) methanol]. Membranes were blocked with 0.5% skimmed milk in TBST [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% (w/v) Tween-20] for 1 h at room temperature (RT) and probed with the appropriate primary antibody, followed by horseradish peroxidase-coupled secondary antibody. Signals were visualized by Immobilon (Merck) and detected by EZ capture MG system (ATTO).
2.9. Co-immunoprecipitation
COS-7 cells were co-transfected with constructs expressing FLAG-tagged and HA-tagged proteins. After transfection, the cells were lysed in buffer C [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (w/v) Tween-20, CI]. Supernatants were recovered by centrifugation at 12,000 ×g for 15 min, followed by immunoprecipitation with ANTI-HA affinity agarose (SIGMA). HA-affinity agarose was washed three times with ice-cold buffer C without CI. An appropriate amount of immunoprecipitates was used for Western blotting with either anti-FLAG M2 or anti-HA antibody.
2.10. Gel filtration chromatography
Purified SQSTM1 proteins were loaded onto the gel filtration column (Superose6 10/300), and eluates were fractionated and collected. Elution was carried out at a flow rate of 0.3 ml/min with a fraction volume of 0.5 ml. Fractionated samples were analyzed by Western blotting using anti-human SQSTM1 antibody.
2.11. Immunocytochemistry
Transfected cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (−) for 20 min at RT, and permeabilized with 0.1% (w/v) Triton-X100 and 5% normal goat serum (NGS) in PBS (−). After blocking with 0.05% (w/v) Triton-X100 and 1% NGS in PBS (−), the primary antibody diluted in PBS (−) containing 0.05% (w/v) Triton-X100 and 1% NGS was added and incubated for 2 h at RT. After washing with PBS (−), the secondary antibody diluted in the same solution as the primary antibody was added and incubated for 2 h at RT. Cells were washed with PBS (−) and mounted by VECTASHIELD Mounting Medium containing 4′,6-diamidino-2-phenylindole (DAPI) for nuclear staining. Confocal laser microscope (LSM700; ZEISS) was used to detect fluorescent signals.
2.12. Imaging analysis
For colocalization analysis, we performed Pearson's correlation coefficient analysis using the Coloc 2 plugin in Fiji (ImageJ). We defined SQSTM1-positive structures whose signal intensities ranged from 50 to 255, and its area size was over 0.31 μm2 (8 pixels) as “SQSTM1-body”, and quantified the number and size of them using the Analyze Particles plugin in Fiji (ImageJ). SQSTM1 structures with >7.84 μm2 (200 pixels) of an area size were defined as background staining and were excluded from the analysis. The size of each SQSTM1-body in each cell was measured. We also counted and normalized the number of SQSTM1-body with the adjusted area of cell body (per 1000 μm2). We independently conducted the transfection experiments 5 times, and analyzed 5–7 randomly-selected images/cells for each construct in each experiment.
2.13. Analysis of the autophagic activity
MEFs prepared from Sqstm1-KO mice were transfected with pCIneoFLAG-SQSTM1 constructs. After 24 h, MEFs were treated with Earle's Balanced Salt Solution (EBSS) (SIGMA) lacking FBS for starvation. To monitor the autophagic flux, lysosomal degradation was inhibited by addition of 250 μM chloroquine (CQ) to the medium, and incubated for additional 6 h. Untransfected MEFs from wild-type and Sqstm1-KO mice were also treated in the same manner. Signal intensities of western blots were quantified by using CS Analyzer Version 3.0 (ATTO). The autophagy flux was calculated from the differences between the ratios of (LC3-II/LC3-I + LC3-II) with and without CQ.
2.14. Statistical analysis
All statistical analyses were conducted using Prism 8 (GraphPad Software). Statistical significance was evaluated by one-way ANOVA with Dunnett's multiple comparison post hoc test or with Tukey's multiple comparison post hoc test. We considered p-values <0.05 to be statistically significant.
3. Results
3.1. Expression of ALS-linked SQSTM1 variants does not affect viability of PC12 cells
Since most of SQSTM1 variations observed in patients with ALS/FTD were heterozygote [[5], [6], [7], [8], [9], [10],12], we first examined whether expression of ALS-linked SQSTM1 variants dominantly affected the cell viability. We measured the viabilities of PC12 cells expressing ALS-linked SQSTM1-PB1 variants (A53T and V90M) or SQSTM1-LIR variants (D337E and L341V). We also tested two artificial SQSTM1 mutants, K7A/D69A (indicated as PB1-2A) and D335A/D336A/D337A (indicated as LIR-3A) as positive controls for PB1 and LIR variants, respectively. PB1-2A mutant is defective not only in self-oligomerization but also in aggregate formation with other ubiquitinated proteins [19,32,33], while LIR-3A mutant loses the affinity to LC3 [18]. Levels of overexpressed SQSTM1 proteins were approximately 5-fold higher than that of endogenous one (Fig. S1). We also confirmed that expression level of each SQSTM1 variant was comparable to that of SQSTM1_WT (Fig. S1). Under these conditions, expression of neither SQSTM1_WT nor its variants showed observable effects on cell viabilities (Fig. S2A), indicating that ALS-linked SQSTM1 variations did not instantly exert deleterious effects to cells under normal cultural conditions.
Since oxidative stress was implicated in the pathogenesis of ALS/FTD [1,2], we next assessed whether expression of ALS-linked SQSTM1 variants induced cell death under oxidative stress conditions. We used menadione as an oxidative stress inducer. Treatment of pCIneo-vector-transfected PC12 cells with 10 μM menadione for 4 h reduced the cell viability by approximately 80% compared to those under normal growth conditions (Figs. S2A and S2B), confirming that experimental conditions used were appropriate to monitor either up- or down-regulation of cell viabilities. Expression of neither SQSTM1_WT nor its ALS-linked variants (A53T, V90M, D337E and L341V) affected menadione-induced cell death (Fig. S2B). Notably, expression of PB1-2A and LIR-3A mutants also exhibited no observable effects (Fig. S2B). These results indicate that SQSTM1 variants have neither harmful nor beneficial effects on the menadione-induced oxidative stress signaling pathway.
To further assess whether expression of ALS-linked SQSTM1 variants induced cell death under mitochondrial stress conditions, we used CCCP, a potent chemical inducer of mitochondrial uncoupling. Treatment of pCIneo-vector transfected PC12 cells with 750 μM CCCP for 10 h reduced the cell viability by approximately 70% compared to control cells under normal growth conditions (Figs. S2A and S2C). Under this experimental setting, expression of neither SQSTM1_WT nor its variants/mutants (A53T, V90M, D337E, L341V, PB1-2A and LIR-3A) affected CCCP-induced cell death (Fig. S2C). These results indicate that not only ALS-linked variants (A53T, V90M, D337E and L341V) but also artificial SQSTM1 mutants (PB1-2A and LIR-3A) have no major impacts on mitochondria stress-induced cell death signaling. Taken together, ALS-linked SQSTM1 variants, at least tested in this study, may not possess dominant discernible cytotoxic activity against PC12 cells under normal as well as acutely-stressed conditions.
3.2. Expression of SQSTM1 results in formation of cytoplasmic SQSTM1-body in Sqstm1-deficient mouse embryonic fibroblasts and PC12 cells
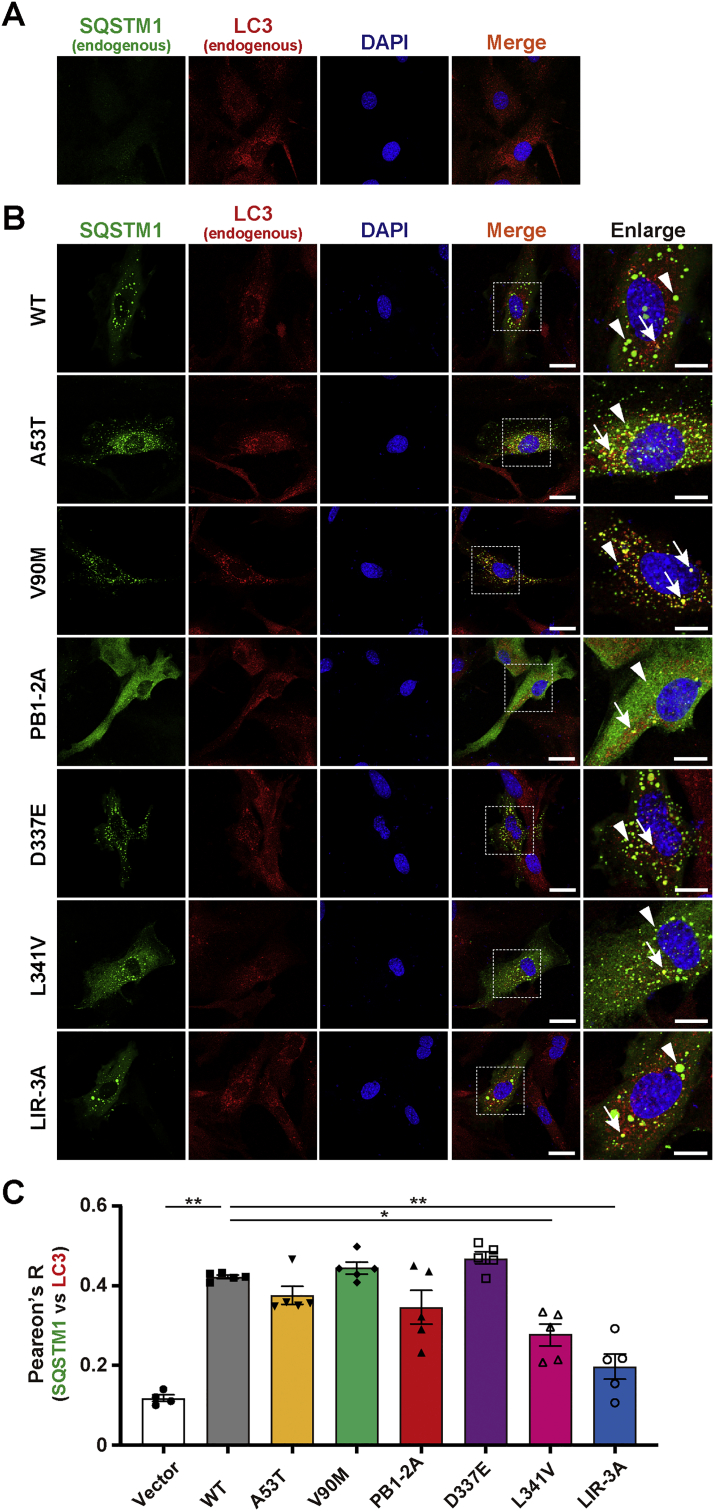
It has previously been reported that the distribution, size and number of SQSTM1-body in cells are altered by the disease-associated and artificial mutations in SQSTM1 [18,28,34]. Thus, we investigated the subcellular distribution of SQSTM1 in PC12 cells. To exclude the possibility that endogenous SQSTM1 affected the distribution of overexpressed SQSTM1 variants/mutants, we also used Sqstm1-KO MEFs. SQSTM1-bodies are visible structures consisting of either membrane-free protein aggregates and/or membrane-confined autophagosome/autolysosomes [28]. SQSTM1_WT was present as numerous round bodies throughout the cytoplasm of both Sqstm1-KO MEFs (Fig. 1A and B) and PC12 cells (Fig. S3). Consistently, some of these, but not all, SQSTM1_WT structures were colocalized with LC3, an autophagosome marker (Figs. 1B and S3), indicating that ectopically-expressed SQSTM1 could form SQSTM1-body in either Sqstm1-KO MEFs or PC12 cells.
Fig. 1.
Subcellular distribution of SQSTM1 mutants in Sqstm1 deficient mouse embryonic fibroblast. Sqstm1 deficient mouse embryonic fibroblasts (Sqstm1-KO MEFs) were transfected with FLAG-tagged pCIneo_SQSTM1 constructs. (A) Representative images for the control experiment (Vector). (B) Representative images for SQSTM1_WT (WT), PB1-domain ALS-linked variants (A53T and V90M), PB1-domain artificial mutant (PB1-2A), LIR ALS-linked variants (D337E and L341V) and LIR artificial mutant (LIR-3A)-expressing cells. Images for SQSTM1 and LC3 were obtained by staining with anti-SQSTM1 (p62-C) and anti-LC3 antibodies, respectively. Nuclei were counterstained with DAPI. High-resolution magnified images are shown on the right. Arrows and arrowheads indicate LC3-positive and -negative SQSTM1-bodies, respectively. Scale bars for merged and enlarged images indicate 20 μm and 10 μm, respectively. (C) Pearson's correlation coefficient analysis of the colocalization between SQSTM1 and LC3. Values of Pearson's R are expressed as mean ± S.E.M. (n = 5 biological replicates). Individual data points are also shown. Statistical significances were evaluated by one-way ANOVA with Tukey's multiple comparison test. The results of comparisons between WT and other conditions are only shown (*p < 0.05, **p < 0.01).
3.3. ALS-linked variations in PB1 domain of SQSTM1 do not alter intracellular distribution of SQSTM1 in Sqstm1-deficient mouse embryonic fibroblasts and PC12 cells
To investigate whether ALS-linked variations in the PB1 domain of SQSTM1 could alter the intracellular distribution of SQSTM1 itself, we tested PB1-2A mutant as a positive control. PB1-2A was diffusely distributed throughout the cytoplasm and was rarely localized to vesicle puncta (Figs. 1B and S3), consistent with previous findings showing that the PB1 domain of SQSTM1 was required not only for its autophagosome localization but also for SQSTM1-body formation [19,20,34]. We next examined two ALS-linked PB1 variants, A53T and V90M. We revealed that both variants were present as LC3-positive as well as -negative SQSTM1 structures in either Sqstm1-KO MEFs or PC12 cells, which were indistinguishable from SQSTM1_WT (Figs. 1B and S3). The results suggest that A53T and V90M variations do not disrupt the self-oligomerizing function of the PB1 domain [32,33].
To verify this, we prepared cell lysates from COS-7 cells co-expressing FLAG-tagged and HA-tagged SQSTM1, and conducted co-immunoprecipitation using HA-affinity beads. FLAG-SQSTM1_WT and HA-SQSTM1_WT were efficiently co-immunoprecipitated, indicating that SQSTM1_WT formed self-oligomer (Fig. S4A). Similarly, both V90M and A53T retained the self-oligomerization ability. Unexpectedly, PB1-2A mutant was less-efficiently but apparently self-oligomerized (Fig. S4A). To further confirm a higher structure of SQSTM1-PB1 variants, we performed gel filtration analysis. Purified SQSTM1-WT, PB1-2A, A53T and V90M were fractionated by Superose6 10/300 column, and resulting fractions were analyzed by western blotting. Not only SQSTM1-WT but also its PB1 variants including PB1-2A mutant were eluted in fractions with an apparent molecular mass of over 975 kDa (Fig. S4B), which was consistent with the result from previous study [20], suggesting that not only SQSTM1-WT and ALS-linked PB1 variants (A53T and V90M) but also PB1-2A mutant can form a huge complex, possibly, through domains other than PB1. Taken together, these results raise the possibility that self-oligomerization is not the sole determinant at least of the intracellular distribution of SQSTM1.
3.4. ALS-linked variations in LIR of SQSTM1 do not alter intracellular distribution of SQSTM1 in Sqstm1-deficient mouse embryonic fibroblasts and PC12 cells
To examine whether ALS-linked variations in the LIR of SQSTM1 could alter the intracellular distribution of SQSTM1, we observed Sqstm1-KO MEFs and PC12 cells expressing either LIR-3A or ALS-linked LIR variants; D337 and L341V. All the LIR variants including D337E, L341V and LIR-3A resembled SQSTM1_WT, in which they were localized to both LC3-positive and -negative SQSTM1 structures (Figs. 1B and S3). Although it has previously been demonstrated that L341V variant disturbs autophagic process [26], our results indicate that ALS-linked variations in the LIR do not have a major impact at least on the formation of SQSTM1-bodies.
3.5. ALS-linked SQSTM1 variations do not alter the size and in number of SQSTM1-bodies in Sqstm1-deficient mouse embryonic fibroblasts
To verify whether ALS-linked SQSTM1 variations affected the size and number of SQSTM1-bodies, we quantified them in Sqstm1-KO MEFs expressing either SQSTM1-WT, ALS-linked variants (A53T, V90M, D337E and L341V) or artificial mutants (PB1-2A and LIR-3A). Both the average size and number of SQSTM1-bodies of ALS-linked variants were mostly comparable to those of SQSTM1_WT (Figs. S5A and S5B). It has previously been reported that loss of the LIR function causes less efficient formation of SQSTM1-bodies in cells [35]. However, LIR-3A mutation did not alter the size and number of SQSTM1-bodies (Figs. S5A and S5B). By contrast, PB1-2A mutation drastically reduced not only the size but also the number of SQSTM1-bodies (Figs. S5A and S5B), consistent with its diffused intracellular distribution (Figs. 1B and S3). Thus, at least ALS-linked SQSTM1 variations tested in this study do not have major impacts on the structural phenotypes of SQSTM1-bodies in cells.
3.6. ALS-linked L341V variant exhibits impaired colocalization with LC3 in Sqstm1-deficient mouse embryonic fibroblasts
It has previously been reported that the ALS-linked L341V variation in the LIR reduces the affinity of SQSTM1 to LC3 in vitro [26]. To verify this, and at the same time, to evaluate the colocalization of the signals between SQSTM1 and LC3, we conducted Pearson's correlation coefficients analysis in Sqstm1-KO MEFs expressing either ALS-linked LIR variant or LIR-3A mutant. As expected, degrees of LC3 colocalization with both L341V variant and LIR-3A mutant were significantly decreased (Fig. 1C). However, another LIR variant, D337E, rather showed the higher level of colocalization, which was comparable to SQSTM1_WT (Fig. 1C). There were no observable effects of other ALS-linked variations on the colocalization between SQSTM1 and LC3 (Fig. 1C). Thus, at least some, but not all, of the amino acid substitutions in the LIR may have a discernible effect on the association between SQSTM1 and LC3.
3.7. Expression of ALS-linked SQSTM1 variants marginally affect the autophagic activity in Sqstm1-deficient mouse embryonic fibroblasts
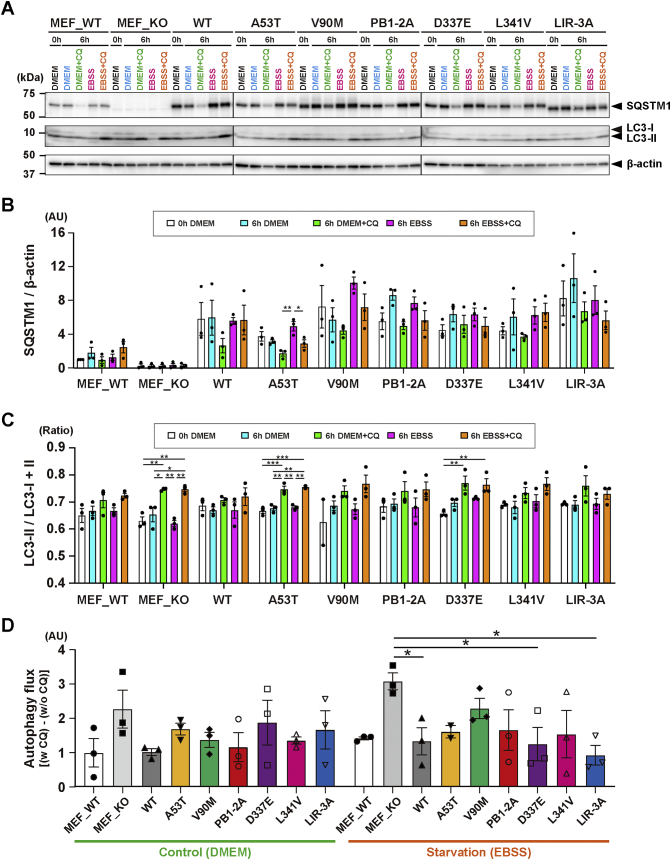
Since some SQSTM1 variations could alter the physical association between SQSTM1 and LC3, we finally investigated whether expression of ALS-linked SQSTM1 variants affected the autophagic activity in cells. MEFs derived from wild-type and Sqstm1-KO mice were subjected to autophagy flux analysis, in which changes of LC3-II in the presence or absence of lysosomal inhibitor CQ was used as an indicator (Fig. 2). While the levels of LC3-II were kept low under both unstarved (DMEM) and starved (EBSS) conditions, they were increased when the cells were treated with CQ (Fig. 2A). Quantification analysis revealed that the responses to CQ were significant in Sqstm-1 KO MEFs and those expressing A53T or D337E variant (Fig. 2C). Interestingly, under starved conditions, the apparent level of the autophagy flux in Sqstm1-KO was significantly increased, and such increased flux was reverted to normal levels by ectopic expression of either D337E variant or LIR-3A mutant, but not by others (Fig. 2D). These results suggest that although some ALS-linked SQSTM1 variants are indeed implicated in the autophagic processes, most variations may have a weaker effect on autophagy. Incidentally, we speculate that the apparent increase in the autophagy flux in Sqstm1-KO MEFs may be due to the combinatorial effects of the decreased basal level of autophagy and the increased compensatory functions of other autophagic adaptor proteins such as optineurin and NDP52 [36].
Fig. 2.
Effects of ectopic expression of SQSTM1 mutants on the autophagic activity in mouse embryonic fibroblast. (A) Western blot analysis of SQSTM1 and LC3. MEFs prepared from Sqstm1-KO mice were transfected with pCIneoFLAG-SQSTM1 constructs. After 24 h, the cells were incubated in DMEM or EBSS with or without 250 μM chloroquine (CQ) for additional 6 h. Untransfected MEFs from wild-type (MEF_WT) and Sqstm1-KO (MEF_KO) mice were also treated in the same manner. Two μg of total proteins from each sample was subjected to SDS-PAGE and analyzed by immunoblotting using anti-SQSTM1 (p62-C) and anti-LC3 antibodies. β-actin was used for a loading control. Representative images are shown. Positions of SQSTM1, LC3-I and -LC3-II are shown on the right. Size-markers are shown on the left. (B) Quantification of the signal intensities of SQSTM1 in western blots. Values of SQSTM1/β-actin (in arbitrary unit; AU) are expressed as mean relative to MEF_WT at 0 h [± S.E.M. (n = 3 biological replicates)]. Individual data points are also shown. (C) Quantification of the signal intensities of LC3 in western blots. Values of the ratio of LC3-II/LC3-I + LC3-II are expressed as mean ± S.E.M. (n = 3 biological replicates). Individual data points are also shown. (D) Autophagy flux in MEFs expressing SQSTM1 variant/mutant. The autophagy flux was defined as; [(the ratio with CQ) – (the ratio without CQ)]. Values (in arbitrary unit; AU) are expressed as mean relative to MEF_WT in DMEM [± S.E.M. (n = 3 biological replicates)]. Individual data points are also shown. Statistical significances were evaluated by one-way ANOVA with Tukey's multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.001) (B and C) or with Dunnett's multiple comparison test (*p < 0.05) (D).
4. Discussion
In this study, to obtain some clues to understanding the pathogenesis of ALS/FTD that is linked to SQSTM1 mutations, we have focused on several SQSTM1 variations, which are originally identified in Japanese and Chinese sporadic ALS patients. We investigated the primary effects of ALS-linked SQSTM1 variant expression on cellular phenotypes including the viability under normal and stressed conditions, intracellular distribution of SQSTM1-bodies and the autophagic activities.
Since most of SQSTM1 variations identified in patients with ALS/FTD were heterozygote [[5], [6], [7], [8], [9], [10],12], we first tested whether expression of ALS-linked SQSTM1 variants dominantly affected the cell viability. Contrary to our expectations, expression of these variants had no major impact on the viability in PC12 cells even under stressed conditions. Thus, expression of ALS-linked SQSTM1 variants, at least tested in this study, do not dominantly exert a deleterious effect on cell survival. Nonetheless, at this stage, we cannot formally exclude the possibility that long-term expression of these variants in cells, which may much recapitulate pathological conditions in motor neurons, could reduce the viability, as we merely adopted experimental settings with short-term exposure to stress-inducing agents and observation in this study.
Previously, it has been demonstrated that both the PB1 domain and the LIR of SQSTM1 have crucial roles on autophagy [18,19]. In addition, it has been reported that under pathological conditions, enlarged granular SQSTM1-positive structures, i.e., SQSTM1-bodies, frequently emerge in the brain of patients with ALS/FTD due to dysfunctional autophagy [2]. Thus, the autophagic degradation of SQSTM1 might be affected by expression of ALS-linked SQSTM1-PB1 and/or -LIR variants. In fact, 3 mutations; i.e., V90M, D337E and L341V, are predicted to be a “possibly damaging substitution” by using PolyPhen-2 prediction tool (http://genetics.bwh.harvard.edu/pph2/), while A53T is a “benign” one. To test this, we investigated the morphology of autophagosomes including SQSTM1-bodies in cells. Although expression of wild-type SQSTM1 as well as its pathogenic variants induced the formation of SQSTM1-bodies that were partially colocalized with LC3, there were no discernible differences in their intracellular distributions both in PC12 cells and Sqstm1-KO MEFs. Furthermore, impacts of ectopic expression of ALS-linked SQSTM1 variants on the autophagic activities were marginal or below detection levels. These results indicate that the process of autophagy may not be largely affected by expression of ALS-linked SQSTM1 variants.
Nonetheless, our quantitative colocalization analysis of SQSTM1-bodies with LC3 in Sqstm1-KO MEFs expressing ALS-linked SQSTM1 variants revealed that ALS-linked LIR variant L341V, but not others, exhibited a significantly decreased colocalization compared to wild-type, supporting the notion that L341V variation reduces the affinity to LC3 [26]. Artificial LIR-3A mutant also showed a decreased level of colocalization with LC3, consistent with the previous findings [34]. The LIR consists of an acidic cluster and hydrophobic residues (DDD and WxxL) [18,34], both of which are essential for the interaction to multiple sites of LC3 [18,34]. Since two ALS-linked LIR variants; D337E and L341V, retain acidic as well as hydrophobic residues within the LIR, it is likely that they still preserve the different levels of binding affinity to LC3. In fact, L341V variant exhibited slightly-higher colocalization with LC3 than did LIR-3A, while another LIR variant, D337E, showed the normal level like wild-type SQSTM1 (Fig. 1C). Notably, it has recently been demonstrated that SQSTM1 can be incorporated into membraneless liquid compartments, which are generated through liquid-liquid phase separation (LLPS) [37,38], by changing its binding partners in a context-dependent manner [15,16,39]. Thus, it is possible that the decreased levels of colocalization of SQSTM1 varinats with LC3 without changing the apparent number of SQSTM1-bodies (Fig. S5) may be explained by the increased number of LC3-negative LLPS-associated SQSTM1-bodies.
One question that has yet to be answered include as to whether and/or how ALS-linked SQSTM1 variations are linked to the pathogenesis of ALS/FTD. In this study, we sought to clarify the pathogenic effects of ectopic expression of ALS-linked SQSTM1 variants using the cell culture system. Despite the fact that the expression levels of SQSTM1 variants are much higher than that of endogenous one, no clear abnormal phenotypes that are linked to SQSTM1 variations have been detected thus far. It is usually appreciated that the transient expression of proteins in conjunction with short-term observation study may have limitations to properly detect chronic and long-term weaker effects of mutant proteins. Therefore, the use of longer-term studies with fully-developed neuronal cultures that are stably expressing SQSTM1 mutants will be required.
Another fundamental issue that has yet to be addressed is as to whether the accumulation of SQSTM1-bodies in cells are actually associated with disease onset and/or progression. Like TAR DNA-binding protein 43 kDa (TDP-43) [40], SQSTM1 can accumulate in the brain and/or spinal cord of ALS/FTD patients irrespective of the presence or absence of mutations in the SQSTM1 gene [14], suggesting that SQSTM1accumulation may not be a causative but consequence of diseases. However, it has also been shown that loss of the SQSTM1 function results in exacerbation of disease phenotypes in a number of animal models for motor neuron diseases including ALS [21] and spinal and bulbar muscular atrophy (SBMA) [41]. Thus, it is reasonable to speculate that malfunction of SQSTM1 may be associated with the acceleration of disease progression and/or aggravation of diseases. However, as there are no direct evidences to support these notions thus far, further detailed studies will be required.
5. Conclusions
This study reveals that ALS-linked SQSTM1 variants have marginal effects on cellular phenotypes at least under transiently-expressed conditions. However, among the ALS patient-derived SQSTM1 variations, L341V variation indeed causes a decreased colocalization of SQSTM1 with LC3, despite of its limited effects on the autophagic activity in cells. Thus, it is still possible that long-term and persistent expression of such ALS-linked SQSTM1 variants could impair normal cellular functions, ultimately leading to neuronal death and neurodegeneration.
CRediT authorship contribution statement
Masahisa Nozaki: Methodology, Formal analysis, Investigation, Data curation, Writing - original draft, Visualization. Asako Otomo: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing - original draft, Writing - review & editing, Visualization. Shun Mitsui: Validation, Investigation, Data curation, Writing - review & editing. Suzuka Ono: Validation, Investigation, Data curation, Writing - review & editing, Visualization. Ryohei Shirakawa: Methodology, Investigation, Data curation, Visualization. YongPing Chen: Methodology, Resources, Data curation. Yutaro Hama: Methodology, Investigation, Data curation, Writing - review & editing. Kai Sato: Validation, Writing - review & editing. XuePing Chen: Resources. Toshiyasu Suzuki: Supervision, Writing - review & editing. Hui-Fang Shang: Conceptualization, Resources, Writing - review & editing, Funding acquisition, Supervision. Shinji Hadano: Conceptualization, Writing - review & editing, Visualization, Project administration, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
We thank Drs. Tetsuro Ishii and Toru Yanagawa at University of Tsukuba for generous gift of Sqstm1-KO mouse. We are grateful to all members of our laboratory at Tokai University for helpful discussion, and also to all members of Support Center for Medical Research and Education at Tokai University for their technical help. This study was supported by a Grant-in-Aid for Scientific Research (B) (26290018 and 19H03551 to SH) from the Japanese Society for Promotion of Science (JSPS), and partly by the National Natural Science Foundation of China (NSFC) and JSPS Bilateral Joint Research Project (to HFS and SH).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ensci.2020.100301.
Appendix A. Supplementary data
Supplementary material
References
- 1.van Es M.A., Hardiman O., Chio A., Al-Chalabi A., Pasterkamp R.J., Veldink J.H., van den Berg L.H. Amyotrophic lateral sclerosis. Lancet. 2017;390(10107):2084–2098. doi: 10.1016/S0140-6736(17)31287-4. [DOI] [PubMed] [Google Scholar]
- 2.Hardiman O., Al-Chalabi A., Chio A., Corr E.M., Logroscino G., Robberecht W., Shaw P.J., Simmons Z., van den Berg L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers. 2017;3:17071. doi: 10.1038/nrdp.2017.71. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen H.P., Van Broeckhoven C., van der Zee J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 2018;34(6):404–423. doi: 10.1016/j.tig.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Ragagnin A.M.G., Shadfar S., Vidal M., Jamali M.S., Atkin J.D. Motor neuron susceptibility in ALS/FTD. Front. Neurosci. 2019;13:532. doi: 10.3389/fnins.2019.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fecto F., Yan J., Vemula S.P., Liu E., Yang Y., Chen W., Zheng J.G., Shi Y., Siddique N., Arrat H., Donkervoort S., Ajroud-Driss S., Sufit R.L., Heller S.L., Deng H.X., Siddique T. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 11, 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 6.Rubino E., Rainero I., Chio A., Rogaeva E., Galimberti D., Fenoglio P., Grinberg Y., Isaia G., Calvo A., Gentile S., Bruni A.C., St George-Hyslop P.H., Scarpini E., Gallone S., Pinessi L., T.S. Group SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 15, 2012;79:1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano M., Nakamura Y., Saigoh K., Sakamoto H., Ueno S., Isono C., Miyamoto K., Akamatsu M., Mitsui Y., Kusunoki S. Mutations in the gene encoding p62 in Japanese patients with amyotrophic lateral sclerosis. Neurology. 2013;80(5):458–463. doi: 10.1212/WNL.0b013e31827f0fe5. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu H., Toyoshima Y., Shiga A., Yokoseki A., Arakawa K., Sekine Y., Shimohata T., Ikeuchi T., Nishizawa M., Kakita A., Onodera O., Takahashi H. Sporadic ALS with compound heterozygous mutations in the SQSTM1 gene. Acta Neuropathol. 2013;126(3):453–459. doi: 10.1007/s00401-013-1150-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Zheng Z.Z., Chen X., Huang R., Yang Y., Yuan L., Pan L., Hadano S., Shang H.F. SQSTM1 mutations in Han Chinese populations with sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2014;35(3) doi: 10.1016/j.neurobiolaging.2013.09.008. 726 e7-9. [DOI] [PubMed] [Google Scholar]
- 10.Kwok C.T., Morris A., de Belleroche J.S. Sequestosome-1 (SQSTM1) sequence variants in ALS cases in the UK: prevalence and coexistence of SQSTM1 mutations in ALS kindred with PDB. EJHG. 2014;22(4):492–496. doi: 10.1038/ejhg.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rea S.L., Majcher V., Searle M.S., Layfield R. SQSTM1 mutations--bridging Paget disease of bone and ALS/FTLD. Exp. Cell Res. 2014;325(1):27–37. doi: 10.1016/j.yexcr.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., Tang L., Zhang N., Pan L., Hadano S., Fan D. Six SQSTM1 mutations in a Chinese amyotrophic lateral sclerosis cohort. Amyotrop. Lat. Scler. Front. Degen. 2015;16(5–6):378–384. doi: 10.3109/21678421.2015.1009466. [DOI] [PubMed] [Google Scholar]
- 13.Yilmaz R., Muller K., Brenner D., Volk A.E., Borck G., Hermann A., Meitinger T., Strom T.M., Danzer K.M., Ludolph A.C., Andersen P.M., Weishaupt J.H., A.L.S.N.M.N.D.N.E.T. German SQSTM1/p62 variants in 486 patients with familial ALS from Germany and Sweden. Neurobiology of aging. 2020;87 doi: 10.1016/j.neurobiolaging.2019.10.018. 139 e9-139 e15. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Martin P., Komatsu M. p62/SQSTM1 - steering the cell through health and disease. J. Cell Sci. 2018;131(21) doi: 10.1242/jcs.222836. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Martin P., Saito T., Komatsu M. p62/SQSTM1: 'Jack of all trades' in health and cancer. FEBS J. 2019;286(1):8–23. doi: 10.1111/febs.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran A., Amanchy R., Linares J.F., Joshi J., Abu-Baker S., Porollo A., Hansen M., Moscat J., Diaz-Meco M.T. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 2011;44(1):134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraft L.J., Dowler J., Manral P., Kenworthy A.K. Size, organization, and dynamics of soluble SQSTM1 and LC3-SQSTM1 complexes in living cells. Autophagy. 2016;12(9):1660–1674. doi: 10.1080/15548627.2016.1199299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 33, 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 19.Itakura E., Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 2011;192(1):17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurzer B., Zaffagnini G., Fracchiolla D., Turco E., Abert C., Romanov J., Martens S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4 doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadano S., Mitsui S., Pan L., Otomo A., Kubo M., Sato K., Ono S., Onodera W., Abe K., Chen X., Koike M., Uchiyama Y., Aoki M., Warabi E., Yamamoto M., Ishii T., Yanagawa T., Shang H.F., Yoshii F. Functional links between SQSTM1 and ALS2 in the pathogenesis of ALS: cumulative impact on the protection against mutant SOD1-mediated motor dysfunction in mice. Hum. Mol. Genet. 2016;25(15):3321–3340. doi: 10.1093/hmg/ddw180. [DOI] [PubMed] [Google Scholar]
- 22.Mitsui S., Otomo A., Nozaki M., Ono S., Sato K., Shirakawa R., Adachi H., Aoki M., Sobue G., Shang H.F., Hadano S. Systemic overexpression of SQSTM1/p62 accelerates disease onset in a SOD1(H46R)-expressing ALS mouse model. Mol. Brain. 2018;11(1):30. doi: 10.1186/s13041-018-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 24.Jain A., Lamark T., Sjottem E., Larsen K.B., Awuh J.A., Overvatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 29, 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goode A., Rea S., Sultana M., Shaw B., Searle M.S., Layfield R. ALS-FTLD associated mutations of SQSTM1 impact on Keap1-Nrf2 signalling. Mol. Cell. Neurosci. 2016;76:52–58. doi: 10.1016/j.mcn.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goode A., Butler K., Long J., Cavey J., Scott D., Shaw B., Sollenberger J., Gell C., Johansen T., Oldham N.J., Searle M.S., Layfield R. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy. 2016;12(7):1094–1104. doi: 10.1080/15548627.2016.1170257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Z., Lim J., Wang Q., Purtell K., Wu S., Palomo G.M., Tan H., Manfredi G., Zhao Y., Peng J., Hu B., Chen S., Yue Z. ALS-FTLD-linked mutations of SQSTM1/p62 disrupt selective autophagy and NFE2L2/NRF2 anti-oxidative stress pathway. Autophagy. 2020;16(5):917–931. doi: 10.1080/15548627.2019.1644076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 30.Hadano S., Ikeda J.E. Purification and functional analyses of ALS2 and its homologue. Methods Enzymol. 2005;403:310–321. doi: 10.1016/S0076-6879(05)03026-0. [DOI] [PubMed] [Google Scholar]
- 31.Hadano S., Otomo A., Kunita R., Suzuki-Utsunomiya K., Akatsuka A., Koike M., Aoki M., Uchiyama Y., Itoyama Y., Ikeda J.E. Loss of ALS2/Alsin exacerbates motor dysfunction in a SOD1-expressing mouse ALS model by disturbing endolysosomal trafficking. PLoS One. 2010;5(3):e9805. doi: 10.1371/journal.pone.0009805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamark T., Perander M., Outzen H., Kristiansen K., Overvatn A., Michaelsen E., Bjorkoy G., Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 36, 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 33.Wilson M.I., Gill D.J., Perisic O., Quinn M.T., Williams R.L. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell. 2003;12(1):39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 34.Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 33, 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 35.Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M., Peterbauer T., Sztacho M., Trapannone R., Tarafder A.K., Sachse C., Martens S. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018;37(5) doi: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamano K., Kikuchi R., Kojima W., Hayashida R., Koyano F., Kawawaki J., Shoda T., Demizu Y., Naito M., Tanaka K., Matsuda N. Critical role of mitochondrial ubiquitination and the OPTN-ATG9A axis in mitophagy. J. Cell Biol. 2020;219(9) doi: 10.1083/jcb.201912144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun D., Wu R., Zheng J., Li P., Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28(4):405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176(3):419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linares J.F., Duran A., Reina-Campos M., Aza-Blanc P., Campos A., Moscat J., Diaz-Meco M.T. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade. Cell Rep. 2015;12(8):1339–1352. doi: 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pamphlett R., Luquin N., McLean C., Jew S.K., Adams L. TDP-43 neuropathology is similar in sporadic amyotrophic lateral sclerosis with or without TDP-43 mutations. Neuropathol. Appl. Neurobiol. 2009;35(2):222–225. doi: 10.1111/j.1365-2990.2008.00982.x. [DOI] [PubMed] [Google Scholar]
- 41.Doi H., Adachi H., Katsuno M., Minamiyama M., Matsumoto S., Kondo N., Miyazaki Y., Iida M., Tohnai G., Qiang Q., Tanaka F., Yanagawa T., Warabi E., Ishii T., Sobue G. p62/SQSTM1 differentially removes the toxic mutant androgen receptor via autophagy and inclusion formation in a spinal and bulbar muscular atrophy mouse model. J. Neurosci. 18, 2013;33:7710–7727. doi: 10.1523/JNEUROSCI.3021-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material