Highlights
-
•
Biomarkers of response to ibrutinib + nivolumab were analyzed in diffuse large B-cell lymphoma (DLBCL), follicular lymphoma and Richter transformation.
-
•
DLBCL patients with elevated PD-L1 by immunohistochemistry tended to have better response and survival.
-
•
Whole exome sequencing identified gene mutations in alternate B-cell receptor pathways linked to response in DLBCL.
-
•
Enriched pathways by gene expression profiling were related to immune activation in responders and proliferation/replication in nonresponders.
-
•
This preliminary work may help to generate hypotheses on genetically defined subsets of patients most likely to benefit from ibrutinib + nivolumab.
Keywords: Ibrutinib, Nivolumab, Non-hodgkin's lymphoma, Biomarkers, Phase I/II trial
Abstract
We analyzed potential biomarkers of response to ibrutinib plus nivolumab in biopsies from patients with diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), and Richter's transformation (RT) from the LYM1002 phase I/IIa study, using programmed death ligand 1 (PD-L1) immunohistochemistry, whole exome sequencing (WES), and gene expression profiling (GEP). In DLBCL, PD-L1 elevation was more frequent in responders versus nonresponders (5/8 [62.5%] vs. 3/16 [18.8%]; p = 0.065; complete response 37.5% vs. 0%; p = 0.028). Overall response rates for patients with WES and GEP data, respectively, were: DLBCL (38.5% and 29.6%); FL (46.2% and 43.5%); RT (76.5% and 81.3%). In DLBCL, WES analyses demonstrated that mutations in RNF213 (40.0% vs. 6.2%; p = 0.055), KLHL14 (30.0% vs. 0%; p = 0.046), and LRP1B (30.0% vs. 6.2%; p = 0.264) were more frequent in responders. No responders had mutations in EBF1, ADAMTS20, AKAP9, TP53, MYD88, or TNFRSF14, while the frequency of these mutations in nonresponders ranged from 12.5% to 18.8%. In FL and RT, genes with different mutation frequencies in responders versus nonresponders were: BCL2 (75.0% vs. 28.6%; p = 0.047) and ROS1 (0% vs. 50.0%; p = 0.044), respectively. Per GEP, the most upregulated genes in responders were LEF1 and BTLA (overall), and CRTAM (germinal center B-cell–like DLBCL). Enriched pathways were related to immune activation in responders and resistance-associated proliferation/replication in nonresponders. This preliminary work may help to generate hypotheses regarding genetically defined subsets of DLBCL, FL, and RT patients most likely to benefit from ibrutinib plus nivolumab.
Introduction
Among novel targeted therapies for the treatment of B-cell malignancies, ibrutinib, a first-in-class, oral, covalent inhibitor of Bruton's tyrosine kinase (BTK), improved clinical outcomes in randomized trials in patients with treatment-naive or relapsed/refractory non-Hodgkin's lymphoma (NHL) [1], [2], [3], [4], [5], [6], [7], [8], [9] leading to approval of ibrutinib in the United States and Europe for the treatment of adult B-cell malignancies, and also for chronic graft versus host disease (cGVHD) [10,11].
Ibrutinib is an investigational therapy for diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), and Richter's transformation (RT), and responses have been observed in phase I/II studies in patients with treatment-naive [12] and relapsed/refractory disease [13], [14], [15]. However, the prognosis for patients with relapsed disease remains poor [16], and the synergistic antitumor activity of ibrutinib combined with other novel agents is currently being investigated as an approach to improve long-term outcomes.
Nivolumab is a fully human immunoglobulin G4 monoclonal antibody that blocks interaction between the programmed death 1 (PD-1) receptor and its ligands, PD-L1 and PD-L2, and augments antitumor activity of T cells [17]. High expression of PD-L1 in solid tumors and lymphomas is generally thought to be related to greater response to anti-PD-1 therapy, but the results from clinical trials are conflicting [17,18]. Based on data from a phase II study [19], nivolumab was approved in the United States for the treatment of classic Hodgkin's lymphoma [20] and has been investigated as monotherapy in DLBCL and FL [21,22], and in combination with ibrutinib in RT [23].
The phase I/IIa LYM1002 study (NCT02329847) evaluated the efficacy and safety of ibrutinib plus nivolumab in 141 patients with relapsed/refractory B-cell malignancies [24]. Safety was consistent with that reported for single-agent ibrutinib or nivolumab. The overall response rate (ORR) was 22/36 (61%) for patients with chronic lymphocytic leukemia/small lymphocytic lymphoma CLL/SLL (including patients with del17p/del11q), 13/40 (33%) for FL, 16/45 (36%) for DLBCL, and 13/20 (65%) for RT. Response to ibrutinib plus nivolumab in RT was high; historically, these patients have had poor outcomes with chemotherapy [25] or single-agent ibrutinib [26].
Biomarker analyses can enhance the efficacy of molecularly targeted therapies by improving rational combinations and identifying patients most likely to benefit from the therapies. Whole genome or exome sequencing studies have identified the spectrum of mutations in genes known to be functionally relevant in DLBCL [27,28], and revealed mutations driving initiation and progression of FL (CREBBP, EZH2, KMT2D, EBF1, MYD88, TNFAIP3) [29] and RT (MYC, BCL2, CDKN2A,TP53, TNFRSF14, TNFSF9) [30]. Some gene mutations, including CD79B, TP53, CARD11, MYD88, EZH2, KMT2D, TNFRSF14, BTG1, MEF2B, and GNA13, have been implicated in the pathogenesis of DLBCL [27]. Exploration of gene variants that may impact response to ibrutinib therapy in NHL (such as BCR and MYD88 pathway mutations in DLBCL and CARD11 mutations in DLBCL and FL) [13,14] is limited and requires further examination.
This analysis evaluated the associations between response to ibrutinib plus nivolumab and a variety of biomarkers including PD-L1 expression by immunohistochemistry (IHC), DNA exome sequencing, and gene expression profiling (GEP), including pathway analyses. Analyses were performed using biopsy samples collected at baseline or before start of treatment from patients with DLBCL (including subtypes), FL, and RT enrolled in the LYM1002 study.
Methods
Patients and study design
Detailed methodology for the LYM1002 study (NCT02329847) was published previously; the study was approved by an independent ethics committee, and all patients provided written informed consent [24]. Briefly, this nonrandomized, open-label phase I/IIa study enrolled adult patients with NHL who received intravenous nivolumab (3 mg/kg) once per 14-day cycle combined with oral ibrutinib 420 mg or 560 mg once daily. Key eligibility criteria were histologically confirmed relapsed/refractory CLL/SLL (with del17p or del11q), DLBCL, FL, or RT (transformation from CLL/SLL only), ≥1 prior systemic therapy (≥2 for FL) but no more than four prior lines of treatment, Eastern Cooperative Oncology Group (ECOG) performance status ≤2, measurable disease, and no prior ibrutinib or anti-PD-1 therapy. Patients were excluded for (a) having major surgery within 4 weeks of the first dose of ibrutinib, (b) getting diagnosed or treated for malignancies other than the indication under study, or (c) requiring treatment with either strong CYP3A inhibitors or warfarin (including equivalent vitamin K antagonists). Biomarker analyses presented herein were conducted in patients with DLBCL, FL, and RT for whom tumor tissue samples were available.
Assessments
DLBCL subtyping
DLBCL subtyping, based on the Wright et al. classification algorithm [31], was conducted in the R software environment using MAS5-normalized (affy v1.48.0, Bioconductor) baseline formalin-fixed paraffin-embedded (FFPE) biopsy GEP data (GeneChip Human Genome U133 Plus 2.0 Array; Affymetrix, Santa Clara, CA, USA).
Treatment response and survival outcomes
Preliminary activity and clinical response to treatment were evaluated by radiological assessments every five cycles for the first 15 months and every 12 cycles thereafter until disease progression, at the end of treatment, and every 6 months during the follow-up period for patients who had not progressed while on therapy and did not start subsequent therapy. Response was assessed per Lugano Classification by Cheson for DLBCL, FL, and RT [32]. For calculation of ORR, responders were defined as patients who achieved complete response (CR) or partial response (PR) by investigator assessment. Progression-free survival (PFS) and overall survival (OS) were estimated using the Kaplan-Meier method and log-rank tests were performed to assess significance.
Biomarker analyses and correlation with clinical outcome
PD-L1 expression
PD-L1 expression as a predictive biomarker for clinical outcomes was evaluated by IHC from baseline tissue biopsies. PD-L1 levels were assessed by IHC as the percentage of tumor cells demonstrating plasma membrane PD-L1 staining of any intensity in a minimum of 100 evaluable tumor cells using the Dako PD-L1 IHC 28–8 pharmDx assay (Agilent Technologies, Glostrup, Denmark). PD-L1 elevation was defined as expression in ≥5% of tumor cells. Kaplan-Meier survival probability with response or survival endpoints was calculated for patients with elevated or nonelevated PD-L1 subgroups with DLBCL, FL, and RT. The association of PD-L1 with clinical response was assessed using Fisher's exact test.
DNA sequence analyses
Exome data were generated from FFPE samples, each from a different patient at baseline. Raw Illumina FASTQ files were processed as follows on the DNAnexus platform (DNAnexus Inc; https://www.dnanexus.com/) using a custom exome analysis workflow: quality was assessed using FastQC 1.0.0, sequences were aligned to the hs37d5 genome build using the BWA-MEM algorithm in BWA Software Package 0.5.9, alignments were recalibrated with the GATK 3.5 Exome Pipeline, variants were annotated with MuTect 1.1.7, and annotations were made with both SnpEff 4.2 (using the GRCh37.75 database and dbNSFP 3.4c) and GEMINI 0.20.0 (modified by using “non-TCGA” gnomAD and ExAC references).
Mutation analysis was performed on a set of cancer-related genes of interest (n = 1742), including those from DLBCL-associated genes (ie, activated B-cell–like [ABC]/germinal center B-cell–like [GCB] discriminating genes, genes used to discriminate between four recently defined subtypes, genes predicted as hypermutated in DLBCL) [33], and genes previously identified from ibrutinib studies, focusing on nonsynonymous single-nucleotide variants that are likely to be somatic based on a set of defined criteria (Supplementary Figure S1). A number of variant filters were used to reduce the likelihood of incorporating sequencing artifacts and germline variants into the analyses.
The significance of variant frequency differences between treatment responders (CR + PR) versus nonresponders (no response or stable disease [SD] + progressive disease [PD]), and between patients with durable responses (PFS >24 months) versus those with PFS ≤24 months, were examined gene-by-gene using Fisher's exact test (no adjustments for multiple hypothesis testing). Patients with no response data were not included in the analyses. Differences in tumor mutational burden (TMB), calculated by dividing the number of inferred somatic mutations in each sample by 30 Mb (the approximate size of the whole exome), were assessed using the Wilcoxon rank sum test.
For patients with DLBCL, mutations were examined in terms of functional groupings by tying mutations in particular genes to potential dysregulation of certain pathways and counting the number of patients with such mutations. Supplementary Table S1 shows which genes were chosen and the functional groups to which they were assigned.
Gene expression analyses
Gene expression microarray data were generated from baseline FFPE biopsy samples using the GeneChip Human Genome U133 Plus 2.0 Array. CEL files were processed and analyzed for DGE and DLBCL subtyping using the R software environment. Raw data were prepared for DGE analyses by Robust Multichip Average normalization (affy v1.48.0, Bioconductor) and annotation with the University of Michigan BrainArray CDF (hgu133plus2hsentrezgcdf, v20.0.0); DGE analyses were performed with empirical Bayes moderation (limma v3.40.6, Bioconductor) and resulting p values were adjusted using the Benjamini-Hochberg false discovery rate-controlling method for multiple hypothesis testing. Gene set enrichment analyses were performed to assess enrichment of canonical pathways from the C2 collection of gene sets from mSigDB. Genes were preranked according to log FC values (from aforementioned DGE results) and analyzed using the Java-based application gsea2–2.2.0.jar with default parameters.
Results
Patients and treatment
Baseline demographics and primary efficacy and safety results for LYM1002 have been reported previously [24]. Briefly, between March 12, 2015 and April 11, 2017, 141 patients were enrolled and treated with daily oral ibrutinib (420 mg or 560 mg) plus intravenous nivolumab (3 mg/kg every 2 weeks): relapsed/refractory CLL/SLL (n = 36; del17p n = 19, del11q n = 17), DLBCL (n = 45), FL (n = 40), and RT (n = 20). At the time of clinical cutoff on October 10, 2017, 35/141 (25%) patients remained on treatment (13 with CLL/SLL, 9 with DLBCL, 7 with FL, and 6 with RT). The most common reasons for treatment discontinuation in all patients were progressive disease or relapse (39%) and adverse events (28%). Median age was 65 years (interquartile range [IQR] 54.0–71.0), 87 (62%) patients were male, 130 (93%) had an ECOG performance status of 0 to 1, and 68 (48%) had bulky disease (≥5 cm). The median number of prior lines of treatment was three. Median follow-up for patients included in this analysis was 18.4 months (IQR 14.8–19.4) for DLBCL, 19.6 months (IQR 14.1–20.7) for FL, and 8.7 months (IQR 6.5–12.1) for RT.
DLBCL subtyping
Twenty-eight patients with DLBCL were evaluable for subtyping using the GEP microarray method; most (19/28) had the GCB subtype, five had the ABC subtype, and four were unclassified.
Treatment responses
Of 70 patients (DLBCL + FL + RT) who had GEP data, 66 were evaluable for response. ORRs were 47.0% (31/66) for all patients, 29.6% (8/27) for DLBCL, 33.3% (6/18) for GCB DLBCL, and 43.5% (10/23) for FL. ORR was highest for patients with RT (81.3%; 13/16) (Table 1). CRs were reported in four patients with DLBCL (two with GCB and two with ABC), three with FL, and two with RT.
Table 1.
Responses in Patients with DLBCL, FL, and RT who had GEP and WES Data.
| GEP Data Set | Total | DLBCLa |
FL | RT | ||
|---|---|---|---|---|---|---|
| N = 66 | All n = 27 |
ABC n = 5 |
GCB n = 18 |
n = 23 | n = 16 | |
| ORR (CR + PR), n (%) | 31 (47.0) | 8 (29.6) | 2 (40.0) | 6 (33.3) | 10 (43.5) | 13 (81.3) |
| CR, n (%) | 9 (13.6) | 4 (14.8) | 2 (40.0) | 2 (11.1) | 3 (13.1) | 2 (12.5) |
| PR, n (%) | 22 (33.3) | 4 (14.8) | 0 | 4 (22.2) | 7 (30.4) | 11 (68.8) |
| Nonresponders, n (%) | 35 (53.0) | 19 (70.4) | 3 (60.0) | 12 (66.7) | 13 (56.5) | 3 (18.8) |
| No response or SD, n (%) | 10 (15.2) | 4 (14.8) | 0 | 2 (11.1) | 6 (26.1) | 0 |
| PD, n (%) | 25 (37.9) | 15 (55.6) | 3 (60.0) | 10 (55.6) | 7 (30.4) | 3 (18.8) |
| WES Data Set | Total | DLBCL |
FL | RT | ||
|---|---|---|---|---|---|---|
| N = 69 | All n = 26 |
ABC n = 4 |
GCB n = 16 |
n = 26 | n = 17 | |
| ORR (CR + PR), n (%) | 35 (50.7) | 10 (38.5) | 2 (50.0) | 6 (37.5) | 12 (46.2) | 13 (76.5) |
| CR, n (%) | 10 (14.5) | 5 (19.2) | 2 (50.0) | 2 (12.5) | 3 (11.5) | 2 (11.8) |
| PR, n (%) | 25 (36.2) | 5 (19.2) | 0 | 4 (25.0) | 9 (34.6) | 11 (64.7) |
| Nonresponders, n (%) | 34 (49.3) | 16 (61.5) | 2 (50.0) | 10 (62.5) | 14 (53.8) | 4 (23.5) |
| No response or SD, n (%) | 12 (17.4) | 4 (15.4) | 0 | 2 (12.5) | 8 (30.8) | 0 |
| PD, n (%) | 22 (31.9) | 12 (46.2) | 2 (50.0) | 8 (50.0) | 6 (23.1) | 4 (23.5) |
All DLBCL patient set also includes patients with unclassified and/or transformed DLBCL not outlined in the Table.
ABC, activated B-cell–like; CR, complete response; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; GCB, germinal center B-cell–like; GEP, gene expression profiling; NHL, non-Hodgkin's lymphoma; ORR, overall response rate; PD, progressive disease; PR, partial response; PRL, partial response with lymphocytosis; RT, Richter's transformation; SD, stable disease; WES, whole exome sequencing.
Of 72 patients who had WES data, 69 were evaluable for response. ORRs were 50.7% (35/69) for all patients, 38.5% (10/26) for DLBCL, 37.5% (6/16) for GCB DLBCL, 46.2% (12/26) for FL, and 76.5% (13/17) for RT (Table 1). CRs were reported in five patients with DLBCL (two with GCB, two with ABC, and 1 with unclassified DLBCL), three with FL, and two with RT (Table 1).
Clinical outcome analyses by biomarker
PD-L1 expression
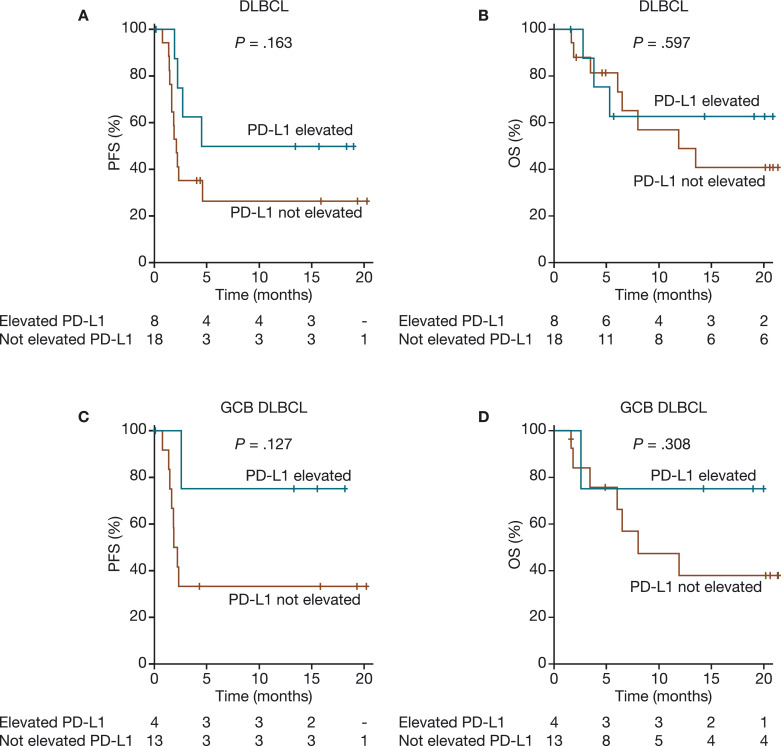
Twenty-six patients with DLBCL, 25 with FL, and 15 with RT had IHC-based PD-L1 data available for analysis. Because only 1 of 25 patients with FL had PD-L1 elevation (≥5% tumor cells by IHC), this analysis focused on patients with DLBCL and RT. Among the 26 patients with DLBCL who had IHC data, 18 patients had PD-L1 expression <5% (range 0–2%) and 8 patients had PD-L1 expression ≥5% (range 5–95%), with the mean (standard deviation [SD]) of 9.6 (21.6)% (Supplementary Figure S2). Of the 8 patients with elevated PD-L1, three had a CR and two had PRs. Of the 26 DLBCL patients with IHC data, 24 also had GEP-based subtyping calls, with six (25.0%) having elevated PD-L1; among these patients, four had GCB DLBCL (one CR, two PRs, and one SD), one had ABC DLBCL (PD), and one was unclassified (PD) (Supplementary Table S2). Based on the IHC analysis, elevated PD-L1 in DLBCL (≥5% tumor cells positive for PD-L1) was observed more frequently in responders versus nonresponders (62.5% [5/8] vs. 18.8% [3/16]; p = 0.065) and was significantly associated with CR (37.5% [3/8] vs. 0% [0/16]; p = 0.028). There was a trend toward improved PFS and OS in patients with DLBCL or GCB DLBCL who had elevated PD-L1 (Fig. 1). In RT, 3/15 (20.0%) patients with available IHC data had elevated PD-L1; all three had PRs with durable PFS and OS. At study closure, two of the three patients with elevated PD-L1 had not progressed, and all three were alive, but no significant correlation could be established because of the small number of patients.
Fig. 1.
PFS and OS by IHC-based PD-L1 expression in patients with DLBCL (A and B) and GCB DLBCL (C and D). The numbers below the X-axis depict patients at risk of progression who had elevated PD-L1 and those who did not have elevated PD-L1. DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B-cell–like; IHC, immunohistochemistry; OS, overall survival; PD-L1, programmed death ligand 1; PFS, progression-free survival.
Exome analyses
Responders versus nonresponders
Exome and response data were available for 26 patients with DLBCL (ORR 38.5%; 10 responders [5 CR, 5 PR], 16 nonresponders), 16 with GCB DLBCL (ORR 37.5%; 6 responders [2 CR, 4 PR], 10 nonresponders), 26 with FL (ORR 46.2%; 12 responders [3 CR, 9 PR], 14 nonresponders), and 17 with RT (ORR 76.5%; 13 responders [2 CR, 11 PR], 4 nonresponders).
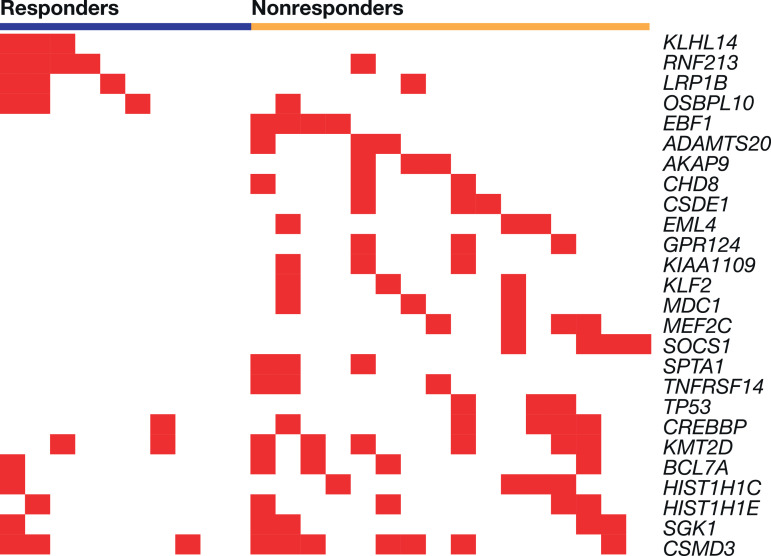
DLBCL responders versus nonresponders were more likely to have mutations in RNF213 (4/10 [40.0%] vs. 1/16 [6.2%]), KLHL14 (3/10 [30.0%] vs. 0/16), LRP1B (3/10 [30.0%] vs. 1/16 [6.2%]), and OSBPL10 (3/10 [30.0%] vs. 1/16 [6.2%]) (Table 2, Fig. 2). The difference in expression between responders and nonresponders was highest for KLHL14 (p = 0.046) Fig. 2 is a heatmap of the frequently occurring mutations from the gene set of interest occurring in patients with DLBCL by responder and nonresponder status, sorted by p values within the responder gene block (top four genes) and the nonresponder gene block (all other genes). DLBCL nonresponders commonly had variants in EBF1, ADAMTS20, and AKAP9. Mutations in the BCR pathway (TNFRSF14, NFKB1B), epigenetic modifiers (CREBBP, KMT2D), and other signaling pathways were also observed (Table 2, Fig. 3). Mutations in HIST1H1C, BCL7A, and SGK1 were reported in both responders and nonresponders but were generally more frequent in nonresponders (Table 2).
Table 2.
Frequent Differentially Mutated Genes between Responders and Nonresponders in Diffuse Large B-Cell Lymphoma (Occurring in at Least Three Patients in Either Group).
| Gene | Responders (n = 10) |
Nonresponders (n = 16) |
Odds Ratio (95% CI) | p Value |
|---|---|---|---|---|
| Mutations more frequent in responders | ||||
| KLHL14 | 3 (30.0%) | 0 (0.0%) | Inf (0.730–Inf) | 0.046 |
| RNF213 | 4 (40.0%) | 1 (6.2%) | 9.053 (0.711–522.371) | 0.055 |
| LRP1B | 3 (30.0%) | 1 (6.2%) | 5.950 (0.397–358.476) | 0.264 |
| OSBPL10 | 3 (30.0%) | 1 (6.2%) | 5.950 (0.397–358.476) | 0.264 |
| Mutations more frequent in nonresponders | ||||
| EBF1 | 0 (0.0%) | 4 (25.0%) | 0.000 (0.000–2.304) | 0.136 |
| ADAMTS20 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| AKAP9 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| CHD8 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| CSDE1 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| EML4 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| GPR124 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| KIAA1109 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| KLF2 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| MDC1 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| MEF2C | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| SOCS1 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| SPTA1 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| TNFRSF14 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| TP53 | 0 (0.0%) | 3 (18.8%) | 0.000 (0.000–3.825) | 0.262 |
| CREBBP | 1 (10.0%) | 5 (31.2%) | 0.257 (0.005–2.940) | 0.352 |
| KMT2D | 2 (20.0%) | 6 (37.5%) | 0.430 (0.034–3.353) | 0.42 |
| BCL7A | 1 (10.0%) | 4 (25.0%) | 0.346 (0.006–4.350) | 0.617 |
| HIST1H1C | 1 (10.0%) | 4 (25.0%) | 0.346 (0.006–4.350) | 0.617 |
| HIST1H1E | 1 (10.0%) | 4 (25.0%) | 0.346 (0.006–4.350) | 0.617 |
| SGK1 | 1 (10.0%) | 4 (25.0%) | 0.346 (0.006–4.350) | 0.617 |
| CSMD3 | 3 (30.0%) | 7 (43.8%) | 0.564 (0.068–3.754) | 0.683 |
CI, confidence interval.
Fig. 2.
Diffuse large B-cell lymphoma gene-level mutation heatmap showing variants from panel of interest occurring in at least three patients, by responder status. Each vertical column represents an individual patient.
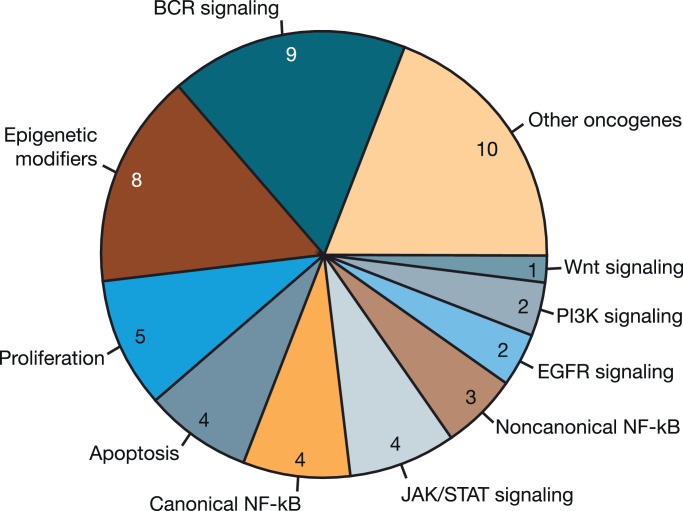
Fig. 3.
Functional groups of mutated genes in patients with diffuse large B-cell lymphoma. The pie chart shows the number of patients (N = 52) with somatic mutations in ≥1 gene representing the associated functional grouping.
Gene mutations implicated in the pathogenesis of DLBCL [27] were generally more frequent in nonresponders (Supplementary Table S2). None of the DLBCL responders had mutations in TP53, MYD88, GNA13, and TNFRSF14, while frequency of these mutations in nonresponders ranged from 12.5% to 18.8%. Of note, there were two mutations in MYD88 (both in nonresponders) and one in CARD11 (in a responder). In DLBCL, a trend toward more BCR pathway–associated mutations was observed in nonresponders, with 10% (1/10) of responders and 50% (8/16) of nonresponders having mutations associated with BCR signaling (p = 0.087).
In the GCB DLBCL subset, there were no gene variants that reached significance between responders and nonresponders (Supplementary Table S3). The most differentially expressed gene mutations were more frequent in nonresponders versus responders: CSMD3 (5/10 [50%] vs. 0/6 [0%], p = 0.093); BCL2 (6/10 [60.0%] vs. 1/6 [16.7%], p = 0.145); KMT2D (6/10 [60.0%] vs. 1/6 [16.7%], p = 0.145); CREBBP, EBF1, and SGK1 (all 4/10 [40.0%] vs. 0/6 [0%], p = 0.234).
Mutation frequencies differing for responders versus nonresponders in FL and RT, respectively, were in BCL2 (9/12 [75.0%] vs. 4/14 [28.6%]; p = 0.047) and ROS1 (0/13 vs. 2/4 [50.0%]; p = 0.044). No major differences were observed in overall TMB between responders and nonresponders with DLBCL (p = 0.215), FL (P = .899), or RT (p = 1.000); however, in GCB DLBCL, TMB was substantially lower for responders (p = 0.003) (Supplementary Figure S3A-D).
PFS >24 versus ≤24 months in DLBCL
In DLBCL, RNF213 (3/7 [42.9%]), NBPF1 (3/7 [42.9%]), KLHL14 (2/7 [28.6%]), and LRP1B (2/7 [28.6%]) variants were more often seen in patients with PFS >24 months. In patients with PFS ≤24 months, the most common variants included KMT2D (8/20 [40.0%]), CREBBP (6/20 [30.0%]), HIST1H1E (5/20 [25.0%]), EP400 (4/20 [20.0%]), and PDE4DIP (6/20 [30.0%]), all of which are involved in chromatin structure; EBF1 (4/20 [20.0%]), CD79B, TP53, ADAMTS20, AKAP9, and TNFRSF14 (all 3/20 [15.0%]) variants were also seen (Table 3). The TMB was substantially lower in DLBCL patients with PFS >24 versus ≤24 months (p = 0.0288) (Supplementary Figure S3E).
Table 3.
Frequent Differentially Mutated Genes Between Patients with PFS >24 versus ≤24 Months in Diffuse Large B-Cell Lymphoma (Occurring in at Least Three Patients Overall).
| Gene | PFS >24 Months (n = 7) |
PFS ≤24 Months (n = 20) |
Odds Ratio (95% CI) | p Value |
|---|---|---|---|---|
| Mutations more frequent in patients with PFS >24 months | ||||
| RNF213 | 3 (42.9%) | 2 (10.0%) | 6.147 (0.527–97.903) | 0.091 |
| KLHL14 | 2 (28.6%) | 1 (5.0%) | 6.889 (0.303–469.371) | 0.156 |
| LRP1B | 2 (28.6%) | 2 (10.0%) | 3.398 (0.200–58.569) | 0.269 |
| TRIO | 2 (28.6%) | 2 (10.0%) | 3.398 (0.200–58.569) | 0.269 |
| NBPF1 | 3 (42.9%) | 4 (20.0%) | 2.863 (0.298–26.744) | 0.328 |
| MEF2B | 2 (28.6%) | 3 (15.0%) | 2.190 (0.145–25.665) | 0.580 |
| SGK1 | 2 (28.6%) | 3 (15.0%) | 2.190 (0.145–25.665) | 0.580 |
| IGLL5 | 2 (28.6%) | 4 (20.0%) | 1.571 (0.111–15.591) | 0.633 |
| BCL2 | 3 (42.9%) | 6 (30.0%) | 1.712 (0.191–14.078) | 0.653 |
| Mutations more frequent in patients with PFS <24 months | ||||
| CREBBP | 0 (0.0%) | 6 (30.0%) | 0.000 (0.000–2.315) | 0.155 |
| PDE4DIP | 0 (0.0%) | 6 (30.0%) | 0.000 (0.000–2.315) | 0.155 |
| HIST1H1E | 0 (0.0%) | 5 (25.0%) | 0.000 (0.000–3.101) | 0.283 |
| KMT2D | 1 (14.3%) | 8 (40.0%) | 0.261 (0.005–2.854) | 0.363 |
| ADAMTS20 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| AKAP9 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| ANK3 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| CD79B | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| CHD8 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| CSDE1 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| EML4 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| EP400 | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| GPR124 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| HIST1H2AC | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| KIAA1109 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| KLF2 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| MAP3K1 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| MDC1 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| MEF2C | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| SF3A1 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| SLC4A5 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| SPEN | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| SPTA1 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| SPTAN1 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| TET2 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| TNFRSF14 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| TP53 | 0 (0.0%) | 3 (15.0%) | 0.000 (0.000–7.204) | 0.545 |
| BTG1 | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| DCC | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| EBF1 | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| EP400 | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| MUC17 | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| NRG1 | 0 (0.0%) | 4 (20.0%) | 0.000 (0.000–4.440) | 0.545 |
| CSMD3 | 2 (28.6%) | 8 (40.0%) | 0.611 (0.047–4.995) | 0.678 |
CI, confidence interval; PFS, progression-free survival.
Gene expression profiling analysis
Differential gene expression
Gene expression and response data were available for 66 patients. The 20 most common genes that were differentially expressed in responders versus nonresponders (all increased in responders) are listed in Table 4. LEF1 and BTLA were the most upregulated genes in responders for all patients included in this analysis (DLBCL + FL + RT). The top 20 genes upregulated and downregulated in responders with DLBCL, FL, and RT are summarized in Supplementary Tables S4, S5, and S6. In GCB DLBCL, CRTAM was a top gene upregulated (2.25-fold) in the responder group.
Table 4.
The Top 20 Genes Differentially Expressed Between Responders and Nonresponders to Ibrutinib plus Nivolumab Among All Patients with Non-Hodgkin's Lymphomaa.
| Gene | Description | Log FC | Adjusted p Value |
|---|---|---|---|
| Genes upregulated in the responder group | |||
| LEF1 | Lymphoid enhancer-binding factor 1 | 1.058 | 0.001 |
| BTLA | B and T lymphocyte associated | 1.095 | 0.001 |
| PATL2 | Protein associated with topoisomerase II homolog 2 (yeast) | 1.111 | 0.003 |
| SIDT1 | SID1 transmembrane family, member 1 | 0.872 | 0.015 |
| PYHIN1 | Pyrin and HIN domain family, member 1 | 1.261 | 0.016 |
| L3MBTL3 | l(3)mbt-like 3 (Drosophila) | 0.982 | 0.028 |
| FAM114A2 | Family with sequence similarity 114, member A2 | 0.545 | 0.028 |
| TBC1D15 | TBC1 domain family, member 15 | 0.400 | 0.028 |
| LEPROTL1 | Leptin receptor overlapping transcript-like 1 | 0.817 | 0.028 |
| SACS | Sacsin molecular chaperone | 0.905 | 0.028 |
| LMBRD1 | LMBR1 domain containing 1 | 0.808 | 0.028 |
| GCLC | Glutamate-cysteine ligase, catalytic subunit | 1.150 | 0.028 |
| APOL3 | Apolipoprotein L, 3 | 0.946 | 0.028 |
| SRBD1 | S1 RNA binding domain 1 | 0.658 | 0.028 |
| TIGIT | T cell immunoreceptor with Ig and ITIM domains | 0.562 | 0.032 |
| CCL4 | Chemokine (C—C motif) ligand 4 | 0.697 | 0.032 |
| MDM1 | Mdm1 nuclear protein | 0.725 | 0.032 |
| SUB1 | SUB1 homolog, transcriptional regulator | 0.850 | 0.032 |
| FCMR | Fc fragment of IgM receptor | 1.378 | 0.032 |
| GMFG | Glia maturation factor, gamma | 0.547 | 0.035 |
| Genes downregulated in the responder group | |||
| C5orf66 | Chromosome 5 open reading frame 66 | –0.503 | 0.053 |
| PAPPA | Pregnancy-associated plasma protein A, pappalysin 1 | –0.442 | 0.056 |
| CCDC120 | Coiled-coil domain containing 120 | –0.331 | 0.061 |
| SOX9 | SRY (sex determining region Y)-box 9 | –1.061 | 0.066 |
| STS | Steroid sulfatase (microsomal), isozyme S | –0.421 | 0.067 |
| LYPD6B | LY6/PLAUR domain containing 6B | –0.573 | 0.075 |
| ARR3 | Arrestin 3, retinal (X-arrestin) | –0.497 | 0.076 |
| LMOD1 | Leiomodin 1 (smooth muscle) | –0.328 | 0.081 |
| LOC102546294 | Uncharacterized LOC102546294 | –0.427 | 0.085 |
| BAIAP2L1 | BAI1-associated protein 2-like 1 | –0.472 | 0.085 |
| SPINK2 | Serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) | –0.545 | 0.090 |
| FGF14 | Fibroblast growth factor 14 | –0.509 | 0.092 |
| HMCN1 | Hemicentin 1 | –0.895 | 0.093 |
| ATG9B | Autophagy related 9B | –0.432 | 0.095 |
| ESRP2 | Epithelial splicing regulatory protein 2 | –0.554 | 0.099 |
| CASC15 | Cancer susceptibility candidate 15 (non-protein coding) | –0.472 | 0.103 |
| ZFHX3 | Zinc finger homeobox 3 | –0.340 | 0.103 |
| CLDN1 | Claudin 1 | –0.826 | 0.103 |
| SLC1A1 | Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | –0.430 | 0.103 |
| LARGE-AS1 | LARGE antisense RNA 1 | –0.494 | 0.103 |
Diffuse large B-cell lymphoma, follicular lymphoma, and Richter's transformation.
Pathway-level gene set enrichment analysis
Among all patients included in this analysis, pathway enrichment results were available for 41 ibrutinib plus nivolumab responders and 37 nonresponders. Among various histologies, responder and nonresponder results, respectively, were available for 8 and 19 patients with DLBCL, 10 and 13 patients with FL, and 13 and 3 patients with RT. Overall, pathways upregulated in responders to ibrutinib plus nivolumab related mostly to RNA translation/metabolism, IL-12 signaling, TCR signaling, IFN-gamma signaling, cytokine/chemokine activity, and general immune activation (Table 5). Nonresponders had enriched activity in pathways related to the extracellular matrix (ECM) processing (eg, glycoproteins, collagen, ECM organization, and general matrisome activity; Table 5). The most enriched pathways in responders and nonresponders with DLBCL, FL, and RT are summarized in Supplementary Tables S7, S8, and S9.
Table 5.
Most Enriched Pathways in Responders and Nonresponders to Ibrutinib plus Nivolumab among Patients with Non-Hodgkin's Lymphomaa.
| Pathway Name | NES | FDR pValue |
FWER pValue |
|---|---|---|---|
| Responders | |||
| Translationb | 2.98 | 0.000 | 0.000 |
| Metabolism of RNAb | 2.93 | 0.000 | 0.000 |
| SRP-dependent co-translational protein targeting to membraneb | 2.88 | 0.000 | 0.000 |
| IL12_2 pathwayc | 2.80 | 0.000 | 0.000 |
| HIV infectionb | 2.79 | 0.000 | 0.000 |
| Influenza life cycleb | 2.79 | 0.000 | 0.000 |
| Metabolism of mRNAb | 2.78 | 0.000 | 0.000 |
| Adaptive immune systemb | 2.73 | 0.000 | 0.000 |
| 3UTR-mediated translational regulationb | 2.71 | 0.000 | 0.000 |
| Host interactions of HIV factorsb | 2.71 | 0.000 | 0.000 |
| Processing of capped intron containing pre-mRNAb | 2.68 | 0.000 | 0.000 |
| CD8 TCR pathwayc | 2.68 | 0.000 | 0.000 |
| TCR signalingb | 2.67 | 0.000 | 0.000 |
| Interferon signalingb | 2.66 | 0.000 | 0.000 |
| Respiratory electron transport ATP synthesis by chemiosmotic coupling and heat production by uncoupling proteinsb | 2.66 | 0.000 | 0.000 |
| TCA cycle and respiratory electron transportb | 2.64 | 0.000 | 0.000 |
| Interferon gamma signalingb | 2.63 | 0.000 | 0.000 |
| TCR pathwayc | 2.60 | 0.000 | 0.000 |
| Signaling by the BCR | 2.59 | 0.000 | 0.000 |
| Antiviral mechanism by IFN-stimulated genesb | 2.59 | 0.000 | 0.000 |
| Nonresponders | |||
| Core matrisomed | –2.92 | 0.000 | 0.000 |
| Integrin 1 pathwayc | –2.77 | 0.000 | 0.000 |
| ECM glycoproteinsd | –2.74 | 0.000 | 0.000 |
| Cytochrome P450 arranged by substrate typeb | –2.58 | 0.000 | 0.000 |
| ECM receptor interactione | –2.56 | 0.000 | 0.000 |
| ECM organizationb | –2.48 | 0.000 | 0.000 |
| Phase 1 functionalization of compoundsb | –2.48 | 0.000 | 0.000 |
| Regulation of IGF activity by IGF-binding proteinsb | –2.48 | 0.000 | 0.000 |
| Cell-cell junction organizationb | –2.41 | 0.000 | 0.000 |
| Collagen formationb | –2.41 | 0.000 | 0.000 |
| A tetrasaccharide linker sequence is required for GAG synthesisb | –2.38 | 0.000 | 0.000 |
| ECM regulatorsd | –2.38 | 0.000 | 0.000 |
| Olfactory signaling pathwayb | –2.35 | 0.000 | 0.000 |
| Linoleic acid metabolisme | –2.32 | 0.000 | 0.002 |
| Avb3 integrin pathwayc | –2.30 | 0.000 | 0.003 |
| Collagensd | –2.29 | 0.000 | 0.003 |
| Tight junction interactionsb | –2.27 | 0.000 | 0.004 |
| Bile acid and bile salt metabolismb | –2.25 | 0.000 | 0.009 |
| Cell junction organizationb | –2.24 | 0.000 | 0.009 |
| Integrin 3 pathwayc | –2.23 | 0.000 | 0.009 |
Diffuse large B-cell lymphoma, follicular lymphoma, and Richter's transformation.
Databases:
REACTOME.
PID.
NABA.
KEGG
FDR, false discovery rate; FWER, family-wise error rate; NES, normalized enrichment score.
The pathways with greater activation in the DLBCL nonresponders were predominantly related to cell cycle, DNA replication, and RNA splicing/processing/metabolism (Supplementary Table S7). The responders with FL had enrichment of activity in pathways related to RNA splicing/processing/metabolism (Supplementary Table S8).
Discussion
Reliable disease subtype identification in patients receiving novel therapies is an important step toward personalizing treatment for patients with relapsed/refractory B-cell malignancies, who have limited options for achieving durable responses. Further, biomarker analyses can help identify patients most likely to benefit from molecularly targeted therapies. This analysis evaluated several potential biomarkers of response to combined treatment with ibrutinib and nivolumab in patients with DLBCL (subtyped using GEP and HTG methods), FL, and RT, using data from the primary LYM1002 phase I/IIa study.
Among patients who had both GEP and response data, good responses to treatment were noted in all cohorts, with the highest response rate in RT (81.3%). Patients with DLBCL had an ORR of 29.6%. Responses were more frequent in patients with the GCB subtype (6/18 [33.3%]), which is in contrast to the ORR of 5% reported previously for single-agent ibrutinib [13]. These results, with the caveat of limited sample size and no central review of GCB status, suggest that ibrutinib in combination with nivolumab may have increased efficacy in this patient population.
PD-L1 expression was investigated in various solid tumors treated with nivolumab, using the definition of PD-L1 positivity as ≥5% cell membrane staining of any intensity [34]. DLBCL tumors often express PD-L1 [35], [36], [37], [38], and analyzing expression of PD-L1 in DLBCL using biopsy samples and a 5% threshold for PD-L1 positivity reported varying percentages of PD-L1–positive DLBCL, ranging from 11% to 49% [35,37,38]. In this study, approximately 30% of patients with DLBCL (including the GCB and ABC subtypes) had elevated PD-L1 expression by IHC. Studies in large groups of patients suggest that non-GCB DLBCL is more commonly associated with PD-L1 expression, although it is observed within both GCB and ABC subtypes [35,36]. Patients with DLBCL and PD-L1 elevation showed a trend toward improved response rates and prolonged survival with ibrutinib and nivolumab, with statistical significance reached for the association between PD-L1 expression and CR (p = 0.028). These results are promising, particularly as PD-L1 positivity in tumor cells has been associated with poor outcomes (particularly OS) to rituximab, cyclophosphamide, doxorubicin vincristine, and prednisone (R-CHOP) or R-CHOP–based regimens in DLBCL [35,36]. All three patients with RT and elevated PD-L1 achieved PRs and none of them experienced PD during the course of the study. However, meaningful correlations between PD-L1 status and RT outcome were not possible due to the small sample size. Only one patient with FL had PD-L1 elevation; previous research indicates that PD-L1 expression is rare in this malignancy [39,40]. In a recent phase II study, nivolumab showed very limited activity in relapsed/refractory FL (ORR 4%), and no correlation between PD-1 or PD-L1 expression and response was noted [22]. In view of this result, the high response in FL observed in our study (43.5%) might have been driven mostly by ibrutinib, consistent with single-agent ibrutinib achieving ORR of 37.5% in a phase II study in relapsed/refractory FL [14].
Further investigation of the combination of BTK and PD-L1 inhibitor therapy is ongoing in B-cell malignancies and could help identify histologies for which this treatment strategy is likely to be most beneficial. Ongoing phase I/II studies are evaluating the safety and efficacy of ibrutinib combined with nivolumab (NCT02420912, NCT02940301) or pembrolizumab (NCT03153202, NCT03514017, NCT03204188, NCT02332980, NCT03204188) in NHL or classic Hodgkin's lymphoma.
Exome analyses were aimed at identifying gene mutations enriched in patients who responded versus those who did not respond to treatment with ibrutinib plus nivolumab. In DLBCL, the top three gene mutations more frequently mutated in responders versus nonresponders included KLHL14 (p = 0.046), RNF213 (p = 0.055), and LRP1B (p = 0.264). KLHL14 belongs to the Kelch-like family of proteins that can serve as subunits of Cullin-RING ubiquitin ligase complex [41] highly expressed in B cells, promoting B1a but suppressing B1b cell development in mice, and thus revealing a role in controlling B cell differentiation [42]. KLHL14 is frequently mutated in ABC DLBCL cells [43], and recent in vitro data have highlighted involvement of KLHL14 in BCR-dependent NF-κB activation [44]. Mutations in RNF213, the Moyamoya disease gene product and an E3 ligase, have been reported in liver cancers [45] and RNF213-ALK fusion has been described in anaplastic large cell lymphoma [46]. In HER2+ breast cancer cells, RNF213 was uncovered to be a substrate for the protein-tyrosine phosphatase PTP1B, and both proteins were required for survival of HER2+ breast cancer in the hypoxic tumor microenvironment[47] Mutations in LRP1B are frequent in melanoma and an association with response to PD-1 blockade has been reported[48].
DLBCL nonresponders commonly had mutations in EBF1, ADAMTS20, and AKAP9 genes generally involved in tumor initiation/proliferation [49], [50], [51]. It has been suggested that mutations of CARD11 (another gene implicated in NF-κB pathway activation downstream of BTK) predict lack of response to ibrutinib in DLBCL and FL [13,14]. In our analyses of patients with DLBCL, nonresponders were more likely to have mutations in genes involved in alternate BCR pathways converging on NF-κB, such as TNFRSF14, MYD88, and NFKB1B, which are among the genes implicated in the pathogenesis of DLBCL and recurrent in refractory disease [27,28]. Notably, none of the DLBCL responders had mutations in TP53, MYD88, and TNFRSF14. Other gene variants occurred frequently but were not linked to response, such as CSMD3, BCL2, and NBPF1.
As mentioned, ibrutinib plus nivolumab had an unexpectedly high antitumor activity in GCB DLBCL, emphasizing the clinical benefit of adding nivolumab to ibrutinib. However, overall TMB was lower in responders with GCB DLBCL and in DLBCL patients with PFS >24 versus ≤24 months, contrary to previous reports in non–small-cell lung carcinoma and melanoma linking higher mutational burden with greater effectiveness of immune checkpoint blockade therapy [52,53]. On the other hand, non–small-cell lung carcinoma and melanoma have a very high average mutational burden, unlike the relatively low average mutational burden in DLBCL [54], suggesting that factors other than TMB may be linked to the antitumoral immune response in DLBCL. Validation of these results in larger patient samples is warranted to fully understand their clinical and biological significance.
In FL, BCL2 mutations were more frequent in responders (75% vs. 28.6% in nonresponders). It has been proposed that BCL2 mutations in FL may be a surrogate for genomic instability triggered by activation-induced cytidine deaminase (AID) [55]. In FL, a significant association between risk and TMB (also often used as a proxy for genomic instability) has been previously demonstrated [56]. The lack of a distinct anticorrelation between TMB and response in the FL cohort of this study (compared with DLBCL) may indicate that there is some degree of benefit being derived from ibrutinib plus nivolumab therapy in genomically unstable patients with high TMB FL. This is supported by the fact that the patients with ≥30 mutated cancer-related genes in the set examined in this work (n = 1742) all had BCL2 mutations and a response rate of 75% (3/4; top two patients with the most mutations were responders), while the patients with response data who had <10 mutated genes of interest had a 0% (0/2) response rate (neither had BCL2 mutations). CREBBP and KMT2D mutations were also frequent in FL, though not significantly associated with response. Longitudinal analyses had previously identified CREBBP and KMT2D variants as early driver mutations in chromatin regulator genes [29]. The mutational spectrum in RT in this study was quite different than that observed in the other histologies. ROS1 was more frequent in RT nonresponders (2/4 vs. 0/13 in responders, p = 0.044); both ROS1 and BCL2 are involved in the NF-κB pathway.
Gene expression analyses uncovered several genes and pathways differentially expressed in responders versus nonresponders to treatment with ibrutinib plus nivolumab. The top genes upregulated in all patients who responded to ibrutinib plus nivolumab included LEF1 and BTLA. LEF1 is expressed at early stages of B-cell differentiation and is essential for cell survival and proliferation. Overexpression of LEF1 is associated with poor prognosis and disease progression in CLL [57], and its presence here may reflect the contribution of ibrutinib to the response. BTLA is a lymphocyte inhibitory receptor that is expressed on Th1 but not Th2 cells [58]. Increased BTLA expression in ibrutinib-responding patients with NHL is consistent with the finding that ibrutinib can drive Th1- versus Th2 T-cell immunity [59]. Moreover, BTLA, TIGIT, and CCL4 are associated with T-cell exhaustion and tumor response to checkpoint blockade [60,61]. Therefore, the fact that each of these genes is highly upregulated in tumor tissue of responding patients suggests that the PD-1 blockade was also a significant part of the clinical efficacy in these patients.
In GCB DLBCL, CRTAM was the most upregulated gene in the responders. CRTAM is expressed on CD8+ T-cells, especially during late-stage activation, and aids in maintaining activated T-cell populations within lymph nodes [62]. A high level of CRTAM expression likely correlates with an immunologically active phenotype, potentially providing a benefit for patients undergoing immune therapy for cancer.
Overall, pathways upregulated in responders to ibrutinib plus nivolumab were mostly related to RNA translation/metabolism, TCR, IL-12, IFN-gamma signaling, cytokine/chemokine activity, and general immune activation; similar trends are seen with DLBCL and RT alone. These results are consistent with studies reporting that PD-1 targets TCR signaling to inhibit functional T-cell activation [63], and anti-PD-1 therapy reduces this inhibition. Moreover, successful activation of T cells depends on a cross-talk between T cells and dendritic cells that involves IL-12 and IFN-gamma signaling [64]. IL-12 and IFN-gamma are also associated with Th1 differentiation [65], and we have previously shown increased secretion of these proteins in ibrutinib responders in FL [66]. Based on two separate analyses in all patients and a subgroup of patients with DLBCL, our results suggest that patients showing high activity of these immune-related pathways may be more responsive to anti-PD-1 agents in combination with ibrutinib. Pathways related to BCR signaling were also enriched in responders with FL, consistent with the mechanism of action of ibrutinib.
Among all patients, nonresponders had enriched activity in pathways related to the ECM processing. A recently published study identified a distinct set of ECM genes upregulated in cancer and correlated with poor prognosis [67]. Transcriptional program dysregulation of these genes was linked to the activation of TGF-beta signaling in cancer-associated fibroblasts and subsequent immunosuppression [67]. The pathways activated in the DLBCL nonresponders related mostly to cell cycle, DNA replication, and RNA splicing/processing/metabolism. These pathways may represent resistance mechanisms because they can drive cancer survival or progression using mechanisms that are not affected by ibrutinib or nivolumab. Interestingly, some of the pathways enriched in responders with FL appeared to be implicated in RNA metabolism or translational regulation, possibly indicating high AID activity in responders. As discussed previously, responder-associated BCL2 mutations in FL may serve as a proxy for AID-mediated genomic instability. The DNA mutator activity of AID, which can ultimately serve to increase neoantigen presentation and thereby response to checkpoint inhibition, is regulated by the RNA exosome complex, meaning that RNA processing could be related to the mechanism behind many FL-associated BCL2 mutations and genomic instability [68].
In conclusion, there was a trend toward improved survival with ibrutinib and nivolumab in patients with DLBCL with elevated PD-L1, although the differences did not reach statistical significance. We have identified several gene mutations, as well as differentially expressed genes and enriched signaling pathways, in patients with DLBCL, FL, and RT who responded versus those who did not respond to treatment with ibrutinib plus nivolumab. The preliminary analyses reported herein may highlight some of the mechanisms involved in the action of this therapeutic combination and help identify patients with B-cell malignancies who are most likely to benefit from treatment with ibrutinib combined with an anti-PD-1/PD-L1 agent, which may represent a salvage therapy for patients with relapsed/refractory NHL.
Role of the funding source
Writing assistance was provided by Ewa Wandzioch and Liqing Xiao of Parexel and funded by Janssen Global Services, LLC.
Data-sharing statement
The data in this publication will be available from the corresponding author upon request.
Author contributions (per CRediT)
Brendan P. Hodkinson: Conceptualization, Formal Analysis, Investigation, Writing – Review & Editing; Michael Schaffer: Formal Analysis, Investigation, Writing – Review & Editing; Joshua D. Brody: Conceptualization, Formal Analysis, Investigation, Writing – Review & Editing; Wojciech Jurczak: Formal Analysis, Investigation, Writing – Review & Editing; Cecilia Carpio: Investigation, Writing – Review & Editing; Dina Ben-Yehuda: Investigation, Writing – Review & Editing; Irit Avivi: Investigation, Writing – Review & Editing; Ann Forslund: Formal Analysis, Investigation, Writing – Review & Editing; Muhit Özcan: Investigation, Writing – Review & Editing; John Alvarez: Investigation, Writing – Review & Editing; Rob Ceulemans: Investigation, Writing – Review & Editing; Nele Fourneau: Formal Analysis, Investigation, Writing – Review & Editing; Sriram Balasubramanian: Conceptualization, Investigation, Formal Analysis, Writing – Original Draft, Writing – Review & Editing; Anas Younes: Conceptualization, Investigation, Formal Analysis, Writing – Original Draft, Writing – Review & Editing.
Declaration of Competing Interest
A. Younes received research funding from Novartis, Janssen, and Curtis and honoraria from Bayer, Merck, Bristol Myers Squibb, Celgene, Incyte, Janssen, Sanofi, Seattle Genetics, and Takeda Millennium. W. Jurczak has served as a consultant or in an advisory role for Janssen-Cilag, Acerta Pharma, Sandoz-Novartis, Celltrion, MEI Pharma, Roche, and Gilead Sciences and has received research funding from Janssen-Cilag, Acerta Pharma, Merck, Gilead Sciences, TG Therapeutics, Pfizer, Incyte, Bayer HealthCare Pharmaceuticals, Sandoz-Novartis, Roche, Celltrion, Takeda Pharmaceuticals, Affimed Therapeutics, and Epizyme. M. Özcan has received research funding from Archigen, AbbVie, Novartis, Bayer, Roche, MSD, Janssen, Celgene, and Takeda and honoraria and travel funding from Takeda, Bristol Myers Squibb, JAZZ, Sanofi, Abdi İbrahim, and Roche. JD Brody received research funding from Janssen, Merck, Bristol Myers Squibb, Seattle Genetics, Takeda, Kite, Acerta, Celgene, and Morphosys. A. Forslund is an employee of Bristol Myers Squibb and owns stock in the company. B. Hodkinson, M. Schaffer, N. Fourneau, J. Alvarez, R. Ceulemans, and S. Balasubramanian are employees of the Janssen Pharmaceutical Companies of Johnson & Johnson. S. Balasubramanian owns stock in Pharmacyclics/AbbVie. C. Carpio, I. Avivi, and D. Ben-Yehuda have nothing to disclose.
Acknowledgments
Acknowledgements
The authors would like to thank all the patients who participated in this study, as well as the study investigators.
Funding
Sponsored by Janssen Research & Development, LLC.
Footnotes
Congress Presentation
Part of the results was presented at the American Association for Cancer Research 2019 Meeting in Atlanta, GA, USA, March 29–April 3, 2019.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100977.
Appendix. Supplementary materials
References
- 1.Byrd J.C., Brown J.R., O'Brien S., Barrientos J.C., Kay N.E., Reddy N.M. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N. Engl. J. Med. 2014;371(3):213–223. doi: 10.1056/NEJMoa1400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Brien S., Furman R.R., Coutre S., Flinn I.W., Burger J.A., Blum K. Single-agent ibrutinib in treatment-naive and relapsed/refractory chronic lymphocytic leukemia: a 5-year experience. Blood. 2018;131(17):1910–1919. doi: 10.1182/blood-2017-10-810044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr P.M., Robak T., Owen C., Tedeschi A., Bairey O., Bartlett N.L. Sustained efficacy and detailed clinical follow-up of first-line ibrutinib treatment in older patients with chronic lymphocytic leukemia: extended phase 3 results from RESONATE-2. Haematologica. 2018;103(9):1502–1510. doi: 10.3324/haematol.2018.192328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanan-Khan A., Cramer P., Demirkan F., Fraser G., Silva R.S., Grosicki S. Ibrutinib combined with bendamustine and rituximab compared with placebo, bendamustine, and rituximab for previously treated chronic lymphocytic leukaemia or small lymphocytic lymphoma (HELIOS): a randomised, double-blind, phase 3 study. Lancet Oncol. 2016;17(2):200–211. doi: 10.1016/S1470-2045(15)00465-9. [DOI] [PubMed] [Google Scholar]
- 5.Noy A., de V.S., Thieblemont C., Martin P., Flowers C.R., Morschhauser F. Targeting Bruton tyrosine kinase with ibrutinib in relapsed/refractory marginal zone lymphoma. Blood. 2017;129(16):2224–2232. doi: 10.1182/blood-2016-10-747345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M.L., Rule S., Martin P., Goy A., Auer R., Kahl B.S. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 2013;369(6):507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M.L., Blum K.A., Martin P., Goy A., Auer R., Kahl B.S. Long-term follow-up of MCL patients treated with single-agent ibrutinib: updated safety and efficacy results. Blood. 2015;126(6):739–745. doi: 10.1182/blood-2015-03-635326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger J.A., Tedeschi A., Barr P.M., Robak T., Owen C., Ghia P. Ibrutinib as initial therapy for patients with chronic lymphocytic leukemia. N. Engl. J. Med. 2015;373(25):2425–2437. doi: 10.1056/NEJMoa1509388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treon S.P., Tripsas C.K., Meid K., Warren D., Varma G., Green R. Ibrutinib in previously treated Waldenstrom's macroglobulinemia. N. Engl. J. Med. 2015;372(15):1430–1440. doi: 10.1056/NEJMoa1501548. [DOI] [PubMed] [Google Scholar]
- 10.Janssen Biotech, Inc., Horsham, PA, USA; Pharmacyclics LLC, Sunnyvale, CA, USA: 2019. IMBRUVICAⓇ (ibrutinib) [prescribing information] [Google Scholar]
- 11.Janssen-Cilag International NV; Beerse, Belgium: 2019. IMBRUVICA (ibrutinib) [summary of Product Characteristics]https://www.ema.europa.eu/en/documents/product-information/imbruvica-epar-product-information_en.pdf [Internet]. [cited April 2, 2020]. Available from: [Google Scholar]
- 12.Fowler N.H., Nastoupil L., De Vos S., Knapp M., Flinn I.W., Chen R. The combination of ibrutinib and rituximab demonstrates activity in first-line follicular lymphoma. Br. J. Haematol. 2020;189(4):650–660. doi: 10.1111/bjh.16424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson W.H., Young R.M., Schmitz R., Yang Y., Pittaluga S., Wright G. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat. Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartlett N.L., Costello B.A., LaPlant B.R., Ansell S.M., Kuruvilla J.G., Reeder C.B. Single-agent ibrutinib in relapsed or refractory follicular lymphoma: a phase 2 consortium trial. Blood. 2018;131(2):182–190. doi: 10.1182/blood-2017-09-804641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaglowski S.M., Jones J.A., Nagar V., Flynn J.M., Andritsos L.A., Maddocks K.J. Safety and activity of BTK inhibitor ibrutinib combined with ofatumumab in chronic lymphocytic leukemia: a phase 1b/2 study. Blood. 2015;126(7):842–850. doi: 10.1182/blood-2014-12-617522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galaznik A., Huelin R., Stokes M., Guo Y., Hoog M., Bhagnani T. Systematic review of therapy used in relapsed or refractory diffuse large B-cell lymphoma and follicular lymphoma. Future Sci OA. 2018;4(7):FSO322. doi: 10.4155/fsoa-2018-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi M., Jiao D., Xu H., Liu Q., Zhao W., Han X. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu-Monette Z.Y., Zhou J., Young K.H. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood. 2018;131(1):68–83. doi: 10.1182/blood-2017-07-740993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Younes A., Santoro A., Shipp M., Zinzani P.L., Timmerman J.M., Ansell S. Nivolumab for classical Hodgkin's lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016;17(9):1283–1294. doi: 10.1016/S1470-2045(16)30167-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasamon Y.L., de Claro R.A., Wang Y., Shen Y.L., Farrell A.T., Pazdur R. FDA approval summary: nivolumab for the treatment of relapsed or progressive classical Hodgkin lymphoma. Oncologist. 2017;22(5):585–591. doi: 10.1634/theoncologist.2017-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesokhin A.M., Ansell S.M., Armand P., Scott E.C., Halwani A., Gutierrez M. Nivolumab in patients with relapsed or refractory hematologic malignancy: preliminary results of a phase Ib study. J. Clin. Oncol. 2016;34(23):2698–2704. doi: 10.1200/JCO.2015.65.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armand P., Janssens A.M., Gritti G., Radford J., Timmerman J.M., Pinto A. Efficacy and safety results from CheckMate 140, a phase 2 study of nivolumab for relapsed/refractory follicular lymphoma. Blood. 2020 doi: 10.1182/blood.2019004753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain N., Ferrajoli A., Basu U., P.A. T., Burger J.A. A phase II trial of nivolumab combined with ibrutinib for patients with Richter transformation. Blood. 2018;32(Supplement 1):296. [Google Scholar]
- 24.Younes A., Brody J., Carpio C., Lopez-Guillermo A., Ben-Yehuda D., Ferhanoglu B. Safety and activity of ibrutinib in combination with nivolumab in patients with relapsed non-Hodgkin lymphoma or chronic lymphocytic leukaemia: a phase 1/2a study. Lancet Haematol. 2019;6(2):e67–e78. doi: 10.1016/S2352-3026(18)30217-5. [DOI] [PubMed] [Google Scholar]
- 25.Jain P., O'Brien S. Richter's transformation in chronic lymphocytic leukemia. Oncology. 2012;26(12):1146–1152. [PubMed] [Google Scholar]
- 26.Tsang M., Shanafelt T.D., Call T.G., Ding W., Chanan-Khan A., Leis J.F. The efficacy of ibrutinib in the treatment of Richter syndrome. Blood. 2015;125(10):1676–1678. doi: 10.1182/blood-2014-12-610782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohr J.G., Stojanov P., Lawrence M.S., Auclair D., Chapuy B., Sougnez C. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U. S. A. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park H.Y., Lee S.B., Yoo H.Y., Kim S.J., Kim W.S., Kim J.I. Whole-exome and transcriptome sequencing of refractory diffuse large B-cell lymphoma. Oncotarget. 2016;7(52):86433–86445. doi: 10.18632/oncotarget.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okosun J., Bodor C., Wang J., Araf S., Yang C.Y., Pan C. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chigrinova E., Rinaldi A., Kwee I., Rossi D., Rancoita P.M., Strefford J.C. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122(15):2673–2682. doi: 10.1182/blood-2013-03-489518. [DOI] [PubMed] [Google Scholar]
- 31.Wright G., Tan B., Rosenwald A., Hurt E.H., Wiestner A., Staudt L.M. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2003;100(17):9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheson B.D., Fisher R.I., Barrington S.F., Cavalli F., Schwartz L.H., Zucca E. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J. Clin. Oncol. 2014;32(27):3059–3068. doi: 10.1200/JCO.2013.54.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz R., Wright G.W., Huang D.W., Johnson C.A., Phelan J.D., Wang J.Q. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novotny J.F., Jr., Cogswell J., Inzunza H., Harbison C., Horak C., Averbuch S. Establishing a complementary diagnostic for anti-PD-1 immune checkpoint inhibitor therapy. Ann. Oncol. 2016;27(10):1966–1969. doi: 10.1093/annonc/mdw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L.Y., Xu X.L., Rao H.L., Chen J., Lai R.C., Huang H.Q. Expression and clinical value of programmed cell death-ligand 1 (PD-L1) in diffuse large B cell lymphoma: a retrospective study. Chin. J. Cancer. 2017;36(1):94. doi: 10.1186/s40880-017-0262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiyasu J., Miyoshi H., Hirata A., Arakawa F., Ichikawa A., Niino D. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood. 2015;126(19):2193–2201. doi: 10.1182/blood-2015-02-629600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andorsky D.J., Yamada R.E., Said J., Pinkus G.S., Betting D.J., Timmerman J.M. Programmed death ligand 1 is expressed by non-Hodgkin lymphomas and inhibits the activity of tumor-associated T cells. Clin. Cancer Res. 2011;17(13):4232–4244. doi: 10.1158/1078-0432.CCR-10-2660. [DOI] [PubMed] [Google Scholar]
- 38.Chen B.J., Chapuy B., Ouyang J., Sun H.H., Roemer M.G., Xu M.L. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin. Cancer Res. 2013;19(13):3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vranic S., Ghosh N., Kimbrough J., Bilalovic N., Bender R., Arguello D. PD-L1 status in refractory lymphomas. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panjwani P.K., Charu V., DeLisser M., Molina-Kirsch H., Natkunam Y., Zhao S. Programmed death-1 ligands PD-L1 and PD-L2 show distinctive and restricted patterns of expression in lymphoma subtypes. Hum. Pathol. 2018;71:91–99. doi: 10.1016/j.humpath.2017.10.029. [DOI] [PubMed] [Google Scholar]
- 41.Shi X., Xiang S., Cao J., Zhu H., Yang B., He Q. Kelch-like proteins: physiological functions and relationships with diseases. Pharmacol. Res. 2019;148 doi: 10.1016/j.phrs.2019.104404. [DOI] [PubMed] [Google Scholar]
- 42.Li S., Liu J., Min Q., Ikawa T., Yasuda S., Yang Y. Kelch-like protein 14 promotes B-1a but suppresses B-1b cell development. Int. Immunol. 2018;30(7):311–318. doi: 10.1093/intimm/dxy033. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J., Grubor V., Love C.L., Banerjee A., Richards K.L., Mieczkowski P.A. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110(4):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J., Phelan J.D., Wright G.W., Haupl B., Huang D.W., Shaffer A.L., 3rd Regulation of B cell receptor-dependent NF-kappaB signaling by the tumor suppressor KLHL14. Proc. Natl. Acad. Sci. U. S. A. 2020;117(11):6092–6102. doi: 10.1073/pnas.1921187117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Xu W., Kang W., Wong S.H., Wang M., Zhou Y. Genomic analysis of liver cancer unveils novel driver genes and distinct prognostic features. Theranostics. 2018;8(6):1740–1751. doi: 10.7150/thno.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Krogt J.A., Bempt M.V., Ferreiro J.F., Mentens N., Jacobs K., Pluys U. Anaplastic lymphoma kinase-positive anaplastic large cell lymphoma with the variant RNF213-, ATIC- and TPM3-ALK fusions is characterized by copy number gain of the rearranged ALK gene. Haematologica. 2017;102(9):1605–1616. doi: 10.3324/haematol.2016.146571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banh R.S., Iorio C., Marcotte R., Xu Y., Cojocari D., Rahman A.A. PTP1B controls non-mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nat. Cell Biol. 2016;18(7):803–813. doi: 10.1038/ncb3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson D.B., Frampton G.M., Rioth M.J., Yusko E., Xu Y., Guo X. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol. Res. 2016;4(11):959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S., Rao N., Ge R. Emerging roles of ADAMTSs in angiogenesis and cancer. Cancers. 2012;4(4):1252–1299. doi: 10.3390/cancers4041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol. Cancer Res. 2009;7(12):1893–1901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo Y.S., Kim M.S., Yoo N.J., Lee S.H. Frameshift mutations of AKAP9 gene in gastric and colorectal cancers with high microsatellite instability. Pathol. Oncol. Res. 2016;22(3):587–592. doi: 10.1007/s12253-016-0042-0. [DOI] [PubMed] [Google Scholar]
- 52.Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934-49 e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zarogoulidis P., Papadopoulos V., Maragouli E., Papatsibas G., Sardeli C., Man Y.G. Nivolumab as first-line treatment in non-small cell lung cancer patients-key factors: tumor mutation burden and PD-L1 ≥50. Transl. Lung Cancer Res. 2018;7(Suppl 1):S28–S30. doi: 10.21037/tlcr.2018.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Correia C., Schneider P.A., Dai H., Dogan A., Maurer M.J., Church A.K. BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood. 2015;125(4):658–667. doi: 10.1182/blood-2014-04-571786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukamoto T., Nakano M., Sato R., Adachi H., Kiyota M., Kawata E. High-risk follicular lymphomas harbour more somatic mutations including those in the AID-motif. Sci. Rep. 2017;7(1):14039. doi: 10.1038/s41598-017-14150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erdfelder F., Hertweck M., Filipovich A., Uhrmacher S., Kreuzer K.A. High lymphoid enhancer-binding factor-1 expression is associated with disease progression and poor prognosis in chronic lymphocytic leukemia. Hematol Rep. 2010;2(1):e3. doi: 10.4081/hr.2010.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe N., Gavrieli M., Sedy J.R., Yang J., Fallarino F., Loftin S.K. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat. Immunol. 2003;4(7):670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- 59.Dubovsky J.A., Beckwith K.A., Natarajan G., Woyach J.A., Jaglowski S., Zhong Y. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122(15):2539–2549. doi: 10.1182/blood-2013-06-507947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji R.R., Chasalow S.D., Wang L., Hamid O., Schmidt H., Cogswell J. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol. Immunother. 2012;61(7):1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X., Harden K., Gonzalez L.C., Francesco M., Chiang E., Irving B. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009;10(1):48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 62.Takeuchi A., Itoh Y., Takumi A., Ishihara C., Arase N., Yokosuka T. CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J. Immunol. 2009;183(7):4220–4228. doi: 10.4049/jimmunol.0901248. [DOI] [PubMed] [Google Scholar]
- 63.Mizuno R., Sugiura D., Shimizu K., Maruhashi T., Watada M., Okazaki I.M. PD-1 primarily targets TCR signal in the inhibition of functional T cell activation. Front. Immunol. 2019;10:630. doi: 10.3389/fimmu.2019.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garris C.S., Arlauckas S.P., Kohler R.H., Trefny M.P., Garren S., Piot C. Successful anti-PD-1 Cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-gamma and IL-12. Immunity. 2018;49(6):1148-61 e7. doi: 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Athie-Morales V., Smits H.H., Cantrell D.A., Hilkens C.M. Sustained IL-12 signaling is required for Th1 development. J. Immunol. 2004;172(1):61–69. doi: 10.4049/jimmunol.172.1.61. [DOI] [PubMed] [Google Scholar]
- 66.Gopal A.K., Schuster S.J., Fowler N.H., Trotman J., Hess G., Hou J.Z. Ibrutinib as treatment for patients with relapsed/refractory follicular lymphoma: results from the open-label, multicenter, phase II DAWN study. J. Clin. Oncol. 2018;36(23):2405–2412. doi: 10.1200/JCO.2017.76.8853. [DOI] [PubMed] [Google Scholar]
- 67.Chakravarthy A., Khan L., Bensler N.P., Bose P., De Carvalho D.D. TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun. 2018;9(1):4692. doi: 10.1038/s41467-018-06654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laffleur B., Basu U., Lim J. RNA exosome and non-coding RNA-coupled mechanisms in AID-mediated genomic alterations. J. Mol. Biol. 2017;429(21):3230–3241. doi: 10.1016/j.jmb.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.