Abstract
During cochlear development, the Notch ligand JAGGED 1 (JAG1) plays an important role in the specification of the prosensory region, which gives rise to sound-sensing hair cells and neighboring supporting cells (SCs). While JAG1's expression is maintained in SCs through adulthood, the function of JAG1 in SC development is unknown. Here, we demonstrate that JAG1 is essential for the formation and maintenance of Hensen's cells, a highly specialized SC subtype located at the edge of the auditory epithelium. Using Sox2CreERT2/+::Jag1loxP/loxP mice of both genders, we show that Jag1 deletion at the onset of differentiation, at embryonic day 14.5, disrupted Hensen's cell formation. Similar loss of Hensen's cells was observed when Jag1 was deleted after Hensen's cell formation at postnatal day (P) 0/P1 and fate-mapping analysis revealed that in the absence of Jag1, some Hensen's cells die, but others convert into neighboring Claudius cells. In support of a role for JAG1 in cell survival, genes involved in mitochondrial function and protein synthesis were downregulated in the sensory epithelium of P0 cochlea lacking Jag1. Finally, using Fgfr3-iCreERT2::Jag1loxP/loxP mice to delete Jag1 at P0, we observed a similar loss of Hensen's cells and found that adult Jag1 mutant mice have hearing deficits at the low-frequency range.
SIGNIFICANCE STATEMENT Hensen's cells play an essential role in the development and homeostasis of the cochlea. Defects in the biophysical or functional properties of Hensen's cells have been linked to auditory dysfunction and hearing loss. Despite their importance, surprisingly little is known about the molecular mechanisms that guide their development. Morphologic and fate-mapping analyses in our study revealed that, in the absence of the Notch ligand JAGGED1, Hensen's cells died or converted into Claudius cells, which are specialized epithelium-like cells outside the sensory epithelium. Confirming a link between JAGGED1 and cell survival, transcriptional profiling showed that JAGGED1 maintains genes critical for mitochondrial function and tissue homeostasis. Finally, auditory phenotyping revealed that JAGGED1's function in supporting cells is necessary for low-frequency hearing.
Keywords: Claudius cells, cochlea development, Hensen's cells, Jagged1, Notch, supporting cells
Introduction
Canonical Notch signaling, which plays essential roles in the development of sensory structures in the vertebrate inner ear is initiated by the binding of a DSL (Delta/Serrate/LAG-2)-type membrane-bound ligand to the transmembrane Notch receptor located on neighboring cells. Ligand binding leads to the release of the intercellular domain of the Notch receptor, which as part of the RBPJ-Mastermind transcription complex, activates transcription of downstream target genes. Mammals have four Notch receptors (Notch1-4) and five DSL-type ligands [Delta-like (Dll) 1, 3, and 4 and Jagged (Jag) 1 and 2] (Kopan and Ilagan, 2009).
In the inner ear, canonical Notch signaling is best known for its roles in lateral inhibition and lateral induction (Eddison et al., 2000; Daudet and Żak, 2020). Notch-mediated lateral induction, a positive feedback mechanism in which the signaling and signal-receiving cells acquire the same cell fate, first defines and maintains the prosensory domains that give rise to specialized sensory epithelia composed of hair cells (HCs) and surrounding supporting cells (SCs) (Daudet and Lewis, 2005; Hartman et al., 2010; Pan et al., 2010; Neves et al., 2011). The Notch ligand JAGGED1 (JAG1) and the downstream Notch effector RBPJ are essential for the early role of Notch signaling in prosensory development. Early loss of JAG1 or RBPJ abolishes, or greatly reduces, the pool of inner ear prosensory progenitors (Brooker et al., 2006; Kiernan et al., 2006; Basch et al., 2011; Yamamoto et al., 2011; Petrovic et al., 2014).
As prosensory progenitor cells differentiate into HCs and SCs during late embryogenesis, Notch signaling plays a key role in limiting the number of HCs that form in a process termed lateral inhibition. Activated by ATOH1, a transcriptional activator and master regulator of HC formation (Bermingham et al., 1999; Chen et al., 2002; Woods et al., 2004), nascent HCs express the Notch ligands Dll1, Dll3, and Jag2 (Adam et al., 1998; Lanford et al., 1999; Morrison et al., 1999; Hartman et al., 2007). Subsequent DLL1/JAG2-mediated activation of Notch1 receptor signaling in adjacent prosensory progenitor cells limits these cells to an SC fate (Lanford et al., 1999; Kiernan et al., 2005; Brooker et al., 2006).
Important effectors of this Notch-mediated HC-repressive function are members of the HES/HEY family. HES/HEY proteins are transcriptional repressors, which antagonize HC formation by repressing Atoh1 expression and ATOH1 activity (Zheng and Gao, 2000; Zine et al., 2001; S. Li et al., 2008; Doetzlhofer et al., 2009; Tateya et al., 2011).
We recently uncovered that canonical Notch signaling, in addition to its role in HC fate repression, is required for the differentiation and survival of cochlear SCs (Campbell et al., 2016). However, the Notch ligand(s) and receptor(s) involved in this process are unknown. A potential candidate for this function is the Notch ligand JAG1. JAG1, which initially is expressed by cochlear and vestibular prosensory progenitors, continues to be highly expressed in SCs (Morrison et al., 1999). While other Notch ligands are downregulated during the first postnatal week, as cochlear HCs and SCs mature, SC-specific JAG1 expression continues throughout adulthood where its function is unknown (Murata et al., 2006; Hartman et al., 2007; Oesterle et al., 2008; Maass et al., 2015).
In the present study, we investigated the role of JAG1 in differentiating and maturing SCs. Our analysis revealed that deletion of Jag1 at the onset of cochlear differentiation alters the patterning and cellular morphology of SCs and leads to a downregulation of genes involved in mitochondrial function and protein synthesis within the sensory epithelium. Furthermore, using morphologic analyses and fate-mapping, we show that Jag1 deletion before and after cochlear differentiation resulted in loss of a specialized SC subtype called Hensen's cells. Functional analysis of adult Jag1 mutant animals after Jag1 deletion at the perinatal stage revealed mild hearing deficits at low frequencies. Together, our results suggest that Jag1-mediated Notch signaling in cochlear SCs is critical for the formation, maintenance, and survival of Hensen's cells.
Materials and Methods
Animals
Sox2CreERT2/+ mice (Arnold et al., 2011) (stock #17593; RRID:IMSR_JAX:017593) and Rosa26loxP-stop-loxP-tdTomato (Rosa26tdTomato) mice (Madisen et al., 2010) (stock #7914; RRID:IMSR_JAX:007914) were purchased from The Jackson Laboratory. Fgfr3-iCreERT2 mice (Rivers et al., 2008; Young et al., 2010) were provided by William Richardson (University College, London) and are now available from The Jackson Laboratory (stock #25809; RRID:IMSR_JAX:025809). Two different Jag1loxP/loxP (Jag1fx/fx) mouse lines were used in this study. For Jag1 deletion using Sox2CreERT2/+ mice, Jag1fx/fx mice (Brooker et al., 2006) (RRID:MGI:3623344) were provided by Julian Lewis (Cancer Research UK London Research Institute). In this line, loxP sites were inserted to flank exons 4 and 5, resulting in a frame shift that generates a stop codon after the start of exon 6 following Cre-mediated recombination. For Jag1 deletion using Fgfr3-iCreERT2 mice and fate-mapping with Sox2CreERT2/+ mice, Jag1fx/fx mice made by Thomas Gridley's laboratory (Kiernan et al., 2006) (MGI: 3692444; stock# 10618; RRID:IMSR_JAX:010618) were purchased from The Jackson Laboratory. In this line, loxP sites were inserted to flank exon 4, resulting in a nonfunctional JAG1 protein after Cre-mediated recombination. Genotyping was performed in house using PCR as previously described for each line or by Transnetyx. All animal work was performed in accordance with approved animal protocols from the Institutional Animal Care and Use Committees at Johns Hopkins University or Southern Illinois University School of Medicine.
Substances given to animals
Sox2CreERT2/+::Jag1fx/fx mice: Pregnant dams time-mated to embryonic day (E) 14.5, received two intraperitoneal injections of tamoxifen (0.125 mg/g body weight, catalog #T5648, Sigma Millipore) and progesterone (0.125 mg/g body weight, catalog #P3972 Sigma Millipore), given 8 h apart. Neonatal pups were injected with (Z)-4-hydroxytamoxifen (0.139 mg per pup, catalog #H7904-5, Sigma Millipore) at postnatal day (P) 0 and P1. Controls included Cre-negative littermates, as well as Sox2CreERT2/+::Jag1fx/fx and Sox2CreERT2/+ mice that did not receive tamoxifen to control for Sox2 haploinsufficiency. Fgfr3-iCreERT2::Jag1fx/fx mice and Sox2CreERT2/+:: Jag1fx/fx::Rosa26tdTomato mice were injected with tamoxifen (0.075 mg/g body weight, catalog #T5648, Sigma Millipore) at P0 and P1, or at P6 and P7. Controls were iCre-negative littermates or Sox2CreERT2/+::Rosa26tdTomato littermates that also received tamoxifen.
Tissue processing
Late embryonic and early postnatal animals were staged using the EMAP eMouse Atlas Project (http://www.emouseatlas.org) Theiler staging criteria. Temporal bones were collected and fixed in 4% PFA (Polysciences) in 10 mm PBS for 2 h at room temperature. Cochleae were then dissected using the whole-mount method as previously described (Montgomery and Cox, 2016). For cryo-sectioning, PFA-fixed inner ears were put through a sucrose gradient (10% sucrose for 30 min, 15% sucrose for 30 min, and 30% sucrose overnight), submerged in OCT (Tissue-Tek, Sakura Finetek), and flash frozen. Sections cut at 14 µm thickness were collected on SuperFrost Plus slides (Thermo Fisher Scientific).
Immunostaining
Cochlear samples were immunostained following standard procedures (Montgomery and Cox, 2016). Briefly, tissue was blocked and permeabilized in 10 mm PBS containing 10% normal horse serum or NGS (Vector Labs), 1% BSA (Thermo Fisher Scientific), and 1% Triton-X-100 (Sigma Millipore) for 1 h at room temperature. Then samples were incubated with primary antibodies diluted in 10 mm PBS containing normal horse serum or NGS (5%), BSA (1%), and Triton-X-100 (0.1%) overnight at 4°C or 37°C. The following primary antibodies were used: rabbit anti-fatty acid binding protein 7 (FABP7) (1:200, catalog #ab32423, Abcam; RRID:AB_880078), rat anti-CD44 (1:500, catalog #550538, BD Biosciences; RRID:AB_393732), rabbit anti-cleaved caspase3 (1:1000, catalog #9661, Cell Signaling Technology; RRID:AB_2341188), goat anti-JAG1 (1:500, catalog #sc-6011, Santa Cruz Biotechnology; RRID:AB_649689), rabbit anti-myosin VIIa (1:200, catalog #25-6790, Proteus BioSciences; RRID:AB_10015251), mouse anti-parvalbumin (1:1000, catalog #P3088, Sigma Millipore; RRID:AB_477329), rabbit anti-S100a1 (1:400, catalog #ab868, Abcam; RRID:AB_306716), and goat anti-SOX2 (1:400, catalog #sc-17 320, Santa Cruz Biotechnology; RRID:AB_2286684). The next day, samples were incubated in corresponding AlexaFluor-conjugated secondary antibodies (1:1000, Invitrogen) for 2 h at room temperature. Nuclei were stained with Hoechst 33342 (1:2000, catalog #H1399, Thermo Fisher Scientific) for 20 min at room temperature, followed by mounting on slides in Prolong Gold (catalog #P36930, Thermo Fisher Scientific). To increase the signal-to-noise ratio, some samples stained with anti-SOX2 antibodies were pretreated with signal enhancer (catalog #136933, Invitrogen) for 30 min at room temperature before the blocking and permeabilization step. Samples were imaged using an LSM 800 confocal microscope and processed with Zen Blue and ZEN 2.3 SP1 software (Carl Zeiss).
Cell counts
Confocal z stacks through the HC layer and corresponding SC layer were taken at two representative regions in each turn of the cochlear (apex, middle, and base). Labeled inner HCs (IHCs), outer HCs (OHCs), and different SC subtypes were quantified in two 150 or 200 µm representative regions in each turn of the cochlea. The length of the imaged segment (150 or 200 µm) was analyzed using ImageJ (http://imagej.nih.gov/ij) or Zen Blue software. HCs and SCs were manually counted in Photoshop CS5 (Adobe) or in Zen Blue software.
Validation of Jag1 deletion
Immunostaining with anti-JAG1 antibodies was used to validate deletion of Jag1 from SCs using the Jag1fx/fx allele that was provided by Julian Lewis (Brooker et al., 2006). However, the Jag1fx/fx allele purchased from The Jackson Laboratory produces a truncated protein that was still detected by anti-JAG1 antibodies. Therefore, we used RT-PCR to validate the deletion of Jag1 from SCs in these mice. Total RNA was extracted from P7 Jag1fx/fx and Fgfr3-iCreERT2::Jag1fx/fx cochleae using tri-reagent (catalog #T9424, Sigma Millipore). RNA was then precipitated with isopropanol (catalog #327930010, Acros Organics) overnight at −20°C, purified with DNase treatment (catalog #AM1906, Thermo Fisher Scientific), and reprecipitated with sodium acetate (catalog #BP333, Thermo Fisher Scientific). Reverse transcription into cDNA was performed using Thermo Scientific Maxima First Strand cDNA synthesis kit (catalog #K1641, Thermo Fisher Scientific). cDNA was amplified with primers that flanked exon 4 of Jag1 (forward: CGACCGTAATCGCATCGTAC, reverse: AGTCCCACAGTAATTCAGATC) (Kiernan et al., 2006), separated on a 1% agarose gel, and visualized with GelRed nucleic acid stain (catalog #41001, Biotium) using a Syngene G:Box Chemi imager (Syngene).
Microarray and qRT-PCR experiments
For microarray and qPCR validation experiments, control (Jag1fx/fx) and experimental animals (Sox2CreERT2/+::Jag1fx/fx) were harvested at P0 after receiving tamoxifen and progesterone at stage E14.5. Cochlear epithelia from individual animals were isolated using dispase/collagenase digest as previously described (Doetzlhofer et al., 2009). Total RNA was extracted using the RNeasy Micro Kit (catalog #74004, QIAGEN). For the microarray experiment, total RNA was labeled with the Affymetrix GeneChip WT PLUS Reagent Kit using the manufacturer's protocol. Labeled RNA was hybridized onto Clariom D mouse microarray (Affymetrix), and chips were scanned and analyzed according to the manufacturer's manuals. Affymetrix CEL files were extracted and data normalized with the Partek GS 6.6 platform (Partek). Partek's extended meta-probe set was used with RMA normalization to create quantile-normalized log2 transcript signal values, which were used in subsequent ANOVAs. The microarray data were deposited in the Gene Expression Omnibus database, accession number GSE148009. Results were validated using qPCR experiments where mRNA was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (catalog #1708891, Bio-Rad). SYBR Green-based qPCR was performed on a CFX-Connect Real Time PCR Detection System (Bio-Rad) using Fast SYBR Green Master Mix reagent (catalog #4385616, Applied Biosystems) and gene-specific primers. Relative gene expression was analyzed using the comparative CT method (Schmittgen and Livak, 2008). The ribosomal gene Rpl19 was used as a reference gene and early postnatal cochlear tissue from WT mice was used as a calibrator. The sequences of qPCR primers used are listed in Table 1.
Table 1.
Primers used for the qPCR validation of microarray data
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Apod | GGCTTCAACGGTGTTTGTTT | GAGGGTGGGATGTTCTCTGA |
| Gpx1 | GTCCACCGTGTATGCCTTCT | CTCCTGGTGTCCGAACTGAT |
| Hey1 | CACTGCAGGAGGGAAAGGTTAT | CCCCAAACTCCGATAGTCCAT |
| Fgfr3 | GGGCTCCTTATTGGACTCG | GCTCCCCTCGGAATTCTTT |
| Igfbp3 | AACCTGCTCCAGGAAACATCAGT | GCTTTCCACACTCCCAGCAT |
| Jag1 | TGTGCAAACATCACTTTCACCTTT | GCAAATGTGTTCGGTGGTAAGAC |
| Mgst3 | TGAGAACGGGCATATGTTCA | GCTTGCTAGGGTCTCCTGTG |
| Rab3b | TTCCGCTATGCTGATGACAC | CCACGGTAGTAGGCTGTGGT |
| Rpl19 | GGTCTGGTTGGATCCCAATG | CCCGGGAATGGACAGTCA |
| S100a1 | TGGATGTCCAGAAGGATGCA | CCGTTTTCATCCAGTTCCTTCA |
| Stox1 | TGCACGCAGAGGATAAGACA | CACTGTGACTCAAAGCCACC |
| Timm13 | GCCATTATGGAGCAGGTGAA | C GCTGCAGTCGAGAGTTGTA |
| Trpa1 | AACACGGCTTTGATGTCCAC | GGGCTGGCTTTCTTGTGATT |
Gene ontology analysis
Gene ontology (GO) enrichment analysis was conducted using the Functional Annotation Tool in Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al., 2009). A curated list of downregulated genes (p ≤ 0.05, log2 (FC) ≤ −2σ, mean log2 Ctrl ≥ 5) was uploaded into the DAVID bioinformatics suite 6.8 and GO terms related to biological process (GOTERM BP DIRECT), molecular function (GOTERM MF DIRECT), cellular compartment (GOTERM CC DIRECT), and KEGG pathway term (KEGG PATHWAY) were obtained using default parameters.
Auditory brainstem response (ABR) measurements
To assess hearing after Jag1 deletion, ABR was measured at P30 in Fgfr3-iCreERT2::Jag1fx/fx mice and iCre-negative littermates injected with tamoxifen at P0/P1. Before ABR recordings, mice were anesthetized with Avertin (250-500 mg/kg, i.p., Sigma Millipore), and kept on a heating pad in a sound-attenuated booth (Industrial Acoustic) throughout the whole procedure. One subdermal stainless-steel recording electrode was inserted at the vertex of the skull, with a second reference electrode inserted under the pinna of the left ear. A ground electrode was inserted at the base of the tail. An electrostatic speaker (EC1, Tucker Davis Technologies System III) was fitted to a tube and inserted into the left ear canal. Acoustic signals were generated using a 16-bit D/A converter (RX6, Tucker Davis Technologies System III), controlled by a customized Auditory Neurophysiology Experiment Control Software (ANECS, Blue Hills Scientific). To generate ABRs, pure tone bursts at 4, 8, 12, 16, and 22 kHz were presented at a rate of 19/s, with a 5 ms duration, and repeated 512 times. Sound intensity at each frequency decreased from 80 to 5 dB, with 5 dB steps. Two blinded reviewers determined the ABR thresholds, which were the lowest sound intensity that produced a visually distinct response in wave I or II.
Experimental design and statistical analysis
The sample size (n) represents the number of animals analyzed per group and is reported in the figure legends for each experiment. Mice of both genders were used in all studies. Animals (biological replicates) were allocated into control or experimental groups based on genotype and/or type of treatment. To avoid bias, masking was used during data analysis. All comparisons were made using a two-way ANOVA followed by a Bonferroni post hoc test, except for data in Figure 2. Here, the determination of genes that were upregulated or downregulated after Jag1 deletion was made using a one-way ANOVA, and the qPCR validation of gene changes was analyzed using a two-tailed Student's t test. p values ≤ 0.05 were considered significant. Data were analyzed using GraphPad Prism 6.02 and 8.0 (GraphPad Software).
Figure 2.
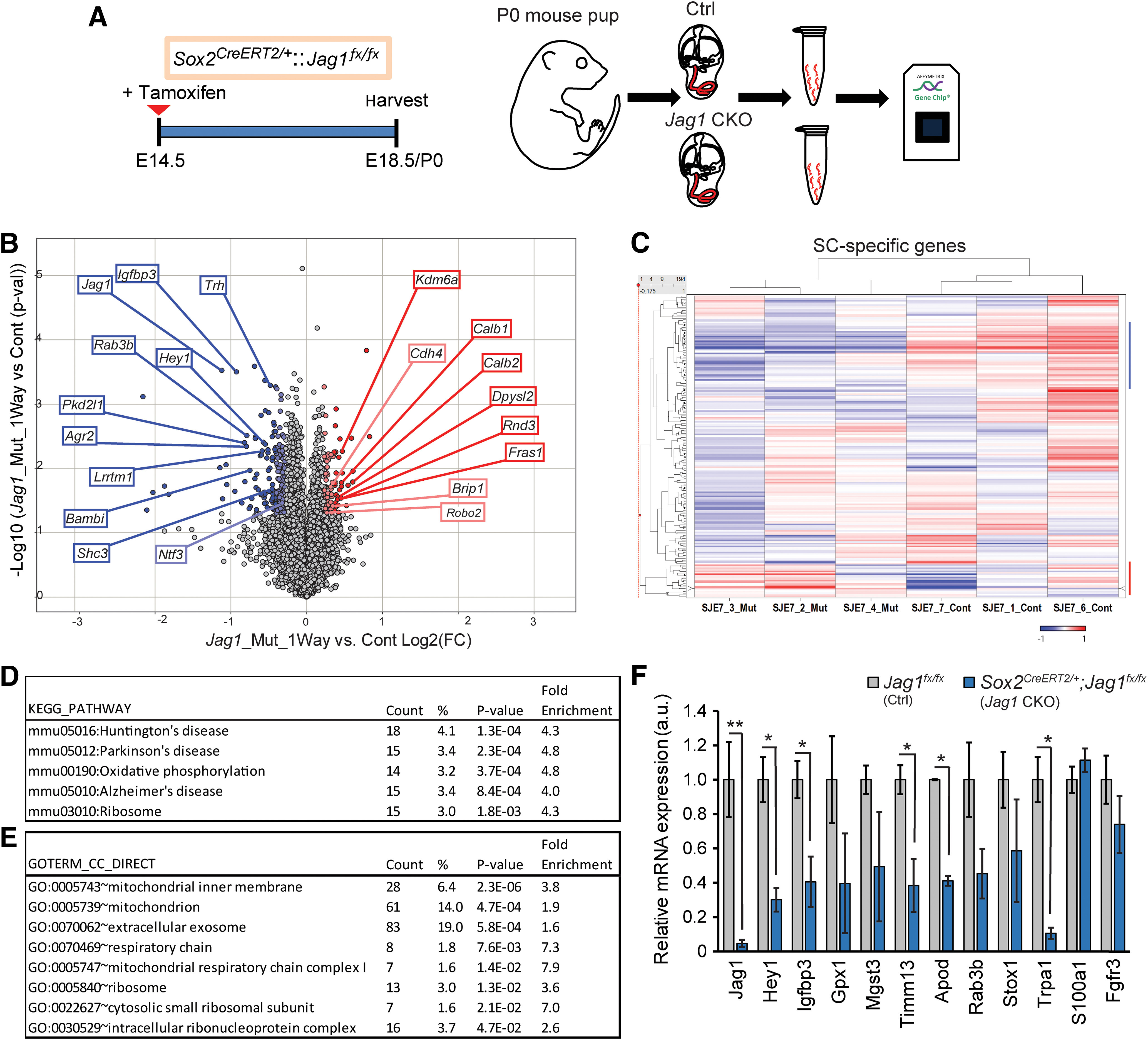
Loss of JAG1 leads to the downregulation of genes involved in mitochondrial function and protein synthesis. A, Experimental strategy. Timed pregnant dams were injected with tamoxifen and progesterone at E14.5, and gene expression in cochlear epithelia from E18.5/P0 Jag1 CKO, (Sox2CreERT2/+::Jag1fx/fx) mice, and control (Ctrl, Jag1fx/fx) littermates was analyzed using Clariom D mouse arrays. B, Volcano plot presents the one-way ANOVA of differential gene expression in Jag1 CKO versus control samples (n = 3 animals/group). Plotted as log2 fold-change (FC) (x axis) versus −log10 p value (y axis). Dark blue (log2 (FC) ≤ −6σ) and light blue (log2 (FC) ≤ −3σ) dots represent downregulated genes. Dark red (log2 (FC) ≥ 6σ) and light red dots (log2 (FC) ≥ 3σ) represent upregulated genes. C, Heat cluster map illustrating JAG1-mediated effects on a list of 200 SC-specific genes curated from published data. D, E, GO analysis of genes downregulated (mean log2 control ≤ 5, p ≤ 0.05, log2 (FC) ≤ −2σ) in P0 Jag1 CKO versus control (Ctrl) cochlear epithelia. Listed are significantly enriched pathways (D) and cellular component (E) GO terms. p values were calculated using Benjamini correction. F, qRT-PCR-based validation of genes of interest that were downregulated at P0 in Jag1 CKO cochlear epithelia compared with Ctrl. SC-specific marker genes S100a1 and Fgfr3 served as controls and no difference between groups was seen. Data are mean ± SEM; minimum of n = 3 animals/group. *p < 0.05; **p ≤0.001; two-tailed Student's t test.
Results
The mammalian auditory sensory epithelium houses several distinct SC subtypes, including Hensen's cells, Deiters' cells, inner and outer pillar cells, inner phalangeal cells, and border cells (Fig. 1A). To investigate the function of JAG1 in differentiating SCs, we made use of Jag1fx/fx mice, previously established by Julian Lewis's group (Brooker et al., 2006) and Sox2CreERT2/+ mice (Arnold et al., 2011). In the cochlea, Sox2CreERT2/+ is highly expressed in HC and SC precursors; and by one week of age, its expression becomes restricted to terminally differentiated SCs (Walters et al., 2015; Gu et al., 2016). We timed-mated Sox2CreERT2/+::Jag1fx/fx with Jag1fx/fx mice and administered tamoxifen to pregnant dams at stage E14.5 (Fig. 1B), which bypasses the earlier requirement for JAG1 in cochlear prosensory progenitor specification and maintenance (Brooker et al., 2006; Kiernan et al., 2006; Hao et al., 2012; Campbell et al., 2016). To control for Sox2 haploinsufficiency (Atkinson et al., 2018), we also collected animals that did not receive tamoxifen treatment (untreated). We harvested control (Jag1fx/fx treated, Jag1fx/fx untreated, and Sox2CreERT2/+::Jag1fx/fx untreated) and Sox2CreERT2/+::Jag1fx/fx treated [Jag1 conditional knockout (CKO)] embryos at E18.5/P0.
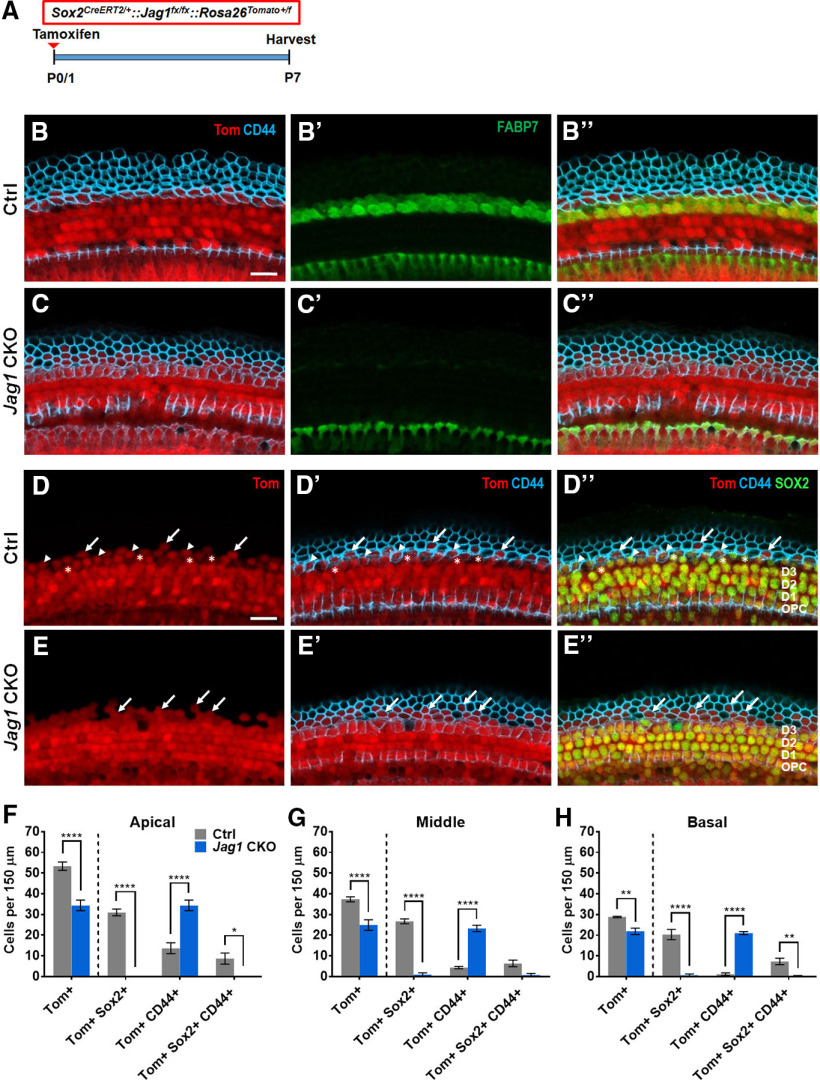
Figure 1.
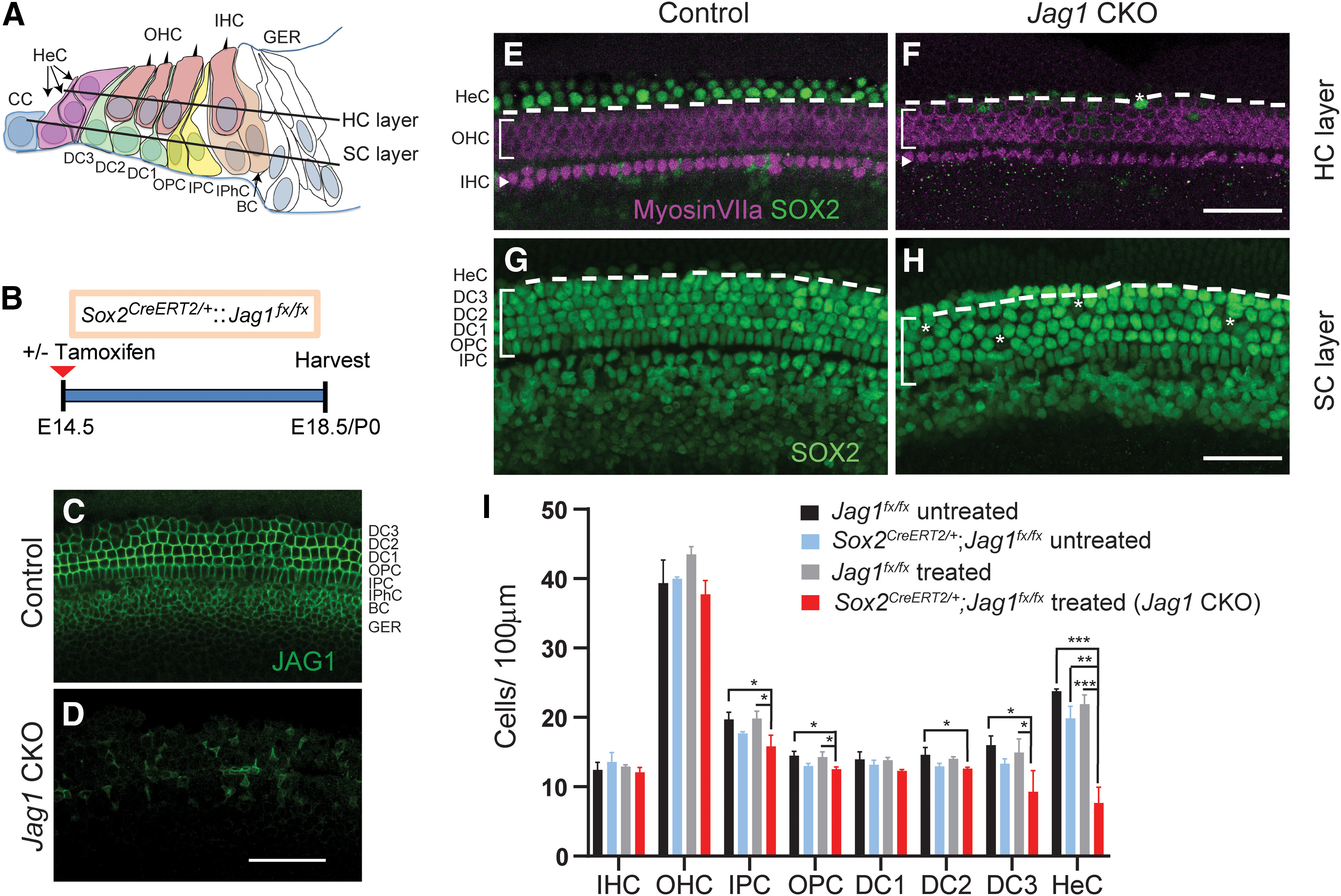
Deletion of Jag1 from stage E14.5 progenitor cells disrupts Hensen's cell formation. A, Schematic of the cellular composition of the neonatal auditory sensory epithelium. BC, Border cell; CC, Claudius cell; DC1, Deiters' cell row 1; DC2, Deiters' cell row 2; DC3, Deiters' cell row 3; GER, greater epithelial ridge; HeC, Hensen's cell, IHC, inner hair cell; IPC, inner pillar cell; IPhC, inner phalangeal cell; OHC, outer hair cell; OPC, outer pillar cell. B, Experimental strategy. Timed pregnant dams were injected with tamoxifen and progesterone at E14.5. Jag1 CKO (Sox2CreERT2/+::Jag1fx/fx treated) mice and control (Jag1fx/fx treated) littermates were analyzed at E18.5/P0. SOX2 immunostaining labels SC nuclei, including Hensen's cells, while myosin VIIa immunostaining labels IHCs and OHCs. Representative confocal images of the cochlear mid-turn are shown. To control for Sox2 haploinsufficiency, E18.5/P0 Sox2CreERT2/+::Jag1fx/fx mice and Jag1fx/fx littermates that did not receive tamoxifen and progesterone (untreated) were also analyzed. C, D, Validation of Jag1 deletion. SC nuclear layer immunostained for JAG1 (green), in Jag1 CKO (D) and control (C, treated, Jag1fx/fx) mice. E, F, HC and Hensen's cell phenotype in P0 control (E, Sox2CreERT2/+::Jag1fx/fx untreated) and P0 Jag1 CKO (F) mice. Control mice (E) contain 1 or 2 rows of SOX2+ (green) Hensen's cells lateral to the myosin VIIa+ (magenta) OHCs (atop dashed white line) (see schematic in A), whereas Jag1 mutant mice contain only few scattered Hensen's cells (white asterisk). G, H, SC phenotype in E18.5 control (G, Jag1fx/fx treated) and E18.5 Jag1 CKO (H) mice. SOX2+ Deiters' cell and Hensen's cell nuclei (atop dashed white line) (see schematic in A) are enlarged and misaligned (white asterisks) in Jag1 CKO mice. I, Quantification of HC and SC subtypes in control and Jag1 CKO mice. Data are mean ± SD; n = 3/group. *p < 0.05; **p ≤ 0.001; ***p ≤ 0.0001; two-way ANOVA, with Bonferroni correction. Scale bar, 50 μm.
First, we analyzed JAG1 expression in control and Jag1 CKO cochlear tissue to validate Cre-mediated deletion of Jag1. In control samples, JAG1 protein was localized at the surface of SCs, except for Hensen's cells (Fig. 1C). By contrast, the majority of SCs in Jag1 CKO animals lacked JAG1 protein expression (Fig. 1D). Next, we analyzed whether Jag1 deletion disrupted the formation or patterning of SCs and/or HCs. To label HCs, we used immunostaining against myosin VIIa, which labels both IHCs and OHCs (Hasson et al., 1995). To visualize SCs, we immunostained against SOX2, which at E18.5/P0 is highly expressed in the nucleus of all SC subtypes, including Hensen's cells, and is expressed at a lower level in the nucleus of IHCs and OHCs (Kempfle et al., 2016). We found that Jag1 deletion had no effect on the number (density) of IHCs or OHCs (Fig. 1E,F,I; two-way ANOVA × genotype F(1,8) = 0.07679, p = 0.7887 for IHCs, F(1,8) = 3.436, p = 0.1009 for OHCs; per treatment F(1,8) = 0.9619, p = 0.3554 for IHCs, F(1,8) = 0.4693, p = 0.5127 for OHCs). Our analysis of the SC phenotype, however, revealed a very significant reduction in the number of Hensen's cells in Jag1 CKO mice compared with all three controls (Fig. 1E–I; two-way ANOVA × genotype F(1,6) = 68.45, p = 0.002; × treatment F(1,6) = 41.49, p = 0.0007). In cochlear tissue of control animals, SOX2+ Hensen's cells were located between the third row of Deiters' cells and Claudius cells, with their nuclei residing in both the HC and SC layers, and with two or three Hensen's cells sitting on top of each other (Fig. 1A,E,G). By contrast, Jag1 CKO cochlear tissue contained either no or only a few scattered Hensen's cells within the HC and SC layers (Fig. 1F,H).
In addition, Deiters' cells in Jag1 CKO mice had enlarged nuclei compared with control mice, and their arrangement appeared to be disorganized, suggesting defects in Deiters' cell differentiation (Fig. 1G,H). Decreased numbers of Deiters' cells in the second and third row were observed in Jag1 CKO samples compared with the Jag1fx/fx treated and/or Jag1fx/fx untreated control groups (Fig. 1I; two-way ANOVA × genotype F(1,8) = 15.20 p = 0.0046 for Deiters' cells row 2, F(1,8) = 12.50, p = 0.0077 for Deiters' cells row 3), but not when compared with Sox2CreERT2/+::Jag1fx/fx untreated controls (Fig. 1I; two way ANOVA × treatment F(1,8) = 1.319, p = 0.2839 for Deiters' cells row 2, F(1,8) = 4.579. p = 0.0648 for Deiters' cells row 3). A similar result was observed for inner and outer pillar cells (Fig. 1I; two-way ANOVA × genotype F(1,8) = 23.35, p = 0.0013 for outer pillar cells, F(1,8) = 18.33, p = 0.0027 for inner pillar cells; × treatment F(1,8) = 0.9287, p = 0.3634 for outer pillar cells, F(1,8) = 1.613, p = 0.2398 for inner pilllar cells), suggesting that Jag1 deficiency combined with Sox2 haploinsufficiency negatively affects the differentiation of pillar and Deiters' cells. Unfortunately, we were unable to address how Jag1 deficiency combined with Sox2 haploinsufficiency may impact pillar and Deiters' cell maturation as conditional deletion of Jag1 at E14.5 resulted in early postnatal lethality. In summary, our analysis demonstrates that JAG1's function is essential for the formation of Hensen's cells in the mammalian cochlea.
To gain insight into how JAG1 may influence Hensen's cell development, we characterized JAG1-mediated gene expression changes in the developing auditory sensory epithelium using whole-genome microarrays. We administered tamoxifen to timed pregnant dams at stage E14.5 and harvested Jag1 CKO (Sox2CreERT2/+::Jag1fx/fx) and control (Jag1fx/fx) mice at stage E18.5/P0 (Fig. 2A). This breeding strategy controls for tamoxifen-induced changes in gene expression, but not for changes because of Sox2 haploinsufficiency (Atkinson et al., 2018). We extracted RNA from enzymatically purified control and Jag1 CKO cochlear sensory epithelia, and analyzed RNA abundance using the GeneChip Mouse Clariom D Arrays (Fig. 2A). Using a one-way ANOVA model, we determined genes that were significantly upregulated (red) or downregulated (blue) in control versus Jag1 CKO cochlear epithelial cells. After removing pseudogenes, imprinted, misaligned, and low expressing genes (average mean log2 (control, Jag1 CKO) < 5.5), a total of 227 genes were differentially expressed between control and Jag1 CKO samples, with 55 genes being significantly upregulated (red dots, p ≤ 0.05, log2(FC) ≥ 3σ) and 172 genes being downregulated (blue dots, p ≤ 0.05, log2(FC) ≤ −3σ) (Fig. 2B). To identify SC-specific genes that are regulated by JAG1-Notch signaling, we used published data to generate a high confidence list of 200 SC-specific genes (Burns et al., 2015; Maass et al., 2016). Our analysis revealed that 41 of these 200 SC-specific genes (21%) were differentially regulated by JAG1-Notch signaling, with 32 being downregulated and 9 being upregulated in Jag1 CKO samples compared with control (Fig. 2C; Extended Data Figs. 2-1, 2-2, genes marked $). Furthermore, the list of downregulated genes included SC-specific genes previously reported to be positively regulated by Notch signaling (Agr3, F2rl1, Hey1, Igfbp3, Lrrtm1, Lsamp, Ntf3, Rab3b, Shc3, and Trh; Fig. 2B; Extended Data Fig. 2-1, genes marked #) (Campbell et al., 2016; Maass et al., 2016). Conversely, the list of upregulated genes included a small subset of genes previously shown to be negatively regulated by Notch signaling (Cdh4, Dpysl2, Robo2, and Cpa6; Extended Data Fig. 2-2, genes marked #) (Maass et al., 2016). However, deletion of Jag1 in the differentiating cochlea did not increase the expression of Atoh1 or other early HC-specific transcription factors (Extended Data Fig. 2-3). This is consistent with our finding that Jag1 deletion at E14.5 does not result in ectopic HC formation (Fig. 1F,I). To uncover the biological processes associated with JAG1-regulated genes, we performed GO enrichment analysis using DAVID. We found that the list of downregulated genes (mean log2 control ≥ 5, p ≤ 0.05, log2(FC) ≤ −2σ) was enriched for genes linked to mitochondrial function (oxidative phosphorylation) and mitochondrial dysfunction (Alzheimer, Parkinson, and Huntington) as well as protein synthesis (ribosome) (Fig. 2D). Furthermore, consistent with such function, the list of downregulated genes was enriched for gene products that are localized to the mitochondrion (mitochondrial inner membrane, respiratory chain, mitochondrial respiratory chain complex I) and ribosomes (cytosolic small ribosomal unit, intracellular ribonucleoprotein complex) (Fig. 2E). In addition, the list of downregulated genes was enriched for gene products that are part of extracellular exosomes, indicating a possible role of JAG1 in the biogenesis of exosomes and/or function of exosome mediated cell–cell communication (Fig. 2E; Extended Data Fig. 2-4).
List of top-ranked downregulated genes in Jag1 mutant cochlear epithelia. Genes are ranked based on fold change. Shown are genes that are downregulated (p ≤ 0.05, log2 (FC) ≤3σ) in Sox2-CreERT2/+::Jag1fx/fx (Jag1 mut) samples compared with Jag1fx/fx (Control, Ctrl) samples. Tamoxifen and progesterone were injected at E14.5 and cochlear epithelia were collected at P0. $ marks SC-specific/enriched genes (Burns et al., 2015; Maass et al., 2016) and # marks genes previously found to be positively regulated in cochlear epithelial cells by Notch signaling (Campbell et al., 2016; Maass et al., 2016). Note that only transcripts with mean log2 values of 5.5 or more in control samples are listed. Download Figure 2-1, DOCX file (38.4KB, docx) .
List of top-ranked upregulated genes in Jag1 mutant cochlear epithelia. Genes are ranked based on fold change (FC). Shown are genes that are upregulated (p ≤ 0.05, log2 (FC) ≥3σ) in Sox2-CreERT2/+::Jag1fx/fx (Jag1 mut) samples compared with Jag1fx/fx (Control, Ctrl) samples. Tamoxifen and progesterone were injected at E14.5 and cochlear epithelia were collected at P0. $ marks SC-specific/enriched genes (Burns et al., 2015; Maass et al., 2016) and # marks genes previously found to be negatively regulated in cochlear epithelial cells by Notch signaling (Campbell et al., 2016; Maass et al., 2016). Note that only transcripts with mean log2 values in Jag1 mutant samples of 5.5 or more are listed. Download Figure 2-2, DOCX file (24.8KB, docx) .
Expression of HC-specific transcription factors is unchanged in Jag1 mutant cochlear epithelia. HC-specific transcription factor expression in Sox2-CreERT2/+::Jag1fx/fx (Jag1 mut) samples compared with Jag1fx/fx (Control, Ctrl) samples. Tamoxifen and progesterone were injected at E14.5 and cochlear epithelia were collected and analyzed at P0. Genes are listed in alphabetical order. Download Figure 2-3, DOCX file (14.2KB, docx) .
List of genes and their associated pathways and cellular components downregulated in Jag1 mutant cochlear epithelia. Gene ontology analysis on genes that were downregulated (mean log2 control ≤5, p ≤ 0.05, log2 (FC) ≤-2σ) in P0 Jag1 CKO versus control (Ctrl) cochlear epithelia. Download Figure 2-4, DOCX file (18.7KB, docx) .
Next, we used qPCR to confirm Jag1 deletion and validate a subset of SC-specific/enriched genes found to be downregulated in Jag1 mutant cochlear epithelia. The genes were selected based on their specific expression in Hensen's cells (Trpa1) (Corey et al., 2004), and their association with biological processes of interest, including oxidative stress and cell death [Gpx1 (Ohlemiller et al., 2000), Apod (Ganfornina et al., 2008), Mgst3 (Lu et al., 2016)], vesicle biogenesis and neurotransmitter release (Rab3b) (Schlüter et al., 2004), mitochondrial inner membrane transport (Timm13) (Rothbauer et al., 2001), and prosensory progenitor cell proliferation (Stox1) (Nie et al., 2015). We also included in our analysis a subset of genes that have been reported to be positively regulated by Notch signaling in SC precursors and SCs (Hey1, Igfbp3) (Campbell et al., 2016; Maass et al., 2016). As controls, we also examined the expression of SC-specific genes Fgfr3 and S100a1, which, based on the microarray data, remained unchanged after the loss of Jag1. Consistent with the microarray data, qPCR analysis revealed that Fgfr3 and S100a1 expression was unaffected by the loss of Jag1, while the expression of Jag1, Hey1, Igfbp3, Timm13, Apod, and Trpa1 was significantly reduced (Fig. 2F). Furthermore, as predicted by the microarray data, we found that on average the expression of Gpx1, Mgst3, Rab3b, and Stox1 was 1.5- to 2.5-fold lower in Jag1 mutant cochlear epithelia compared with control cochlear epithelia (Fig. 2F). However, because of high variability across samples, the reduction in the expression of these genes was not significant. Together, our gene expression analysis confirms the observed defect in Hensen's cell formation and suggests a role for JAG1 in tissue homeostasis and metabolism.
In mice, HCs and SCs are not functional at birth and undergo a two-week-long process of postnatal maturation (Legendre et al., 2008; Lelli et al., 2009; Szarama et al., 2012). To address whether JAG1 function is required during postnatal maturation, we deleted Jag1 after the completion of HC and SC differentiation. To that end, we injected Jag1 CKO mice and their Jag1fx/fx littermates with 4-hydroxy-tamoxifen at P0/P1. To control for Sox2 haploinsufficiency, we also collected Sox2CreERT2/+::Jag1fx/fx and Jag1fx/fx mice that did not receive 4-hydroxy-tamoxifen treatment (untreated) (Fig. 3A). Control (Jag1fx/fx treated, Jag1fx/fx untreated, and Sox2CreERT2/+::Jag1fx/fx untreated) and Jag1 CKO mice were harvested and analyzed at P5 or P7. JAG1 immunostaining confirmed the successful deletion of Jag1 in cochlear tissue obtained from Jag1 CKO mice (Fig. 3B,C). Next, we quantified HC and SC subtypes at stage P7 in Jag1 CKO and control mice using the HC and SC markers myosin VIIa and SOX2, respectively. Quantification of SC numbers revealed a very significant reduction in the number of Hensen's cells in Jag1 CKO mice compared with control mice (Fig. 3J; two-way ANOVA × genotype F(1,10) = 20.43, p = 0.0011 and × treatment F(1,10) = 12.98, p = 0.0048). A recent report found that Sox2 haploinsufficiency at the neonatal stage stimulates pillar cell proliferation and the production of ectopic IHCs (Atkinson et al., 2018). Consistent with these recent findings, quantification of HC numbers in control and Jag1 CKO mice uncovered a mild increase in the number of IHC in the two groups of mice that contained the Sox2CreERT2/+ allele compared with the Jag1fx/fx treated and Jag1fx/fx untreated controls (Fig. 3J; two-way ANOVA × genotype F(1,7) = 34.20, p = 0.0006 for IHCs, F(1,7) = 0.6557, p = 0.4447 for OHCs; × treatment F(1,7) = 0.7362, p = 0.4193 for IHCs, F(1,7) = 1.175, p = 0.3143 for OHCs). Also, we observed a mild, but significant, increase in the number of inner pilllar cells in Jag1 CKO mice compared with Sox2CreERT2/+::Jag1fx/fx untreated control animals (Fig. 3J; two-way ANOVA × treatment F(1,10) = 6.004, p = 0.0342 for inner pilllar cells).
Figure 3.
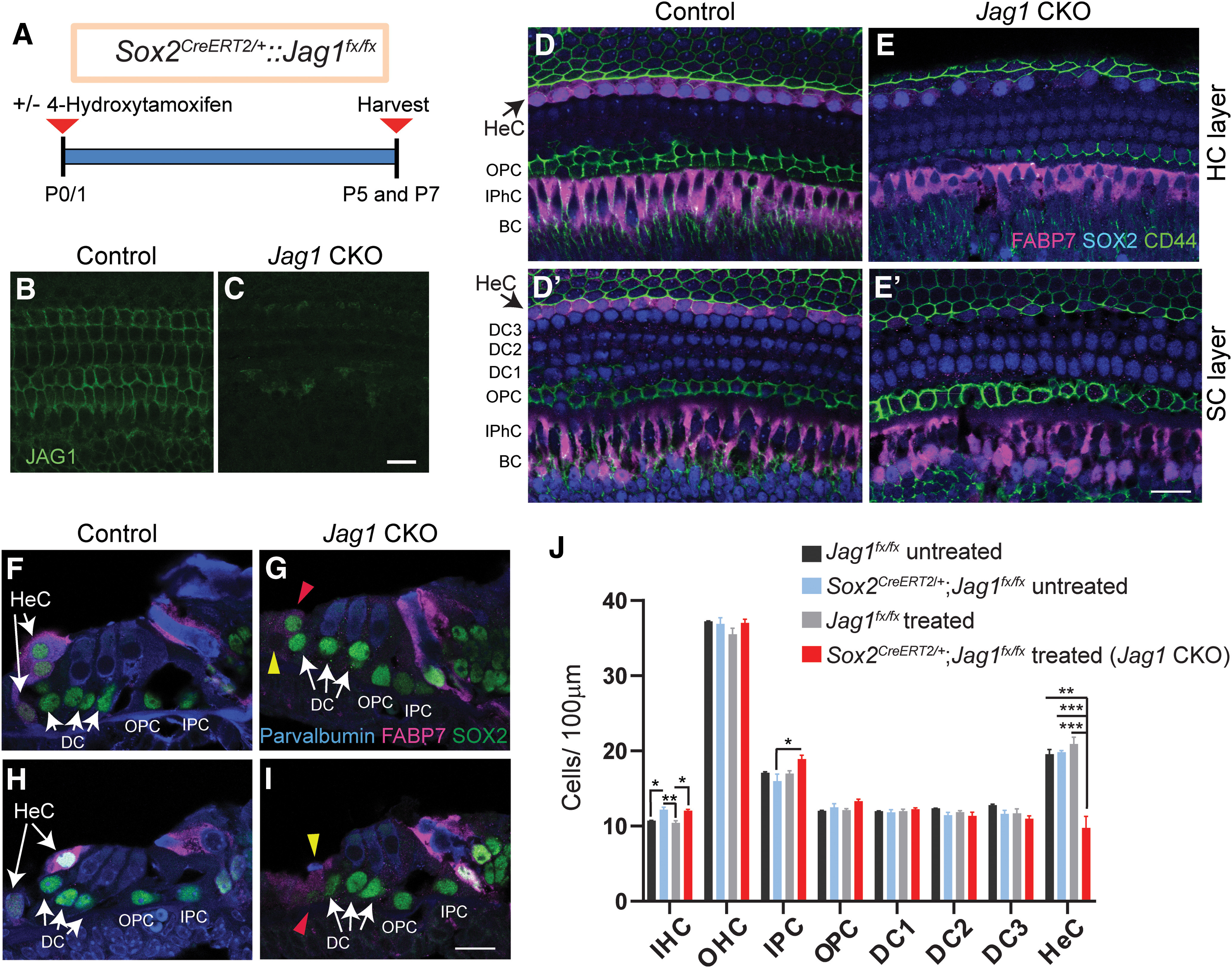
Deletion of Jag1 from neonatal SCs results in the loss of Hensen's cells. A, Experimental strategy. Sox2CreERT2/+::Jag1fx/fx and Jag1fx/fx were treated with 4-hydroxy-tamoxifen at P0/P1, and their cochleae were analyzed at P5 or P7. Untreated control mice (Sox2CreERT2/+::Jag1fx/fx untreated and Jag1fx/fx, untreated) were also used. B, C, Validation of conditional Jag1 deletion. Shown is the SC nuclear layer, immunostained for JAG1 (green), in Jag1 CKO (C) and control (B, Jag1fx/fx treated) mice. D–I, SOX2 immunostaining (D–E′, blue; F–I, green) labels SC nuclei, including Hensen's cells. FABP7 (magenta) labels Hensen's cells, inner phalangeal cells, and border cells. Parvalbumin (F–I, blue) labels HCs, and CD44 (D–E′, green) labels Claudius cells and outer pillar cells. Shown are representative single-plane confocal images of the cochlear mid-turn. D–E′, Top down view of the HC (D,E) and SC nuclear layers (D′,E′) in Jag1 CKO (E,E′) and control (D,D′, Sox2CreERT2/+::Jag1fx/fx untreated) mice. Black arrows indicate the location of Hensen's cells residing within the HC (D) and SC layer (D′) in control mice. F–I, Cochlear cross-sections of Jag1 CKO (G,I) and control mice (F,H, Sox2CreERT2/+::Jag1fx/fx, untreated). White arrows indicate 2 or 3 Hensen's cell nuclei that are stacked on top of each other in control mice. Red arrowheads indicate SOX2+ Hensen's cell-like cells (G,I). Yellow arrowheads indicate dying cell (I) and missing Hensen's cell (G) in Jag1 CKO mice. J, Quantification of HC and SC subtypes in control and Jag1 CKO mice. Data are mean ± SD; n = 3/group. *p < 0.05; **p ≤ 0.001; ***p ≤ 0.0001; two-way ANOVA, with Bonferroni correction. DC1, Deiters' cell row 1; DC2, Deiters' cell row 2; DC3, Deiters' cell row 3; HeC, Hensen's cell; IPC, inner pillar cell; IPhC, inner phalageal cell; OPC, outer pillar cell. Scale bar, 20 μm.
To confirm the observed Hensen's cell loss in Jag1 CKO mice, we conducted additional experiments in which we used a combination of FABP7, SOX2, and CD44 immunostaining to distinguish between Hensen's cells, Deiters' cells, and Claudius cells. In the postnatal cochlea, FABP7 is highly expressed in the cytoplasm of Hensen's cells, inner phalangeal cells, and border cells (Saino-Saito et al., 2010), whereas CD44 is selectively expressed on the cell membranes of outer pillar cells and Claudius cells (Hertzano et al., 2010) (Fig. 3D,D′). In P5 control mice, two or three Hensen's cell nuclei (SOX2+, FABP7+) are stacked on top of each other (Fig. 3F,H), with a single row of Hensen's cells within the SC nuclear layer and a row of stacked Hensen's cells residing in the HC layer (Fig. 3D,D′). By contrast, the cochlea of Jag1 CKO mice only contained a few scattered SOX2+ Hensen's cell-like nuclei within the HC layer and SC layer (Fig. 3E,E′,G,I). These Hensen's cell-like cells expressed FABP7 at a much-reduced level than Hensen's cells in control mice, and some SOX2+ Hensen's cell-like cells in Jag1 CKO mice appeared to coexpress CD44, suggesting that these cells may undergo a cell-fate conversion and become Claudius cells (Fig. 3E′).
To further investigate the Hensen's cell-like nuclei with CD44 expression, we performed fate-mapping experiments using Sox2CreERT2/+::Jag1fx/fx::Rosa26tdTomato/+ and Sox2CreERT2/+::Rosa26tdTomato/+ controls that were injected with tamoxifen at P0/P1 (Fig. 4A). In control samples collected at P7, all SOX2+ SC subtypes, including Hensen's cells that were colabeled with FABP7, expressed tdTomato (Tom; Fig. 4B-B′′), and a small number of CD44+ Claudius cells also expressed Tom (Fig. 4D-D′′). In samples with Jag1 deletion, there was a loss of FABP7 expression in the Hensen's cell region and there appeared to be an increased number of Tom+ cells expressing CD44 (Fig. 4E-E′′). Quantification of Tom+ cells lateral to the third row of Deiters' cells at P7 showed a decrease in the number of Tom+ cells and Tom+, SOX2+ cells in all turns of the cochlea when Jag1 was deleted (Fig. 4D–H; two-way ANOVA × genotype F(1,5) = 29.81, p = 0.028 for apex, F(1,5) = 15.68, p = 0.0107 for middle, and F(1,5) = 13.67, p = 0.014 for base), which suggests that Jag1 is important for the survival of some Hensen's cells. In support of these findings, we frequently noted missing cells (Fig. 3G) as well as cellular debris (Fig. 3I) within the Hensen's cell region in Jag1 CKO mice. However, staining with a marker of apoptosis, cleaved caspase-3, revealed no caspase3+ Hensen's cells in the cochlea of Jag1 CKO mice between P1 and P4. There was also an approximately twofold increase in the number of cells labeled with both Tom and CD44 in Jag1 CKO cochleae (Fig. 4D–H). Thus, some Hensen's cells appear to lose SOX2 expression and convert into Claudius cells.
Figure 4.
Fate-mapping analysis reveals that Hensen's cells die or convert into Claudius cells after deletion of Jag1. A, Experimental strategy. Sox2CreERT2/+::Jag1fx/fx::Rosa26tdTomato/+ (Jag1 CKO) and Sox2CreERT2/+::Rosa26tdTomato/+ (Ctrl) mice were injected with tamoxifen at P0/P1, and cochleae were analyzed at P7. B–C′′, Representative confocal images from Ctrl (B–B′′) and Jag1 CKO (C–C′′) mice in the apical turn, showing expression of FABP7 (a Hensen's cell marker, green), CD44 (a Claudius cell marker, blue), and tdTomato (Tom, red). D–E′′, Representative confocal images of Ctrl (D–D′′) and Jag1 CKO (E–E′′) mice, showing expression of SOX2 (a SC marker, green), CD44 (blue), and Tom (red). Arrows indicate Tom+, CD44+ cells. Asterisks indicate Tom+, SOX2+ cells. Arrowheads indicate Tom+, SOX2+, CD44+ cells. F–H, Quantification of Tom+ cells lateral to the third row of Deiters' cells that also express CD44 and/or SOX2 in the apical (F), middle (G), and basal (H) turns of the cochleae in Ctrl and Jag1 CKO mice. Data are mean ± SEM; n = 3 or 4/group. *p < 0.05; **p < 0.01; ****p < 0.0001; two-way ANOVA followed by a Bonferroni post hoc test. Scale bar, 20 μm.
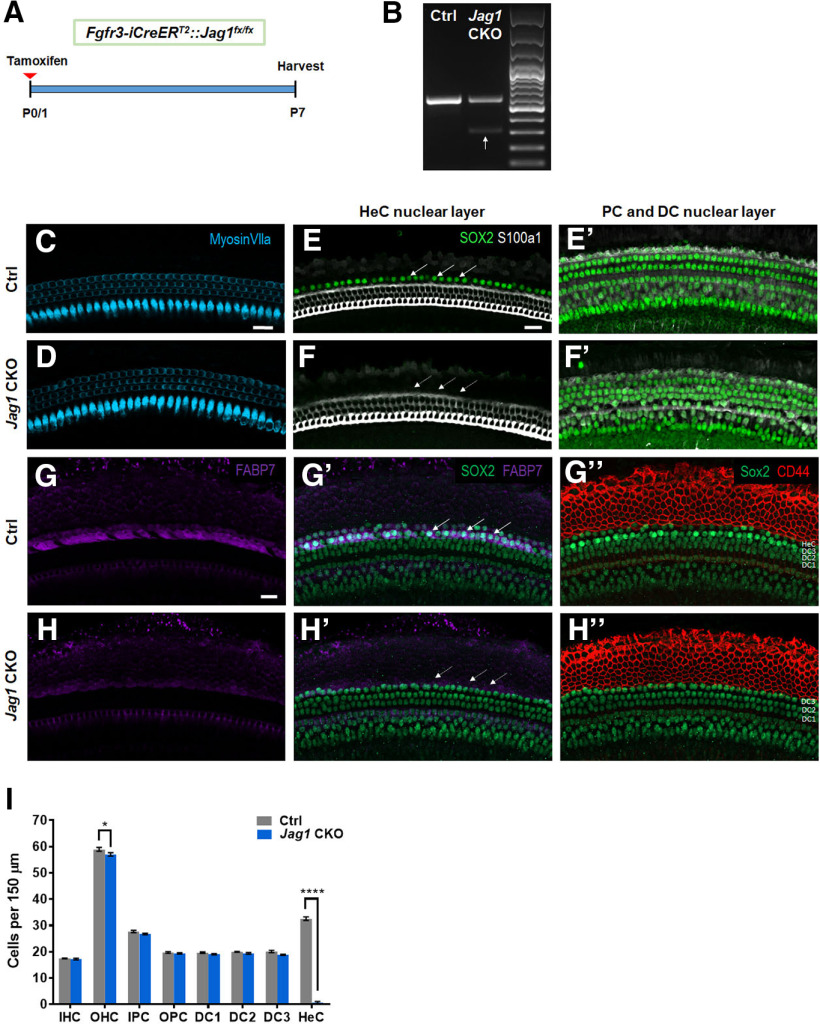
An even more severe loss of Hensen's cells was observed when Jag1 was deleted in the neonatal cochlea using the transgenic Fgfr3-iCreERT2 line. The Fgfr3-iCreERT2 line has been used extensively in the auditory field to disrupt gene function in pillar and Deiters' cells and has normal Sox2 expression levels (Kirjavainen et al., 2015; Atkinson et al., 2018). Fgfr3-iCreERT2 mice injected with tamoxifen at P0/P1 have CreER expression in ∼100% of pillar and Deiters' cells, as well as in varying amounts of OHCs depending on the cochlear turn (Cox et al., 2012; McGovern et al., 2017). Fgfr3-iCreERT2::Jag1fx/fx and iCre-negative control mice were injected with tamoxifen at P0 and P1 to delete Jag1, which was confirmed by RT-PCR showing a second smaller band that lacks exon 4 in Fgfr3-iCreERT2::Jag1fx/fx mice (Fig. 5A,B). We quantified HCs and individual SC subtypes at P7 using myosin VIIa to label HCs and SOX2 to label SC nuclei (Fig. 5C–F′) and observed a significant loss of Hensen's cells in Fgfr3-iCreERT2::Jag1fx/fx mice (Fig. 5I; two-way ANOVA × genotype F(1,6) = 191, p < 0.0001). To confirm that Hensen's cells are missing, we used two methods: (1) S100a1 immunostaining to label the cytoplasm and nuclei of pillar and Deiters' cells (White et al., 2006; Buckiova and Syka, 2009); and (2) FABP7 immunostaining to label the cytoplasm of Hensen's cells (Saino-Saito et al., 2010). In control samples, there are two rows of SOX2+, FABP7+ cells located lateral to the S100a1+ third row of Deiters' cells (Fig. 5E,E′,G,G′). However, after Jag1 deletion in Fgfr3-iCreERT2::Jag1fx/fx mice, there were no SOX2+ cells located lateral to the third row of Deiters' cells (Fig. 5F,F′,H′), and there was a reduction in FABP7 expression (Fig. 5G,H). We also performed immunostaining with CD44, to label Claudius cells, which are located lateral to Hensen's cells. In Fgfr3-iCreERT2::Jag1fx/fx mice, the CD44 expression pattern appeared similar to the control; however, the CD44 staining was immediately adjacent to the SOX2+ Deiters' cell row 3 nuclei (Fig. 5G′′,H′′). Together, these results confirm that the missing SOX2+ cells are Hensen's cells. In addition, there was a slight decrease in the number of OHCs, but no changes in the number of IHCs in Fgfr3-iCreERT2::Jag1fx/fx mice at P7 (Fig. 5C,D,I), which rules out the possibility of Hensen's cells converting into HCs. Together, these results indicate that Sox2 haploinsufficiency is not responsible for the loss of Hensen's cells after Jag1 deletion.
Figure 5.
Massive loss of Hensen's cells at P7 after neonatal deletion of Jag1 from pillar and Deiters' cells. A, Experimental strategy. Fgfr3-iCreERT2::Jag1fx/fx (Jag1 CKO) and Jag1fx/fx (Ctrl) mice were injected with tamoxifen at P0/P1, and cochleae were analyzed at P7. B, RT-PCR results using RNA extracted from P7 Ctrl and Jag1 CKO whole cochlea. The upper band (541 bp) indicates the WT allele containing exon 4. The lower band (286 bp, white arrow) confirms deletion of Jag1 exon 4 in Jag1 CKO mice. The upper band is still present in Jag1 CKO mice since JAG1 is expressed in all SCs and Fgfr3-iCreERT2 only targets pillar and Deiters' cells. C, D, Representative confocal slice images of HC, labeled with myosin VIIa (blue) in P7 Ctrl (C) and Jag1 CKO (D) mice. E–F′, Representative confocal slice images of SC nuclear layers at the level of Hensen's cell nuclei (E,F) and at the level of pillar and Deiters' cell nuclei (E′,F′) showing SOX2 (an SC marker, green) and S100a1 (a pillar and Deiters' cell marker, white) expression in P7 Ctrl (E–E′) and Jag1 CKO (F–F′) mice. Arrows indicate the location of Hensen's cells (E,F). G–H′′, Representative confocal slice images of SOX2 (green), FABP7 (a Hensen's cell marker, magenta), and CD44 (a Claudius cell marker, red) expression in P7 Ctrl (G–G′′) and Jag1 CKO (H–H′′) mice. Arrows indicate the location of Hensen's cells (G′,H′). I, Quantification of IHCs, OHCs, and individual SC subtypes (inner pillar cells to Hensen's cells) in Ctrl and Jag1 CKO mice. Data are mean ± SEM; n = 4/group. *p < 0.05; ****p < 0.0001; two-way ANOVA followed by a Bonferroni post hoc test. DC1, Deiters' cell row 1; DC2, Deiters' cell row 2; DC3, Deiters' cell row 3; HeC, Hensen's cell; IPC, inner pillar cell; OPC, outer pillar cell. Scale bar, 20 μm.
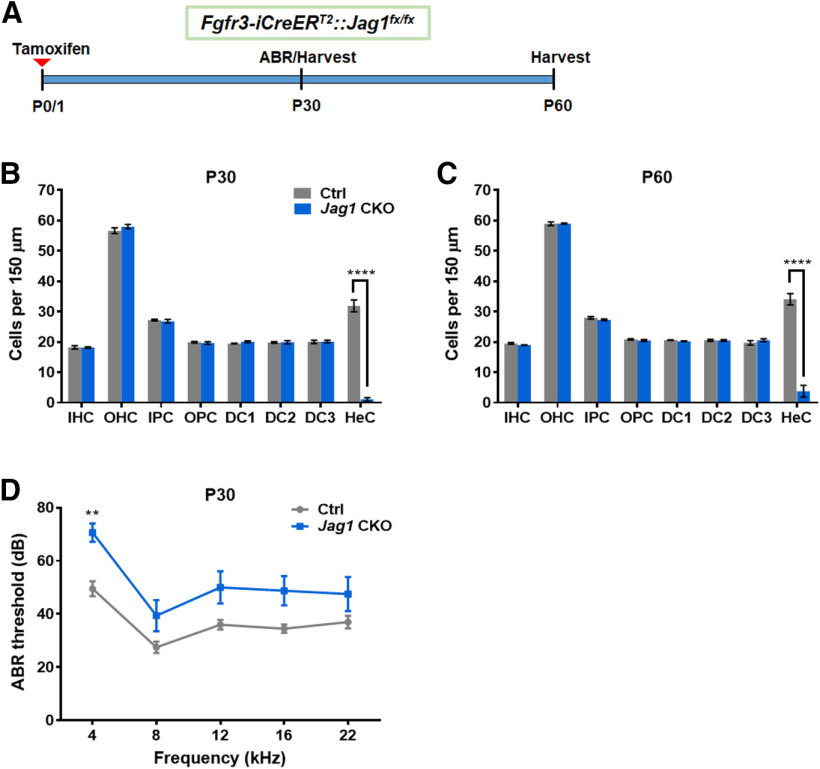
We next examined Fgfr3-iCreERT2/+::Jag1fx/fx mice at older ages to investigate whether SC loss progressed to other cell types. After tamoxifen treatment at P0 and P1, the number of HCs and individual SC subtypes were quantified in Fgfr3-iCreERT2::Jag1fx/fx cochlea and iCre-negative controls at P30 and P60 (Fig. 6A). Similar to the changes we observed at P7, there was no difference in number of IHCs or OHCs at P30 or P60 compared with the control samples (Fig. 6B,C). For SCs, the only significant difference observed at P30 and P60 was a decrease in Hensen's cells (Fig. 6B,C; two-way ANOVA × genotype F(1,6) = 59.25, p = 0.0003 for P30 and F(1,6) = 73.68, p = 0.0001 for P60). Thus, neonatal Jag1 deletion induces Hensen's cell loss that occurs by P7 and does not affect other cell types or cause further morphologic changes in adulthood.
Figure 6.
Neonatal Jag1 deletion from pillar and Deiters' cells caused hearing deficits in the low-frequency range. A, Experimental strategy. Fgfr3-iCreERT2::Jag1fx/fx (Jag1 CKO) and Jag1fx/fx (Ctrl) mice were injected with tamoxifen at P0/P1, followed by ABR at P30, and analysis of the cochlea at P30 or P60. B, C, Quantification of IHCs, OHCs, and individual SC subtypes (inner pillar cells to Hensen's cells) in Ctrl and Jag1 CKO mice at P30 (B) and P60 (C). Data are mean ± SEM; n = 4/group. ****p < 0.0001. D, ABR thresholds of Ctrl and Jag1 CKO at P30. Data are mean ± SEM; n = 8-10/group. **p < 0.01; two-way ANOVA followed by a Bonferroni post hoc test. DC1, Deiters' cell row 1; DC2, Deiters' cell row 2; DC3, Deiters' cell row 3; HeC, Hensen's cell; IPC, inner pillar cells; OPC, outer pillar cells.
To assess whether Hensen's cell loss affected hearing, we performed ABR at P30 using Fgfr3-iCreERT2::Jag1fx/fx mice and iCre-negative controls that were injected with tamoxifen at P0 and P1 (Fig. 6A). While there was a main effect of genotype (two-way ANOVA F(1,16) = 11.44, p = 0.038), the only significant increase in ABR thresholds in Fgfr3-iCreERT2::Jag1fx/fx mice was observed at 4 kHz (Fig. 6D).
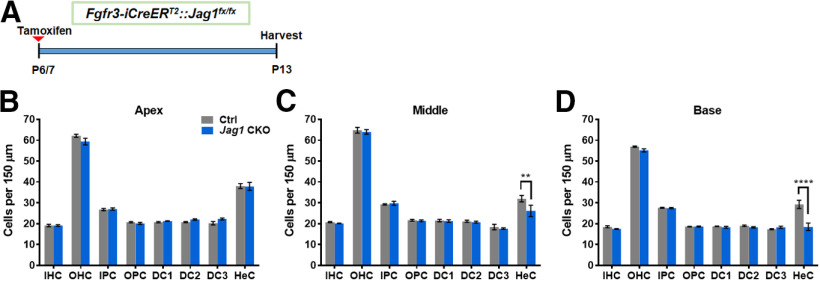
To investigate the function of Jag1 at an older age when HCs and SCs are more mature, we injected Fgfr3-iCreERT2::Jag1fx/fx mice with tamoxifen at P6 and P7 and quantified HCs and individual SC subtypes one week later, at P13 (Fig. 7A). Again, the numbers of IHCs and OHCs remained unchanged (Fig. 7B–D). However, unlike the neonatal Jag1 deletion that resulted in massive Hensen's cell loss across all three cochlear turns, deletion of Jag1 at P6/P7 induced a mild loss of Hensen's cells that exhibited a gradient across cochlear turns. In the apical turn of Fgfr3-iCreERT2::Jag1fx/fx mouse cochleae, no Hensen's cell loss was observed (Fig. 7B; two-way ANOVA × genotype F(1,6) = 0.01205, p = 0.9162). In the middle turn, there was a ∼20% loss of Hensen's cells (Fig. 7C; two-way ANOVA × genotype F(1,6) = 1.479, p = 0.2696); and in the basal turn, Jag1 deletion at P6/P7 caused a larger ∼40% loss of Hensen's cells (Fig. 7D; two-way ANOVA × genotype F(1,6) = 10.81, p = 0.0166). Thus, there appears to be a critical period for Jag1's function in SCs.
Figure 7.
Mild loss of Hensen's cells after deletion of Jag1 from pillar and Deiters' cells at 1 week of age. A, Experimental strategy. Fgfr3-iCreERT2::Jag1fx/fx (Jag1 CKO) and Jag1fx/fx (Ctrl) mice were injected with tamoxifen at P6/P7, and cochleae are analyzed at P13. B–D, Quantification of IHCs, OHCs, and individual SC subtypes (inner pillar cells to Hensen's cells) in the apical (B), middle (C), and basal (D) turns of the cochleae in Ctrl and Jag1 CKO mice at P13. Data are mean ± SEM; n = 4/group. **p < 0.01; ****p < 0.0001; two-way ANOVA followed by a Bonferroni post hoc test. DC1, Deiters' cell row 1; DC2, Deiters' cell row 2; DC3, Deiters' cell row 3; HeC, Hensen's cell; IPC, inner pillar cells; OPC, outer pillar cells.
Discussion
JAG1 has a well-established role in specifying the prosensory region during inner ear development via Notch-mediated lateral induction (Kiernan, 2013). However, its later expression in SCs is without a defined function. Here, we show that JAG1 is essential for the formation of Hensen's cells and is required for Hensen's cell survival and cell-fate maintenance during the early phase of cochlear maturation and that Jag1 loss leads to low-frequency hearing deficits.
Disruption of Notch signaling during cochlear differentiation and early maturation triggers the conversion of SC precursors and newly formed SCs into HCs (Tang et al., 2006; Yamamoto et al., 2006; Takebayashi et al., 2007; Doetzlhofer et al., 2009). Thus, it could be reasoned that Hensen's cells are lost because of conversion into OHCs. However, we did not observe any increase in the number of HCs in response to Jag1 deletion, indicating that JAG1 is dispensable for HC fate repression and that the loss of Hensen's cells was not because of their conversion into OHCs. Instead, we observed morphologic evidence of dead and dying Hensen's cells in P6/7 Jag1 mutant cochlear tissue, and fate-mapping experiments indicate that approximately one-third of Hensen's cells are lost because of cell death.
There is precedence for Notch signaling promoting cell survival. NOTCH1 and NOTCH3 are essential for the survival of neural stem/progenitor cells (Androutsellis-Theotokis et al., 2006; Mason et al., 2006), and loss of Notch1, Dll1, or Jag1 induces apoptosis in glioma stem cells (Purow et al., 2005). Furthermore, laterally located cochlear prosensory cells and later laterally located SCs, in particular Deiters' cells, are lost in the absence of RPBJ, indicating an essential role for canonical Notch signaling in their survival (Basch et al., 2011; Yamamoto et al., 2011; Campbell et al., 2016).
Mechanistic work indicates that Notch signaling promotes cell survival through activation of nuclear factors, such as STAT3, MYC, and NFKB, as well as though activation of cytoplasmic signals, including Akt and mTOR, resulting in increased oxidative metabolism and transcription of prosurvival genes (Oishi et al., 2004; Androutsellis-Theotokis et al., 2006; Chan et al., 2007; Perumalsamy et al., 2010; Hossain et al., 2018). Consistent with a positive link between cochlear JAG1/Notch signaling and Akt-mTOR signaling, our gene expression analysis revealed that loss of Jag1 in cochlear SC precursors resulted in the downregulation of genes associated with oxidative phosphorylation and protein synthesis. However, the majority of these metabolic genes are broadly expressed within the auditory sensory epithelium, and further studies are needed to establish a causative link between the death of Hensen's cells and potential changes in the metabolism of Hensen's cells or SCs in general.
Alternatively, it is possible that JAG1 promotes Hensen's cell survival through other mechanisms. We found that loss of Jag1 in SC precursors caused the downregulation of genes whose proteins localize to extracellular exosomes. Exosomes are endosome-derived microvesicles that enable the transfer of protective proteins/RNAs between exosome-secreting and exosome-receiving cells (McGough and Vincent, 2016). In the utricle, heat shock-induced exosomes have been found to protect HCs from aminoglycoside-induced HC death (Breglio et al., 2020). Thus, the observed Hensen's cell loss in Jag1 mutant cochlear tissue may be the product of defective or abnormal exosome signaling. A limitation of the microarray experiment is that we used Jag1 mutant cochlear tissue, which lacked one copy of Sox2 while controls did not; therefore, some of the observed differences in gene expression may be because of Sox2 haploinsufficiency (Atkinson et al., 2018). Future studies will address whether there is a regulatory link between JAG1 and SC-mediated exosome signaling and whether JAG1 positively regulates prosurvival pathways, such as AKT-mTOR signaling in SCs, including Hensen's cells (Saxton and Sabatini, 2017).
Our fate-mapping experiments revealed that, in response to neonatal Jag1 deletion, two-thirds of Hensen's cells converted into CD44+, SOX2– Claudius cell-like cells. What triggered the upregulation of CD44 and downregulation of SOX2 in a subset of Hensen's cells is unclear. JAG1's role in repressing a Hensen's cell-to-Claudius cell-fate switch is reminiscent of its early role in vestibular prosensory cell maintenance. There, JAG1-Notch signaling prevents the conversion of prosensory progenitors into nonsensory cells through maintaining the expression of the transcription factor SOX2 and through antagonizing the transcription factor LMX1A (Mann et al., 2017; Brown et al., 2020). A similar antagonistic relationship between Claudius cell-specific genes and JAG1-SOX2 signaling may be at play, and future investigations into the molecular mechanisms of Claudius cell-fate determination are warranted.
Hensen's cells are a SC subtype located between the third row of Deiters' cells and Claudius cells, with no direct contact with HCs. Thus, Hensen's cells may be uniquely vulnerable to Jag1 deletion since they only receive Notch signals from neighboring Deiters' cells through the JAG1 ligand. By contrast, the Notch receptors located on pillar and Deiters' cells are also activated by the Notch ligands DLL1 and JAG2, expressed by HCs (Lanford et al., 1999; Kiernan et al., 2005; Brooker et al., 2006). Thus, Jag1 deletion using Sox2CreERT2/+ or Fgfr3-iCreERT2 removes all Notch signaling from Hensen's cells, whereas pillar and Deiters' cells would still receive Notch signaling from DLL1 and JAG2.
The nature of the Notch receptor(s) through which JAG1 signals in SCs is currently unknown. Differentiating and maturing SCs, including Hensen's cells, highly express Notch1 and Notch3 (Maass et al., 2015; Kolla et al., 2020), making these receptors premier candidates to mediate JAG1's function in SCs. JAG1 has been shown to interact with Notch receptors within the same cell, attenuating the ability of the cell to send or receive Notch signals (Henrique and Schweisguth, 2019). However, it is unlikely that JAG1 function in Hensen's cells is mediated through a cis-signaling mechanism because differentiating and maturing Hensen's cells lack Jag1 transcript or protein (Morrison et al., 1999; Kolla et al., 2020). Furthermore, our Fgfr3-iCreERT2::Jag1fx/fx data indicate that defects in Hensen's cell maintenance and survival are observed, even in the absence of Hensen's cell-specific Jag1 deletion.
Hensen's cells' dependence on JAG1 appears to have a critical period as Jag1 deletion at P6 produced a less severe phenotype where only 20%-40% of Hensen's cells were lost in the middle and basal turns of the cochlea, and no loss in the apical turn. This tonotopic difference is unlikely caused by differences in maturation since SCs first form in the base and SCs in the apical turn are less mature than those in the middle and basal turns (Lim and Anniko, 1985). Instead, the milder Hensen's cell phenotype at P6 may be because of a lower Cre-mediated recombination efficiency and/or an increase in JAG1 protein stability compared with P0. The latter may explain why the apex, which expresses JAG1 at a twofold higher levels than other regions of the cochlea, shows no loss of Hensen's cells (Son et al., 2012).
Jag1 deletion during cochlear differentiation not only resulted in a loss of Hensen's cells, but caused cellular patterning defects within the sensory epithelium. A recent study proposed that Hensen's cells are involved in the patterning of OHCs during cochlear differentiation, where Hensen's cell division and migration lateral to OHCs produce a shearing motion that facilitates the arrangement of OHCs and Deiters' cells into the final checkerboard-like pattern (Cohen et al., 2020). Thus, the HC and SC patterning defects we observed may be the result of missing Hensen's cells. Alternatively, the defects in cellular patterning in Jag1 mutant cochlea may be because of altered cell adhesion and/or polarity properties of SCs and HCs.
We also observed a decrease in the number of Deiters', inner pillar, and outer pillar cells when Jag1 was deleted at E14.5 using Sox2CreERT2/+ mice compared with controls that had normal Sox2 expression, but not compared with controls that had only one copy of Sox2, which suggests that JAG1 and SOX2 cooperate in pillar and Deiters' cell differentiation. Deletion of Jag1 at P0 using Sox2CreERT2/+ mice resulted in an opposite phenotype, with a mild, but significant, increase in the number of inner pillar cells compared with controls with one copy of Sox2, but not compared with controls with normal Sox2 expression, suggesting that, in the early postnatal cochlea, SOX2 and JAG1 may have opposing functions.
When Fgfr3-iCreERT2 mice were used to delete Jag1 at P0, there was no change in the number of inner pillar cells. This different result could be because Fgfr3-iCreERT2 mice have both copies of Sox2, or that inner pillar cells still receive JAG1 signaling since the Fgfr3-iCreERT2 line does not target the SC subtypes located on the medial side of inner pillar cells. These results emphasize the need for controls of various genotypes and the benefit of using complementary approaches to study gene function.
We found that Jag1 deletion at P0 using Fgfr3-iCreERT2 resulted in mild but significant hearing deficits at 4 kHz. The reason for the decrease in low-frequency hearing sensitivity is unknown. However, it is intriguing to speculate that it is causally linked to diminished Hensen's cell function. Hensen's cells have a role in the formation of the tectorial membrane (Rau et al., 1999; Kim et al., 2019), they participate in recycling potassium ions from the base of OHCs to the endolymph through gap junction connections with Deiters' cells (Zwislocki et al., 1992; Sato and Santos-Sacchi, 1994; Kikuchi et al., 1995; Lautermann et al., 1998), and they are involved in maintaining osmotic balance through their expression of aquaporin4 (Takumi et al., 1998; J. Li and Verkman, 2001). Moreover, a recent study provides evidence that, in the cochlear apex, cells located lateral to OHCs (Hensen's cells and Claudius cells) possess passive, yet sharply tuned, vibratory characteristics that may tune OHC force generation (Gao et al., 2014). Deregulation of any of these processes could alter hearing sensitivity, with the latter altering low-frequency sensitivity.
Footnotes
This work was supported by National Institute on Deafness and Other Communication Disorders Grants R01DC011571 to A.D. and R01DC014441 to B.C.C., and the Office of the Assistant Secretary of Defense for Health Affairs Grant W81XWH-15-1-0475 to B.C.C. The Southern Illinois University School of Medicine Research Imaging Facility is supported by Office of Naval Research Grant N00014-15-1-2866. We thank Dr. William Richardson (University College London) for providing the Fgfr3-iCreERT2 mouse line; and Dr. Julian Lewis (Cancer Research UK London Research Institute) for providing one of the Jag1fx/fx mouse lines.
B.C.C. is a consultant for Turner Scientific, LLC, and Otonomy, Inc. The remaining authors declare no competing financial interests.
References
- Adam J, Myat A, Le Roux I, Eddison M, Henrique D, Ish-Horowicz D, Lewis J (1998) Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development 125:4645–4654. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD (2006) Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442:823–826. 10.1038/nature04940 [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K (2011) Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9:317–329. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson PJ, Dong Y, Gu S, Liu W, Najarro EH, Udagawa T, Cheng AG (2018) Sox2 haploinsufficiency primes regeneration and Wnt responsiveness in the mouse cochlea. J Clin Invest 128:1641–1656. 10.1172/JCI97248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Ohyama T, Segil N, Groves AK (2011) Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjκ. J Neurosci 31:8046–8058. 10.1523/JNEUROSCI.6671-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY (1999) Math1: an essential gene for the generation of inner ear hair cells. Science 284:1837–1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Breglio AM, May LA, Barzik M, Welsh NC, Francis SP, Costain TQ, Wang L, Anderson DE, Petralia RS, Wang YX, Friedman TB, Wood MJ, Cunningham LL (2020) Exosomes mediate sensory hair cell protection in the inner ear. J Clin Invest 130:2657–2672. 10.1172/JCI128867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J (2006) Notch ligands with contrasting functions: jagged1 and Delta1 in the mouse inner ear. Development 133:1277–1286. 10.1242/dev.02284 [DOI] [PubMed] [Google Scholar]
- Brown RM, Nelson JC, Zhang H, Kiernan AE, Groves AK (2020) Notch-mediated lateral induction is necessary to maintain vestibular prosensory identity during inner ear development. Dev Biol 462:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckiova D, Syka J (2009) Calbindin and S100 protein expression in the developing inner ear in mice. J Comp Neurol 513:469–482. 10.1002/cne.21967 [DOI] [PubMed] [Google Scholar]
- Burns JC, Kelly MC, Hoa M, Morell RJ, Kelley MW (2015) Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat Commun 6:8557. 10.1038/ncomms9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DP, Chrysostomou E, Doetzlhofer A (2016) Canonical Notch signaling plays an instructive role in auditory supporting cell development. Sci Rep 6:19484. 10.1038/srep19484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ (2007) Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood 110:278–286. 10.1182/blood-2006-08-039883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N (2002) The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129:2495–2505. [DOI] [PubMed] [Google Scholar]
- Cohen R, Amir-Zilberstein L, Hersch M, Woland S, Loza O, Taiber S, Matsuzaki F, Bergmann S, Avraham K, Sprinzak D (2020) Mechanical forces drive ordered patterning of hair cells in the mammalian inner ear. Nat Comm 11:5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DP, Garcia-Anoveros J, Holt JR, Kwan KY, Lin SY, Vollrath MA, Amalfitano A, Cheung EL, Derfler BH, Duggan A, Geleoc GS, Gray PA, Hoffman MP, Rehm HL, Tamasauskas D, Zhang DS (2004) TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature 432:723–730. 10.1038/nature03066 [DOI] [PubMed] [Google Scholar]
- Cox BC, Liu Z, Lagarde MM, Zuo J (2012) Conditional gene expression in the mouse inner ear using Cre-loxP. J Assoc Res Otolaryngol 13:295–322. 10.1007/s10162-012-0324-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudet N, Lewis J (2005) Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development 132:541–551. 10.1242/dev.01589 [DOI] [PubMed] [Google Scholar]
- Daudet N, Żak M (2020) Notch signalling: the multitask manager of inner ear development and regeneration. Adv Exp Med Biol 1218:129–157. 10.1007/978-3-030-34436-8_8 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N (2009) Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell 16:58–69. 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddison M, Le Roux I, Lewis J (2000) Notch signaling in the development of the inner ear: lessons from Drosophila. Proc Natl Acad Sci USA 97:11692–11699. 10.1073/pnas.97.22.11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganfornina MD, Do Carmo S, Lora JM, Torres-Schumann S, Vogel M, Allhorn M, Gonzalez C, Bastiani MJ, Rassart E, Sanchez D (2008) Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 7:506–515. 10.1111/j.1474-9726.2008.00395.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SS, Wang R, Raphael PD, Moayedi Y, Groves AK, Zuo J, Applegate BE, Oghalai JS (2014) Vibration of the organ of Corti within the cochlear apex in mice. J Neurophysiol 112:1192–1204. 10.1152/jn.00306.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R, Brown RM 2nd, Hsu CW, Cai T, Crowder AL, Piazza VG, Vadakkan TJ, Dickinson ME, Groves AK (2016) Lineage tracing of Sox2-expressing progenitor cells in the mouse inner ear reveals a broad contribution to non-sensory tissues and insights into the origin of the organ of Corti. Dev Biol 414:72–84. 10.1016/j.ydbio.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Koesters R, Bouchard M, Gridley T, Pfannenstiel S, Plinkert PK, Zhang L, Praetorius M (2012) Jagged1-mediated Notch signaling regulates mammalian inner ear development independent of lateral inhibition. Acta Otolaryngol 132:1028–1035. 10.3109/00016489.2012.690533 [DOI] [PubMed] [Google Scholar]
- Hartman BH, Hayashi T, Nelson BR, Bermingham-McDonogh O, Reh TA (2007) Dll3 is expressed in developing hair cells in the mammalian cochlea. Dev Dyn 236:2875–2883. 10.1002/dvdy.21307 [DOI] [PubMed] [Google Scholar]
- Hartman BH, Reh TA, Bermingham-McDonogh O (2010) Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci USA 107:15792–15797. 10.1073/pnas.1002827107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS (1995) Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci USA 92:9815–9819. 10.1073/pnas.92.21.9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Schweisguth F (2019) Mechanisms of Notch signaling: a simple logic deployed in time and space. Development 146:dev172148 10.1242/dev.172148 [DOI] [PubMed] [Google Scholar]
- Hertzano R, Puligilla C, Chan SL, Timothy C, Depireux DA, Ahmed Z, Wolf J, Eisenman DJ, Friedman TB, Riazuddin S, Kelley MW, Strome SE (2010) CD44 is a marker for the outer pillar cells in the early postnatal mouse inner ear. J Assoc Res Otolaryngol 11:407–418. 10.1007/s10162-010-0211-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain F, Sorrentino C, Ucar DA, Peng Y, Matossian M, Wyczechowska D, Crabtree J, Zabaleta J, Morello S, Del Valle L, Burow M, Collins-Burow B, Pannuti A, Minter LM, Golde TE, Osborne BA, Miele L (2018) Notch signaling regulates mitochondrial metabolism and NF-kappaB activity in triple-negative breast cancer cells via IKKalpha-dependent non-canonical pathways. Front Oncol 8:575. 10.3389/fonc.2018.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kempfle JS, Turban JL, Edge AS (2016) Sox2 in the differentiation of cochlear progenitor cells. Sci Rep 6:23293. 10.1038/srep23293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE. (2013) Notch signaling during cell fate determination in the inner ear. Semin Cell Dev Biol 24:470–479. 10.1016/j.semcdb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T (2006) The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet 2:e4. 10.1371/journal.pgen.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T (2005) The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development 132:4353–4362. 10.1242/dev.02002 [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Kimura RS, Paul DL, Adams JC (1995) Gap junctions in the rat cochlea: immunohistochemical and ultrastructural analysis. Anat Embryol (Berl) 191:101–118. 10.1007/BF00186783 [DOI] [PubMed] [Google Scholar]
- Kim DK, Kim JA, Park J, Niazi A, Almishaal A, Park S (2019) The release of surface-anchored alpha-tectorin, an apical extracellular matrix protein, mediates tectorial membrane organization. Sci Adv 5:eaay6300. 10.1126/sciadv.aay6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirjavainen A, Laos M, Anttonen T, Pirvola U (2015) The Rho GTPase Cdc42 regulates hair cell planar polarity and cellular patterning in the developing cochlea. Biol Open 4:516–526. 10.1242/bio.20149753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, Palermo AT, So KS, Mays JC, Orvis J, Burns JC, Hertzano R, Driver EC, Kelley MW (2020) Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun 11:2389. 10.1038/s41467-020-16113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW (1999) Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet 21:289–292. 10.1038/6804 [DOI] [PubMed] [Google Scholar]
- Lautermann J, ten Cate WJ, Altenhoff P, Grummer R, Traub O, Frank H, Jahnke K, Winterhager E (1998) Expression of the gap-junction connexins 26 and 30 in the rat cochlea. Cell Tissue Res 294:415–420. 10.1007/s004410051192 [DOI] [PubMed] [Google Scholar]
- Legendre K, Safieddine S, Kussel-Andermann P, Petit C, El-Amraoui A (2008) alphaII-betaV spectrin bridges the plasma membrane and cortical lattice in the lateral wall of the auditory outer hair cells. J Cell Sci 121:3347–3356. 10.1242/jcs.028134 [DOI] [PubMed] [Google Scholar]
- Lelli A, Asai Y, Forge A, Holt JR, Geleoc GS (2009) Tonotopic gradient in the developmental acquisition of sensory transduction in outer hair cells of the mouse cochlea. J Neurophysiol 101:2961–2973. 10.1152/jn.00136.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Verkman AS (2001) Impaired hearing in mice lacking aquaporin-4 water channels. J Biol Chem 276:31233–31237. 10.1074/jbc.M104368200 [DOI] [PubMed] [Google Scholar]
- Li S, Mark S, Radde-Gallwitz K, Schlisner R, Chin MT, Chen P (2008) Hey2 functions in parallel with Hes1 and Hes5 for mammalian auditory sensory organ development. BMC Dev Biol 8:20. 10.1186/1471-213X-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DJ, Anniko M (1985) Developmental morphology of the mouse inner ear: a scanning electron microscopic observation. Acta Otolaryngol Suppl 422:1–69. [PubMed] [Google Scholar]
- Lu L, Pandey AK, Houseal MT, Mulligan MK (2016) The genetic architecture of murine glutathione transferases. PLoS One 11:e0148230 10.1371/journal.pone.0148230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass JC, Gu R, Basch ML, Waldhaus J, Lopez EM, Xia A, Oghalai JS, Heller S, Groves AK (2015) Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front Cell Neurosci 9:110. 10.3389/fncel.2015.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass JC, Gu R, Cai T, Wan YW, Cantellano SC, Asprer JS, Zhang H, Jen HI, Edlund RK, Liu Z, Groves AK (2016) Transcriptomic analysis of mouse cochlear supporting cell maturation reveals large-scale changes in notch responsiveness prior to the onset of hearing. PLoS One 11:e0167286. 10.1371/journal.pone.0167286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann ZF, Gálvez H, Pedreno D, Chen Z, Chrysostomou E, Żak M, Kang M, Canden E, Daudet N (2017) Shaping of inner ear sensory organs through antagonistic interactions between Notch signalling and Lmx1a. Elife 6:e33323 10.7554/eLife.33323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HA, Rakowiecki SM, Gridley T, Fishell G (2006) Loss of notch activity in the developing central nervous system leads to increased cell death. Dev Neurosci 28:49–57. 10.1159/000090752 [DOI] [PubMed] [Google Scholar]
- McGough IJ, Vincent JP (2016) Exosomes in developmental signalling. Development 143:2482–2493. 10.1242/dev.126516 [DOI] [PubMed] [Google Scholar]
- McGovern MM, Brancheck J, Grant AC, Graves KA, Cox BC (2017) Quantitative analysis of supporting cell subtype labeling among CreER lines in the neonatal mouse cochlea. J Assoc Res Otolaryngol 18:227–245. 10.1007/s10162-016-0598-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SC, Cox BC (2016) Whole mount dissection and immunofluorescence of the adult mouse cochlea. J Vis Exp 107:53561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Hodgetts C, Gossler A, Hrabe de Angelis M, Lewis J (1999) Expression of Delta1 and Serrate1 (Jagged1) in the mouse inner ear. Mech Dev 84:169–172. 10.1016/s0925-4773(99)00066-0 [DOI] [PubMed] [Google Scholar]
- Murata J, Tokunaga A, Okano H, Kubo T (2006) Mapping of notch activation during cochlear development in mice: implications for determination of prosensory domain and cell fate diversification. J Comp Neurol 497:502–518. 10.1002/cne.20997 [DOI] [PubMed] [Google Scholar]
- Neves J, Parada C, Chamizo M, Giraldez F (2011) Jagged 1 regulates the restriction of Sox2 expression in the developing chicken inner ear: a mechanism for sensory organ specification. Development 138:735–744. 10.1242/dev.060657 [DOI] [PubMed] [Google Scholar]
- Nie X, Zhang K, Wang L, Ou G, Zhu H, Gao WQ (2015) Transcription factor STOX1 regulates proliferation of inner ear epithelial cells via the AKT pathway. Cell Prolif 48:209–220. 10.1111/cpr.12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR (2008) Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol 9:65–89. 10.1007/s10162-007-0106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, McFadden SL, Ding DL, Lear PM, Ho YS (2000) Targeted mutation of the gene for cellular glutathione peroxidase (Gpx1) increases noise-induced hearing loss in mice. J Assoc Res Otolaryngol 1:243–254. 10.1007/s101620010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Kamakura S, Isazawa Y, Yoshimatsu T, Kuida K, Nakafuku M, Masuyama N, Gotoh Y (2004) Notch promotes survival of neural precursor cells via mechanisms distinct from those regulating neurogenesis. Dev Biol 276:172–184. 10.1016/j.ydbio.2004.08.039 [DOI] [PubMed] [Google Scholar]
- Pan W, Jin Y, Stanger B, Kiernan AE (2010) Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc Natl Acad Sci USA 107:15798–15803. 10.1073/pnas.1003089107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumalsamy LR, Nagala M, Sarin A (2010) Notch-activated signaling cascade interacts with mitochondrial remodeling proteins to regulate cell survival. Proc Natl Acad Sci USA 107:6882–6887. 10.1073/pnas.0910060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic J, Formosa-Jordan P, Luna-Escalante JC, Abelló G, Ibañes M, Neves J, Giraldez F (2014) Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development 141:2313–2324. 10.1242/dev.108100 [DOI] [PubMed] [Google Scholar]
- Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, Maric D, Eberhart CG, Fine HA (2005) Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res 65:2353–2363. 10.1158/0008-5472.CAN-04-1890 [DOI] [PubMed] [Google Scholar]
- Rau A, Legan PK, Richardson GP (1999) Tectorin mRNA expression is spatially and temporally restricted during mouse inner ear development. J Comp Neurol 405:271–280. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD (2008) PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci 11:1392–1401. 10.1038/nn.2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Hofmann S, Muhlenbein N, Paschen SA, Gerbitz KD, Neupert W, Brunner M, Bauer MF (2001) Role of the deafness dystonia peptide 1 (DDP1) in import of human Tim23 into the inner membrane of mitochondria. J Biol Chem 276:37327–37334. 10.1074/jbc.M105313200 [DOI] [PubMed] [Google Scholar]
- Saino-Saito S, Suzuki R, Tokuda N, Abe H, Kondo H, Owada Y (2010) Localization of fatty acid binding proteins (FABPs) in the cochlea of mice. Ann Anat 192:210–214. 10.1016/j.aanat.2010.06.007 [DOI] [PubMed] [Google Scholar]
- Sato Y, Santos-Sacchi J (1994) Cell coupling in the supporting cells of Corti's organ: sensitivity to intracellular H+ and Ca+2. Hear Res 80:21–24. 10.1016/0378-5955(94)90004-3 [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 169:361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Schmitz F, Jahn R, Rosenmund C, Südhof TC (2004) A complete genetic analysis of neuronal Rab3 function. J Neurosci 24:6629–6637. 10.1523/JNEUROSCI.1610-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Son EJ, Wu L, Yoon H, Kim S, Choi JY, Bok J (2012) Developmental gene expression profiling along the tonotopic axis of the mouse cochlea. PLoS One 7:e40735. 10.1371/journal.pone.0040735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarama KB, Gavara N, Petralia RS, Kelley MW, Chadwick RS (2012) Cytoskeletal changes in actin and microtubules underlie the developing surface mechanical properties of sensory and supporting cells in the mouse cochlea. Development 139:2187–2197. 10.1242/dev.073734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S, Yamamoto N, Yabe D, Fukuda H, Kojima K, Ito J, Honjo T (2007) Multiple roles of Notch signaling in cochlear development. Dev Biol 307:165–178. 10.1016/j.ydbio.2007.04.035 [DOI] [PubMed] [Google Scholar]
- Takumi Y, Nagelhus EA, Eidet J, Matsubara A, Usami S, Shinkawa H, Nielsen S, Ottersen OP (1998) Select types of supporting cell in the inner ear express aquaporin-4 water channel protein. Eur J Neurosci 10:3584–3595. 10.1046/j.1460-9568.1998.00360.x [DOI] [PubMed] [Google Scholar]
- Tang LS, Alger HM, Pereira FA (2006) COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development 133:3683–3693. 10.1242/dev.02536 [DOI] [PubMed] [Google Scholar]
- Tateya T, Imayoshi I, Tateya I, Ito J, Kageyama R (2011) Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev Biol 352:329–340. 10.1016/j.ydbio.2011.01.038 [DOI] [PubMed] [Google Scholar]
- Walters BJ, Yamashita T, Zuo J (2015) Sox2-CreER mice are useful for fate mapping of mature, but not neonatal, cochlear supporting cells in hair cell regeneration studies. Sci Rep 5:11621. 10.1038/srep11621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N (2006) Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441:984–987. 10.1038/nature04849 [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW (2004) Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci 7:1310–1318. 10.1038/nn1349 [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Tanigaki K, Tsuji M, Yabe D, Ito J, Honjo T (2006) Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J Mol Med (Berl) 84:37–45. 10.1007/s00109-005-0706-9 [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Chang W, Kelley MW (2011) Rbpj regulates development of prosensory cells in the mammalian inner ear. Dev Biol 353:367–379. 10.1016/j.ydbio.2011.03.016 [DOI] [PubMed] [Google Scholar]
- Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, Richardson WD (2010) An Fgfr3-iCreER(T2) transgenic mouse line for studies of neural stem cells and astrocytes. Glia 58:943–953. 10.1002/glia.20976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ (2000) Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci 3:580–586. 10.1038/75753 [DOI] [PubMed] [Google Scholar]
- Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, Kageyama R, de Ribaupierre F (2001) Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci 21:4712–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwislocki JJ, Slepecky NB, Cefaratti LK, Smith RL (1992) Ionic coupling among cells in the organ of Corti. Hear Res 57:175–194. 10.1016/0378-5955(92)90150-l [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of top-ranked downregulated genes in Jag1 mutant cochlear epithelia. Genes are ranked based on fold change. Shown are genes that are downregulated (p ≤ 0.05, log2 (FC) ≤3σ) in Sox2-CreERT2/+::Jag1fx/fx (Jag1 mut) samples compared with Jag1fx/fx (Control, Ctrl) samples. Tamoxifen and progesterone were injected at E14.5 and cochlear epithelia were collected at P0. $ marks SC-specific/enriched genes (Burns et al., 2015; Maass et al., 2016) and # marks genes previously found to be positively regulated in cochlear epithelial cells by Notch signaling (Campbell et al., 2016; Maass et al., 2016). Note that only transcripts with mean log2 values of 5.5 or more in control samples are listed. Download Figure 2-1, DOCX file (38.4KB, docx) .
List of top-ranked upregulated genes in Jag1 mutant cochlear epithelia. Genes are ranked based on fold change (FC). Shown are genes that are upregulated (p ≤ 0.05, log2 (FC) ≥3σ) in Sox2-CreERT2/+::Jag1fx/fx (Jag1 mut) samples compared with Jag1fx/fx (Control, Ctrl) samples. Tamoxifen and progesterone were injected at E14.5 and cochlear epithelia were collected at P0. $ marks SC-specific/enriched genes (Burns et al., 2015; Maass et al., 2016) and # marks genes previously found to be negatively regulated in cochlear epithelial cells by Notch signaling (Campbell et al., 2016; Maass et al., 2016). Note that only transcripts with mean log2 values in Jag1 mutant samples of 5.5 or more are listed. Download Figure 2-2, DOCX file (24.8KB, docx) .
Expression of HC-specific transcription factors is unchanged in Jag1 mutant cochlear epithelia. HC-specific transcription factor expression in Sox2-CreERT2/+::Jag1fx/fx (Jag1 mut) samples compared with Jag1fx/fx (Control, Ctrl) samples. Tamoxifen and progesterone were injected at E14.5 and cochlear epithelia were collected and analyzed at P0. Genes are listed in alphabetical order. Download Figure 2-3, DOCX file (14.2KB, docx) .
List of genes and their associated pathways and cellular components downregulated in Jag1 mutant cochlear epithelia. Gene ontology analysis on genes that were downregulated (mean log2 control ≤5, p ≤ 0.05, log2 (FC) ≤-2σ) in P0 Jag1 CKO versus control (Ctrl) cochlear epithelia. Download Figure 2-4, DOCX file (18.7KB, docx) .