Abstract
Cryptosporidium species subtypes are generally identified via DNA sequencing of the gp60 gene tandem repeat motif region. Due to the immunogenic nature of its glycoprotein products, gp60 is subject to host selective pressures, genetic recombination and evolutionary processes that drive extensive polymorphism at this locus. The elucidation of the polymorphic nature of this gene has led to the current mainstay in Cryptosporidium subtyping nomenclature.
This study aimed to develop a real-time polymerase chain reaction based method utilising a post-PCR application, high resolution melting (HRM) analysis, in conjunction with the abovementioned gp60 nomenclature system, in order to differentiate between Cryptosporidium parvum gp60 subtypes. Subtype differentiation is based on the difference between the melting temperatures of individual subtypes conferred by variations in the polymorphic region of gp60.
• Nested gp60 primers were designed to amplify a target region of <200 base pairs for effective HRM analysis
• This method presents a rapid, sensitive, cost effective alternative to conventional sequencing.
• This method is highly flexible and may be applied to other loci in order to facilitate multi-locus analysis and improve the discriminative abilities of the method.
Keywords: Molecular epidemiology, Cryptosporidium, Enteric parasitology, DNA sequencing, Real-time PCR, High resolution melting(HRM) analysis, gp60 gene, Infectious disease
Graphical abstract
Specifications table
| Subject Area: | Immunology and Microbiology |
| More specific subject area: | Molecular Parasitology |
| Method name: | 60 kDa glycoprotein gene based differentiation of Cryptosporidium parvum subtype isolates via high resolution melting (HRM) analysis. |
| Name and reference of original method: | N/A |
| Resource availability: | N/A |
Method details
Method background
Gastrointestinal and diarrhoeal disease contribute significantly to global morbidity and mortality, accounting for more than 1.3 million deaths in 2015 [1]. Cryptosporidium, a globally ubiquitous, protozoan parasite, is an aetiology to which diarrhoea induced death is commonly attributed. Over 90% of cases of cryptosporidiosis are attributable to two species, Cryptosporidium parvum and Cryptosporidium hominis; although nearly 20 species and genotypes have been reported in humans [2]. First reported as an agent of human disease in the 1976 [[3],[4]], cryptosporidiosis, in addition to disproportionately affecting immunocompetent children under the age of five, can result in protracted or recurrent bouts of potentially fatal diarrhoeal illness in immunocompromised individuals.
Previously, diagnostic analysis of this enteric pathogen was predominantly limited to microscopy via fluorescent or acid-fast staining. However, microscopic analysis of oocysts precluded thorough epidemiological analyses, lacking in the ability to distinguish between species of Cryptosporidium morphologically. However, since the release of both the C. parvum and C. hominis genomes in the early 2000s [5], [6], [7], a host of molecular methods have been developed to distinguish between Cryptosporidium species and subspecies, with species identification largely achieved through polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of various genes and DNA sequencing or real-time PCR based analysis of the small subunit ribosomal RNA (SSU rRNA or 18S), a method which has become the mainstay in differentiating C. parvum and C. hominis [8], [9], [10], [11]. Subtype identification is conducted through DNA sequence analysis of the 60 kDa glycoprotein gene (gp60) in accordance with nomenclature developed by Strong et al. and modified by Sulaiman et al. [12,13]. Within this system, subtype assignment is based on the number of TCA repeats (represented by the letter A), TCG repeats (represented by the letter G), TCT repeats (represented by the letter T) and other repetitive sequences, such as the ACATCA (represented by the letter R) associated with the C. parvum IIa allele family, within the gp60 tandem repeat motif region. Subtype families are named as Ia, Ib, Ic, Id, Ie, If, etc. for C. hominis and IIa, IIb, IIc, IId, etc. for C. parvum, with further species families named in ascending order.
The ability to analyse Cryptosporidium species and subtypes is critical as both inter- and intra-species variation in host range and/or virulence/transmissibility have been described [2]. Additionally, Cryptosporidium species exhibit regional variation; with both C. parvum and C. hominis predominance found to vary between industrialised countries, while C. hominis tends to predominate in developing countries [14]. It is worth noting that C. parvum is the predominant species in several European countires [15]. Within Cryptosporidium species, gp60 subtypes also exhibit regional variation, with C. parvum IIa subtypes the most prevalent case of infection in industrialised nations [16]. Indeed, gp60 is one of the most polymorphic genes in the Cryptosporidium genome, subject to selective pressures and genetic recombination. Consequently, Cryptosporidium populations are often panmictic in structure, with this structure particularly common within C. parvum [17,18]. Thus, analysis of the regional variation exhibited by various Cryptosporidium gp60 subspecies is also key to understanding and elucidating the population genetics.
The development of real-time PCR, a rapid, highly sensitive, fluorescence based technology has revolutionised clinical microbiology, with myriad potential applications in molecular parasitology. Real-time PCR also boasts a suite of advanced post-PCR applications, one of which, high resolution melting (HRM) analysis, is capable of interrogating DNA sequence variations via determination of the relationship between melting temperature and DNA fragment denaturation [19]. HRM analysis is sensitive enough to resolve a single base pair difference between amplicons, and has been applied to single nucleotide polymorphism (SNP) and tandem-repeat number based genotyping and determination of DNA methylation status [19]. Recently HRM analysis has been applied to the differentiation of Cryptosporidium species from other apicomplexan parasites, Toxoplasma gondii, Sarcocystis species, Neospora species, and to differentiate between Cryptosporidium species [20], [21], [22]. HRM analysis has also been successfully applied to the intra-species differentiation of Cryptosporidium cuniculus gp60 subtypes Va and Vb [23].
In this study, following confirmation of isolate bank species and subtype designations via a real-time PCR based 18S rRNA gene speciation method and DNA sequencing of the gp60 gene, respectively, HRM analysis was applied to the differentiation of C. parvum gp60 subtypes of allele family IIa. This subtype family is responsible for the majority of C. parvum cases in Europe [15]. Additionally, it has been established that analysis of multiple loci is required for reliable and sufficiently discriminatory genotyping of Cryptosporidium subtypes [24]. This study was conducted in order to develop a HRM analysis based method capable of rapid, detailed epidemiological analyses using the locus of interest, that was also capable of being expanded upon for future development of a multi-locus variable number tandem repeat analysis (MLVA) tool. Such a tool would improve the discriminatory power of the method and would also be applicable to any future pan- European multi-locus genotyping schemes that may be introduced.
Ethical statement
Full ethical approval was obtained from the Cork Institute of Technology (CIT) Research Ethics Committee prior to study commencement (reference number MF-C-JOL12/2014).
Clinical sample acquisition and total nucleic acid extraction
Samples were acquired through collaboration with the Clinical Microbiology Department of Cork University Hospital (CUH). Over a period of three years, from August 2015 to August 2018, inclusive, 163 Cryptosporidium positive isolates were amassed during routine molecular enteric pathogen screening. Sample acceptance for routine molecular enteric pathogen screening was limited to samples from patients presenting with diarrhoea, with an acceptance criterion requiring samples to be graded as type 5 or higher on the Bristol Stool Form Chart (BSFC).
The EntericBio GastroPanel II (Serosep, Limerick, Ireland) one-step, heat treatment extraction protocol, a preliminary stage in the routine molecular enteric pathogen screening process, was utilised, as per manufacturer instructions, for sample DNA extraction.
-
•
EntericBio Stool Preparation Solution (SPS) was inoculated with a sample coated FloqSwab (Copan, Italy) .
-
•
The sample inoculated SPS tube was vortexed for 30 s.
-
•
The inoculated SPS tube was then heated at 103 °C for 30 min to achieve DNA extraction.
18S rRNA gene based species identification
Clinical enteric pathogen screening was limited to genus level identification. Consequently, a previously published method by Mary et al., employing real-time polymerase chain reaction (PCR) based amplification of an 18S rRNA gene target, was used to rapidly differentiate between C. parvum and Cryptosporidium hominis isolates [25]. This method, as described by Mary et al. [25], employed a pan-Cryptosporidium specific primer pair directed at a conserved region of the18S rRNA gene, while two varying minor groove binding fluorescent probes differentiated between C. parvum and C. hominis, respectively. The following modifications to the method were performed: the VIC fluorescent dye bound to the minor groove binding probe for C. parvum was replaced with HEX, in order to align with the LightCycler96 (LC96) (Roche Molecular Diagnostics, Germany) detection specifications; on-site optimisation of the method on the LC96 instrument indicated optimal probe concentrations to be 50 nM, rather than the 100 nM as specified by Mary et al. [25].
Multiplex real-time PCR step-wise description:
-
•
All reactions were carried out at a final volume of 20 µl, with mastermix constituent volumes and concentrations outlined in Table 1. These volumes were multiplied by the desired number of reactions plus an additional 10%, to account for pipetting error.
-
•
15 µl of the prepared mastermix was added to each reaction tube.
-
•
Lastly, 5 µl of genomic template DNA was added to the relevant reaction tube.
-
•
Real-time PCR reactions were conducted on a Roche LC96 thermocycler under the following cycling conditions: initial denaturation at 94 °C for 10 min, subsequent 3-step amplification for 45 cycles, including denaturation at 94 °C for 10 s, annealing at 54 °C for 30 s and extension at 72 °C for 10 s.
Table 1.
Multiplex 18S rRNA gene real-time PCR mastermix components.
| Reagent | Volume (μl) | Final Concentration |
|---|---|---|
| LightCycler 480 High Resolution Melting Master | 4 | 1x |
| Molecular grade water | 10 | N/A |
| Forward primer (10 µM) (‘5 – CATGGATAACCGTGGTAAT – 3′) | 0.4 | 200 nM |
| Reverse primer (10 µM) (‘5 –TACCCTACCGTCTAAAGCTG,– 3′) | 0.4 | 200 nM |
| C. parvum probe (10 µM) (HEX-ATCACATTAAATGT-MGBBHQ) | 0.1 | 50 nM |
| C. hominis probe (10 µM) (FAM-ATCACAATTAATGT-MGB-BHQ) | 0.1 | 50 nM |
| Template | 5 | N/A |
Additional notes:
-
•
The concentration of genomic Cryptosporidium DNA present in each sample could not be determined, as any quantified value would represent the total nucleic acid concentration derived from all microorganisms present in the original faecal sample, given the general extraction protocol employed.
gp60 primer design
MUSCLE software (https://www.ebi.ac.uk/Tools/msa/muscle/) was utilised to conduct multiple sequence alignment on subtype sequences from C. parvum gp60 allele families prevalent in Europe [15], specifically the IIa-IIj families (GenBank accession numbers: AB242224-AB242229, AY382675, AY738185, AY738186, AY738188-AY7381889, AY873780-AY873782, AY738191, AY738195, DQ192502- DQ192508, DQ630514-DQ630515, DQ630517, DQ630519, DQ648531-DQ648537, EU140508), in order to identify homologous regions circumscribing the tandem repeat region of gp60 between the various subspecies.
The online primer designing tool, Primer-Blast (www.ncbi.nlm.nih.gov/tools/primer-blast/), was used to design both outer and inner primers within these homologous regions. The characteristics of the resultant primers are outlined in Table 2. Due to the presence of subtype dependant variation in the number of tandem repeats present in the targeted region, amplicon sizes vary slightly.
Table 2.
gp60 gene targeting primer pairs designed for use in gp60 subtyping and HRM analysis.
| Primer name | Target gene | Annealing temperature (°C) | Sequence (5′ – 3′) | Fragment size (bp) |
|---|---|---|---|---|
| gp60Outer | 60 kDa glycoprotein gene | 65 | F: TCTCCGTTATAGTCTCCGCTGT R: TGCGGGATCTGTTTGGTCTT |
462 - 498 |
| gp60Inner | 60 kDa glycoprotein gene | 60 | F: CCTTCCGTTATAGTCTCCGCT R: CTTCTCCGCCATCTGCTTCT |
141 – 177 |
First round real-time PCR, amplicon purification and sample sequencing
All first round real-time PCR amplifications, employing the gp60Outer primer pair, were conducted using the LC96 instrument (Roche, Basel, Switzerland). The reagent volumes necessary for a single reaction are outlined in Table 3. These volumes were multiplied by the desired number of reactions plus an additional 10%, to account for pipetting error.
Table 3.
First round gp60 real-time PCR mastermix components.
| Reagent | Volume (μl) | Final Concentration |
|---|---|---|
| FastStart Essential DNA Green Master (Roche, Basel, Switzerland) | 10 | 1x |
| Molecular grade water | 4.2 | N/A |
| Gp60Outer forward primer (10 µM) | 0.4 | 200 nM |
| Gp60Outer reverse primer (10 µM) | 0.4 | 200 nM |
| Template | 5 | N/A |
Step-wise description:
-
•
During reaction mastermix preparation, DNA was withheld from the mastermix.
-
•
Reactions were conducted at a volume of 20 µl.
-
•
15 µl of the prepared mastermix was added to each reaction tube.
-
•
5 µl of genomic template DNA was subsequently added to the relevant reaction tube.
-
•
Real-time PCR reactions were conducted under the following cycling conditions: initial denaturation at 95 °C for 10 min, subsequent 3-step amplification for 45 cycles, including denaturation at 95 °C for 30 s, annealing at 65 °C for 30 s and extension at 72 °C for 40 s.
-
•
All resulting clinical isolate amplicons were purified using the High Pure PCR product purification kit (Roche Molecular Diagnostics, Germany).
-
•
Samples were sequenced bidirectionally off-site via Sanger sequencing (Eurofins, Cologne, Germany).
Additional notes:
-
•
Amplicon purification was conducted as per the manufacturer's instructions, with one minor modification. At the elution phase of the purification protocol, 20 µl elution buffer was added to the column, instead of the indicated 50 µl, and an additional incubation step at 35 °C for 5 min was included prior to centrifugation, in order to improve DNA yield.
gp60 subtype identification
Sequence data were subsequently analysed and gp60 subtype designations were successfully determined for 149 of the 163 samples (91.4%).
Overall, 12 C. parvum subtypes were identified amongst the clinical samples, with one of each sample of each subtype selected for further analysis. An additional 6 reference samples were also included in the study, giving a total of 18 C. parvum family IIA subtypes selected for HRM analysis, as outlined in Table 4.
Table 4.
Provenance of gp60 subtyped C. parvum isolates included in the HRM analysis study.
| Species | gp60 Subspecies Designation | Source | Country of origin |
|---|---|---|---|
| C. parvum | IIaA10G2R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA15G1R2 | Clinical Isolate | Ireland |
| C. parvum | IIaA15G2R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA16R1 | Reference Sample | United Kingdom |
| C. parvum | IIaA16G1R1 | Reference Sample | United Kingdom |
| C. parvum | IIaA17G1R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA17G2R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA17G3R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA17G4R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA18G1R1 | Reference Sample | United Kingdom |
| C. parvum | IIaA18G3R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA19G3R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA19G4R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA20G1R1 | Reference Sample | United Kingdom |
| C. parvum | IIaA20G2R1 | Reference Sample | United Kingdom |
| C. parvum | IIaA20G3R1 | Clinical Isolate | Ireland |
| C. parvum | IIaA21G1R1 | Reference Sample | United Kingdom |
| C. parvum | IIaA21G3R1 | Clinical Isolate | Ireland |
Nested real-time PCR and HRM curve acquisition
Second round, inner gp60 primer amplification reactions were conducted on the LC96 analyser (Roche Molecular Diagnostics, Germany). The reagent volumes necessary for a single reaction are outlined in Table 5. These volumes were multiplied by the desired number of reactions plus an additional 10%, to account for pipetting error.
Table 5.
Second round gp60 real-time PCR mastermix components.
| Reagent | Volume (μl) | Final Concentration |
|---|---|---|
| LightCycler 480 High Resolution Melting Master (Roche) | 10 | 1x |
| Molecular grade water | 4.4 | N/A |
| Gp60Inner forward primer (10 µM) | 0.6 | 300 nM |
| Gp60Inner reverse primer (10 µM) | 0.6 | 300 nM |
| MgCl2 (25 mM) | 2.4 | 3.0 mM |
| Template | 2 | N/A |
Step-wise description:
-
•
First round reaction PCR products were diluted 1:100 in molecular grade water. During reaction mastermix preparation, DNA was withheld from the mastermix.
-
•
Reactions were conducted at a volume of 20 µl.
-
•
18 µl of the prepared mastermix was added to each reaction tube.
-
•
2 µl of genomic template DNA was subsequently added to the relevant reaction tube.
-
•
Real-time PCR reactions were conducted under the following cycling conditions: initial denaturation at 95 °C for 10 min, subsequent 3-step amplification for 35 cycles, including denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 40 s.
-
•
Incorporated into the second round real-time PCR cycling conditions, HRM was conducted immediately post-PCR amplification. HRM conditions were as follows: DNA was initially denatured by heating at 95 °C for 60 s, followed by cooling at 40 °C for 60 s and subsequently increasing the temperature by 2.2 °C/s from 65 °C to 97 °C, taking 15 continuous readings/ °C, in order to monitor the change in fluorescence.
Additional notes:
-
•
MgCl2 was added as a separate mastermix constituent for second round amplification reactions. LightCycler 480 High Resolution Melting Master Mix(Roche, Basel, Switzerland) does not contain MgCl2 to allow for optimisation of MgCl2 concentrations, ensuring specific amplification for optimal HRM analysis.
High Resolution Melting (HRM) analysis of inner gp60 amplicons
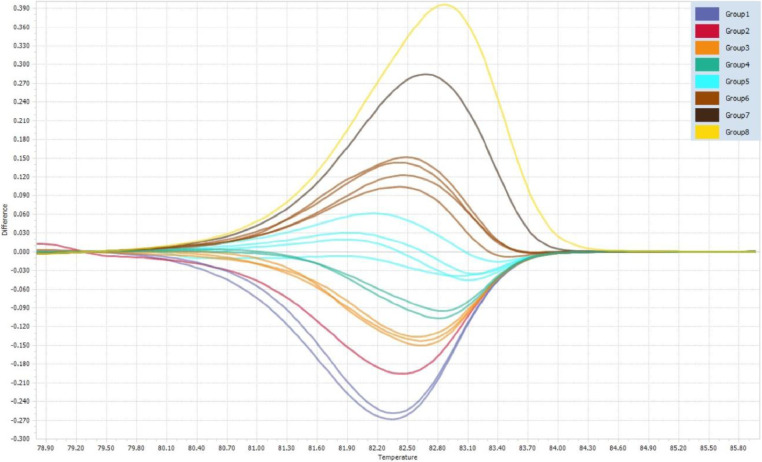
Following high resolution melting-curve acquisition, the resulting data were analysed and a normalisation region of 79 to 86 °C was applied for analysis within the LC96 software (Roche Molecular Diagnostics, Basel, Switzerland). The positive/negative threshold was set to the default 0.05 relative fluorescence units (RFU). Delta melting temperature (Tm) discrimination and curve shape discrimination parameters were set to 100%. Normalised melting curve, differential melting peak, and difference plots were generated and analysed in order to determine the precise melting temperature of each C. parvum subtype. Analysis was conducted on all duplicate runs.
This method successfully differentiated the studied subtypes into 8 subtype groupings, as specified in Table 6 and highlighted in Fig. 1. This method also has the potential to be expanded upon to include other tandem repeat loci for improved C. parvum subtype discrimination, adopting a multi-locus genotyping approach that is commonly employed in routine MLVA and population genetics analyses of Cryptosporidium species.
Table 6.
Intra- and inter-assay reproducibility of melting temperatures of C. parvum gp60 subtypes exhibited by HRM analysis.
| gp60 subtype | Subtype grouping | Intra-assay Reproducibility |
Inter-assay Reproducibility |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Tm* 1 | Tm* 2 | Mean Tm*±SD⁎⁎ | %CV⁎⁎⁎ | Tm* 1 | Tm* 2 | Mean Tm*±SD⁎⁎ | %CV⁎⁎⁎ | ||
| IIaA21G1R1 | 1 | 82.02 | 81.96 | 81.99±0.04 | 0.052 | 81.94 | 81.88 | 81.91±0.04 | 0.052 |
| IIaA20G1R1 | 1 | 82.02 | 82.02 | 82.02±0.00 | 0.000 | 82 | 81.94 | 81.97±0.04 | 0.052 |
| IIaA16R1 | 2 | 82.16 | 82.22 | 82.19±0.04 | 0.052 | 82.14 | 82.08 | 82.11±0.04 | 0.052 |
| IIaA20G2R1 | 3 | 82.29 | 82.29 | 82.29±0.00 | 0.000 | 82.21 | 82.21 | 82.21±0.00 | 0.000 |
| IIaA17G1R1 | 3 | 82.24 | 82.29 | 82.27±0.04 | 0.043 | 82.34 | 82.27 | 82.31±0.05 | 0.060 |
| IIaA18G1R1 | 3 | 82.29 | 82.35 | 82.32±0.04 | 0.052 | 82.28 | 82.21 | 82.25±0.05 | 0.060 |
| IIaA16G1R1 | 4 | 82.42 | 82.42 | 82.42±0.00 | 0.000 | 82.4 | 82.41 | 82.41±0.01 | 0.010 |
| IIaA15G1R2 | 4 | 82.48 | 82.42 | 82.45±0.04 | 0.051 | 82.41 | 82.34 | 82.38±0.05 | 0.060 |
| IIaA17G2R1 | 5 | 82.55 | 82.62 | 82.59±0.05 | 0.060 | 82.6 | 82.54 | 82.57±0.04 | 0.051 |
| IIaA21G3R1 | 5 | 82.49 | 82.55 | 82.52±0.04 | 0.051 | 82.4 | 82.4 | 82.40±0.00 | 0.000 |
| IIaA20G3R1 | 5 | 82.55 | 82.55 | 82.55±0.00 | 0.000 | 82.54 | 82.47 | 82.51±0.05 | 0.060 |
| IIaA19G3R1 | 5 | 82.62 | 82.68 | 82.65±0.04 | 0.051 | 82.54 | 82.47 | 82.51±0.05 | 0.060 |
| IIaA15G2R1 | 6 | 82.81 | 82.8 | 82.81±0.01 | 0.010 | 82.8 | 82.73 | 82.77±0.05 | 0.060 |
| IIaA18G3R1 | 6 | 82.81 | 82.81 | 82.81±0.00 | 0.000 | 82.73 | 82.67 | 82.70±0.04 | 0.051 |
| IIaA17G3R1 | 6 | 82.75 | 82.75 | 82.75±0.00 | 0.000 | 82.79 | 82.73 | 82.76±0.04 | 0.051 |
| IIaA19G4R1 | 6 | 82.81 | 82.88 | 82.85±0.05 | 0.060 | 82.8 | 82.8 | 82.80±0.00 | 0.000 |
| IIaA17G4R1 | 7 | 83.14 | 83.14 | 83.14±0.00 | 0.000 | 83.12 | 83.06 | 83.09±0.04 | 0.051 |
| IIaA10G2R1 | 8 | 83.32 | 83.29 | 83.31±0.02 | 0.025 | 83.26 | 83.26 | 83.26±0.00 | 0.000 |
Melting temperature.
Standard deviation.
Coefficient of variance.
Fig. 1.
Difference plot of C. parvum subtype groupings.
Method validation
Reproducibility of HRM analysis
In order to assess the intra-experimental reproducibility, duplicate reactions were analysed of the subtype samples within a single PCR run. Inter-experimental variation was assessed in separate, duplicate runs of identical subtype sample composition.
Based on the data obtained from intra- and inter-experimental reproducibility experiments, the averages, standard deviations, and coefficients of variation of Tm peaks generated by the HRM software were calculated and indicated high levels of reproducibility, as outlined in Table 6.
Limit of detection evaluation
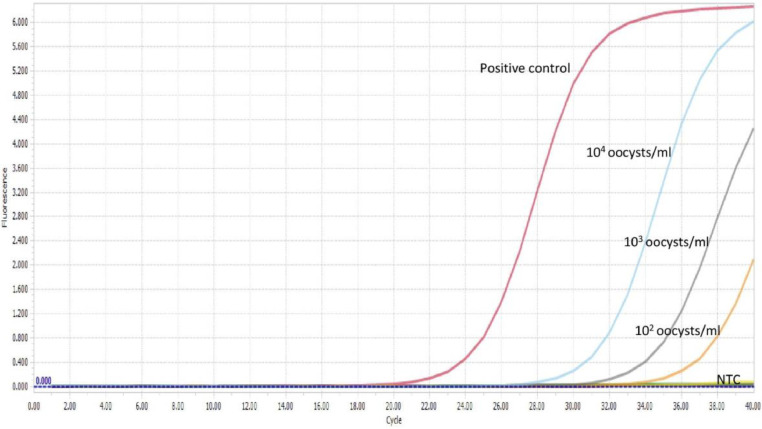
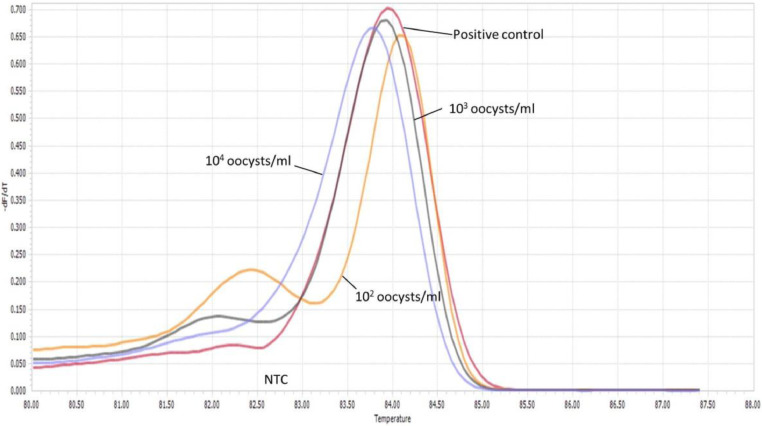
The limit of detection (LoD) for both gp60 outer and inner primer pairs was evaluated using Cryptosporidium DNA extracted from a semi-purified (via salt floatation) faecal sample containing Cryptosporidium parvum oocysts. Prior to extraction, oocysts within a semi-purified faecal sample were microscopically enumerated in triplicate with KOVA Glasstic Slides (Medical Supply Company, Dublin, Ireland), with the average oocyst concentration determined to be approximately 1 × 104 oocysts per ml. DNA was extracted from oocyst samples, following the same SPS extraction protocol outlined previously for clinical samples. A 10-fold serial dilution, ranging from 1 × 104 oocysts per ml to 1 × 10−2 oocysts per ml was prepared from the C. parvum genomic extract.
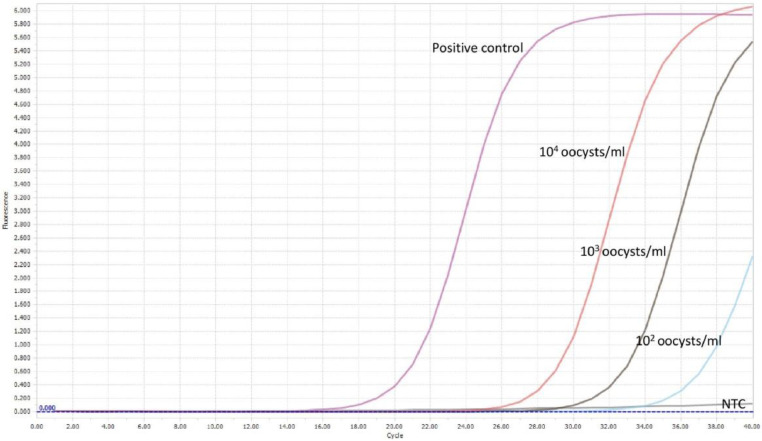
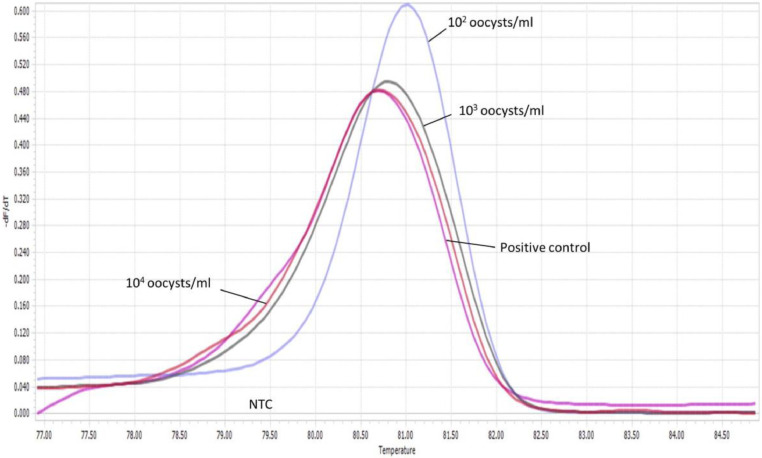
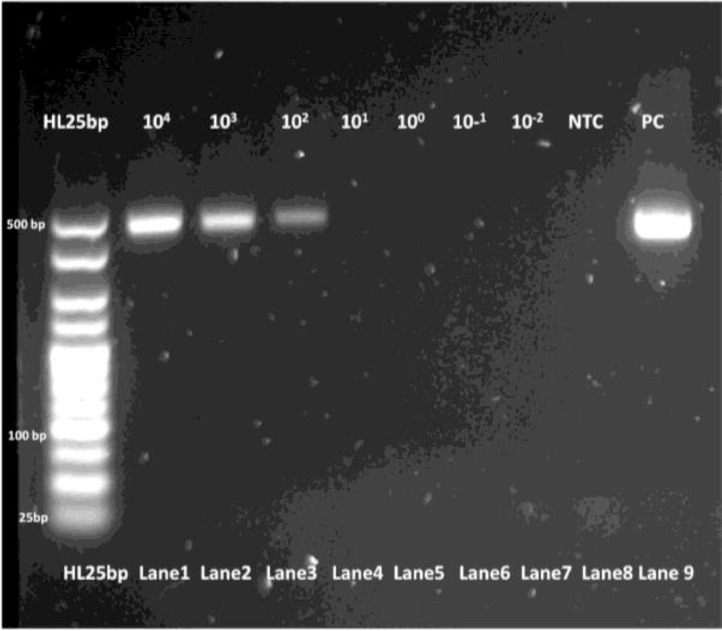
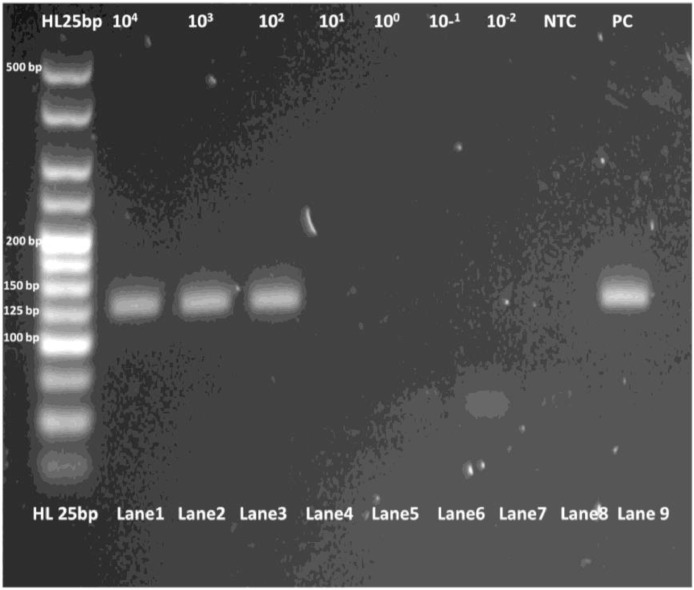
The limit of detection for both C. parvum gp60 outer and inner primer pairs was assessed employing the respective amplification conditions outlined previously, and gel electrophoresis. All reactions were conducted in duplicate. For clarity, single reactions are shown in Figs. 2, 3, 5 and 6. The limit of detection for both outer and inner primer pairs was 1 × 102 oocysts/ml, as seen in Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7.
Fig. 2.
Real-time PCR amplification curves of C. parvum outer gp60 primer pair sensitivity evaluation.
Fig. 3.
HRM analysis melting peaks of C. parvum outer primer pair sensitivity evaluation.
Fig. 5.
Real-time PCR amplification curves of C. parvum inner gp60 primer pair sensitivity evaluation.
Fig. 6.
HRM analysis melting peaks of C. parvum inner primer pair sensitivity evaluation.
Fig. 4.
PCR products of C. parvum outer gp60 primer pair sensitivity evaluation visualized via gel electrophoresis.
Fig. 7.
PCR products of C. parvum inner gp60 primer pair sensitivity evaluation visualized via gel electrophoresis.
Specificity evaluation
The specificity of both C. parvum outer and inner gp60 primers sets was also evaluated, using DNA extracts from varying enteric pathogens, of various genera, both bacterial and parasitic in nature. Salmonella, Shigella, Campylobacter, Verotoxigenic Escherichia coli (VTEC) and G. lamblia positive faecal DNA extracts detected during routine molecular enteric screening in CUH were tested, in addition to genomic DNA extracted from Blastocystis hominis cysts (ATCC, United States of America). Salmonella, Shigella, Campylobacter, Verotoxigenic Escherichia coli (VTEC) and G. lamblia were extracted on-site in the Medical Microbiology Department of CUH, via the EntericBio SPS based protocol as outlined previously. DNA was extracted from B. hominis cysts employing the DNeasy Blood and Tissue Kit (Qiagen, German), as per manufacturer's instructions for DNA extraction from Gram-negative bacteria.
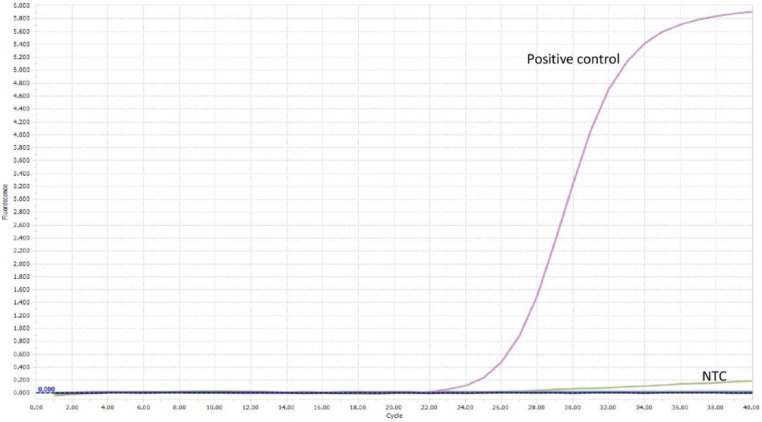
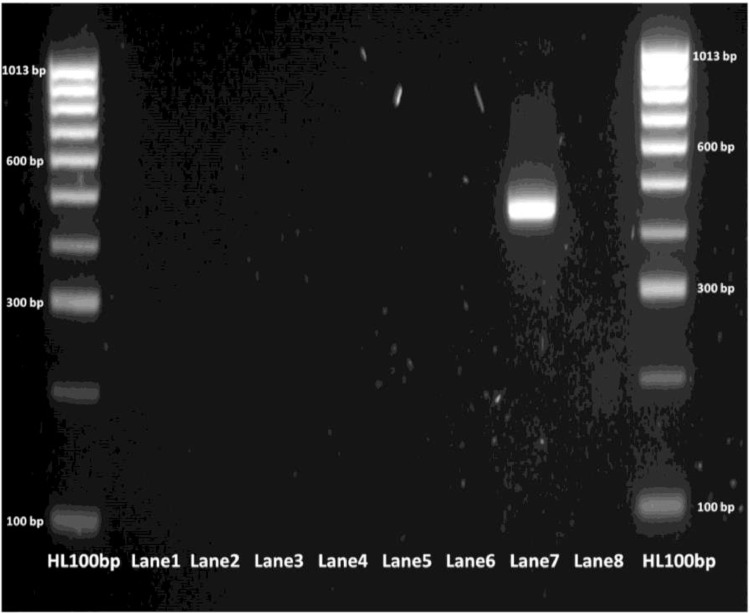
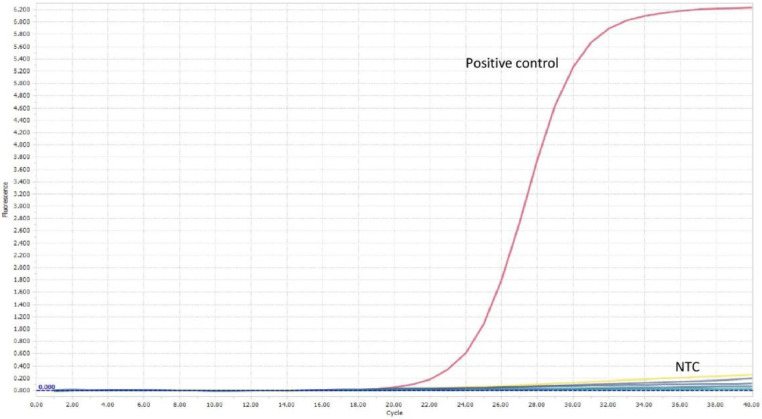
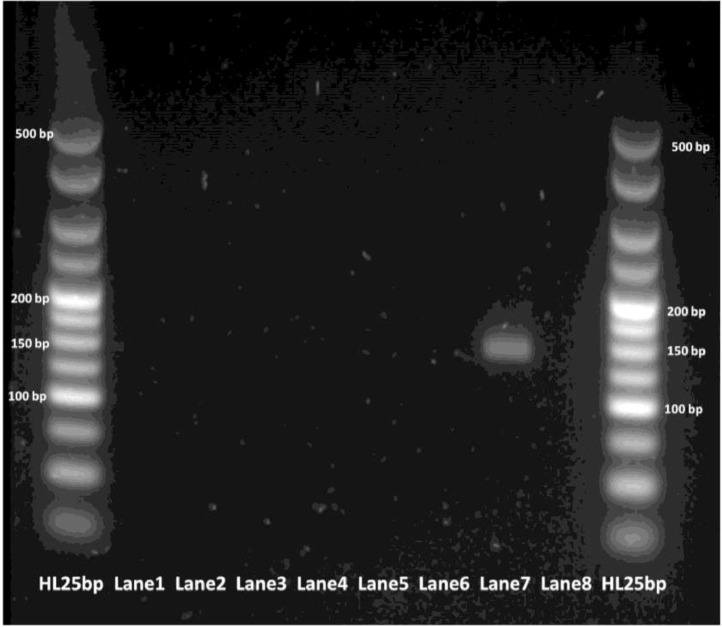
Extracted DNA was amplified under the real-time PCR conditions highlighted previously for both gp60 primer sets, respectively. All reactions were conducted in duplicate. Amplification of the C. parvum template DNA alone was observed for both primer pairs, as seen in Fig. 8, Fig. 9, Fig. 10, Fig. 11.
Fig. 8.
Real-time PCR amplification curves of C. parvum outer gp60 primer pair specificity evaluation.
Fig. 9.
PCR products of C. parvum outer gp60 primer pair specificity evaluation visualized via gel electrophoresis. Lane 1 - Salmonella.; Lane 2 - Shigella; Lane 3 - Campylobacter; Lane 4 - VTEC; Lane 5 - G. lamblia; Lane 6 - B. hominis; Lane 7 - C. parvum (positive control); Lane 8 - NTC.
Fig. 10.
Real-time PCR amplification curves of C. parvum inner gp60 primer pair specificity evaluation.
Fig. 11.
PCR products of C. parvum inner gp60 primer pair sensitivity evaluation visualized via gel electrophoresis. Lane 1 - Salmonella.; Lane 2 - Shigella; Lane 3 - Campylobacter; Lane 4 - VTEC; Lane 5 - G. lamblia; Lane 6 - B. hominis; Lane 7 - C. parvum (positive control); Lane 8 - NTC.
Declaration of Competing Interest
There are no conflicts of interest, of which we are aware, relating to this body of work.
Acknowledgements
The authors would like to thank the Irish Research Council (IRC) for their financial support of this research through the Government of Ireland Postgraduate Scholarship Programme [GOIPG/2014/918]. We would also like to express our sincere gratitude to the entire staff of the Clinical Microbiology Department of Cork University Hospital for their support of this work. The authors also wish to extend thanks to Dr Rachel Chalmers and Dr Kristin Elwin of the Cryptosporidium Reference Unit, Swansea, Wales, for their scientific support and expertise.
References
- 1.Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Reiner R.C. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan U., Hijjawi N. New developments in Cryptosporidium research. Int. J. Parasitol. 2015;45:367–373. doi: 10.1016/j.ijpara.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Meisel J.L., Perera D.R., Meligro C., Rubin C.E. Overwhelming watery diarrhea associated with a cryptosporidium in an immunosuppressed patient. Gastroenterology. 1976;70:1156–1160. [PubMed] [Google Scholar]
- 4.Nime F.A., Burek J.D., Page D.L., Holscher M.A., Yardley J.H. Acute enterocolitis in a human being infected with the protozoan Cryptosporidium. Gastroenterology. 1976;70:592–598. [PubMed] [Google Scholar]
- 5.Abrahamsen M.S., Templeton T.J., Enomoto S., Abrahante J.E., Zhu G., Lancto C.A. Complete Genome Sequence of the Apicomplexan, Cryptosporidium parvum. Science (80-) 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 6.Xu P., Widmer G., Wang Y., Ozaki L.S., Alves J.M., Serrano M.G. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 7.Bankier A.T., Spriggs H.F., Fartmann B., Konfortov B.A., Madera M., Vogel C. Integrated mapping, chromosomal sequencing and sequence analysis of Cryptosporidium parvum. Genome Res. 2003;13:1787–1799. doi: 10.1101/gr.1555203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peralta R.H.S., Velásquez J.N., Cunha F.de S., Pantano M.L., Sodré F.C., da Silva S. Genetic diversity of Cryptosporidium identified in clinical samples from cities in Brazil and Argentina. Mem. Inst. Oswaldo Cruz. 2016;111:30–36. doi: 10.1590/0074-02760150303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leone A., Ripabelli G., Sammarco M.L., Grasso G.M. Detection of Cryptosporidium spp. from human faeces by PCR-RFLP, cloning and sequencing. Parasitol. Res. 2009;104:583–587. doi: 10.1007/s00436-008-1233-8. [DOI] [PubMed] [Google Scholar]
- 10.Abu Samra N., Thompson P.N., Jori F., Frean J., Poonsamy B., du Plessis D. Genetic Characterization of Cryptosporidium spp. in Diarrhoeic Children from Four Provinces in South Africa. Zoonoses Public Health. 2013;60:154–159. doi: 10.1111/j.1863-2378.2012.01507.x. [DOI] [PubMed] [Google Scholar]
- 11.Insulander M., Silverlås C., Lebbad M., Karlsson L., Mattsson J.G., Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol. Infect. 2013;141:1009–1020. doi: 10.1017/S0950268812001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulaiman I.M., Hira P.R., Zhou L., Al-Ali F.M., Al-Shelahi F.A., Shweiki H.M. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strong W.B., Gut J., Nelson R.G. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y., Ryan U.M., Xiao L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Cacciò S.M., Chalmers R.M. Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. 2016;22:471–480. doi: 10.1016/j.cmi.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Torres E., Li N., Wang L., Bowman D., Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int. J. Parasitol. 2013;43:1141–1147. doi: 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 17.De Waele V., Van den Broeck F., Huyse T., McGrath G., Higgins I., Speybroeck N. Panmictic Structure of the Cryptosporidium parvum Population in Irish Calves: Influence of Prevalence and Host Movement. Appl. Environ. Microbiol. 2013;79:2534–2541. doi: 10.1128/AEM.03613-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herges G.R., Widmer G., Clark M.E., Khan E., Giddings C.W., Brewer M. Evidence that cryptosporidium parvum populations are panmictic and unstructured in the upper midwest of the united states. Appl. Environ. Microbiol. 2012;78:8096–8101. doi: 10.1128/AEM.02105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong S.Y.C., Giffard PM. Microbiological applications of high-resolution melting analysis. J. Clin. Microbiol. 2012;50:3418–3421. doi: 10.1128/JCM.01709-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chelbi H., Essid R., Jelassi R., Bouzekri N., Zidi I., Ben Salah H. High-resolution melting-curve (HRM) analysis for C. meleagridis identification in stool samples. Microb. Pathog. 2018;115:332–337. doi: 10.1016/j.micpath.2017.12.070. [DOI] [PubMed] [Google Scholar]
- 21.Fehlberg H.F., Maciel B.M., Albuquerque G.R. Identification and discrimination of Toxoplasma gondii, Sarcocystis spp., Neospora spp., and Cryptosporidium spp. by righ-resolution melting analysis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0174168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pangasa A., Jex A.R., Campbell B.E., Bott N.J., Whipp M., Hogg G. High resolution melting-curve (HRM) analysis for the diagnosis of cryptosporidiosis in humans. Mol. Cell. Probes. 2009;23:10–15. doi: 10.1016/j.mcp.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Hadfield S.J., Chalmers RM. Detection and characterization of Cryptosporidium cuniculus by real-time PCR. Parasitol. Res. 2012;111:1385–1390. doi: 10.1007/s00436-012-2874-1. [DOI] [PubMed] [Google Scholar]
- 24.Chalmers R.M., Pérez-Cordón G., Cacció S.M., Klotz C., Robertson L.J. participants of the Cryptosporidium genotyping workshop (EURO-FBP). Cryptosporidium genotyping in Europe: The current status and processes for a harmonised multi-locus genotyping scheme. Exp. Parasitol. 2018;191:25–30. doi: 10.1016/j.exppara.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Mary C., Chapey E., Dutoit E., Guyot K., Hasseine L., Jeddi F. Multicentric evaluation of a new real-time PCR assay for quantification of Cryptosporidium spp. and identification of Cryptosporidium parvum and Cryptosporidium hominis. J. Clin. Microbiol. 2013;51:2556–2563. doi: 10.1128/JCM.03458-12. [DOI] [PMC free article] [PubMed] [Google Scholar]