Abstract
The diverse viruses in the phylum Nucleocytoviricota (also known as NLCDVs, Nucleo-cytoplasmic Large DNA Viruses) typically possess large icosahedral virions. However, in several families of Nucleocytoviricota, the icosahedral capsid was replaced by irregular particle shapes, most notably, the amphora-like virions of pandoraviruses and pithoviruses, the largest known virus particles in the entire virosphere. Pandoraviruses appear to be the most highly derived viruses in this phylum because their evolution involved not only the change in the virion shape, but also, the actual loss of the gene encoding double-jelly roll major capsid protein (DJR MCP), the main building block of icosahedral capsids in this virus assemblage. Instead, pandoravirus virions are built of unrelated abundant proteins. Here we show that the second most abundant virion protein of pandoraviruses, major virion protein 2 (MVP2), evolved from an inactivated derivative of a bacterial glycoside hydrolase of the GH16 family. The ancestral form of MVP2 was apparently acquired early in the evolution of the Nucleocytoviricota, to become a minor virion protein. After a duplication in the common ancestor of pandoraviruses and molliviruses, one of the paralogs displaces DJR MCP in pandoraviruses, conceivably, opening the way for a major increase in the size of the virion and the genome. Exaptation of a carbohydrate-binding protein for the function of the MVP is a general trend in virus evolution and might underlie the transformation of the virion shape in other groups of the Nucleocytoviricota as well.
Keywords: giant viruses, glycoside hydrolase family 16, exaptation, capsid protein, virus evolution, Pandoraviridae
1. Introduction
Nucleo-cytoplasmic large DNA viruses (NCLDVs), recently officially classified into a phylum Nucleocytoviricota (Koonin et al. 2020), represent an expansive and highly diverse group of eukaryotic viruses (Koonin and Yutin 2019). They are associated with most major eukaryotic lineages and emerge as important players in ecosystems across Earth’s biomes (Moniruzzaman et al. 2020; Schulz et al. 2020). Analysis of marine microbial metagenomes suggested that there is more phylogenetic diversity in a single family of NCLDVs than currently known in bacteria and archaea, combined (Mihara et al. 2018). Officially, phylum Nucleocytoviricota includes seven virus families, namely, Phycodnaviridae, Mimiviridae, Ascoviridae, Iridoviridae, Marseilleviridae, Asfarviridae, and Poxviridae. However, many family level groups remain unclassified, most notably, including pandoraviruses (Philippe et al. 2013; Legendre et al. 2018), molliviruses (Legendre et al. 2015; Christo-Foroux et al. 2020), pithoviruses (Legendre et al. 2014), cedratviruses (Andreani et al. 2016; Bertelli et al. 2017), orpheoviruses (Andreani et al. 2017a), faustoviruses (Reteno et al. 2015; Benamar et al. 2016), pacmanviruses (Andreani et al. 2017b), mininucleoviruses (Subramaniam et al. 2020), and medusaviruses (Yoshikawa et al. 2019).
These viruses span a range of genome sizes. The smallest among NCLDVs are mininucleoviruses with genomes of about 74 kb (Subramaniam et al. 2020). On the other side of the spectrum are pandoraviruses, the so far undisputed champions of the virosphere in terms of genome size, which have genomes of up to 2.5 Mb, larger than the genomes of many cellular organisms (Philippe et al. 2013). Pandoraviruses are not the only true giants among the NCLDVs: some members of at least two other virus groups, mimiviruses and orpheovirus, have genomes larger than 1 Mb (Guglielmini et al. 2019; Koonin and Yutin 2019). Mapping of the genome size distribution on the phylogeny of NCLDVs and reconstruction of gene gain and loss indicate that gigantism has evolved on several independent occasions and that giant viruses have evolved from viruses with considerably smaller genomes (Guglielmini et al. 2019; Koonin and Yutin 2019). However, the mechanisms and particular adaptations permitting the dramatic expansion of virus genomes remain unclear.
Most members of the Nucleocytoviricota have icosahedral capsids constructed from the characteristic double-jelly roll (DJR) major capsid proteins (MCPs) and single jelly roll minor capsid proteins, an architecture shared with several groups of smaller eukaryotic and prokaryotic viruses (Krupovic and Bamford 2008). This feature has been used as the basis for unification of all DJR MCP viruses, including Nucleocytoviricota, into the kingdom Bamfordvirae, within the realm Varidnaviria (Koonin et al. 2020). However, departure from the canonical icosahedral capsid has occurred on several independent occasions in the history of Nucleocytoviricota (Fig. 1A). Perhaps, the most extensively studied and hence best understood case is that of poxviruses, in which the homolog of the DJR MCP instead of performing a role of capsid protein functions as a scaffolding protein and is proteolytically removed from the mature virions which assume a brick-shaped morphology (Condit, Moussatche, and Traktman 2006). Non-icosahedral virions are also characteristic of ascoviruses, pithoviruses, cedratviruses, orpheovirus, pandoraviruses and, potentially, molliviruses. Virions of ascoviruses, depending on the species, are either bacilliform, ovoidal, or allantoid in shape and have complex symmetry (Asgari et al. 2017). Pithoviruses, cedratviruses, orpheovirus, and pandoraviruses also have ovoid virions, with certain differences in the terminal ‘cork-like’ structures (Philippe et al. 2013; Legendre et al. 2014; Andreani et al. 2016, 2017a; Silva et al. 2018). In contrast, mollivirus virions are spherical with a thick coat or tegument covered with fibers (Quemin et al. 2019). In phylogenetic analyses, molliviruses and pandoraviruses form a clade that is lodged deep within the family Phycodnaviridae (Yutin and Koonin 2013), an expansive family of algal viruses with complex icosahedral virions (Fang et al. 2019; Van Etten, Agarkova, and Dunigan 2019). Despite the diversity of virion morphologies, all but one group of Nucleocytoviricota viruses with non-icosahedral virions encode orthologs of the DJR MCP, suggesting that, following the poxvirus paradigm, these proteins retain roles in virion assembly or/and structure. Pandoraviruses represent the only apparent exception, even though in molliviruses, the closest relatives of pandoraviruses, DJR MCP is one of the major components of the virion (Legendre et al. 2015). Instead, pandoravirus virions (1 μm in length and 0.5 μm in diameter) contain two major virion proteins (MVP) with molecular masses of ∼60 kDa which are unrelated to the MCPs of other known viruses (Philippe et al. 2013). The thick, electron-dense tegument layer of pandoravirus virions has been reported to contain cellulose which could be stained with cellulose-specific fluorescent dye and enzymatically degraded by cellulase (Brahim Belhaouari et al. 2019). Thus, pandoraviruses appear to represent the most radical departure from the paradigmatic virion architecture common to other viruses in the Nucleocytoviricota. However, how this change has occurred and what is the source of the pandoravirus MVPs, remains unknown.
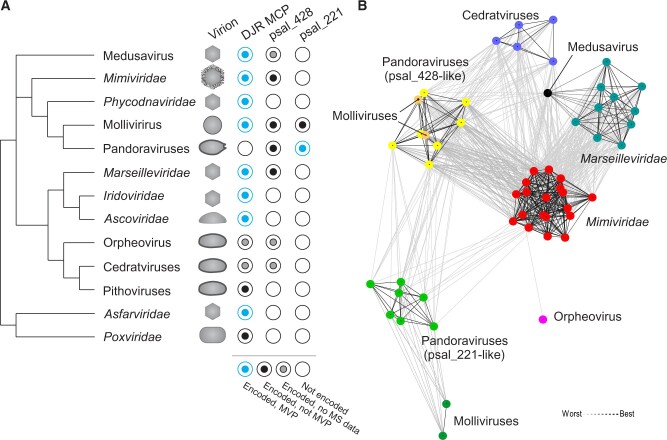
Figure 1.
Conservation of pandoravirus MVP2 (psal_221) homologs among giant viruses. (A) The conservation of DJR MCPs, psal_221 and psal_428 as well as virion morphologies are overlaid on a schematic tree reflecting the evolutionary relationships between different groups of giant viruses. MVP, major virion protein. (B) Clustering of the MVP2 homologs based on their sequence similarity. Lines connect nodes (sequences) with P-value ≤ 1e−20.
Here, we investigated the provenance of the MVPs of pandoraviruses and show that one of the two most abundant proteins evolved from an inactivated derivative of a bacterial glycoside hydrolase of family 16 (GH16). We propose a scenario for the gradual evolution of pandoraviruses from icosahedral ancestor. These results provide a striking example of exaptation of a cellular protein for an MVP function.
2. Results and discussion
2.1 Homologs of the pandoravirus MVPs in other viruses
We analyzed the provenance of the two major, most abundant virion proteins of pandoraviruses that ranked 1 and 2 in proteomic analyses (Philippe et al. 2013). To this end, sequences of the corresponding Pandoravirus salinus proteins, MVP1 (psal_273; YP_008436917) and MVP2 (psal_221; YP_008436815), were used as queries in homology searches. Homologs of MVP1 were restricted to pandoraviruses, and sensitive profile–profile comparisons did not reveal similarity to any other known proteins, consistent with the original assessment (Philippe et al. 2013). In contrast, numerous homologs of MVP2 were readily identifiable in other viruses by BLASTP (n = 54; E < 5e−03). In particular, MVP2 homologs were widespread in (but restricted to) members of the class Megaviricetes (Koonin et al. 2020), including pandoraviruses, molliviruses, mimiviruses, marseilleviruses, cedratviruses, orpheovirus, and medusavirus (Fig. 1A). Notably, pandoraviruses and molliviruses encode two MVP2 homologs each (psal_221 and psal_428 in P. salinus), whereas other viruses carry a single MVP2-like gene. Clustering of the viral sequences based on pairwise similarities, followed by community detection analysis using convex clustering algorithm implemented in CLANS (Frickey and Lupas 2004), revealed six clusters (Fig. 1B). From the largest to the smallest, these clusters are: 1, Mimiviridae; 2, Marseilleviridae and Medusavirus; 3, psal_428-like proteins of pandoraviruses and molliviruses; 4, psal_221-like proteins of pandoraviruses; (5) cedratviruses; and 6, psal_221-like proteins of molliviruses. Finally, the orpheovirus sequence was an outlier, loosely connected to the Mimiviridae cluster. The pithoviruses, which form a monophyletic group with orpheovirus and cedratviruses, do not encode a homolog of either psal_221 or psal_428 (Fig. 1A). Given that orpheovirus and cedratvirus homologs show higher sequence similarity to the corresponding proteins of mimiviruses rather than to each other (Fig. 1B), it appears likely that the protein has been acquired independently by the ancestors of the two virus groups. More generally, the connectivity pattern suggests that most viruses encode orthologs of psal_428, whereas genuine orthologs of MVP2 are conserved only in pandoraviruses and molliviruses. The clusters of MVP2-like proteins were generally consistent with the classification of the respective viruses (Fig. 1B), suggesting that these genes have not undergone recent exchange between viruses from different families.
In all viruses, for which proteomics data on the viral particles is available, MVP2 homologs were found to be virion components (Fig. 1A). However, except for the psal_221 orthologs in pandoraviruses, these proteins constitute only minor fraction of the virion proteins. Whereas psal_221 is the second most abundant protein in P. salinus virion, its homolog psal_428 is ranked only at 161 (Legendre et al. 2018). In molliviruses, which are the closest relatives of pandoraviruses, but employ DJR MCP for virion formation, ml_494 and ml_264, the orthologs of P. salinus psal_221 and psal_428, are ranked only 96 and 56, respectively (Legendre et al. 2015). Among members of the Marseilleviridae, MVP2 homologs were identified in virions of Marseillevirus (Boyer et al. 2009), Melbournevirus (Fabre et al. 2017; Okamoto et al. 2018), and Noumeavirus (Fabre et al. 2017). In Melbournevirus, MEL_089, along with the DJR MCP, was among the five proteins highly resistant to urea treatment (Okamoto et al. 2018), suggesting that in certain properties, namely, resilience to denaturing treatment, MVP2-like proteins are similar to the bona fide MCPs. A potential function of an MVP2-like protein has been suggested only in mimiviruses. L829, a homolog of MVP2, has been identified in virions of several mimiviruses, including Acanthamoeba polyphaga mimivirus (Renesto et al. 2006) and Tupanvirus soda lake (Abrahão et al. 2018; Schrad et al. 2020). Spontaneous deletion of a locus in the mimivirus genome, which included the L829 gene, resulted in the loss of the characteristic fibers surrounding the mimivirus virions, and proteomic analysis of the purified fibers further suggested that L829 is one of several components of the fibers (Boyer et al. 2011). However, RNAi-mediated knock down of the L829 gene did not eliminate fibers completely, only making them shorter and less dense, suggesting that L829 is not a major component of these structures (Sobhy et al. 2015). The fiber layer present in mimiviruses has not been observed in other MVP2-encoding viruses, including pandoraviruses and molliviruses, suggesting that the function(s) of MVP2 in these viruses differs from that in the mimivivurses.
2.2 Bacterial origins of MVP2-like proteins
Searches with the psal_221 sequence against the RefSeq database (BLASTP, E < 5e−03) showed that, in addition to viruses, MVP2 homologs are widespread in bacteria, particularly, in actinobacteria, with most annotated as hypothetical proteins. Clustering of the bacterial and viral MVP2 homologs revealed three interconnected major modules of bacterial sequences (Fig. 2A). Actinobacteria dominated all three modules: 88 per cent in Module 1 (n = 92), 60 per cent in Module 2 (n = 31), and 62 per cent in Module 3 (n = 8; Supplementary Table S1). Firmicutes and proteobacteria represented the second largest groups in Modules 2 (29%) and 3 (31%), respectively. All viral homologs clustered with sequences from Module 1 (Fig. 2A). Notably, psal_221 orthologs connect to bacterial sequences through viral homologs, consistent with this group representing divergent paralogs that evolved in the context of giant virus genomes. However, whether all virus proteins are monophyletic or were recruited from bacteria on multiple independent occasions could not be concluded from the clustering analysis due to similar levels of sequence similarity among viral as well as between viral and bacterial homologs, resulting in a dense connectivity between the corresponding clusters.
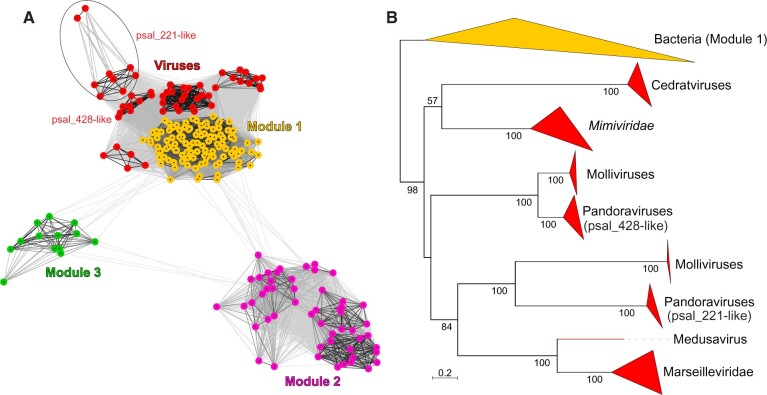
Figure 2.
Relationship between bacterial and viral MVP2 homologs. (A) Clustering of bacterial and viral sequences based on their sequence similarity. Lines connect nodes (sequences) with P-value ≤ 1e−14. The cluster of pandoravirus and mollivirus orthologs of psal_221 is circled. Bacterial Modules 1–3 are indicated with different colors and labeled in the figure. Composition of the three modules can be found in Supplementary Table S1. (B) Maximum likelihood phylogenetic tree of MVP2 homologs encoded by viruses (red clades) and bacteria from Module 1 (yellow clade). Closely related sequences are collapsed to triangles, whose side lengths are proportional to the distances between closest and farthest leaf nodes. The tree was constructed with IQ-Tree (Minh et al. 2020). Numbers at the nodes represent bootstrap supports. The scale bar represents the number of substitutions per site.
To study the relationships between viral and bacterial homologs in Module 1, we performed maximum likelihood phylogenetic analysis using IQ-Tree (Minh et al. 2020). In the mid-point rooted tree shown in Fig. 2B, viral and bacterial sequences form two major clades, suggesting monophyly of all viral sequences. Thus, in all likelihood, the bacterial ancestor of MVP2-like proteins has been introduced into the virus world on a single occasion. Although basal nodes in the phylogeny are poorly resolved (Fig. 2B), phyletic distribution of the protein in giant viruses (Fig. 1A) suggests that the corresponding gene has been disseminated horizontally. Notably, all viruses that encode MVP2 homologs infect amebae, phagocytic protists that feed on bacteria and often harbor bacterial endosymbionts. Accordingly, amebae are considered to be a ‘melting pot’ of gene exchange between bacteria and giant viruses (Boyer et al. 2009). Indeed, none of the giant viruses infecting other hosts encodes this protein. In particular, the absence of MVP2 in the closest relatives of the mimiviruses that infect algae (Santini et al. 2013; Moniruzzaman et al. 2014) and microzooplankton (Fischer et al. 2010; Deeg, Chow, and Suttle 2018) is consistent with the possibility that the evolution of this protein took place in the context of virus–host interactions in ameba, whereby ancestors of the MVP2-encoding viruses have co-infected the same host and exchanged genes.
2.3 Ancestral function of MVP2-like proteins
To gain insights into the potential function of the bacterial proteins ancestral to MVP2 homologs, we performed PaperBLAST which explores the database of 700,000 scientific articles that mention over 400,000 different proteins (last update 24 March 2020; Price and Arkin 2017). However, no relevant information was found. Homology searches against the PDB database also returned no hits, suggesting that none of the proteins in Modules 1–3 were functionally or structurally characterized. Therefore, we performed sensitive profile–profile comparisons with HHpred using as queries profiles separately constructed for each of the three bacterial modules shown in Fig. 2A. In all three cases, HHpred-returned proteins of the GH16 (cd00413) as the best hits (Supplementary Fig. S1), although the significance of the hit for Module 3 was more modest compared with those obtained for the other two modules (probability of 82% vs 96–98%). The GH16 family is an expansive, and taxonomically widespread assemblage of functionally diverse enzymes, including lichenase, xyloglucosyltransferase, agarase, kappa-carrageenase, endo-beta-glucanases, endo-beta-galactosidase, and more (CAZypedia Consortium 2018; Viborg et al. 2019; Linton 2020). These enzymes adopt a common β-jelly roll fold consisting of two closely packed, curved antiparallel β sheets which create a deep channel harboring the catalytic site (Fig. 3A) that consists of the conserved triad of acidic residues in the pattern EXDX(X)E, where X is any amino acid (Viborg et al. 2019). Examination of the alignment of the GH16 representatives, viral MVP2 homologs and bacterial sequences from Modules 1 to 3 (Supplementary Fig. S2) showed that the active site in the latter two groups carries mutations of the conserved glutamate and aspartate residues in the catalytic site (Fig. 3B), suggesting that these proteins are not enzymatically active. Thus, in all likelihood, the bacterial protein acquired by giant viruses was already inactivated.
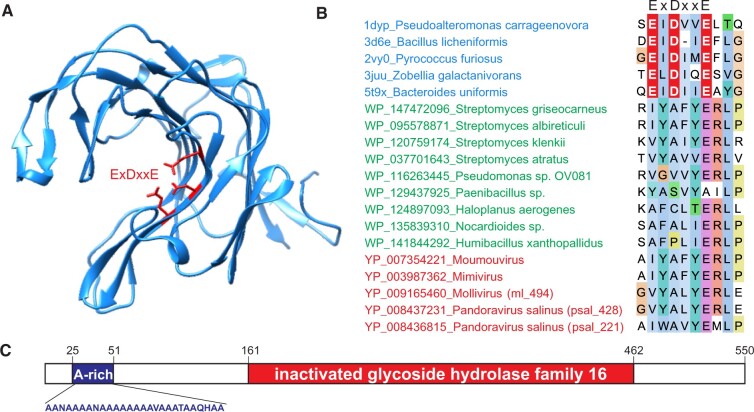
Figure 3.
MVP2 homologs are derived from inactivated GH16. (A) Structure of a representative GH16, kappa-carrageenase of Pseudoalteromonas carrageenovora (PDB ID: 1DYP; Michel et al. 2001). Acidic active site residues are shown using stick representation and are colored red. B. Alignment of representative GH16 (blue) as well as inactivated derivatives from Modules 1 to 3 (green) and viruses (red). Only the region harboring the active site (ExDxxE) of GH16 is shown. Active site triad is shown in white font on the red background. The full alignment can be found in Supplementary Fig. S2. (C) Domain organization of P. salinus MVP2 (psal_221). A-rich, alanine-rich region.
In addition to the inactivated GH16 domain, psal_221 contains an N-terminal region of unclear function in which twenty-one out of twenty-seven amino acid residues are alanines (Fig. 3C). A similar N-terminal Ala-rich region of the Antigen I/IIf from Streptococcus mutans is responsible for binding to collagen, laminin, keratin, and fibronectin (Sciotti et al. 1997). Thus, it appears likely that the Ala-rich region of psal_221 also mediates specific protein–protein interaction. The Ala-rich region is conserved in all psal_221 pandoravirus orthologs but not in other viral MVP2 homologs, including psal_428. Notably, it is also missing in the psal_221 orthologs in molliviruses, suggesting that the region has been acquired following the divergence of pandoraviruses and molliviruses from their common ancestor and might be specifically involved in the formation of the amphora-like pandoravirus virions.
2.4 Evolution of pandoravirus MVP2
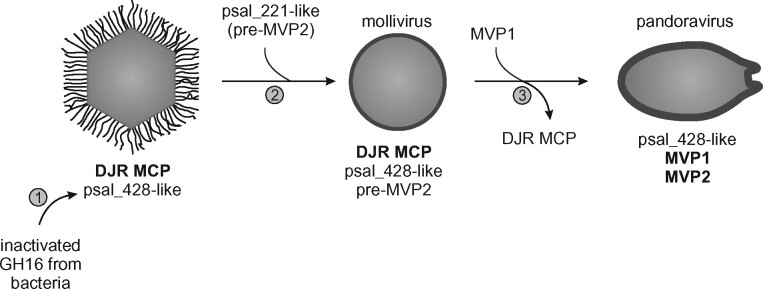
Collectively, our results suggest a scenario for the evolution of major structural proteins of pandoravirus virions (Fig. 4). The roots of the MVP2 ancestor can be traced to a peculiar group of functionally uncharacterized bacterial proteins which have evolved from GH16 family enzymes through inactivating mutation in the active site. Given that GH16 enzymes act on carbohydrates, the inactivated GH16 derivatives, most likely, retained the ability to bind carbohydrates and function in this capacity. In particular, MVP2 might interact with and stabilize the cellulose component of the pandoravirus tegument layer (Brahim Belhaouari et al. 2019). Following the acquisition, the inactivated GH16 gene has been exchanged between different groups of giant ameba-infecting viruses, where they function as minor components of the viral particles. In the ancestor of molliviruses and pandoraviruses, this gene underwent duplication and, in pandoraviruses, one of the copies gained an Ala-rich region, giving rise to pre-MVP2. The advance of pre-MVP2 to the status of an MVP was likely concomitant with the acquisition of MVP1, the provenance of which remains enigmatic, and the loss of the ancestral DJR MCP which is conserved in all other viruses encoding MVP2-like proteins, including molliviruses. The switch from the icosahedral DJR MCP-based capsids to amphora-like virions has likely led to changes in virion assembly mechanisms and potentially precipitated substantial increase in virion and genome sizes that occurred in pandoraviruses.
Figure 4.
Proposed evolutionary scenario for the emergence of pandoraviruses from an icosahedral ancestor. 1, Acquisition from bacteria of an inactivated gene encoding a GH16-family protein, an ancestor of psal_428; 2, Duplication of the psal_428 gene in the ancestor of molliviruses and pandoraviruses yielding pre-MVP2 (psal_221); 3, Displacement of the DJR MCP with a MVP1 of unknown provenance and refunctionalization of MVP2 precipitate the emergence of amphora-shaped pandoravirus virions.
2.5 Concluding remarks
Dramatic changes in virion architecture are common in virus evolution, across the virosphere, from the smallest of viruses to the giants discussed here. This transition is typically accompanied by replacement, in a particular virus lineage, of the ancestral morphogenetic module with a different one, which often comes from other viruses (Caprari et al. 2015; Guardado-Calvo and Rey 2017; Kazlauskas et al. 2017; Wolf et al. 2018; Koonin et al. 2020). Exchange or even loss of morphogenetic modules appears to be more frequent in viruses with smaller genomes. For instance, RNA viruses of the order Tymovirales can have either icosahedral virions (Tymoviridae), flexible filamentous (e.g. Alphaflexiviridae) virions, or no capsids at all (Deltaflexiviridae). Furthermore, capsid protein genes can be exchanged between viruses with RNA and DNA genomes (Diemer and Stedman 2012; Roux et al. 2013). Intuitively, mixing and matching of morphogenetic and genome replication modules should be more efficient when both modules consist of small numbers of genes, as is indeed the case for viruses with small genomes. In contrast, the morphogenetic modules of giant viruses are far more complex, many of them encapsidating in excess of 100 proteins (Renesto et al. 2006; Legendre et al. 2014, 2018; Schrad et al. 2020). Thus, replacement of the key components in such intricate systems is more likely to disrupt the co-evolved dependencies and lead to defective assemblies. Nevertheless, pandoraviruses succeeded in achieving just that: the DJR MCP has been lost and the typical icosahedral capsid turned into a giant amphora-like virion. Our findings discussed here shed light on this remarkable metamorphosis. We show that one of the two MVPs of pandoraviruses has evolved from a pre-existing minor virion component, in itself, a horizontally acquired inactivated bacterial enzyme, suggesting a gradual transformation of a non-capsid protein into a bona fide MVP.
MVP2 of pandoraviruses represents a case of exaptation of a cellular protein for a function in virion assembly and structure. A similar route has been proposed for the origin of the MVPs of certain archaeal, animal, and plant viruses (Krupovic et al. 2015; Krupovic and Koonin 2017). More generally, the evolution of the pandoravirus MVP2 appears to recapitulate the recently proposed scenario for the origin of viruses, under which capsid proteins continuously evolve from functionally diverse cellular proteins (Krupovic, Dolja, and Koonin 2019). The evolution of pandoravirus MVP2 from an inactivated GH16 family glycoside hydrolase mirrors the proposed evolution of the jelly roll fold capsid proteins, the most common type of capsid proteins, from carbohydrate-binding proteins (Krupovic and Koonin 2017). Intriguingly, virions of pithoviruses, superficially similar to those of pandoraviruses, and mature brick-shaped virions of poxviruses do not contain homologs of either MVP1 or MVP2 of pandoraviruses and are instead constructed from unrelated proteins (Resch et al. 2007; Legendre et al. 2014). Similarly, major virions proteins of poxviruses have no obvious homologs among other known viruses. Although sequence similarity searches thus far have not provided insights into the provenance of these proteins, structural studies and/or development of more sensitive homology detection tools should help solving this puzzle.
3. Methods
3.1 Sequence searches and clustering
Viral homologs of pandoravirus proteins psal_273 (YP_008436917) and psal_221 (YP_008436815) were identified using PSI-BLASTP (Altschul et al. 1997) against the non-redundant protein database and NCBI. The bacterial homologs of psal_221 were collected by running two iterations of PSI-BLAST against the RefSeq database restricted to bacterial sequences. The redundancy in the resultant dataset was removed by clustering the sequences with MMseq2 (Steinegger and Söding 2017) to 80 per cent sequence identity over 80 per cent of the alignment length.
Sequences were clustered using CLANS with BLAST option (Frickey and Lupas 2004). CLANS is an implementation of the Fruchterman-Reingold force-directed layout algorithm, which treats protein sequences as point masses in a virtual multidimensional space, in which they attract or repel each other based on the strength of their pairwise similarities (CLANS P-values; Frickey and Lupas 2004). Thus, evolutionarily more closely related sequences gravitate to the same parts of the map, forming distinct clusters. Clusters of viral psal_221 homologs were identified by CLANS convex clustering algorithm at P-value = 1e−20. Whereas clustering of bacterial and viral homologs was performed at CLANS P-value = 1e−14.
3.2 Multiple sequence alignments and remote homology detection
Sequences were aligned using MAFFT in ’Auto’ mode (Katoh, Rozewicki, and Yamada 2019) and visualized with Jalview (Waterhouse et al. 2009). Sequence searches based on profile–profile comparisons were used to detect remote homology. Multiple sequence alignments were used as queries for profile–profile comparisons in HHpred against different protein databases, including Pfam, PDB, CDD, and SCOPe (Zimmermann et al. 2018).
3.3 Phylogenetic analysis
For phylogenetic analysis, uninformative positions we removed using TrimAl with gap threshold of 0.2 (Capella-Gutierrez, Silla-Martinez, and Gabaldon 2009). The final alignment contained 392 positions. The maximum likelihood tree was constructed using IQ-TREE v2 (Minh et al. 2020). The best-fitting substitution model was selected by IQ-TREE and was LG+F+R5. Branch supports were estimated using bootstrap (1,000 replicates). The tree was visualized with iTOL (Letunic and Bork 2019).
Data availability
All the data used in this work are available from the GenBank database and from the Supplementary Material.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
M.K. was supported by l’Agence Nationale de la Recherche Grant ANR-17-CE15-0005-01 (ENVIRA). N.Y. and E.V.K. are supported through the Intramural Research Program of the National Institutes of Health of the USA (National Library of Medicine).
Conflict of interest: None declared.
Supplementary Material
Contributor Information
Mart Krupovic, Department of Microbiology, Archaeal Virology Unit, Institut Pasteur, Paris 75015, France.
Natalya Yutin, National Library of Medicine, National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD 20894, USA.
Eugene Koonin, National Library of Medicine, National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD 20894, USA.
References
- Abrahão J. et al. (2018) ‘Tailed Giant Tupanvirus Possesses the Most Complete Translational Apparatus of the Known Virosphere’, Nature Communications, 9: 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. (1997) ‘Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs’, Nucleic Acids Research, 25: 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J. et al. (2016) ‘Cedratvirus, a Double-Cork Structured Giant Virus, is a Distant Relative of Pithoviruses’, Viruses, 8: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J. et al. (2017. a) ‘Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses’, Frontiers in Microbiology, 8: 2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani J. et al. (2017. b) ‘Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between Asfarviridae and Faustoviruses’, Journal of Virology, 91: e00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S. et al. ; ICTV Report Consortium. (2017) ‘ICTV Virus Taxonomy Profile: Ascoviridae’, Journal of General Virology, 98: 4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benamar S. et al. (2016) ‘Faustoviruses: Comparative Genomics of New Megavirales Family Members’, Frontiers in Microbiology, 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C. et al. (2017) ‘Cedratvirus lausannensis - Digging into Pithoviridae Diversity’, Environmental Microbiology, 19: 4022–34. [DOI] [PubMed] [Google Scholar]
- Boyer M. et al. (2009) ‘Giant Marseillevirus Highlights the Role of Amoebae as a Melting Pot in Emergence of Chimeric Microorganisms’, Proceedings of the National Academy of Sciences of the United States of America, 106: 21848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M. et al. (2011) ‘Mimivirus Shows Dramatic Genome Reduction after Intraamoebal Culture’, Proceedings of the National Academy of Sciences of the America, 108: 10296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahim Belhaouari D. et al. (2019) ‘Evidence of a Cellulosic Layer in Pandoravirus massiliensis Tegument and the Mystery of the Genetic Support of Its Biosynthesis’, Frontiers in Microbiology, 10: 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., Gabaldon T. (2009) ‘trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses’, Bioinformatics, 25: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprari S. et al. (2015) ‘Sequence and Structure Analysis of Distantly-Related Viruses Reveals Extensive Gene Transfer between Viruses and Hosts and among Viruses’, Viruses, 7: 5388–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAZypedia Consortium. (2018) ‘Ten Years of CAZypedia: A Living Encyclopedia of Carbohydrate-Active Enzymes’, Glycobiology, 28: 3–8. [DOI] [PubMed] [Google Scholar]
- Christo-Foroux E. et al. (2020) ‘Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses’, Journal of Virology, 94: e01997–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit R. C., Moussatche N., Traktman P. (2006) ‘In a Nutshell: Structure and Assembly of the Vaccinia Virion’, Advances in Virus Research, 66: 31–124. [DOI] [PubMed] [Google Scholar]
- Deeg C. M., Chow C. T., Suttle C. A. (2018) ‘The Kinetoplastid-Infecting Bodo saltans virus (BsV), a Window into the Most Abundant Giant Viruses in the Sea’, eLife, 7: e33014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer G. S., Stedman K. M. (2012) ‘A Novel Virus Genome Discovered in an Extreme Environment Suggests Recombination between Unrelated Groups of RNA and DNA Viruses’, Biology Direct, 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E. et al. (2017) ‘Noumeavirus Replication Relies on a Transient Remote Control of the Host Nucleus’, Nature Communications, 8: 15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q. et al. (2019) ‘Near-Atomic Structure of a Giant Virus’, Nature Communications, 10: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. G. et al. (2010) ‘Giant Virus with a Remarkable Complement of Genes Infects Marine Zooplankton’, Proceedings of the National Academy of Sciences of the United States of America, 107: 19508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T., Lupas A. (2004) ‘CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity’, Bioinformatics, 20: 3702–4. [DOI] [PubMed] [Google Scholar]
- Guardado-Calvo P., Rey F. A. (2017) ‘The Envelope Proteins of the Bunyavirales’, Advances in Virus Research, 98: 83–118. [DOI] [PubMed] [Google Scholar]
- Guglielmini J. et al. (2019) ‘Diversification of Giant and Large Eukaryotic dsDNA Viruses Predated the Origin of Modern Eukaryotes’, Proceedings of the National Academy of Sciences of the United States of America, 116: 19585–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019) ‘MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization’, Briefings in Bioinformatics, 20: 1160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas D. et al. (2017) ‘Evolutionary History of ssDNA Bacilladnaviruses Features Horizontal Acquisition of the Capsid Gene from ssRNA Nodaviruses’, Virology, 504: 114–21. [DOI] [PubMed] [Google Scholar]
- Koonin E V et al. (2020) ‘Global Organization and Proposed Megataxonomy of the Virus World’, Microbiology and Molecular Biology Reviews, 84: e00061–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Yutin N. (2019) ‘Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism’, Advances in Virus Research, 103: 167–202. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Bamford D. H. (2008) ‘Virus Evolution: How Far Does the Double Beta-Barrel Viral Lineage Extend?’, Nature Reviews Microbiology, 6: 941–8. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Koonin E. V. (2017) ‘Multiple Origins of Viral Capsid Proteins from Cellular Ancestors’, Proceedings of the National Academy of Sciences of the United States of America, 114: E2401–E2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Dolja V. V., Koonin E. V. (2019) ‘Origin of Viruses: Primordial Replicators Recruiting Capsids from Hosts’, Nature Reviews Microbiology, 17: 449–58. [DOI] [PubMed] [Google Scholar]
- Krupovic M. et al. (2015) ‘Evolution of an Archaeal Virus Nucleocapsid Protein from the CRISPR-Associated Cas4 Nuclease’, Biology Direct, 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M. et al. (2014) ‘Thirty-Thousand-Year-Old Distant Relative of Giant Icosahedral DNA Viruses with a Pandoravirus Morphology’, Proceedings of the National Academy of Sciences of the United States of America, 111: 4274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M. et al. (2015) ‘In-Depth Study of Mollivirus sibericum, a New 30,000-y-Old Giant Virus Infecting Acanthamoeba’, Proceedings of the National Academy of Sciences of the United States of America, 112: E5327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M. et al. (2018) ‘Diversity and Evolution of the Emerging Pandoraviridae Family’, Nature Communications, 9: 2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2019) ‘Interactive Tree of Life (iTOL) v4: Recent Updates and New Developments’, Nucleic Acids Research, 47: W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton S. M. (2020) ‘Review: The Structure and Function of Cellulase (Endo-Beta-1,4-Glucanase) and Hemicellulase (Beta-1,3-Glucanase and Endo-Beta-1,4-Mannase) Enzymes in Invertebrates That Consume Materials Ranging from Microbes, Algae to Leaf Litter’, Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 240: 110354. [DOI] [PubMed] [Google Scholar]
- Michel G. et al. (2001) ‘The Kappa-Carrageenase of P. carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases’, Structure, 9: 513–25. [DOI] [PubMed] [Google Scholar]
- Mihara T. et al. (2018) ‘Taxon Richness of “Megaviridae” Exceeds Those of Bacteria and Archaea in the Ocean’, Microbes and Environments, 33: 162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B. Q. et al. (2020) ‘IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era’, Molecular Biology and Evolution, 37: 1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M. et al. (2014) ‘Genome of Brown Tide Virus (AaV), the Little Giant of the Megaviridae, Elucidates NCLDV Genome Expansion and Host-Virus Coevolution’, Virology, 466-467: 60–70. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M. et al. (2020) ‘Dynamic Genome Evolution and Complex Virocell Metabolism of Globally-Distributed Giant Viruses’, Nature Communications, 11: 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K. et al. (2018) ‘Cryo-EM Structure of a Marseilleviridae Virus Particle Reveals a Large Internal Microassembly’, Virology, 516: 239–45. [DOI] [PubMed] [Google Scholar]
- Philippe N. et al. (2013) ‘Pandoraviruses: Amoeba Viruses with Genomes up to 2.5 Mb Reaching That of Parasitic Eukaryotes’, Science, 341: 281–6. [DOI] [PubMed] [Google Scholar]
- Price M. N., Arkin A. P. (2017) ‘PaperBLAST: Text Mining Papers for Information about Homologs’, mSystems, 2: e00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin E. R. et al. (2019) ‘Complex Membrane Remodeling during Virion Assembly of the 30,000-Year-Old Mollivirus sibericum’, Journal of Virology, 93: e00388–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renesto P. et al. (2006) ‘Mimivirus Giant Particles Incorporate a Large Fraction of Anonymous and Unique Gene Products’, Journal of Virology, 80: 11678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W. et al. (2007) ‘Protein Composition of the Vaccinia Virus Mature Virion’, Virology, 358: 233–47. [DOI] [PubMed] [Google Scholar]
- Reteno D. G. et al. (2015) ‘Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae’, Journal of Virology, 89: 6585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S. et al. (2013) ‘Chimeric Viruses Blur the Borders between the Major Groups of Eukaryotic Single-Stranded DNA Viruses’, Nature Communications, 4: 2700. [DOI] [PubMed] [Google Scholar]
- Santini S. et al. (2013) ‘Genome of Phaeocystis globosa Virus PgV-16T Highlights the Common Ancestry of the Largest Known DNA Viruses Infecting Eukaryotes’, Proceedings of the National Academy of Sciences of the United States of America, 110: 10800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrad J. R. et al. (2020) ‘Structural and Proteomic Characterization of the Initiation of Giant Virus Infection’, Cell, 181: 1046–61.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F. et al. (2020) ‘Giant Virus Diversity and Host Interactions through Global Metagenomics’, Nature, 578: 432–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciotti M. A. et al. (1997) ‘The N-Terminal Half Part of the Oral Streptococcal Antigen I/IIf Contains Two Distinct Binding Domains’, FEMS Microbiology Letters, 153: 439–45. [DOI] [PubMed] [Google Scholar]
- Silva L. et al. (2018) ‘Cedratvirus getuliensis Replication Cycle: An in-Depth Morphological Analysis’, Scientific Reports, 8: 4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhy H. et al. (2015) ‘Identification of Giant Mimivirus Protein Functions Using RNA Interference’, Frontiers in Microbiology, 6: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M., Söding J. (2017) ‘MMseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets’, Nature Biotechnology, 35: 1026–8. [DOI] [PubMed] [Google Scholar]
- Subramaniam K. et al. (2020) ‘A New Family of DNA Viruses Causing Disease in Crustaceans from Diverse Aquatic Biomes’, mBio, 11: e02938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J. L., Agarkova I. V., Dunigan D. D. (2019) ‘Chloroviruses’, Viruses, 12: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viborg A. H. et al. (2019) ‘A Subfamily Roadmap of the Evolutionarily Diverse Glycoside Hydrolase Family 16 (GH16)’, Journal of Biological Chemistry, 294: 15973–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M. et al. (2009) ‘Jalview Version 2–a Multiple Sequence Alignment Editor and Analysis Workbench’, Bioinformatics, 25: 1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y. I. et al. (2018) ‘Origins and Evolution of the Global RNA Virome’, mBio, 9: e02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa G. et al. (2019) ‘Medusavirus, a Novel Large DNA Virus Discovered from Hot Spring Water’, Journal of Virology, 93: e02130–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Koonin E. V. (2013) ‘Pandoraviruses Are Highly Derived Phycodnaviruses’, Biology Direct, 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L. et al. (2018) ‘A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core’, Journal of Molecular Biology, 430: 2237–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this work are available from the GenBank database and from the Supplementary Material.