Abstract
Gastric cancer is the third leading cause of cancer‐related deaths worldwide. Novel biomarkers circRNAs can play an important role in the development of gastric cancer as oncogenes or tumor suppressor genes. The purpose of this study was to clarify the relationship between the abnormal expression of multiple circRNAs and their prognostic value in gastric cancer patients through a meta‐analysis. We researched articles reporting the relationship between circRNAs and the prognosis of gastric cancer published in PubMed, Cochrane, Embase, Web of Science, Wanfang, CNKI, and VIP databases before 31 December 2019. Thirty‐five articles were selected for the meta‐analysis, involving 3135 gastric cancer patients. The total HR values (95% CI) of OS and DFS related to highly expressed circRNAs that indicated worse prognosis were 1.83 (1.64‐2.03; p < 0.001) and 1.66 (1.33‐2.07; p < 0.001), respectively. The total HR (95% CI) of OS and DFS related to highly expressed circRNAs that indicated better prognosis was 0.54 (0.45‐0.66; p < 0.001) and 0.58 (0.43‐0.78; p < 0.001), respectively. Two panels of five circRNAs predicted a more considerable HR value (circ_0009910, hsa_circ_0000467, hsa_circ_0065149, hsa_circ_0081143, and circDLST; and circSMARCA5, circLMTK2, hsa_circ_0001017, hsa_circ_0061276, and circ‐KIAA1244). The results of the meta‐analysis were 2.63 (2.08‐3.33; p < 0.001) and 0.39 (0.27‐0.59; p < 0.001) for OS, respectively. The two panels of dysregulated circRNAs can be considered as more suitable potential prognostic tumor biomarkers in patients with gastric cancer because of their larger HR values.
Keywords: circRNAs, gastric cancer, meta‐analysis, prognosis
We obtained the HR value which combined analysis of survival outcomes and time from gastric cancer patients in each clinical trial as a reference to judge the prognostic value of circRNAs in gastric cancer. We found that two panels of five circRNAs showed a greater prognostic value: 2.63(2.08‐3.33), p < 0.001 (circ_00099 10, hsa_circ_0000467, hsa_circ_0065149, hsa_circ_0081143, circDLST); 0.39(0.27‐ 0.59), p < 0.001 (circSMARCA5, circLMTK2, hsa_circ_0001017, hsa_circ_0061276, circ‐KIAA1244).

1. INTRODUCTION
Gastric cancer (GC) is one of the most common cancers globally. In 2018, there were more than 1 million new cases, and the estimated number of deaths reached 783,000 (equivalent to 1 in 12 deaths worldwide). GC is the fifth most common type of cancer worldwide. It is also the third leading cause of cancer‐related deaths. 1 Approximately 70% of new cases are concentrated in developing countries, especially China. According to these studies, China has a high incidence of GC. The incidence of GC in men is second to lung cancer, while the probability of women being diagnosed with GC is only lower than that of breast cancer and lung cancer, ranking third in the incidence of malignant tumors over the same period. 2
The occurrence and development of GC are the results of multiple factors. It has been reported that Helicobacter pylori infection and a family history of GC are the main risk factors. 3 In addition, another study stated that poor eating habits (high‐salt diet, preserved food, fried food), unhealthy lifestyle (alcoholism, cigarettes, excessive overeating), history of stomach surgery, and occupational exposure history (cement, mineral dust, chromium) can promote the development of GC. 4 , 5 However, advanced GC can be caused by extensive infiltration of tumor tissue in multiple organs, leading to a poor prognosis. Identifying accurate, convenient, and economical biomarkers to determine prognosis has always been a hotspot in cancer research. Liquid biomarkers have advantages such as noninvasiveness, painlessness, convenient material acquisition, and continuous testing in vitro. They are not only widely used in clinical applications, but also continue to be extensively researched. In recent years, the study of liquid biomarkers has expanded from the traditional protein level to the molecular level. However, because of the limitations of biological characteristics and experimental conditions, many biomarkers have the characteristics of low experimental sensitivity and low repeatability of detection. Therefore, biomarkers with high sensitivity, excellent specificity, and high repeatability that accurately reflect patient prognosis and have the ability to guide treatment require further investigation.
CircRNAs are a recently identified type of RNA that have a covalent circular structure. With the emergence of high‐throughput sequencing, exoenzyme‐based enrichment tools, and bioinformatics, the detection amount and types of circRNAs have increased rapidly, and a large number of circRNA families have been found in various animals, plants, and humans. Memczak and Hansen have made breakthrough progress in the study of the biological function of circRNAs, and proposed that circRNAs can function as miRNA sponges in gene expression, gene regulation, and posttranscriptional processes for the first time. 6 , 7 Many studies have since shown that circRNAs have an important role in the development of tumors. CircRNAs can affect gene expression and transcription in many diseases including GC through various mechanisms. 8 Biological experiments revealed that many circRNAs contain miRNA binding sites, which can be used as miRNA sponges to inhibit or promote the progression of GC. In addition, circRNAs regulate the transcription of parental genes, participating in protein interaction and translating proteins. 9 A variety of circRNAs can function as oncogenes or tumor suppressor genes. As circRNAs are characterized by their high abundance, high stability, conservatism, and tissue specificity, they are considered to have excellent potential value, especially as biomarkers for diagnosis and prognosis. Treatment targets are also the focus of research. 8 , 10 The dysregulation of specific circRNAs as molecular biomarkers is closely related to the prognosis of GC patients.
However, because single‐center, small‐sample studies with different experimental methods and standards were adopted by different research centers, the ability to evaluate the relationship between the expression of multiple circRNAs and the prognosis of GC patients is limited. To the best of our knowledge, this is the largest study to further elucidate the relationship between the expression of multiple circRNAs and the prognosis of GC, so as to advance our understanding of the prognostic value of circRNAs and promote the development of targeted therapy with circRNAs.
2. METHODS
2.1. Search strategy
We performed searches for studies on the relationship between circRNAs and the prognosis of GC in PubMed, Cochrane, EMBASE, Web of Science, Wanfang, China National Knowledge Infrastructure (CNKI), and China Science and Technology Journal (VIP) databases published before 31 December 2019. The specific retrieval strategies were as follows: #1 “RNA, circular” OR “circular RNA*” OR “circRNA*”; #2 “stomach neoplasms” OR “gastric cancer” OR “stomach cancers” OR “gastric carcinoma” OR “advanced gastric cancer”; and #3 “prognosis” OR “prediction” OR “survival” OR “hazard ratio” OR “mortality” OR “follow up studies.” The search formula was as follows: #1 AND #2 AND #3 (combination of subject words and free words).
2.2. Inclusion and exclusion criteria:
The inclusion criteria were as follows: (a) patients with GC diagnosed pathologically; (b) prognosis of patients with stage I‐IV disease was predicted based on the expression level of circRNAs; (c) expression level of circRNAs can be divided into high expression and low expression; and (d) we could directly obtain the overall survival (OS), disease‐free survival (DFS), hazard ratio (HR), and 95% confidence interval (95% CI) values from the text or extract them from the Kaplan‐Meier curve. The exclusion criteria were as follows: (a) reviews, letters, case reports, and nonclinical related studies; (b) non‐English, non‐Chinese, nonhuman studies, and articles without data; and (c) duplicate articles.
3. DATA EXTRACTION
According to the above inclusion and exclusion criteria, two researchers independently selected articles from a large number of literature and extracted and entered relevant data from the articles that met the criteria. In cases of disagreement, the final results were determined by discussion between the two researchers, and the results were recorded after reaching a consensus. Articles that were clearly unrelated to the research theme were excluded by reading the title/abstract, and then, the full text of the remaining articles was read after excluding such articles. The literature was classified according to inclusion and exclusion criteria and determined as to whether it was eventually included. The content of the data extraction mainly included basic information of the included studies (including first author, type of circRNAs, publication time, country, number of patients, test methods, sample types, cutoff value) and survival data (including follow‐up time, HR, and 95% CI). We used the NOS (Newcastle‐Ottawa Scale) scale for quality evaluation and assigned the articles from 0 to 9 according to the content of the literature and judgment criteria.
3.1. Statistical analysis
Survival analysis is an analysis method that takes the patient's death as the end point of observation and combines the survival time. We mainly obtained the HR value from the survival analysis curve. Our methods of obtaining HR were as follows: (a) If the HR and 95% CI had been given in the literature, it could be collected directly. (b) If the Kaplan‐Meier survival curve was given in the literature, we used Engauge digitizer 4.1 software to read the survival data from the survival curve, and then, calculated the HR and 95% credibility interval using the method described by Tierney. 11
We used the I 2 statistic method to test the heterogeneity between the included studies. If the heterogeneity test results were p > 0.1 and I 2 < 50%, the included studies were considered as having no obvious heterogeneity, and the fixed effects model (FEM) was used to combine the HR value. If p ≤ 0.1 and I2 > 50%, it was considered that there was obvious heterogeneity between the included studies, and a random effects model (REM) was used to merge the HR value. At this time, the source of heterogeneity needed to be further explored and a subgroup analysis was performed to determine whether the heterogeneity could be explained by clinical heterogeneity or methodological heterogeneity. It was also feasible to use a sensitivity analysis to identify the impact of a single research result on the meta‐analysis conclusion. If the heterogeneity was too large or the original data could not be obtained, only a descriptive analysis was performed instead of a meta‐analysis. RevMan 5.3 software was used to combine the effect size and calculate I 2 statistics to judge the heterogeneity.
4. RESULTS
4.1. Search results
After searching for articles according to the specific detection strategy, 292 articles were initially screened. One hundred and thirty‐six duplicate articles were deleted after importing into Endnote Document Manager. Seventy‐eight articles including irrelevant articles, reviews, letters, case reports, and nonclinical related studies were excluded after carefully reading the titles and abstracts. Forty‐three articles including non‐data studies, non‐prognosis studies, nonhuman studies, and noncompliant studies were excluded through intensive reading of the full text. Finally, 35 literature studies with a total of 3135 patients were included in this meta‐analysis. 8 , 10 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 The process is shown in Figure 1.
FIGURE 1.
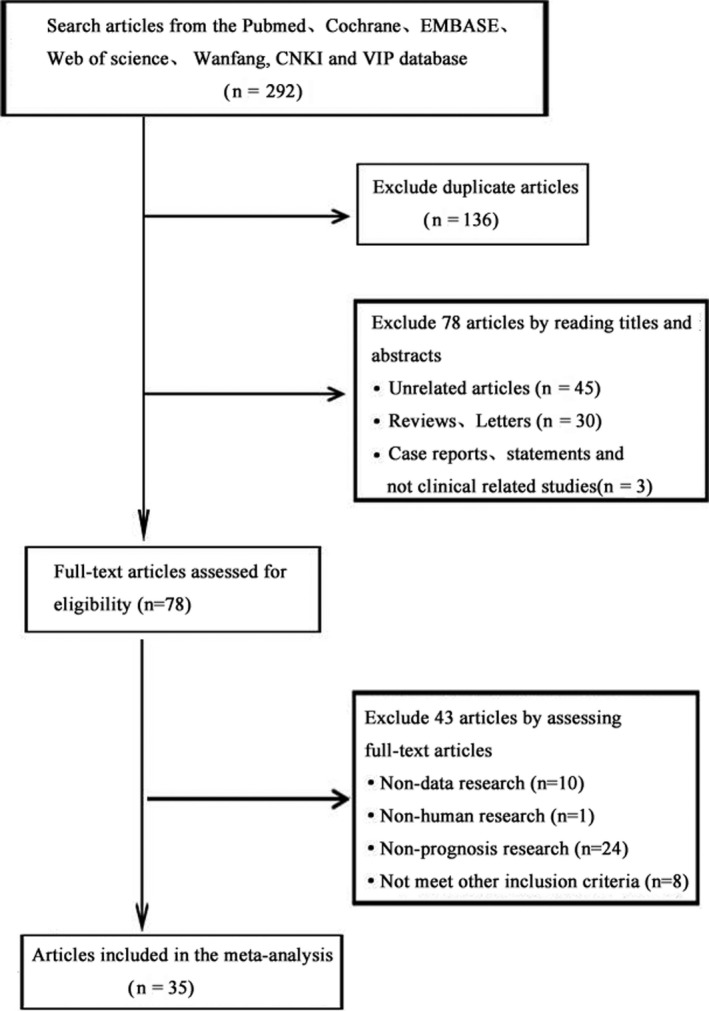
Flow chart of the study selection process
4.2. Characteristics of included studies
The meta‐analysis of 35 articles, involving 3135 people, was the largest sample study for predicting the effect of circRNAs on the prognosis of GC to date. All included studies were from China and were published between 2016 and 2019. HRs and 95% CIs were provided in 13 articles and survival curves were provided in 22 articles. The expression levels of circRNAs included in all studies were detected using qRT‐PCR technology. The literature quality evaluation was independently completed by two researchers by referring to the relevant literature. Cutoff values of high or low circRNA expression were mostly median or mean. The average score of the literature was 7.9. The highest score was 9 and the lowest was 7. The details of these characteristics are shown in Table 1.
TABLE 1.
Main characteristics of circRNAs studies for prognosis analysis of gastric cancer
| References | circRNAs (n = 40) | Year | Nation | Number (n = 3135) | OS | Cutoff value | Sample types | Follow‐up NOS (month) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | |||||||||
| Chen et al. | circPVT1 a | 2016 | China | 187 | 0.59 | 0.41‐0.86 | 2‐fold | Tissue | 100 | 8 |
| Li et al. | hsa_circ_0001017 b | 2017 | China | 278 | 0.41 | 0.16‐0.94 | median | Plasma | 60 | 7 |
| Li et al. | hsa_circ_0061276 b | 2017 | China | 278 | 0.43 | 0.18‐0.93 | median | Plasma | 60 | 7 |
| Pan et al. | ciRS−7 a | 2017 | China | 102 | 2.03 | 1.20‐4.11 | median | Tissue | 60 | 8 |
| Zhang et al. | circRNA_100269 b | 2017 | China | 112 | 0.74 | 0.34‐1.58 | NA | Tissue | 50 | 7 |
| Zhang et al. | Circular RNA_LARP4 | 2017 | China | 80 | 0.53 | 0.24‐0.99 | 2‐fold | Tissue | 150 | 8 |
| Li et al. | circ_0056618 a | 2018 | China | 32 | 1.67 | 0.61‐3.56 | NA | Tissue | 100 | 8 |
| Li et al | circRBMS3 a | 2018 | China | 69 | 1.95 | 1.06‐3.60 | 2‐fold | Tissue | 60 | 8 |
| Liu et al. | circ_0009910 a | 2018 | China | 129 | 2.35 | 1.67‐3.77 | NA | Tissue | 60 | 7 |
| Liu et al. | circular RNA YAP1 b | 2018 | China | 80 | 0.64 | 0.27‐1.53 | 13.37 e | Tissue | 120 | 8 |
| Lu et al. | hsa_circ_0000467 a | 2018 | China | 51 | 2.68 | 1.42‐4.79 | median | Tissue | 50 | 7 |
| Ouyang et al. |
circPDSS1 a |
2018 | China | 20 | 1.65 | 0.50‐3.49 | 2‐fold | Tissue | 60 | 8 |
| Sun et al. | circPVRL3 b | 2018 | China | 62 | 0.44 | 0.20‐0.96 | NA | Tissue | 120 | 8 |
| Tang et al. | circ‐KIAA1244 b | 2018 | China | 87 | 0.41 | 0.08‐0.92 | 1.443 e | Plasma | 40 | 8 |
| Cai et al. | circSMARCA5 b | 2019 | China | 60 | 0.38 | 0.15‐0.95 | median | Tissue | 40 | 8 |
| Cai et al. | circHECTD1 a | 2019 | China | 50 | 1.58 | 0.72‐2.80 | 2‐fold | Tissue | 50 | 8 |
| Du et al. | circ‐PRMT5 a | 2019 | China | 90 | 2.26 | 1.14‐4.47 | median | Tissue | 167 | 8 |
| Ding et al. | circ‐DONSON a | 2019 | China | 142 | 1.80 | 1.01‐3.61 | NA | Tissue | 120 | 8 |
| He et al. | circLMTK2 b | 2019 | China | 111 | 0.36 | 0.17‐0.76 | median | Tissue | 100 | 8 |
| Li et al. | circ‐ERBB2 a | 2019 | China | 58 | 1.57 | 0.57‐4.29 | median | Tissue | 60 | 8 |
| Liu et al. | circHIPK3 a | 2019 | China | 53 | 1.78 | 1.12‐3.22 | NA | Tissue | 80 | 8 |
| Lu et al. | circ‐RanGAP1 a | China | 97 | 1.43 | 1.05‐2.27 | 2‐fold | Tissue | 60 | 8 | |
| Rong et al. | circPSMC3 b | 2019 | China | 106 | 0.59 | 0.30‐0.95 | 2‐fold | Tissue | 50 | 8 |
| Shao et al. | Hsa_circ_0065149 a | 2019 | China | 96 | 3.15 | 1.23‐5.04 | 10.19 e | Tissue | 72 | 8 |
| Tao et al. | hsa_circ_0000419 b | 2019 | China | 96 | 0.81 | 0.18‐1.57 | 8.14 e | Tissue | 60 | 7 |
| Wang et al. | circLMTK2 a | 2019 | China | 121 | 1.75 | 1.02‐3.13 | median | Tissue | 100 | 9 |
| Wang et al. | circOSBPL10 a | 2019 | China | 70 | 1.61 | 0.79‐3.29 | NA | Tissue | 70 | 8 |
| Wang et al. | Five‐circRNAs a , c | 2019 | China | 192 | 1.63 | 1.36‐1.95 | median | Tissue | 160 | 9 |
| Wu et al. | circ‐PRKCI a | 2019 | China | 50 | 1.64 | 0.57‐2.83 | NA | Tissue | 60 | 8 |
| Wu et al. | circZNF609 a | 2019 | China | 80 | 1.58 | 0.78‐2.91 | 1.124 e | Tissue | 200 | 8 |
| Wen et al. | Hsa_circ_0001614 b | 2019 | China | 63 | 0.53 | 0.22‐1.24 | NA | Tissue | NA d | 8 |
| Wu et al. | circ‐DCAF6 a | 2019 | China | 62 | 2.06 | 1.06‐4.01 | mean | Tissue | 60 | 7 |
| Xue et al. | hsa_circ_0081143 a | 2019 | China | 30 | 2.37 | 1.72‐ 4.20 | 2‐fold | Tissue | 60 | 8 |
| Yang et al. | hsa_circ_0005556 b | 2019 | China | 100 | 0.67 | 0.35‐1.28 | NA | Tissue | 72 | 8 |
| Zhang et al. | circDLST a | 2019 | China | 71 | 3.76 | 1.46‐5.67 | NA | Tissue | 100 | 8 |
| Zhang et al. | Hsa_circ_0067997 a | 2019 | China | 48 | 1.47 | 0.51‐4.22 | 1.5‐fold | Tissue | 50 | 8 |
All circRNAs was detected by qRT‐PCR.
Oncogene, Upregulated in gastric cancer tissue or plasma;
Suppressor, Downregulated in gastric cancer tissue or plasma;
hsa_circ_0103398, hsa_circ_0119099, hsa_circ_0121124, hsa_circ_0127859, and hsa_circ_0139915;
NA: Not application;
The cutoff value of ROC curve
4.3. Effect of the expression level of circRNAs on the overall survival of GC
After in‐depth reading of the included 35 articles, we found that 22 articles reported that circRNAs are highly expressed in GC tissues and are negatively correlated with patient prognosis. After quantitative analysis, high expression of these circRNAs indicated poor prognosis for GC patients (vs. low‐expression group: HR = 1.83; 95% CI, 1.64‐2.03; p < 0.001). I 2 = 0 indicated that the research results were highly reliable (Figure 2A); a funnel plot is shown in Figure 2B. At the same time, another 13 articles reported that circRNAs are lowly expressed in GC tissues and have a positive correlation with patient prognosis. The total HR value of the high‐expression group vs. the low‐expression group could be obtained (HR = 0.54; 95% CI, 0.45‐0.66; p < 0.001). There was also no heterogeneity, indicating that the research results were highly reliable (Figure 2C); a funnel plot is shown in Figure 2D. A panel of five circRNAs predicted a greater HR (circ_0009910, hsa_circ_0000467, hsa_circ_0065149, hsa_circ_0081143, and circDLST), which is displayed in Figure 3A. The following five circRNAs showed a smaller HR: circSMARCA5, circLMTK2, hsa_circ_0001017, hsa_circ_0061276, and circ‐KIAA1244 (Figure 3C). The results of the meta‐analysis were 2.63 (2.08‐3.33; p < 0.001) and 0.39 (0.27‐0.59; p < 0.001), respectively. Figure 3B,D show the respective funnel plots. In summary, circRNAs can be used as biological markers to judge the prognosis of GC patients. The roles of the included circRNAs in the development of GC and the specific mechanisms are shown in Table 2.
FIGURE 2.
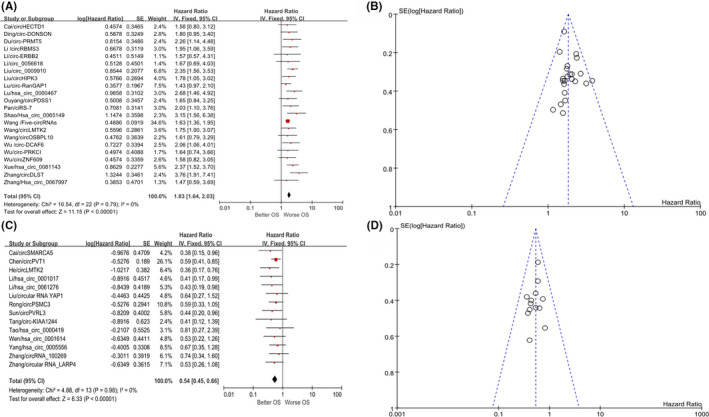
Forest and funnel plots for the prognostic value of circRNAs in overall survival (OS) of gastric cancer patients. High‐expressing circRNAs indicate worse prognosis (A, B) or better prognosis (C, D) for gastric cancer patients
FIGURE 3.
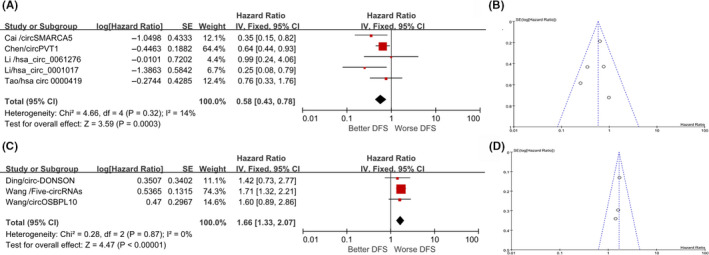
Forest and funnel plots of five circRNAs with maximum (A, B) and minimum (C, D) HR values for gastric cancer
TABLE 2.
The role of circRNAs in the development of gastric cancer
| circRNAs (n = 40) | Gene symbol | Chromosome | Impact on functions of cells | Mechanism |
|---|---|---|---|---|
| circ‐PRMT5 a | PRMT5 | chr14 | Proliferation/apoptosis | Sponging miR−145 and miR−1304 to upregulate MYC |
| circ‐PRKCI a | PRKCI | / | Proliferation/invasion | Binding to microRNA−545 |
| circ‐DONSON a | DONSON | chr21q22.11 | Proliferation/migration/ invasion/ apoptosis | NURF complex dependent activation of transcription factor SOX4 |
| circHECTD1 a | HECTD1 | / | Glutaminolysis/proliferation/migration/ invasion | Targeting miR−1256 and activating β‐catenin/c‐Myc signaling to facilitates glutaminolysis |
| circZNF609 a | ZNF609 | / | Proliferation/ migration | Sponging miR−483‐3p and regulating CDK6 |
| circ_0056618 a | / | / | Proliferation/ metastasis | Regulating miR−206/CXCR4 |
| circRBMS3 a | RBMS3 | chr3 | Proliferation/invasion | Regulating miR−153‐SNAI1 axis |
| circDLST a | DLST | chr 14q24.3 | Proliferation/Colony formation | Sponging miR−502‐5p and activating the NRAS/ MEK1/ ERK1/2 signaling |
| circOSBPL10 a | OSBPL10 | chr3 | Proliferation/invasion/ migration | A miRNA sponge for miR−136‐5P |
| Five‐circRNAs a , c | / | / | / | One carbon pool by folate pathway |
| hsa_circ_0000467 a | SKA3 | chr13 | Proliferation/migration/invasion | Tumor promoter |
| circ‐ERBB2 a | ERBB2 | / | Proliferation/apoptosis, migration/invasion | MiR−503/CACUL1 and miR−637/MMP−19 signaling |
| ciRS−7 a | / | / | Proliferation/metastasis/ colony formation | Abrogates the tumor suppressive effect of miR−7 via PTEN/PI3 K/AKT signaling pathway |
| Hsa_circ_0065149 a | SETD2 | chr3 | / | Harbor hsa‐miR−197‐5p, hsa‐miR−222‐3p, hsa‐miR−330‐5p, and hsa‐miR−486‐3p seed sequences |
| circHIPK3 a | HIPK3 | / | Proliferate/migrate | Regulating the Wnt/β‐catenin pathway |
| Hsa_circ_0067997 a | FNDC3B | / | Viability/colony formation/invasive ability | Inhibition of miR−515‐5p and activation of X chromosome‐linked inhibitor of apoptosis (XIAP) |
| circ_0009910 a | / | / | Proliferation/migration/invasion | Decreased expression of E‐cadherin and increased expression of N‐cadherin and snail |
| circLMTK2 a | LMTK2 | chr7 | Proliferation/metastasis | A sponge of miR−150‐5p |
| circ‐RanGAP1 a | RANGAP1 | chr22q13.2 | Invasion/metastases | Regulates VEGFA expression by targeting miR−877‐3p |
| circ‐DCAF6 a | / | chr1 | Proliferation/apoptosis | Sponging miR−1231 and miR−1256 |
| hsa_circ_0081143 a | / | / | cisplatin resistance | Targeting miR−646/CDK6 pathway |
| circPDSS1 a | PDSS1 | / | apoptosis | Sponging miR−186‐5p and modulating NEK2 |
| circPVT1 a | PVT1 | chr8q24 | Proliferation | A sponge for members of the miR−125 family |
| circular RNA YAP1 b | YAP1 | / | Proliferation/invasion | Regulating the miR−367‐5p/p27Kip1axis |
| circPSMC3 b | PSMC3 | / | Proliferation/metastasis | Acting as a competitive endogenous RNA through sponging miR−296‐5p |
| hsa_circ_0005556 b | NBAS | chr2 | Proliferation/invasion/chemotherapy resistance | CircRNA‐miRNA sponging sites: miR−548c−3p, miR‐ 587, miR−4739, miR−125a−3p, miR−297 |
| circRNA_100269 b | LPHN2 | chr1 | Proliferation | Targeting miR−630 |
| hsa_circ_0000419 b | RAB3IP | chr12 | / | Harbor hsa‐miR−141‐5p and hsa‐miR−589‐3p by matching miRNAs seed sequences |
| circLMTK2 b | LMTK2 | chr7q21.3 | Cell viability /mobility | Bax and cleaved Caspase 3 expression was enhanced significantly accompanying with suppressed Bcl−2 |
| circ‐KIAA1244 b | KIAA1244 | Cellular components/molecular functions/ signaling pathways | / | |
| Circular RNA_LARP4 b | LARP4 | chr12 | Proliferation/invasion | Sponging miR−424‐5p and regulating LATS1 expression |
| Hsa_circ_0001614 b | SENP6 | chr6 | Inflammatory cells | / |
| circSMARCA5 b | SMARCA5 | / | Proliferation/migration/ invasion | / |
| circPVRL3 b | PVRL3 | / | proliferation/ migration | Sponge to 9 miRNAs d and be able to have a binding with AGO2, FUS, LIN28A, PTB, and EIF4A3 |
| hsa_circ_0001017 b | XPO1 | chr2 | / | / |
| hsa_circ_0061276 b | NRIP1 | / | / | / |
Oncogene, Upregulated in gastric cancer tissue or plasma.
Suppressor, Downregulated in gastric cancer tissue or plasma.
hsa_circ_0103398, hsa_circ_0119099, hsa_circ_0121124, hsa_circ_0127859, and hsa_circ_0139915.
miR‐203, miR‐1272, miR‐1283, miR‐31, miR‐638, miR‐496, miR‐485‐3p, miR‐766, and miR‐876‐3p.
4.4. Effect of circRNA expression levels on disease‐free survival of GC
The included seven studies provided the HRs and 95% CIs of the disease‐free survival of GC patients, involving 1025 patients. We performed a quantitative analysis and found that there were four reports of high expression that were positively correlated with the disease‐free survival of patients. Quantitative analysis of total HR for the high‐expression group vs. the low‐expression group could be obtained (HR = 0.58; 95% CI, 0.43‐0.78; p = 0.0003). There was no heterogeneity, indicating that the research results were highly credible (Figure 4A); a funnel plot is shown in Figure 4B. At the same time, the high expression of circRNAs reported by another three reports was negatively correlated with the disease‐free survival of patients. The total HR for the high‐expression group vs. the low‐expression group could be obtained (HR = 1.66; 95% CI, 1.33‐2.07; p < 0.001).The results of the heterogeneity test also indicated that the research results were highly reliable (Figure 4C); a funnel plot is shown in Figure 4D. In summary, circRNAs can be used as biological markers that affect the quality of disease‐free survival and the prognosis of GC patients.
FIGURE 4.
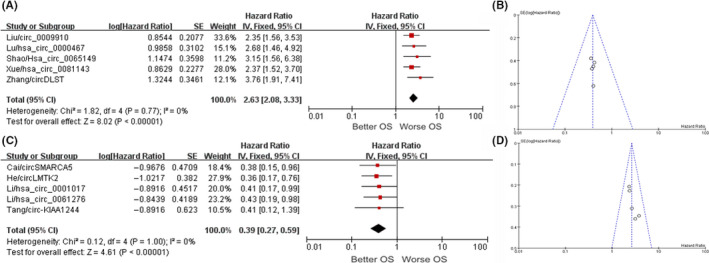
Forest and funnel plots for prognostic value of circRNAs in disease‐free survival (DFS) of gastric cancer patients. High‐expressing circRNAs indicate worse prognosis (A, B) or better prognosis (C, D) for gastric cancer patients
5. DISCUSSION
Because of chemotherapy resistance and tumor metastasis, the prognosis of GC is poor. Although new anticancer drugs continue to emerge, GC still poses a huge threat to human health. In recent years, increasing numbers of studies have focused on the clinical role of circRNAs. Many reports show that circRNAs can be used as molecular biomarkers for the prognosis of GC patients. 10 , 12 , 29 Through a large data meta‐analysis, we did not focus on single circRNAs but provided evidence to elucidate the prognostic value of the abnormal expression of multiple circRNAs in GC patients. Furthermore, two groups of circRNAs with the largest HRs were selected to illustrate their prognostic value and provide ideas for future combined testing. The results of the meta‐analysis showed that the total HR value (95% CI) of OS and DFS was 1.83 (1.64‐2.03; p < 0.01) and 1.66 (1.33‐2.07; p < 0.01), respectively. The total HR (95% CI) of OS and DFS was 0.54 (0.45‐0.66; p < 0.01) and 0.58 (0.43‐0.78; p < 0.01), respectively. In addition, two panels of five circRNAs showed a greater prognostic value: 2.63 (2.08‐3.33; p < 0.01) (circ_0009910, hsa_circ_0000467, hsa_circ_0065149, hsa_circ_0081143, and circDLST); and 0.39 (0.27‐0.59; p < 0.01) (circSMARCA5, circLMTK2, hsa_circ_0001017, hsa_circ_0061276, and circ‐KIAA1244). These results indicated that abnormal expression of circRNAs can be used as tumor biomarkers for the prognosis of GC patients.
CircRNAs can promote or inhibit the process of tumor cell migration, invasion, proliferation, drug resistance, and epithelial‐mesenchymal transition (EMT). CircRNAs can function through different target genes, which is related to prognosis. For example, Li et al. 45 showed that circ_104916 could inhibit the growth of GC. The expression level of circ_104916 in GC tissue and five GC cell lines was downregulated. Further experiments in vitro showed that circ_104916 could inhibit the proliferation, invasion, migration, and EMT of GC cells. The low expression of circ_104916 was related to poor prognosis. Wang et al. 17 reported that circLMTK2, derived from exons 10 and 11 of LMTK2 mRNA, has a negative effect on GC cells by inhibiting the function of miR‐150‐5p such as blocking DNA synthesis and reducing cell proliferation, migration, and invasion ability in GC cells. CircRNA‐circPDSS1 promotes the progression of GC by regulating miR‐186‐5p and NEK2. 21 Wu et al. 37 proved that circ‐DCAF6 is the upstream negative regulator of miR‐1231 and miR‐1256, and its enhancement promotes cell proliferation and indicates a poor prognosis. CircRNA‐100269 can target miR630 to inhibit the proliferation of tumor cells. 23 CircLARP4, derived from exons 9 and exon 10 of LARP4, has high stability (half‐life >24 h), and it is also regarded as a potential biomarker. CircLARP4 can be regarded as the sponge of miR‐424‐5p to inhibit its effect of promoting cell proliferation and invasion. Therefore, the high expression of circLARP4 is related to better prognosis. 46 CircRNAs can also induce apoptosis and inhibit cell proliferation, migration, and invasion in GC through the EMT signaling pathway. 47 Xue et al. 41 suggested that has_circ_0081143 might act as a ceRNA to inhibit miR‐646, increasing the expression of CDK6 and promoting the resistance of GC to cisplatin. Another research group 48 found that the expression of circAKT3 in GC tissues of cisplatin‐resistant patients was higher than that in cisplatin‐sensitive patients. Further mechanistic research showed that circAKT3 promoted DNA damage repair and inhibited cell apoptosis through sponging miR198. The process increased the expression of its downstream target PIK3R1, providing a new treatment prospect for cisplatin‐resistant patients. Shao et al. 49 found that the expression of hsa_circ_0014717 was significantly downregulated in 77.2% of GC tissues. Its expression level in GC tissues was negatively correlated with tumor stage, distant metastasis, carcinoembryonic antigen, and CA19‐9. Therefore, the low expression of hsa_circ_0014717 was associated with poor prognosis. In addition, hsa_circ_0014717 was also detected in body fluids, indicating that hsa_circ_0014717 is a noninvasive biomarker. Another study 29 reported that circPVT1 encoded by PVT1 was upregulated in GC, which inhibited miR‐125 and promoted cell proliferation. By analyzing the expression of circPVT1 in 187 patients with GC and their 100‐month follow‐up results, the researchers found that circPVT1 was an independent prognostic biomarker of OS and DFS in patients with GC. This suggests that circPVT1 not only plays a role in the treatment of GC, but also may be a prognostic marker for GC patients.
An important mechanism of circRNAs in cancer‐related signal transduction is circRNA‐miRNA‐protein. The PTEN/PI3K/AKT/mTOR signaling pathway is the most used research pathway between circRNAs and GC. CircPSMC3 indirectly upregulates PTEN through sponging miRNAs. 19 Cirs7 targets miR7 to inhibit the expression of PTEN, AKT, and MTOR. Therefore, Cirs7 can downregulate PTEN expression and upregulate AKT and MTOR expression. 8 CircOSBPL10 has a carcinogenic role in GC by activating the Wnt/β‐catenin signaling pathway. Overexpression of circOSBPL10 will lead to upregulation of WNT2 expression by sponging mir‐136‐5p. Wnt2 activates the Wnt/β‐catenin signaling pathway and promotes the development of GC. 50 In addition, circHIPK3 and circHECTD1 also promote the development of GC through the Wnt/β‐catenin pathway. 15 , 51 The expression of circ‐ZFR in tumor tissue was significantly lower than in para‐carcinoma tissue, while the expression of circ‐ZFR in GC cell lines HGC‐27, AZ521, and AGS was significantly lower than that in the gastric epithelial cell line GES1. Circ‐ZFR causes cell cycle arrest and apoptosis of GC through sponging miR‐107/miR‐130a, which binds to the 3ʹ untranslated region (UTR) of PTEN. 52 Many studies have shown that PTEN can be targeted and regulated by miR‐107 and miR‐130a, affecting the activity of cancer cells. 53 , 54 These results suggest that the circ‐ZFR‐miR‐107/miR‐130a‐PTEN pathway has an important role in the development of GC. Although many signaling pathways related to GC have been described, the involvement of other signaling pathways such as the Notch pathway is still unclear.
There are still many deficiencies in our research. First, almost all tissue and plasma samples in our study were collected over a long period. Although the samples were preserved at a low temperature, there is still some influence on the degradation of circRNAs. Second, we cannot guarantee that all relevant studies are included in our analysis, and all the included studies with high prognostic value showed positive results. The publication bias reached a consensus. Thus, our results might overestimate the prognostic value of circRNAs among GC patients. Finally, we should perform more experiments to verify the reliability and economy of the analysis results in body fluid samples to judge whether circRNAs can be used as simple, fast, accurate, economic, and noninvasive tumor markers in clinical settings.
Evaluating the prognosis of patients and exploring the mechanism of circRNAs has a critical role in clinical decision‐making and in the development of novel targeted gene therapy. CircRNAs can perform physiological functions by acting as miRNA sponges and translation templates, regulating the expression of parental genes, and acting on RNA binding proteins. Despite many studies, we still know little about circRNAs, and they are far from being used in clinical practice. First, researchers mainly use tumor tissue or tumor cell samples to perform research on cancer‐related circRNAs, which should be extended to the detection of noninvasive samples (such as blood, urine, and saliva). Second, in the future, circRNAs may be considered as a potential therapeutic target. However, how to transport circRNAs to relevant sites to obtain long‐term efficacy and how to achieve nonimmune rejection are urgent problems. Finally, the goal of circRNA research is to apply them safely to the clinical treatment of human diseases, and thus, a large number of clinical studies are needed to complete this process.
6. CONCLUSION
By detecting the expression levels of a panel of circRNAs in tissues or body fluids, we can make appropriate clinical decisions based on different prognoses.
CONFLICT OF INTEREST
The authors have no financial disclosures or conflicts of interest.
AUTHOR CONTRIBUTIONS
H. Chen. and C. Liang had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: X. Wang, Y. Liu; Manuscript drafting and reviewing: C. Ren; Acquisition, analysis, and interpretation of data: all authors; Statistical analysis: C. Liang; Administrative, technical, and material support: H. Chen. H. Chen and C. Liang contributed equally to this work.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (81573220), the Foundation for Young Medical Scholars in Jiangsu Province (QNRC2016325 and QNRC2016333), and the Foundation for Young Medical Scholars in Yangzhou (RCC201835)
Chen H, Liang C, Wang X, et al. The prognostic value of circRNAs for gastric cancer: A systematic review and meta‐analysis. Cancer Med. 2020;9:9096–9106. 10.1002/cam4.3497
Hui Chen and Chengtong Liang contributed equally.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in [repository name e.g., “figshare”] at http://doi.org/[doi], reference number [reference number].
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- 3. Choi IJ, Kim CG, Lee JY, et al. Family history of gastric cancer and helicobacter pylori treatment. N Engl J Med. 2020;382:427–436. [DOI] [PubMed] [Google Scholar]
- 4. Yusefi AR, Bagheri Lankarani K, Bastani P, Radinmanesh M, Kavosi Z. Risk factors for gastric cancer: a systematic review. Asian Pac J Cancer Prev. 2018;19:591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fang X, Wei J, He X, et al. Landscape of dietary factors associated with risk of gastric cancer: a systematic review and dose‐response meta‐analysis of prospective cohort studies. Eur J Cancer. 2015;51:2820–2832. [DOI] [PubMed] [Google Scholar]
- 6. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 7. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 8. Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS‐7 abrogates the tumor suppressive effect of miR‐7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119:440–446. [DOI] [PubMed] [Google Scholar]
- 9. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 10. Lu J, Zhang PY, Xie JW, et al. Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal. 2019;33:e22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time‐to‐event data into meta‐analysis. Trials. 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M, Liu KD, Zhang L, et al. Circ‐0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22:8248–8256. [DOI] [PubMed] [Google Scholar]
- 13. Li HB, Yao GL, Feng B, Lu XL, Fan YG. Circ_0056618 and CXCR4 act as competing endogenous in gastric cancer by regulating miR‐206. J Cell Biochem. 2018;119:9543–9551. [DOI] [PubMed] [Google Scholar]
- 14. Zhang J, Hou LD, Liang R, et al. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR‐502‐5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 2019;18(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cai J, Chen Z, Wang J, et al. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR‐1256 and activating β‐catenin/c‐Myc signaling. Cell Death Dis. 2019;10(8):576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He J, Chen J, Ma B, Jiang L, Zhao GF. CircLMTK2 acts as a novel tumor suppressor in gastric cancer. Biosci Rep. 2019;39(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S, Tang D, Wang W, et al. circLMTK2 acts as a sponge of miR‐150‐5p and promotes proliferation and metastasis in gastric cancer. Mol Cancer. 2019;18(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du WB, Li D, Guo XM, et al. Circ‐PRMT5 promotes gastric cancer progression by sponging miR‐145 and miR‐1304 to upregulate MYC. Artif Cells Nanomed Biotechnol. 2019;47:4120–4130. [DOI] [PubMed] [Google Scholar]
- 19. Rong DW, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR‐296‐5p. Mol cancer. 2019;18(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Li G, Xue M, Yang F, Jin Y, Fan Y, Li W. CircRBMS3 promotes gastric cancer tumorigenesis by regulating miR‐153–SNAI1 axis. J Cell Physiol. 2019;234:3020–3028. [DOI] [PubMed] [Google Scholar]
- 21. Ouyang YM, Li YJ, Huang YG, et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR‐186‐5p and modulating NEK2. J Cell Physiol. 2019;234:10458–10469. [DOI] [PubMed] [Google Scholar]
- 22. Tang WW, Fu K, Sun HD, Rong DW, Wang HJ, Cao HY. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol cancer. 2018;17(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR‐630. Aging. 2017;9:1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cai J, Chen ZQ, Zuo XL. circSMARCA5 functions as a diagnostic and prognostic biomarker for gastric cancer. Dis Markers. 2019;2019:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ding L, Zhao Y, Dang S, et al. Circular RNA circ‐DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol cancer. 2019;18:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu L, Li Y, Xu XM, Zhu X. Circular RNA circ‐PRKCI promotes cell proliferation and invasion by binding to microRNA‐545 in gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:9418–9426. [DOI] [PubMed] [Google Scholar]
- 27. Lu J, Wang Y‐H, Yoon C, et al. Circular RNA circ‐RanGAP1 regulates VEGFA expression by targeting miR‐877–3p to facilitate gastric cancer invasion and metastasis. Cancer lett. 2020;471:38–48. [DOI] [PubMed] [Google Scholar]
- 28. Chen J, Chen J, Wu J, et al. Circular RNA profile identifies circOSBPL10 is unregulated and as a proliferative factor in gastric cancer. Ann Oncol. 2018;29:vii55. [Google Scholar]
- 29. Chen J. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Ann Oncol. 2016;27:ix78. [DOI] [PubMed] [Google Scholar]
- 30. Liu H, Liu Y, Bian ZL, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR‐367‐5p/p27 (Kip1) axis. Mol cancer. 2018;17(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J, Hou LD, Zhu JS. Circular RNA‐LARP4 inhibits growth and invasion of gastric cancer by sponging has‐miR‐424‐5p and upregulating large tumor suppressor kinase 1. J Dig Dis. 2017;18:22. [Google Scholar]
- 32. Wu WD, Wei NX, Shao G, Jiang CN, Zhang SR, Wang LH. circZNF609 promotes the proliferation and migration of gastric cancer by sponging miR‐483‐3p and regulating CDK6. Onco Targets Ther. 2019;12:8197–8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tao X, Shao Y, Lu R, et al. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract. 2020;216(1):152763. [DOI] [PubMed] [Google Scholar]
- 34. Wang F, Li X, Zhao X, Xue Y. Detection of a 5‐circRNA signature to improve prognostic prediction in gastric cancer. J Investig Med. 2020;68(3):762–769. [DOI] [PubMed] [Google Scholar]
- 35. Yang L, Yu Y, Yu X, et al. Downregulated expression of hsa‐circ‐0005556 in gastric cancer and its clinical significance. Dis Markers. 2019;2019:2624586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun HD, Xu ZP, Sun ZQ, et al. Down‐regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8(1):10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu L, Liu D, Yang Y. Enhanced expression of circular RNA circ‐DCAF6 predicts adverse prognosis and promotes cell progression via sponging miR‐1231 and miR‐1256 in gastric cancer. Exp Mol Pathol. 2019;110:104273. [DOI] [PubMed] [Google Scholar]
- 38. Wen J. Expression of hsa_circ_0001614 in gastric cancer and its relationship with prognosis of gastric cancer patients [Master]. China Medical University; 2019. [Google Scholar]
- 39. Shao Y, Tao X, Lu R, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26(3):1475–1482. [DOI] [PubMed] [Google Scholar]
- 40. Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR‐515‐5p and activation of X chromosome‐linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol. 2019;47:308–318. [DOI] [PubMed] [Google Scholar]
- 41. Xue M, Li G, Fang X, Wang L, Jin Y, Zhou Q. Hsa‐circ‐0081143 promotes cisplatin resistance in gastric cancer by targeting miR‐646/CDK6 pathway. Cancer Cell Int. 2019;19:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/p‐catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23:7905–7912. [DOI] [PubMed] [Google Scholar]
- 43. Li XS, He M, Guo JJ, Cao T. Upregulation of circular RNA circ‐ERBB2 predicts unfavorable prognosis and facilitates the progression of gastric cancer via miR‐503/CACUL1 and miR‐637/MMP‐19 signaling. Biochem Biophys Res Commun. 2019;511:926–930. [DOI] [PubMed] [Google Scholar]
- 44. Li TW. Identification and diagnostic value of gastric cancer‐related circular RNA [Master]. Ningbo University; 2017. [Google Scholar]
- 45. Li J, Zhen L, Zhang Y, et al. Circ‐104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR‐424‐5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22:2297–2303. [DOI] [PubMed] [Google Scholar]
- 48. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR‐198 suppression. Mol Cancer. 2019;18:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S, Zhang X, Li Z, et al. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. Oncogene. 2019;38:6985–7001. [DOI] [PubMed] [Google Scholar]
- 51. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/β‐catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23:7905–7912. [DOI] [PubMed] [Google Scholar]
- 52. Liu T, Liu S, Xu Y, et al. Circular RNA‐ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR‐130a/miR‐107 and modulating PTEN. Cancer Res Treat. 2018;50:1396–1417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Wei H, Cui R, Bahr J, et al. miR‐130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017;77:6168–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiong J, Wang D, Wei A, et al. Deregulated expression of miR‐107 inhibits metastasis of PDAC through inhibition PI3K/Akt signaling via caveolin‐1 and PTEN. Exp Cell Res. 2017;361:316–323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in [repository name e.g., “figshare”] at http://doi.org/[doi], reference number [reference number].


