Abstract
Objective: To investigate the difference between trimethylation of monocyte histone H3K4 and DNA methylation in acute rejection (AR) after renal transplantation in rats and reveal the epigenetic mechanism of the AR rats based on metabolomics. Method: Peripheral blood mononuclear cells (PBMCs) were isolated, and CD4 +CD25 + Treg cells were sorted by flow cytometry. The Foxp3 mRNA and protein levels of CD4 +CD25 + Treg cells were detected by real-time RT-PCR and Western blotting, respectively. High-throughput screening was applied to evaluate the H3K4 methylation of monocytes using chromatin immunoprecipitation with DNA microarray (ChIP-chip) and verified by ChIP with real-time PCR (ChIP-qPCR). Methylated DNA immunoprecipitation sequencing was combined with real-time PCR (MeDIP-qPCR) to detect the DNA methylation level of positive genes (ABCC4, Mef2d, Tbx1 and Eif6). Real-time quantitative PCR (qRT-PCR) and Western blotting were used to detect the mRNA and protein levels of positive genes. The difference in lipid metabolism in the blood of (non) acute rejection rats was analysed by HPLC/MS. Results: AR rats showed an apparent increase in BUN and Cr levels, as well as IL-2, IL-10 and IFN-γ. Compared with non-AR rats, the expression of CD4 +CD25 + Treg cells and Foxp3 mRNA and protein were significantly lower in AR rats. AR rats also showed an increase in H3K4 trimethylation of CD4 +CD25 + Treg and decrease in DNA methylation. There were significant differences in the DNA methylation level of four positive genes between AR and non-AR rats (P<0.05), in addition to differences in the expression levels of mRNA and protein. Pathological examination of the transplanted kidney indicated that AR rats had more severe pathological injury of the kidney than the non-AR rats. There were significant increase in the contents of several phosphatidylcholines, lysophosphatidylcholine, free fatty acids and carnitine in AR rats which detected by HPLC/MS. Conclusion: H3K4 trimethylation increased in PBMCs in AR rats, while DNA methylation decreased, indicating the presence of epigenetic differences between AR and non-AR rats. Metabolomic studies indicated a significant increase in AR rats in the contents of several metabolites, such as phosphatidylcholines, lysophosphatidylcholine, free fatty acids and carnitine, suggesting an increasein phospholipase activity and leading to an energy metabolism imbalance with intensification of β-oxidation. DNA methylation may be associated with an increase in phosphatidylcholines, lysophosphatidylcholine and free fatty acids in AR rats, which may further affect energy metabolism by enhancing the tricarboxylic acid cycle in AR rats.
Keywords: Rat, acute rejection, histone methylation, DNA methylation, metabolomics
Introduction
Epigenetics refers to the fact that the DNA sequence of organisms does not change, but the gene expression has undergone hereditary changes. Epigenetics can affect the homeostasis of the immune system, it’s believed to play an important role in the homeostasis of the immune system after kidney transplantation and is a major influential factor in normal immune functions. Histone modification and DNA methylation are two major epigenetic mechanisms [1-3]. Histone methylation is involved in many chromatin-related activities such as gene expression regulation and maintenance of genomic stability, including acetylation, methylation, phosphorylation and ubiquitination. Methylation mainly occurs in the side chains of arginine and lysine. The methylation of Lys (H3K4) in H3 histone is a marker of gene transcription activation. Gene silencing and gene transcription decrease under the catalysis of enzymes when DNA methylation occurs, however, when DNA demethylation occurs, methylated genes can be demethylated, and silenced inactivated genes can be activated, resulting in increased gene transcription and expression. In addition to their respective influence on chromosome structure, gene transcription and replication, histone modification and DNA methylation also interact with each other [4-6]. In acute rejection (AR) following kidney transplantation, metabolic disorders of several phospholipids can be detected in the serum. In an organism, membrane lipid microenvironment consisting of lipids not only plays an important role in intercellular signal transduction but also in energy metabolism. Lipid metabolism disorder can interfere with normal intercellular signal transduction and disrupt the energy metabolism balance, thus further leading to the failure of the transplanted kidney in CKD patients. Some external factors may cause epigenetic dysregulation in the immune response, leading to the abnormal expression of relevant genes and immune imbalance, which in turn leads to autoimmune diseases.
Kidney transplantation is an effective method for the treatment of end-stage kidney disease, but some immune rejection of donor kidney may lead to kidney transplantation failure. Therefore, it is considered that immune response is the key to determine the success of renal transplantation. It will trigger a series of immune activities when received kidney transplantation, and over-strong immune response can produce a variety of cellular immune effects by activating a variety of T lymphocytes, which can lead to acute rejection. This study focused on the regulatory mechanism of Treg cell differentiation in AR following kidney transplantation in rats and analysed the working mechanism of H3K4 trimethylation from the perspective of genetic modification. Moreover, high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS) was used to detect lipid metabolism in blood.
Methods
Laboratory animals and grouping
Male Wistar and SD rats (aged 14 weeks, 250-300 g) were used to construct the renal allograft transplantation model and all rats were fed adaptively for 2 weeks. There were two experimental groups, AR and non-AR. In the non-AR group, Wistar rats were the recipients, and the homologous series Wistar rats were the donors (N=15). In the AR group, Wistar rats were the recipients and SD rats were the donors (N=15). To analyse the metabolomes, a normal control group and sham-operated group were set up. The normal control group consisted of Wistar rats that did not undergo any operation (N=15). The sham-operated group consisted of Wistar rats that underwent laparotomy and suturing only (N=15). Animal anesthesia machine, isoflurane inhalation anesthesia, 3% induction, 1.5% maintenance, flow rate 0.7 L/min, real-time adjustment of flow rate according to the depth of anesthesia. The modelling effect of the AR and non-AR group were verified by the post-operative renal function test and pathological renal biopsy. Rats were fasted for 12 h before surgery with free access to water and no immunosuppressive therapy was administered after surgery. All the rats were killed by intravenous injection of KCl (10%) through caudal vein after surgery 7 days and kidney tissues were obtained. Three rats died during the whole experiment. The first death occurred in the early stage of the experiment. Considering that the anesthesia caused no breathing and pulse after the surgery, the other two died on the 3th and 4th day after the surgery. Speculating that it may be related to the loss of renal function caused by postoperative infection according to the postoperative feeding and dissection.
This study was approved by the Animal Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University (No, IACUC-20140214063).
Inclusion criteria
All included rats had good growth status with normal mobility and surviving transplanted kidneys. Survival of the transplanted kidneys was determined based on the following criteria: (1) the circulation resumed after anastomoses, the kidneys immediately filled and turned red, and the kidneys exhibited elasticity and hardness; (2) there was no exudation from the anastomotic site, the renal artery had normal pulsation, and the renal vein had no distortion or blood stasis and good filling; and (3) ureteral peristaltic activity was observed 2-3 min after the operation, and the transplanted kidneys were considered surviving if urine was observed flowing out of the ureteral orifice.
Exclusion criteria
Rats were removed from the study when any of the following conditions were occurred: (1) post-operative infection; (2) post-operative complications such as the renal artery or vein thrombosis, bleeding or stenosis of the anastomotic site; and (3) stenosis, necrosis and fistula of the ureter in the anastomotic site of the ureter.
Sample collection
Blood sample collection
Blood samples were collected before and on the 7th day after surgery, and they were anticoagulated with heparin sodium. The subpopulation of T cell in fresh blood was determined, sorted and purified by flow cytometry, and the serum was extracted and stored at -80°C.
Collection of kidney samples
Kidneys were harvested at 7 days after surgery. If the rats died before the 7th day after surgery, the kidney samples were harvested upon death and subjected to HE staining.
Detection procedures
Liver and kidney function test
Detection indicators included aspartate transaminase (AST), alanine aminotransferase (ALT), blood urea nitrogen (BUN), and creatinine (Cr), which were measured using a Backman LX 20 chemistry analyser.
Cytokine detection
The cytokines IL-2, IL-10 and IFN-γ were detected using ELISA. The kit was purchased from R&D, America.
Histopathology of the kidney
The pathological diagnosis was based on the Banff’ 97 classification criteria, and pathological changes in the kidneys were observed in the normal control group, AR group and non-AR group.
CD4 +CD25 + Treg sorting and ratio detection
Peripheral blood was collected and red cells were removed with red blood cell lysis buffer; then, the cells were resuspended in 500 µl pre-cooled PBS and supplemented with 5 µl FITC-labelled CD4 antibody and PE-labelled CD25 antibody. After incubation on ice for 30 min in the dark, the cells were centrifuged at 1000 rpm for 5 mins, and the supernatant was discarded. The cells were washed twice with pre-cooled PBS and then resuspended in 1000 µl FACS buffer. CD4 +CD25 + Treg cells were sorted using a flow cytometer (BD, America), and the ratio changes were monitored.
Detection of Foxp3 mRNA and protein expression of CD4 +CD25 + Treg cells
RNA extraction
PBMCs were isolated and added with 1 ml TRIzol, followed by vortex mixing and incubation at room temperature for 15 min. Next, 200 µL chloroform was added to the mixture and mixed by inverting the tube. The cells were incubated at room temperature for 5 min and centrifuged at 4°C, 12000 rpm for 15 min. The supernatant was collected into a new centrifuge tube (1.5 ml). Next, an equal volume of isopropanolwas added and mixed by inverting the tube, followed by incubation at -20°C for 30 min; then, the sample was and centrifuged again at 4°C at 12000 rpm for 15 min. The supernatant was then discarded, and the RNA precipitate was washed with 1 mL 75% ethanol (prepared in DEPC-treated water) and mixed well. After centrifugation for 3 mins at 4°C and 12000 rpm, the supernatant was discarded and the precipitate was collected and dried at room temperature for 2-3 min. RNA was dissolved in RNase-free H2O and detected based on the A260/A280 ratio. The RNA integrity was also assessed.
Reverse transcription and quantitative fluorescence PCR
The DNA removal system was prepared using qualified RNA samples together with 2 µL 5×g DNA buffer, 4 µL total RNA and 4 µL RNase-free ddH2O. After complete mixing and centrifugation, the cells were incubated at 42°C for 3 min and then placed on ice. The reverse transcription system was prepared using 2 µL 10× first-strand buffer, 1 µL RT primer mix, 2 µL FQ-RT primer mix and 5 µL RNase-free ddH2O. The reverse transcription mixture was added to the reaction buffer and mixed well. After incubation at 42°C for 15 min followed by 95°C for 3 min, the cells were placed on ice, and cDNA was synthesized by reverse transcription. The quantitative fluorescence PCR system consisted of the following: Foxp3 primers: 5’-CAGCTAAGCCTATGGCTCCTT-3’, 5’-AGGGTGCCACCATGACTAGG-3’, 228 bp; and β-Actin primers, serving as the internal reference: 5’-CCCATCTATGAGGGTTACGC-3’, 5’-TTTAATGTCACGCACGATTTC-3’, 150 bp. The volume of each primer was 0.4 µL, together with 10 µL SYBR Select Master Mix (2X), 1 µL cDNA template, and 8.2 µL RNase-free water. Foxp3 mRNA expression was detected using an ABI7500 Real-Time PCR system.
Protein extraction and detection
PBMCs were collected, and proteins were isolated; the proteins were subjected to 12% alkyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which was followed by membrane transfer and incubation with the primary and secondary antibodies (Abcam, United States). Next, the ECL Kit (Abcam, USA) was used for colour rendering, and the protein level of Foxp3 was determined based on the relative grey value.
mRNA and protein expression of positive genes and DNA methylation levels
Detection of H3K4 trimethylation using ChIP-chip
Chromatin immunoprecipitation combined chip technology (ChIP-chip) was performed according to the literature [7]. DNA was amplified, and the products were purified. ChIP DNA and Input DNA were labelled with Cy3 or Cy5 dye and hybridized with the 12K CpG island array as fluorescent probes. High-throughput screening was applied to the H3K4 trimethylation of the CD4 +CD25 + Treg cells. The microarrays were scanned with aGenePix 4000B Array Scanner (Axon Instruments). The raw data were analysed using Genespring software.
Verification by ChIP-qPCR
As described for ChIP-chip, DNA amplification was performed on an ABI7700 real-time PCR system. Primers for positive genes were designed using Primer 5.0 and are as follows: Abcc4 primers: 5’-CGCTAGGGTTTGGGGAGATG-3’, 5’-GGCCAGGCGGTGTATTATCT-3’, 150 bp; Mef2d primers: 5’-TGGTGGCATAGGTTTGATGGT-3’, 5’-GCAGAGTAGGGTGGCATGTAT-3’, 143 bp; Eif6 primers: 5’-TGGTGGCATAGGTTTGATGGT-3’, 5’-GCAGAGTAGGGTGGCATGTAT-3’, 146 bp; and Tbx1 primers: 5’-GAGACGAAAGTGCGCAGGAA-3’, 5’-CAGGGGAGTGGGAAAACCAG-3’, 120 bp. Standard and melting curves were plotted to ensure the accuracy and specificity of the amplification. The relative content of template in the samples was expressed by the Ct (threshold cycle). The relative expression of target genes was calculated using the 2-ΔΔCt method.
MeDIP-qPCR detection of gene methylation levels
The DNA methylation level was measured for positive genes using the SssI methyl acceptance assay according to the literature. Real-time PCR was applied to the target genes and control for each sample. Primers for positive genes were as follows: Abcc4 5’-CCTTCAGGGTGGTGCTTAGG-3’, 5’-GAAAGGAGGCTGGGGAAAGG-3’, 150 bp; Mef2d 5’-GATTAGAGAACGCGATCTGGC-3’, 5’-TGGCAGGGGTCTAAGGGTAAAT-3’, 111 bp; Eif6 5’-CTAGTAACCATCGAGTCCGCC-3’, 5’-CTAGCTTTCCGTT TCCGCCTGTAT-3’, 112 bp; and Tbx1 5’-CTGGGCTTCGTATCCTGTGC-3’, 5’-TGGGCCTCAAGACACCACTA-3’, 116 bp. Standard and melting curves were plotted to ensure the accuracy and specificity of the amplification. The relative content of the template in the samples was expressed by the Ct (threshold cycle). The relative expression of target genes was calculated using the 2-ΔΔCt method.
qRT-PCR and Western blot detection of the mRNA and protein of positive genes
Total RNA extraction was performed using TRIzol reagent. The mRNA expression of positive genes was detected. cDNA was synthesized using a reverse transcription kit. Primers were designed for positive genes and the internal reference (β-Actin) and are as follows: Abcc4 primers: 5’-AGGCAAGTCGTCCCTGTTG-3’, 5’-ATTGCTCCTCACGGTTCCTG-3’, 130 bp; Mef2d primers: 5’-GCAACGGCCTAAACAAGGTC-3’, 5’-CTTTGCCTCCCTGGGAAGTG-3’, 120 bp; Eif6 primers: 5’-GATCTGAGGGGTTTGAGAGGG-3’, 5’-TGTTCTCGAACGACGCTCTG-3’, 140 bp; Tbx1 primers: 5’-GATACCAGCCCCGTTTCCAT-3’, 5’-CTGCGTGATCCGGTGATTCT-3’, 143 bp; and β-Actin primers: 5’-CCCATCTATGAGGGTTACGC-3’, 5’-TTTAATGTCACGCACGATTTC-3’, 150 bp. Standard and melting curves were plotted to ensure the accuracy and specificity of the amplification. The relative content of template in the samples was expressed using the Ct (threshold cycle). The relative expression of target genes was calculated using the 2-ΔΔCt method.
The protein levels of related positive genes were similar to that of Foxp3 protein.
HPLC/MS detection of differential metabolites in blood samples
Metabolic fingerprints were obtained from the plasma using HPLC-qTOF-MS for AR rats. Differential metabolites were analysed by PLS-DA in AR rats. HPLC-PDA with a high peak capacity and sensitivity and MS with high resolution were used. During detection, hydrophilic interaction chromatography and reversed-phase chromatography were used. Blood samples were detected for AR and non-AR rats in positive and negative ionization modes.
The above procedures were undertaken by the CAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software. For each sample, quantitative fluorescence PCR was repeated three times. The relative expression of target genes was expressed as the mean ± standard deviation (x̅ ± s). Intergroup comparisons were conducted using independent-samples and paired t-tests, One way ANOVA method is used to compare multiple data sets. The comparison of multiple data between the same indicators is performed by Bonferroni in Post Hoc Multiple Comparisons model. P<0.05 indicated a significant difference. Metabolomics data were analysed by principal component analysis and the least squares method.
Results
The effect of AR on liver and kidney function
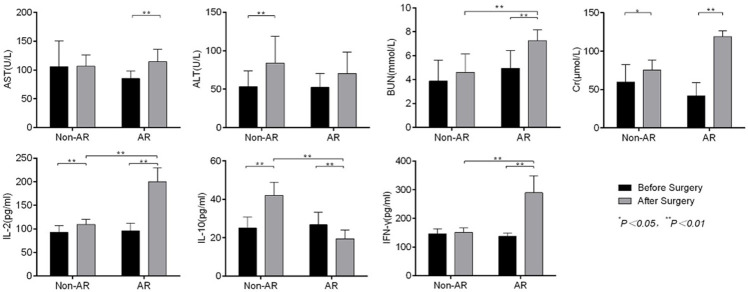
The AR group presented with a significant increase in ALT, BUN and Cr levels compared with the condition before surgery (P<0.05). The BUN and Cr level were higher in the AR group compared with the non-AR group, but there was no statistically significant difference (P>0.05) (Figure 1).
Figure 1.
Comparison of liver (AST, ALT), kidney functions (BUN, Cr) and related cytokines (IL-2, IL-10 and IFN-γ) before and after surgery. AST, ALT, BUN and Cr increased significantly in the AR group, which indicated that liver and kidney functions were abnormal, respectively. IL-2, IL-10 and INF-γ were significantly higher than in the non-AR group.
Cytokine levels in PBMCs in AR rats
Non-AR rats showed a significant increase in IL-2 and IL-10 levels compared with before surgery, while IL-2, IL-10 and IFN-γ also increased significantly after surgery in AR rats (P<0.01). Compared with the non-AR group, IL-2, IL-10 and IFN-γ levels also increased significantly after surgery in the AR group (P<0.01) (Figure 1).
Histopathological changes of the kidney
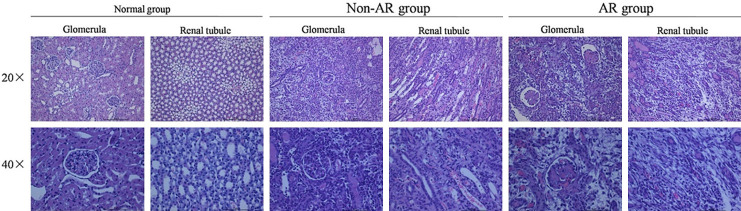
Compared with the normal group and non-AR group, pathological damage was more severe in the AR group. AR rats showed an increase in glomerular volume, mesangial widening, mesangial basement and cell hyperplasia, and capillary hyperaemia. Neutrophil granulocytes and nuclear fragments were observed in a few capillaries. Renal tubules were partially necrotic, predominantly in the medulla. We also observed renal interstitial oedema, vasodilation and congestion of the interstitial tissues, neutrophil infiltration, and infiltration by lymphocytes, plasmacytes and neutrophils in the interstitial tissues. Endothelial exfoliation was significant in renal blood cells (Figure 2).
Figure 2.
Pathological changes in glomeruli and renal tubules in the normal group, AR group and non-AR group. The glomeruli and renal tubules were significantly damaged in the renal tissue in the non-AR group compared with the normal group, and the renal interstitial showed oedema and inflammatory cell infiltration that was accompanied by endothelial cell damage. All the above changes were more pronounced in the renal tissue of the AR group.
Changes in the ratio of CD4 +CD25 + Treg cells and relative Foxp3 expression
The ratio of CD4 +CD25 + Treg cells in the AR group was significantly lower than that of the non-AR group. Foxp3 mRNA and protein expression of CD4 +CD25 + Treg cells were also much lower compared with the non-AR group (Figure 3).
Figure 3.
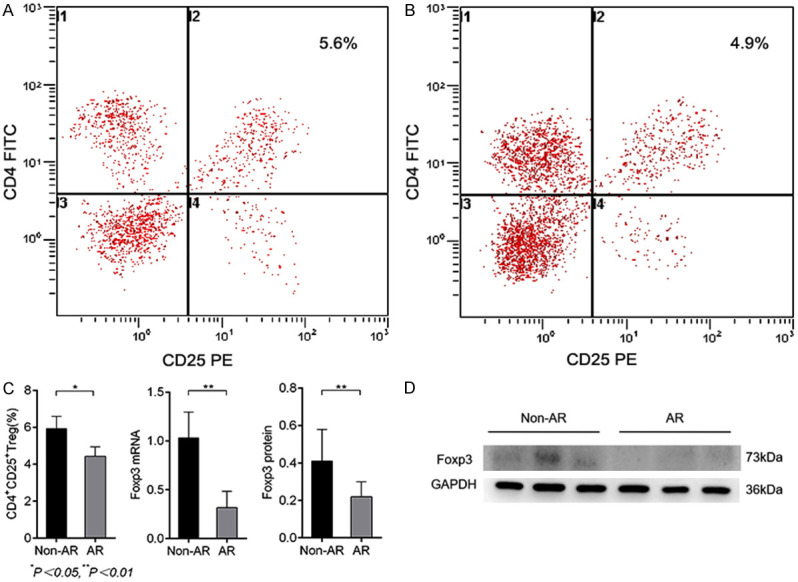
The level of CD4 +CD25 + Treg cells and Foxp3 mRNA between the Non-AR and AR groups. A, B: CD4 +CD25 + Treg ratio detected by flow cytometry. C: Comparison of CD4 +CD25 + Treg ratios, Foxp3 and protein expression levels between the AR and non-AR groups. D: Expression of Foxp3 protein in the AR and non-AR groups.
mRNA expression of positive genes in CD4 +CD25 + Treg cells and DNA methylation levels
Analysis of the CpG island microarray data
Fc values were calculated for all genes using the built-in software of the GenePix 4000B scanner. All data items with absolute values of Fc equal to or greater than 4 and a P-value less than 0.05 were selected. There was significant difference in H3K4me3 levels in 141 probes, among which 31 probes indicated an increase in H3K4me3 levels and 110 probes a decrease in H3K4me3 levels. Table 1 shows 20 genes with either an increase or decrease in H3K4me3 levels. The H3K4me3 level was compared between the groups. According to the CHIP-chip results, 2 genes with decreased H3K4me3 levels (Abcc4 and Mef2d) and 2 genes with increased H3K4me3 levels (Eif6 and Tbx1) were chosen in the AR group.
Table 1.
H3K4me3 gene with P-value less than 0.05 between AR group and non-AR group in CpG Island microarray
| Gene_name | Fold Change | P value | Gene Expression |
|---|---|---|---|
| Abcc4 | 63.1351351 | 0.0240011 | Up |
| Mef2d | 38.0833333 | 0.0209243 | Up |
| Acad11 | 47.6857142 | 0.0281887 | Up |
| Msra | 36.9047619 | 0.0452636 | Up |
| Phkb | 28.1754386 | 0.0373665 | Up |
| Mapk14 | 28.0000000 | 0.0174344 | Up |
| Nr1h3 | 17.3910256 | 0.0064995 | Up |
| Nfkb2 | 17.3520000 | 0.0484555 | Up |
| Man1c1 | 12.9317269 | 0.0394279 | Up |
| Kat8 | 11.8461538 | 0.0105543 | Up |
| Tbx1 | 0.06083650 | 0.0065373 | Down |
| Eif6 | 0.19382321 | 0.0007147 | Down |
| Dpysl5 | 0.24773413 | 0.0015608 | Down |
| Ly49s6 | 0.24665392 | 0.0238669 | Down |
| Foxred2 | 0.19780219 | 0.0177199 | Down |
| Fbxl17 | 0.19753979 | 0.0024538 | Down |
| Slc2a12 | 0.19597989 | 0.0474165 | Down |
| Tdrp | 0.19512195 | 0.0001020 | Down |
| Nkx1-2 | 0.19242902 | 0.0005184 | Down |
| Eml2 | 0.19178082 | 0.0257578 | Down |
Verification results by Chip-Qpcr
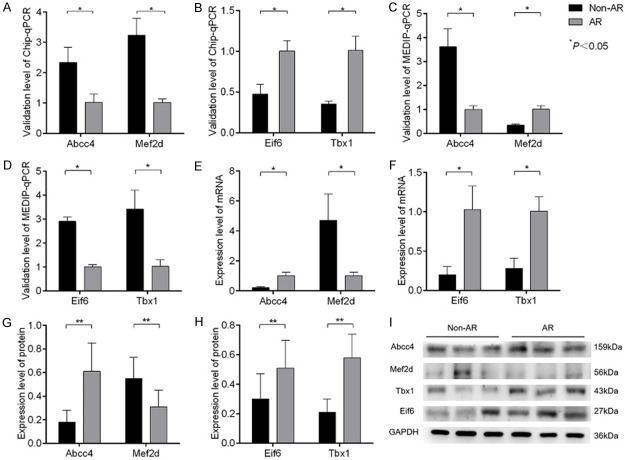
To verify the CpG island microarray results, ChiP-qPCR was applied to the DNA precipitate. There was a significant difference in the quantitative fluorescence PCR results for the four target genes between the AR and non-AR groups, which was consistent with the microarray analysis (Figure 4A, 4B).
Figure 4.
mRNA and protein expressions of positive genes in CD4 +CD25 + Treg cells and DNA methylation levels. A, B: Verification by Chip-qPCR for the four positive genes. C, D: Verification by MEDIP-qPCR of the four positive genes. E, F: Verification of the mRNA expression of the four positive genes. G-I: Protein expression levels of four positive genes.
DNA methylation levels
There was a significant difference in the DNA methylation levels of the four positive genes between the AR and non-AR groups (P<0.05). The methylation level increased for the Mef2d gene but decreased for the Abcc4, Eif6 and Tbx1 genes (Figure 4C, 4D).
mRNA and protein expression of four positive genes
There was a significant difference in the mRNA and protein expression of the four positive genes between the AR and non-AR groups. The mRNA (Figure 4E, 4F) and protein (Figure 4G-I) expression levels were upregulated for the Abcc4, Eif6 and Tbx1 genes but downregulated for the Mef2d gene.
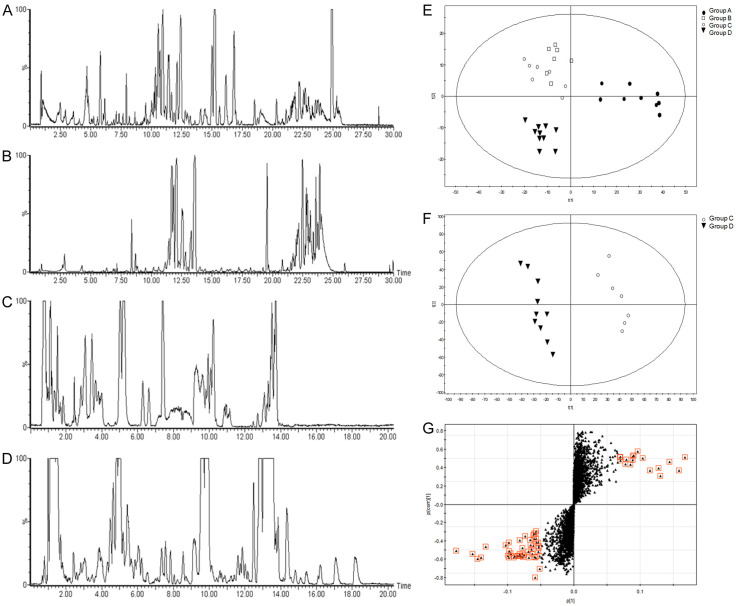
HPLC/MS analysis of metabolites in blood samples
The normal control group and sham operation group were set up for metabolomic analysis. The optimal separation time and mobile phase system were determined after repeated experiments. The metabolic fingerprints were obtained from the plasma using HPLC-qTOF-MS (Figure 5A-D). The pattern recognition method PLS-DA achieved a good discrimination among the four groups. There were neither crossovers nor overlaps in the distribution of the integral value of principal components between the AR and non-AR groups, which indicated a significant difference in metabolic fingerprints between the two groups (Figure 5E). PLS-DA discriminant analysis was further applied to the two groups to obtain the score plot (Figure 5F) and S-plot (Figure 5G).
Figure 5.
HPLC/MS analysis of metabolites in blood samples. A-D: HPLC-ESI-qTOF-MS patterns of plasma from rats after kidney transplantation (A: RP-HPLC MS in positive ionization mode; B: RP-HPLC MS in negative ionization mode; C: HILIC-HPLC MS in positive ionization mode; D: HILIC-HPLC MS in negative ionization mode). E: PLS-DA score plot of the four groups (●: Normal control group, □: Sham operation group, ○: AR group, ▼: Non-AR group). F: PLS-DA score plot of the AR and non-AR groups (▼: Non-AR group, ○: AR group). G: PLS-DA S-plot of the AR and non-AR groups. The compounds in red with the greatest distance from the origin of the S-plot were the main differential metabolites between the AR and non-AR groups.
The compounds shown in red with the greatest distance from the origin of the S-plot were the main differential metabolites between the AR and non-AR groups. To further verify the differential metabolites, the mass number and nuclear-cytoplasmic ratio of compounds in the red block diagram in Figure 5G were introduced into the Metlin metabolite database to search the compounds (the fluctuation range of the mass number was controlled within 0.05). The main differential metabolites were found to be creatinine, several phosphatidylcholines, lysophosphatidylcholine, free fatty acids and carnitine, the contents of which were significantly increased in the AR group. These results indicated higher phospholipase activity (Table 2).
Table 2.
Molecular species identified as potential biomarkers for classifying the acute graft rejection group from the non-acute graft rejection group
| No | Mass number | Compound | Detected by | Change |
|---|---|---|---|---|
| 1 | 126.03 | Taurine | HILIC in the positive ion mode | ↑ |
| 2 | 114.02 | Creatnine | HILIC in the positive ion mode | ↑ |
| 3 | 284.30 | Stearic amide C18:1 | RP in the positive ion mode | ↑ |
| 4 | 310.32 | Eicosanoic amide C20:2 | RP in the positive ion mode | ↑ |
| 5 | 312.33 | Eicosanoic amide C20:1 | RP in the positive ion mode | ↑ |
| 6 | 400.34 | Palmitoyl carnitine C16:0 | RP and HILIC in the positive ion mode | ↑ |
| 7 | 281.26 | FFA 18:1 | RP in the negative ion mode | ↑ |
| 8 | 279.24 | FFA 18:2 | RP in the negative ion mode | ↑ |
| 9 | 506.37 | LPC 18:0 | RP and HILIC in the positive and negative ion modes | ↑ |
| 10 | 524.36 | LPC 18:0 | RP and HILIC in the positive and negative ion modes | ↑ |
| 11 | 538.39 | LPC 19:0 | RP and HILIC in the positive and negative ion modes | ↑ |
| 12 | 542.33 | LPC 20:5 | RP and HILIC in the positive and negative ion modes | ↑ |
| 13 | 482.33 | LPC 15:0 | RP and HILIC in the positive and negative ion modes | ↑ |
| 14 | 734.57 | PC 32:0 | RP and HILIC in the positive and negative ion modes | ↑ |
| 15 | 832.58 | PC 40:7 | RP and HILIC in the positive and negative ion modes | ↑ |
| 16 | 522.39 | LPC 19:0 | RP and HILIC in the positive and negative ion modes | ↑ |
| 17 | 830.61 | PC 36:1 | RP and HILIC in the positive and negative ion modes | ↑ |
| 18 | 826.59 | PC 36:4 | RP and HILIC in the positive and negative ion modes | ↑ |
| 19 | 808.58 | PC 38:5 | RP and HILIC in the positive and negative ion modes | ↑ |
| 20 | 806.57 | PC 38:6 | RP and HILIC in the positive and negative ion modes | ↑ |
| 21 | 834.60 | PC 40:6 | RP and HILIC in the positive and negative ion modes | ↑ |
| 22 | 832.58 | PC 40:7 | RP and HILIC in the positive and negative ion modes | ↑ |
| 23 | 282.31 | Oleic amide C18:2 | RP in the positive ion mode | ↑ |
“↑” represent that the metabolite is up-regulated in the transplanted rats compared with the non-AR group.
Discussion
AR following kidney transplantation is an intense immune activity that can occur in response to receiving a transplanted kidney. If the immune response is too excessive, several T-lymphocytes will be activated to produce different cellular immune responses, leading to failure of the transplanted kidney. In these processes, Treg cells play the dominant role in maintaining immune tolerance of the organism. CD4 +CD25 + Treg cells can inhibit allograft rejection as the main cells involved in immune tolerance. Foxp3 is specifically expressed in CD4 +CD25 + Treg cells and is a member of the Foxhead transcription factor family. The gene encoding Foxp3 is located on the X chromosome. Continuous expression of Foxp3 is needed to maintain Treg functions.
The main hypothesis we wanted to clarify in this study is the molecular mechanism of abnormal lipid metabolism induced by abnormal modification of CD4 +CD25 + Treg cells H3K4me3 in the acute rejection kidney in rat kidney transplantation, which mainly includes the following three components: 1) Abnormal modification of CD4 +CD25 + Treg cell H3K4me3 may lead to AR in renal transplantation. 2) Abnormal lipid metabolism directly affects the biological activity of the biofilm system and can cause AR in renal transplantation by affecting energy metabolism. 3) Abnormal modification of CD4 +CD25 + Treg cell H3K4me3 is correlated with abnormal lipid metabolism, which can lead to AR in renal transplantation. Our results indicated that severe liver (ASL, ALT) and renal (BUN, Cr) injury occur after AR when compared with non-AR rats, and there was an apparent increase in the levels of IL-2, IL-10 and IFN-γ. Histopathological examinations revealed typical injuries of renal glomeruli and tubulointerstitium. The ratio of CD4 +CD25 + Treg cells decreased significantly, as did Foxp3 expression. Under normal conditions, CD4 +CD25 + Treg cells account for 5%-10% of peripheral CD4 + T cells in humans and rats. Secretion of immunosuppressive cytokines such as IL-10 and IL-4 can also produce an immunosuppressive effect. IL-2 and IFN-γ are secreted by Thl cells and can promote the activation of T-lymphocytes and the cellular immune response. In the donor-specific transfusion (DST)-induced immune tolerance model, IL-2 could stabilize the internal environment containing CD4 +CD25 + T cells [8]. It has been reported that a low level of IL-2 can promote the proliferation of CD4 +CD25 + T cells, whereas a high level of IL-2 blocks the immunomodulatory effect of CD4 +CD25 + T cells [9]. CD4 +CD25 + Treg cells can be inactivated by intercellular contact, which is accompanied by the secretion of IL-10. This process finally inhibits syngeneic CD4 + T cells and results in immune tolerance. A study has shown that mutation of the Foxp3 gene can lead to specific autoimmune diseases. The promotor region is the important regulator of Foxp3 expression and the target of epigenetic modification. Epigenetic modification regulates the differentiation of initial T cells into ordinary T cells or Treg cells and plays an important role in controlling Treg-related genes, especially expression of the Foxp3 gene. It is generally believed that renal function following kidney transplantation is also influenced by epigenetics.
Epigenetics is the study of heritable changes in gene function that do not involve changes in the DNA sequence. The main epigenetic mechanisms include histone modification and DNA methylation. Histone modification is involved in numerous chromatin-related activities, such as the regulation of gene expression and maintenance of genomic stability. The catalysis of related enzymes during DNA methylation can silence genes and reduce mRNA transcriptional expression; DNA demethylation can demethylate genes that have been methylated, activate genes that have been silenced, and increase gene transcription and expression. The essence of DNA methylation is the establishment, maintenance and removal of the methylation mechanism. Cysteine residues in the domain of DNA methyltransferase form covalent intermediates with the sixth carbon atom (C6) on the cytosine ring, destroying the aromatic ring configuration of the whole base and, thus, activating the fifth carbon atom (C5), transferring the methyl of S-adenosylmethionine (SAM) to cytosine C5 catalysed by the enzyme, releasing the proton of cytosine C5, and breaking the covalent bond of C6. SAM transforms into S-adenosyl homocysteine (SAH). Studies have shown that methylation of cytosine in the CpG island can inhibit gene transcription by binding to transcription factors or altering the chromatin structure [10]. By using the CpG island array, it was found that the H3K4me3 level was significantly different in 141 probes between the AR and non-AR groups. Among them, H3K4me3 levels increased in 31 probes and decreased in 110 probes in the AR group compared with the non-AR group. After screening with CHIP-chip, it was found that compared with the non-AR group, the methylation level of the Abcc4 and Mef2d genes decreased significantly in the AR group, while that of the Eif6 and Tbx1 genes increased significantly. As indicated by the detection of DNA methylation levels, the DNA methylation of the Mef2d gene in AR rats increased significantly, resulting in a downregulation of its mRNA and protein expression. The DNA methylation level of the Abcc4, Eif6 and Tbx1 genes decreased significantly, resulting in an upregulation of its mRNA and protein expression. The potential underlying mechanisms may be as follows: 1. Abcc4, also known as multidrug resistance-associated protein 4 (MRP4), is an integrin located on the cell membrane and widely found in lungs, kidney, bladder and gallbladder [11]. Abcc4 inhibits the migration of dendritic cells and antigen presentation in vivo [12]. In AR, Abcc4 is upregulated, which results in the failure to produce a normal immune response by inhibiting antigen presentation or weakening or blocking immune signal transduction. This phenomenon further aggravates graft loss. 2. Mef2d (myocyte enhancer factor 2D) is a member of the Mef2 gene family, and Mef2 protein is a member of the MADS transcription factor family [13]. Members can form homologous or heterologous dimers by binding to MADS domain, thus producing a response to various intercellular and intracellular signal pathways [14]. Post-translational modification is the most important pathway regulating Mef4 protein function [15]. Activated T-cells can lead to cell apoptosis via Mef2-mediated activation of the transcription of the Nur77 gene [16-18]. In AR, stress-induced generation of H2O2 can greatly enhance the activity of Cdk5 in the nuclei and cytoplasm [19] and cause phosphorylation of the structural domain of Mef2d. These phenomena will increase the instability of Mef2 and induce its degeneration. Moreover, Cdk5-mediated phosphorylation of Mef2d can activate cleavage of Mef2d by caspase, which also facilitates its degradation and leads to cell apoptosis [19]. 3. Eif6 (eukaryotic initiation factor 6) is a nuclear matrix protein involved in the assembly of the 60S ribosomal subunit, and Eif6 is a rate-limiting factor for translation initiation. Eif6 has an inhibitory effect on kidney fibrosis by inhibiting TGF-βl expression. The kidney is rich in TGF-βl, which is activated by AR. A study has shown that TGF-βl mediates Treg expression and, hence, induces the production of Foxp3. It can stimulate the differentiation of undifferentiated CD4 + T cells into Foxp3+ Treg cells. A high level of Eif6 can further reduce the ratio of CD4 +CD25 + Treg cells and Foxp3 expression by inhibiting TGF-βl, thus leading to AR [20]. 4. Tbx1 or T-box1 is localized to the nuclei and is a member of the T-box transcription factor family. The Tbx1 gene is localized onchromosome 22q11.2, abnormalities of which correlate significantly to renal diseases. Tbx1 mediates the translocation of Smad1 to the nuclei via Hoxd10, thus regulating the downstream gene. Samd1 is a member of the SMADS family and is an intracellular kinase substrate for TGF-β receptor. In diabetic nephropathy, chronic renal failure and AR following kidney transplantation, Samd1 mediates intracellular transduction of TGF-β. This process directly leads to glomerular hypertrophy and promotes the synthesis of mesangial cells and secretion of extracellular matrix while inhibiting extracellular matrix degradation. This is considered the pathological mechanism of glomerular sclerosis.
DNA methylation plays a major role in gene suppression at the transcriptional level. The possible mechanisms are as follows [21]: 1. DNA methylation directly interferes with specific transcription factors, and the major groove of the DNA axis is the site where many protein factors bind to DNA. After cytosine is methylated, 5-mC protrudes into the main groove, thereby interfering with the binding of the transcription factor to DNA (Figure 6Aa1, 6Aa2). 2. Sequence-specific methylated DNA-binding proteins bind to the methylated CpG island in the promoter region, recruiting some proteins to form a transcriptional repressor complex, preventing the binding of transcription factors to the target sequence of the promoter region and, thereby, affecting gene transcription (e.g., methylated cytosine binding proteins 1 and 2 (MEcp1 and MeCP2) and MBDs [22] (Figure 6Bb1-b3). 3. DNA methylation inhibits gene expression by altering the chromatin structure (Figure 6C).
Figure 6.
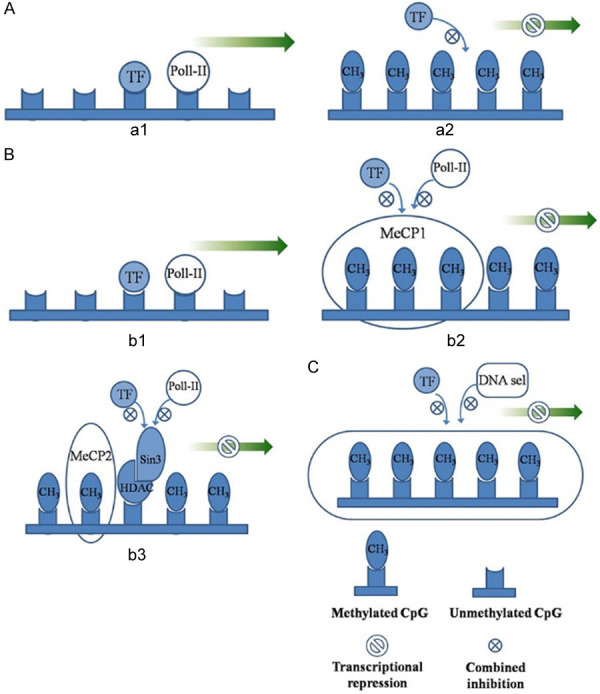
Possible mechanisms of transcriptional inhibition by methylation. A: a1: Activated transcription; a2: Transcriptional factors can not bind, resulting in transcriptional repression; B: b1: Activated transcription; b2: Transcriptional inhibition caused by MeCP1; b3: Transcriptional inhibition caused by MeCP2; C: Transcriptional inhibition due to changes in chromatin structure.
Characteristic screening of immune and rejection markers after renal transplantation can directly reflect the types and levels of different metabolites before and after transplantation. In this study, creatinine, PCs, LPCs, FFA and carnitine were the main metabolites in the two groups. In the AR group, significant changes were found in the levels of various phospholipid molecules, especially PC and LPC. LPCs are a class of PCs derivatives formed by removing FFA from PCs. FFA produces ATP energy mainly through oxidative phosphorylation in vivo, which requires carnitine as a carrier to transfer lipid acyl coenzyme A to mitochondria for further oxidation. The results of this study showed that the contents of PC, LPCs, FFA, carnitine and other metabolites increased significantly in the acute rejection group, suggesting an abnormal metabolism of phospholipids during acute rejection, but the mechanism of the increase of phospholipid metabolites is not very clear. We believe that the mechanism may include the following points. 1. Cell membranes are composed of lipids and membrane proteins. There are many ion channels and lipoprotein receptors embedded between the inner and outer membranes of cell membranes. Phospholipids are an important component of lipid components in cell membranes. Differences in types and levels of phospholipids in cell membranes can directly affect the biological characteristics of cell membranes, thus indirectly affecting substance transport, cell differentiation, receptor-ligand binding, and transmembrane signal transduction, among others. 2. Acute renal allograft rejection is a T lymphocyte-mediated cellular immune response. Immediately after acute rejection, resting T lymphocytes recognize alloantigens (HLA) through direct, indirect and IL-2 receptor channels. The activation of these signalling pathways is closely related to the integrity of cell membrane components. 3. Acute rejection of renal transplantation involves the production of reactive oxygen species (ROS), lipid peroxidation and antioxidation [23-25]. Oxidative stress can stimulate and activate the metabolism of PCs while upregulating the level of LPCs, which play an important role in promoting inflammation, stimulating cell proliferation and tumour cell invasion [26,27]. 4. The generated PCs can be transformed into FFA in vivo, which is positively correlated with the carnitine level. FFA can increase the rate of oxidative phosphorylation of mitochondria, accelerate the efficiency of beta oxidation, increase the rate of CDP-choline formation, and lead to the formation of PCs (Figure 7), and the level of FFA in plasma has been shown to be a sensitive index of lipid peroxidation damage [28,29]. Therefore, the increase in FFA and carnitine levels can also be used as markers of lipid peroxidation progression in acute rejection. In AR rats, a variety of metabolites, such as sphingomyelin, PCs and LPCs, increased significantly, suggesting an increase in phospholipase activity, enhancement of β-oxidation via the tricarboxylic acid cycle (TCA) pathway, and the presence ofa high-load lipid peroxidation rate. The increase in these metabolites may be one of the factors that further lead to graft failure.
Figure 7.
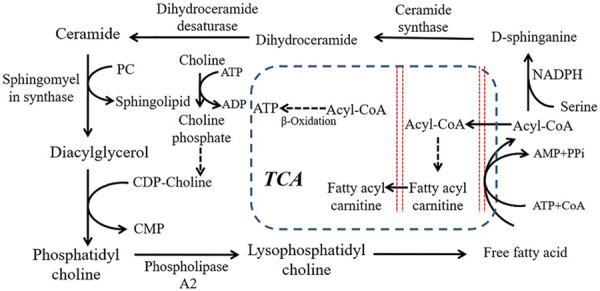
Possible pathways of plasma phospholipid metabolism in acute rejection of renal transplantation. With the increase in FFA level produced by PCs, FFA can peroxide under oxidative stress, increase the rate of oxidative phosphorylation of mitochondria, and accelerate the process of β-oxidation through the TCA pathway to produce ATP to supply energy. Simultaneously, the increased FFA levels can also increase the rate of CDP-choline formation, leading to an increase in PC generation.
In our study, compared with the non-AR group, the histone methylation level of Abcc4 and Mef2d decreased in AR group, as did the DNA methylation of Abcc4. In contrast, the DNA methylation level increased for Mef2d, and the histone methylation level increased for Eif6 and Tbx1, with a decrease in their DNA methylation levels. This phenomenon indicated a synergistic effect between histone methylation and DNA methylation of Mef2d, Eif6 and Tbx1. However, histone methylation and DNA methylation were consistent for Abcc4, probably because H3K4me caused an inhibition of the genomic DNA methylation in the CpG island [30]. In the AR group, we observed a significant increase in the content of metabolites such as PCs, LPCs, FFA and the carnitine and energy metabolism imbalance. The interaction mechanism may be as follows. 1. Methylation affects intercellular signal transduction, immune regulation and energy metabolism by influencing the modification of metabolites, such as amino acids and lipids; 2. PCs are not only related to mutual transformation of phospholipids, but it also has an indirect regulatory role in energy metabolism, especially mitochondrial β-oxidation [31]. 3. PCs can be synthesized in vivo by methylation and the PC pathway. In both eukaryotes and prokaryotes, PCs can be synthesized by trimethylation following the catalysis of phosphatidylethanolamine N-methyltransferase (PEMT), with S-adenosyl methionine (SAM) as the methyl donor and mono-methyl-PET (MMPE) and dimethyl-PET (DMPE) as intermediates. During methylation, SAM is converted into S-adenosyl-L-homocysteine (SAH) [32,33] (Figure 8). However, the regulatory role of PEMT and the mediating role of PCs in energy metabolism remain to be further studied.
Figure 8.
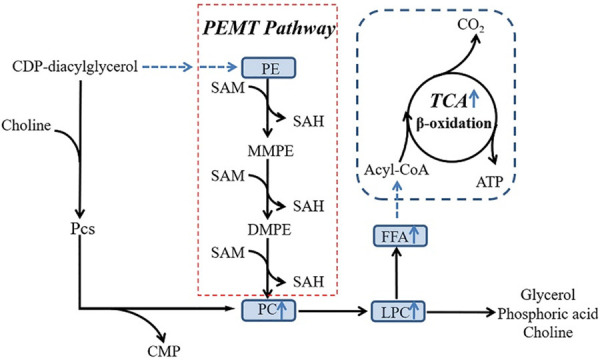
Relationship between lipid metabolism and methylation. PCs not only connect the mutual conversion between phospholipids butalso indirectly regulate the energy metabolism of the body, especially the β oxidation process in mitochondria. PCs can be synthesized by methylation and the PC pathways in vivo. Eukaryotes and prokaryotes can be catalysed by phosphatidylethanolamine N-methyltransferase (PEMT) via S-adenosylmethylsulfide. As a methyl donor, the amino acid is produced by three consecutive methylations of the intermediate monomethyl-PEG (MMPE) and dimethyl-PEG (DMPE). During the methylation process, SAM is converted into S-adenosyl homocysteine (SAH).
The main hypothesis we wanted to clarify in this study was the molecular mechanism of abnormal lipid metabolism induced by abnormal modification of CD4 +CD25 + Treg cell H3K4me3 in acute rejection kidney in rat kidney transplantation. Compared with non-AR rats, the histone methylation level of Abcc4 and Mef2d in AR rats decreased, while the DNA methylation level of Mef2d increased. In contrast, the DNA methylation level of Abcc4 decreased. In the AR group, the histone methylation level of Eif6 and Tbx1 increased, while the DNA methylation level decreased. However, the above difference in mRNA expression between the genes may be not caused by changes in DNA sequence. We speculate that H3K4 methylation, a representative of histone methylation and DNA methylation, can alter the immune status by modifying epigenetics. Furthermore, metabolomic studies indicated the presence of differential metabolites of lipid metabolism between non-AR and AR groups, including LCs and PLCs. This phenomenon supported a lipid metabolism disorder in AR following kidney transplantation, in addition to epigenetic changes. To conclude, specific modification of these gene loci can be performed in AR based on the features of lipid metabolism. The research findings shed new light onto the early diagnosis and treatment of AR following kidney transplantation.
Deficiencies
The main shortcomings of this study are as follows: 1) The number of experimental rats included was too small. Small sample experiments may have some influence on the experimental results due to individual differences. 2) Animal models can not completely simulate the pathophysiological process of the human body. In this study, only animal experiments were carried out, and no blood samples were collected from patients after renal transplantation. Further studies should be carried out using human specimens. 3) In this study, the mechanism of lipid metabolism disorder leading to AR was mostly focused on energy metabolism, which is an important link in the occurrence and development of AR, but combined with the relevant literature, lipid metabolism may also be closely related to apoptosis, organelle dysfunction and abnormal membrane protein composition. We will continue to study other possible mechanisms of AR development in kidney transplantation in the future.
Disclosure of conflict of interest
None.
References
- 1.Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi Y, Wu J, Suzuki K, Martinez-Redondo P, Li M, Liao HK, Wu MZ, Hernández-Benítez R, Hishida T, Shokhirev MN, Esteban CR, Sancho-Martinez I, Belmonte JCI. Integration of CpG-free DNA induces de novo methylation of CpG islands in pluripotent stem cells. Science. 2017;356:503–508. doi: 10.1126/science.aag3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ, Cantley LC. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547:419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–32. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papiernik M, de Moraes ML, Pontoux C, Vasseur F, Pénit C. Regulatory CD4 T cells: expression of IL-2R alpha chain, resistance to clonal deletion and IL-2 dependency. Int Immunol. 1998;10:371–8. doi: 10.1093/intimm/10.4.371. [DOI] [PubMed] [Google Scholar]
- 9.Brinster C, Shevach EM. Bone marrow-derived dendritic cells reverse the anergic state of CD4 +CD25 + T cells without reversing their suppressive function. J Immunol. 2005;175:7332–40. doi: 10.4049/jimmunol.175.11.7332. [DOI] [PubMed] [Google Scholar]
- 10.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 11.Carozzo A, Diez F, Gomez N, Cabrera M, Shayo C, Davio C, Fernández N. Dual role of cAMP in the transcriptional regulation of multidrug resistance-associated protein 4 (MRP4) in pancreatic adenocarcinoma cell lines. PLoS One. 2015;10:e0120651. doi: 10.1371/journal.pone.0120651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Ven R, de Groot J, Reurs AW, Wijnands PG, van de Wetering K, Schuetz JD, de Gruijl TD, Scheper RJ, Scheffer GL. Unimpaired immune functions in the absence of Mrp4 (Abcc4) Immunol Lett. 2009;124:81–7. doi: 10.1016/j.imlet.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–93. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 14.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 15.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–7. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 16.Youn HD, Sun L, Prywes R, Liu JO. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 17.Youn HD, Liu JO. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity. 2000;13:85–94. doi: 10.1016/s1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 18.Swanson BJ, Jäck HM, Lyons GE. Characterization of myocyte enhancer factor 2 (MEF2) expression in B and T cells: MEF2C is a B cell-restricted transcription factor in lymphocytes. Mol Immunol. 1998;35:445–58. doi: 10.1016/s0161-5890(98)00058-3. [DOI] [PubMed] [Google Scholar]
- 19.Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 20.Lan Q, Zhou X, Fan H, Chen M, Wang J, Ryffel B, Brand D, Ramalingam R, Kiela PR, Horwitz DA, Liu Z, Zheng SG. Polyclonal CD4+ Foxp3+ Treg cells induce TGFβ-dependent tolerogenic dendritic cells that suppress the murine lupus-like syndrome. J Mol Cell Biol. 2012;4:409–19. doi: 10.1093/jmcb/mjs040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 22.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller TF, Serkova N, Niemann CU, Freise CE. Influence of donor pretreatment with N-acetylcysteine on ischemia/reperfusion injury in rat kidney grafts. J Urol. 2004;171:1296–300. doi: 10.1097/01.ju.0000103928.64939.6a. [DOI] [PubMed] [Google Scholar]
- 24.Kosieradzki M, Kuczynska J, Piwowarska J, Wegrowicz-Rebandel I, Kwiatkowski A, Lisik W, Michalak G, Danielewicz R, Paczek L, Rowinski WA. Prognostic significance of free radicals: mediated injury occurring in the kidney donor. Transplantation. 2003;75:1221–1227. doi: 10.1097/01.TP.0000065282.46425.87. [DOI] [PubMed] [Google Scholar]
- 25.Oehlschläger S, Albrecht S, Hakenberg OW, Manseck A, Froehner M, Zimmermann T, Wirth MP. Measurement of free radicals and NO by chemiluminescence to identify the reperfusion injury in renal transplantation. Luminescence. 2002;17:130–132. doi: 10.1002/bio.675. [DOI] [PubMed] [Google Scholar]
- 26.Carson MJ, Lo D. The push-me pull-you of T cell activation. Science. 2001;293:618–619. doi: 10.1126/science.1063516. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F, Jia Z, Gao P, Kong H, Li X, Chen J, Yang Q, Yin P, Wang J, Lu X, Li F, Wu Y, Xu G. Metabonomics study of atherosclerosis rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry. Talanta. 2009;79:836–844. doi: 10.1016/j.talanta.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Deiana M, Aruoma OI, Rosa A, Crobu V, Casu V, Piga R, Dessi MA. The effect of ferric-nitrilotriacetic acid on the profile of polyunsaturated fatty acids in the kidney and liver of rats. Toxicol Lett. 2001;123:125–33. doi: 10.1016/s0378-4274(01)00372-1. [DOI] [PubMed] [Google Scholar]
- 29.Niemann CU, Saeed M, Akbari H, Jacobsen W, Benet LZ, Christians U, Serkova N. Close association between the reduction in myocardial energy metabolism and infarct size: dose-response assessment of cyclosporine. J Pharmacol Exp Ther. 2002;302:1123–8. doi: 10.1124/jpet.102.036848. [DOI] [PubMed] [Google Scholar]
- 30.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Wen H, Liu J, Yu C, Zhao X, Shi X, Xu G. Metabonomics study of the acute graft rejection in rat renal transplantation using reversed-phase liquid chromatography and hydrophilic interaction chromatography coupled with mass spectrometry. Mol Biosyst. 2012;8:871–8. doi: 10.1039/c2mb05454j. [DOI] [PubMed] [Google Scholar]
- 32.López-Lara IM, Geiger O. Novel pathway for phosphatidylcholine biosynthesis in bacteria associated with eukaryotes. J Biotechnol. 2001;91:211–21. doi: 10.1016/s0168-1656(01)00331-5. [DOI] [PubMed] [Google Scholar]
- 33.Sohlenkamp C, López-Lara IM, Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog Lipid Res. 2003;42:115–62. doi: 10.1016/s0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]