Abstract
The heterogeneity of diffuse large B-cell lymphoma (DLBCL) acts as a main barrier to identify the genetic basis of the disease and the choice of treatment. Differentially expressed genes (DEGs) from three mRNA expression profile datasets were screened using GEO2R, and bioinformatics analysis was performed on the DEGs. A total of six upregulated and 13 downregulated DEGs were identified. Among these, two hub genes with a high degree of correlation were selected. FBN1 and TIMP1 were identified via STRING analysis and validated by GEPIA. FBN1 and TIMP1 were highly expressed in DLBCL tissues. FBN1 expression was significantly higher in patients of the Ann Arbor stage group (III-IV), with higher IPI score (3-5), and in the non-GCB group. Patients with high TIMP1 expression were more frequently associated with B symptoms, Ann Arbor stage (III-IV), higher IPI score (3-5) and were in the non-GCB group. Furthermore, FBN1 siRNA decreased FBN1 and TIMP1 expression and downregulation of TIMP1 attenuated TIMP1 expression but not of FBN1. Migration of DLBCL cells reduced when treated with either FBN1 or TIMP1 siRNA. Moreover, FBN1 or TIMP1 siRNA decreased the expression of Wnt target genes. Simultaneous overexpression of TIMP1 resulted in an increase in these proteins. This confirmed that both FBN1 and TIMP1 were positively associated with DLBCL progression. Further analysis revealed that FBN1/TIMP1 interaction could improve DLBCL cell migration and regulate the Wnt signaling pathway. Although the underlying mechanisms regarding the interaction between FBN1 and TIMP1 requires further clarification, they might be potential therapeutic targets for DLBCL therapy.
Keywords: Diffuse large B-cell lymphoma, bioinformatics analysis, fibrillin-1, tissue inhibitor of matrix metalloproteinases 1, cell migration
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a subgroup of non-Hodgkin lymphoma (NHL) with high morbidity. Although chemotherapy of DLBCL has greatly advanced in the past 20 years, the overall survival of a large number of patients with DLBCL remains regrettable, especially upon diagnosis of bone marrow metastasis [1]. Therefore, a better understanding of DLBCL and identification of new critical targets for early detection of DLBCL is immediately required.
Data-mining of genes using bioinformatics approach is helpful for identifying novel important genes involved in the pathogenesis of diseases, thereby providing important insights and a foundation for further new research [2]. In the present study, three gene expression profile datasets were obtained from the Gene Expression Omnibus (GEO) database. Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed for the differentially expressed genes (DEGs). Furthermore, we analyzed protein-protein interaction (PPI) networks to explore the interactions and identified certain factors that might be involved in the regulatory mechanisms of DLBCL.
Fibrillin-1 (FBN1) is an essential protein in the extracellular matrix that modulates the tissue microenvironment [3]. FBN1 is associated with vertebral growth and development. Fibrillin microfibrils play a vital role in modulating bone growth [4]. Besides, a murine model with FBN1intragenic duplication presented dramatically expanded lumbar vertebra and extended costal cartilages [5]. A recent study indicated that overexpression of FBN1 might reveal early recurrence of ovarian cancer and chemosensitivity to platinum [6]. However, the influence of FBN1 in DLBCL remains unclear.
Tissue inhibitor of matrix metalloproteinases 1 (TIMP1) belongs to the TIMP family which is comprised of four known members (TIMP1, TIMP2, TIMP3 and TIMP4) [7]. TIMP1 negatively regulates the invasion and metastasis of tumor cells by inhibiting MMP-9 activity [8]. Several studies have validated that overexpression of TIMP1 resulted in increased expression of genes involved in proliferation, apoptosis and signal transduction [9-11]. In addition, TIMP1 was found to decrease tumor cell sensitivity to multiple anticancer drugs [12]. Specifically, TIMP1 could reduce cyclinB1 expression and activate the NF-κB signaling pathway and hence prevent breast cancer cells from undergoing chemotherapy-induced cell death [13]. However, the precise function and potential mechanisms of TIMP1 in DLBCL needs to be elucidated.
In this study, we identified DEGs in DLBCL from GSE2350, GSE32018 and GSE44337 datasets. We then analyzed the PPI networks to examine the interactions and identified two important proteins (FBN1 and TIMP1) which might be associated with the regulatory mechanisms of DLBCL. We found that the expression levels of FBN1 and TIMP1 were associated with clinicopathological characteristics. We further demonstrated that specific siRNAs of FBN1 and TIMP1 could suppress the migration of DLBCL cells. Furthermore, FBN1/TIMP1 interaction could induce DLBCL cell migration by activating the Wnt3/β-catenin signaling pathway. Together, these data highlight new roles of FBN1 and TIMP1 in lymphocyte migration.
Materials and methods
Raw data
A total of three human DLBCL mRNA expression datasets [GSE2350, GSE32018 and GSE44337] were obtained from the National Center of Biotechnology Information (NCBI) GEO (https://www.ncbi.nlm.nih.gov/geo/). All the datasets contained a comparison between 41 DLBCL samples and 19 normal lymphocyte samples.
Identification of DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r), a web tool, can perform sophisticated R-based analysis of GEO data and display the results as a table of DEGs that can be visualized using GEO Profile graphics. The adjusted P-values (adj. P) and Benjamini and Hochberg false discovery rate were optimized to balance the identification of statistically significant genes with the limitations of false-positives. LogFC (fold change) >1 and adj. P<0.05 were considered as statistically significant. The web-based tool VENNY (version 2.1, http://bioinfogp.cnb.csic.es/tool/venny/index.html) was used to acquire the overlapping DEGs in the three datasets [14].
GO and KEGG pathway analysis of DEGs
GO analysis annotates genes and gene products according to their characteristic biological functions based on highthroughput genome or transcriptome data. KEGG is a collection of databases for analyzing genomes and biological pathways. The Database for Annotation Visualization and Integrated Discovery (DAVID: http://david.ncifcrf.gov) (version 6.8), which integrates the information of biological functions of genes with graphical displays, was used to sort out the GO and KEGG pathway identified co-DEGs. A false discovery rate threshold was set at P<0.05. The Search Tool for the Retrieval of Interacting Genes (STRING) database was applied to identify and visualize the functional interactions between proteins.
Patients and data collection
In this study, 110 patients verified clinically and pathologically as DLBCL, and who were enrolled in Shenjing Hospital of China Medical University (Shenyang, China) from January 2013 to January 2018 were chosen as the subjects. Furthermore, 30 tissue specimens of patients, which were pathologically diagnosed as reactive lymphoid hyperplasia during the same period were selected as the controls. The clinicopathological data of patients were procured. The Clinical Ethics Committee of Shenjing Hospital of China Medical University approved this study, and informed consent was obtained from the patients.
Immunohistochemistry
Immunohistochemical staining was performed as previously described [15]. Paraffin-embedded sections of DLBCL tissues were incubated with primary rabbit anti-FBN1 and anti-TIMP1antibodies (ab53076 and ab109125, 1:200, Abcam, Cambridge, MA, USA) and subsequently with secondary horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (KeyGEN BioTECH company, Nanjing, China). Immunohistochemical staining was performed according to the manufacturer’s instructions using DAB solution and counterstained with hematoxylin (KeyGEN BioTECH company, Nanjing, China). Staining intensities and percentage of positive tumor cells were scored independently by two pathologists who were blinded to the patient clinical outcome. Protein expression level was classified based on the percentage of positive cells and the intensity of staining. Immunoreactivity was scored as follows: Staining intensity -/+, <25% positive cells; staining intensity ++, ≥25% and <50% positive cells; staining intensity +++, 50-75% positive cells; and staining intensity ++++, >75% positive cells. Samples with high expression were those with ≥50% (+++ or ++++) of cells staining for FBN1 or TIMP1.
FBN1 and TIMP1 validation by GEPIA
In this study, the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) was used to examine the expression of FBN1 and TIMP1, and to analyze their expression levels in normal and DLBCL tissues and the association of their expression levels with overall survival. We selected P<0.05 as a threshold.
Western blotting
Approximately 40 µg of total protein was used for each treatment to detect target genes. Solubilized proteins were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, USA). Peroxidase-based chemiluminescent detection was performed according to standard laboratory protocols. Antibodies used for immunoblotting were as follows: rabbit anti-FBN1, anti-TIMP1 and anti-GAPDH antibodies (ab231094, ab109125 and ab181602, 1:2000, Abcam, Cambridge, MA, USA), rabbit monoclonal anti-Wnt3a, anti-GSK-3, anti-β-Catenin, anti-c-Myc, anti-cyclin D1 antibodies (#2721, #12456, #8480, #18583 and #55506, 1:2000, Cell Signaling Technology, Danvers, MA, USA).
Cell culture and transfection
The DLBCL cell lines OCI-LY1 and SU-DHL-2 were purchased from BNBIO company (Beijing, China). Cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS, Invitrogen, MO, USA) in a humidified incubator at 37°C with 5% CO2.
FBN1 siRNA, TIMP1 siRNA and control siRNA were purchased from JTS scientific company (Wuhan, China) (Supplementary Table 1). The control siRNA was the scrambled sequence of the gene-specific siRNA. The gene-specific siRNAs were transiently transfected into cells using lipofectamine 3000 (Invitrogen, MD, USA) according to the manufacturer’s instructions. Lentivirus carrying TIMP1-wt construct was synthesized by Genechem Co., LTD (Shanghai, China), and the viral particles were subsequently transduced into DLBCL cells. The cells overexpressing TIMP1 were screened using puromycin (Invitrogen, NY, USA) and confirmed by western blotting.
Cell migration and proliferation assays
The migration of cells was measured using a transmigration chamber assay (Corning, MA, USA). In brief, cells (2×106 cells/ml) were starved in serum-free RPMI-1640 medium for 1 h. Cell suspension was added to the upper chamber with a pore size of 8 µm (Corning, MA, USA) and 600 µl of complete medium was added to the lower chamber. The plate was then incubated for 2-3 h at 37°C, and the number of cells that migrated through the inserts was monitored by the cell migration assay. Cell proliferation was determined using the Cell Counting Kit-8 reagent (CCK-8, Beyotime, Shanghai, China) according to the manufacturer’s instructions. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad, CA, USA). The experiment was performed in triplicate.
Statistical analysis
All data were analyzed using GraphPad Prism 6.0 statistical software (GraphPad Software, Inc., CA, USA) and SPSS 20.0 (IBM Corp., NY, USA). Kaplan-Meier analysis and log-rank test were performed for analysis of overall survival (by GEPIA) and for pathway enrichment analysis (by DAVID). The two-tailed paired Student’s t-test was performed for analyzing the differences between two groups. One-way analysis of variance (ANOVA) and Bonferroni post hoc test were used for evaluating differences among multiple groups. The sensitivity and specificity for the prediction of OS (Overall Survival) and PFS (Progression-Free-Survival) were analyzed by receiver operating characteristic (ROC) curves. The area under the curve (AUC) was used to measure the prognostic or predictive accuracy. Pearson’s test was used for correlation analysis. We compared the two groups using chi-squared (χ2) or Fisher’s exact test for categorical variables. Results with P<0.05 were considered as statistically significant.
Results
Identification of DEGs in DLBCL
Three GEO datasets were analyzed which included 41 DLBCL samples and 19 normal lymphocyte samples (Figure 1A). A total of 584, 275 and 960 DEGs were upregulated and 744, 807 and 644 DEGs were downregulated in the these three datasets, respectively. In total, 19 DEGs presented similar expression pattern in all three datasets (Figure 1B), including 6 upregulated and 13 downregulated in DLBCL samples compared to normal lymphocyte samples (Table 1).
Figure 1.
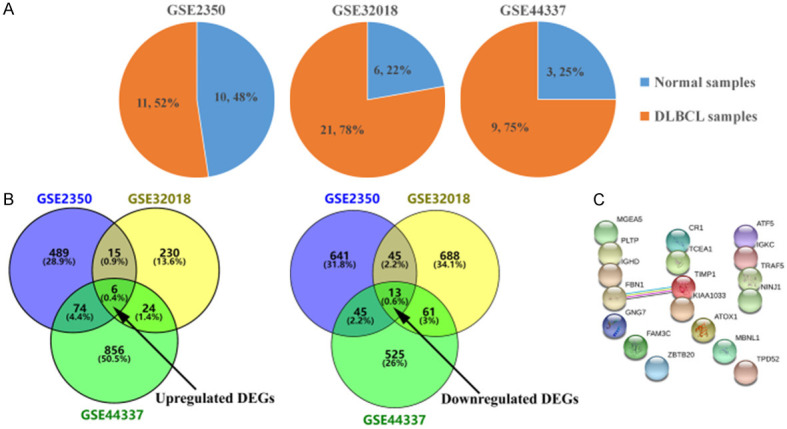
Identification of consistent DEGs between normal lymphocyte and DLBCL samples. A. Statistical analysis of the three microarray datasets obtained from the GEO database. The percentage of normal lymphocyte and DLBCL samples are shown in the three datasets. B. Venn diagrams of common DEGs in the three datasets. The number and percentage of DEGs are displayed. C. The PPI network of DEGs was constructed using the STRING database.
Table 1.
Identification of consistent differentially expressed genes, including 6 upregulated and 13 downregulated genes, from GSE2350, GSE32018 and GSE44337 datasets
| Gene Symbol | Down/Up | GSE2350 | GSE32018 | GSE44337 | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| logFC | adj.P.Val | logFC | adj.P.Val | logFC | adj.P.Val | ||
| PLTP | Up | 1.029311 | 3.64E-02 | 1.409244 | 2.26E-02 | 3.08744 | 3.49E-03 |
| ATOX1 | Up | 1.064798 | 4.03E-04 | 1.410082 | 1.55E-03 | 3.100636 | 3.24E-04 |
| FBN1 | Up | 1.675059 | 3.23E-02 | 1.244236 | 2.29E-02 | 3.639916 | 1.50E-03 |
| ATF5 | Up | 1.871464 | 9.04E-04 | 1.518801 | 2.83E-02 | 3.301105 | 2.31E-03 |
| NINJ1 | Up | 2.585884 | 5.58E-04 | 1.168389 | 3.12E-02 | 1.931035 | 6.21E-03 |
| TIMP1 | Up | 3.5823225 | 1.88E-02 | 2.318486 | 2.54E-05 | 4.860962 | 3.55E-04 |
| IGKC | Down | -3.820658 | 1.33E-06 | -2.626743 | 7.09E-03 | -3.717709 | 6.70E-03 |
| CR1 | Down | -3.177401 | 1.70E-04 | -1.048999 | 1.30E-02 | -1.303208 | 1.77E-03 |
| IGHD | Down | -2.812763 | 2.26E-02 | -1.265743 | 1.33E-02 | -6.193135 | 5.89E-04 |
| FAM3C | Down | -2.764777 | 1.96E-04 | -1.129327 | 2.66E-03 | -3.274943 | 1.53E-03 |
| GNG7 | Down | -2.002193 | 6.69E-07 | -1.253173 | 7.34E-03 | -4.87254 | 5.95E-08 |
| TPD52 | Down | -1.97413 | 2.30E-04 | -1.386452 | 2.97E-02 | -2.13456 | 2.25E-04 |
| IGLV1-44 | Down | -1.716078 | 6.67E-04 | -2.132387 | 5.70E-03 | -1.404451 | 1.54E-03 |
| ZBTB20 | Down | -1.684524 | 1.03E-03 | -2.060043 | 9.75E-03 | -1.35991 | 1.46E-03 |
| MGEA5 | Down | -1.678981 | 5.92E-06 | -1.541935 | 2.22E-03 | -1.298316 | 4.52E-03 |
| TRAF5 | Down | -1.599049 | 1.25E-05 | -1.278165 | 1.04E-02 | -1.340282 | 8.08E-03 |
| KIAA1033 | Down | -1.473975 | 9.09E-04 | -1.061922 | 8.20E-04 | -1.075805 | 3.52E-03 |
| TCEA1 | Down | -1.341718 | 5.15E-05 | -1.236592 | 6.38E-03 | -1.135546 | 1.31E-03 |
| MBNL1 | Down | -1.046436 | 5.42E-04 | -1.759458 | 1.25E-04 | -1.464918 | 2.33E-04 |
Functional enrichment analysis of DEGs and identification of hub genes
GO-BP analysis indicated that the DEGs were mainly involved in ‘complement activation’, ‘positive regulation of B cell activation’, ‘phagocytosis recognition’ and ‘phagocytosis engulfment’ as shown in Table 2. In GO-CC analysis, the DEGs were mainly enriched in ‘extracellular region’, ‘immunoglobulin complex’ and ‘extracellular exosome’. With respect to MF, the DEGs were mainly enriched in ‘antigen binding’ and ‘immunoglobulin receptor binding’. Furthermore, in the KEGG pathway enrichment analysis, the DEGs were mainly enriched in ‘human immunodeficiency virus 1 infection’, ‘human cytomegalovirus infection’ and ‘pathways in cancer’. In addition, the significantly enriched REACTOME pathways were identified as ‘platelet activation, signaling and aggregation’, ‘post-translational protein phosphorylation’, ‘regulation of IGF transport and uptake by IGFBPs’, ‘platelet degranulation’ and so on.
Table 2.
GO enrichment analysis, KEGG and REACTOME pathway analyses of differentially expressed genes associated with DLBCL
| Category | Term | Count | % | P-Value | Genes |
|---|---|---|---|---|---|
| GOTERM_BP | GO: 0006958~complement activation, classical pathway | 4 | 21.0526 | 1.52E-04 | CR1, IGLV1-44, IGHD, IGKC |
| GOTERM_BP | GO: 0050871~positive regulation of B cell activation | 2 | 10.5263 | 0.0275 | IGHD, IGKC |
| GOTERM_BP | GO: 0006910~phagocytosis, recognition | 2 | 10.5263 | 0.0296 | IGHD, IGKC |
| GOTERM_BP | GO: 0006911~phagocytosis, engulfment | 2 | 10.5263 | 0.0369 | IGHD, IGKC |
| GOTERM_CC | GO: 0005576~extracellular region | 6 | 31.5789 | 0.0173 | IGLV1-44, FAM3C, FBN1, IGKC, PLTP, TIMP1 |
| GOTERM_CC | GO: 0042571~immunoglobulin complex, circulating | 2 | 10.5263 | 0.0186 | IGHD, IGKC |
| GOTERM_CC | GO: 0070062~extracellular exosome | 7 | 36.8421 | 0.0471 | CR1, FAM3C, IGHD, FBN1, IGKC, GNG7, TIMP1 |
| GOTERM_MF | GO: 0003823~antigen binding | 3 | 15.7894 | 0.0047 | IGLV1-44, IGHD, IGKC |
| GOTERM_MF | GO: 0034987~immunoglobulin receptor binding | 2 | 10.5263 | 0.0259 | IGHD, IGKC |
| KEGG_PATHWAY | hsa05170: Human immunodeficiency virus 1 infection | 2 | 212 | 0.0017 | GNG7, TRAF5 |
| KEGG_PATHWAY | hsa05163: Human cytomegalovirus infection | 2 | 225 | 0.0019 | GNG7, TRAF5 |
| KEGG_PATHWAY | hsa05200: Pathways in cancer | 2 | 530 | 0.0098 | GNG7, TRAF5 |
| REACTOME_PATHWAY | R-HSA-76002: Platelet activation, signaling and aggregation | 3 | 260 | 5.38E-05 | GNG7|FAM3C|TIMP1 |
| REACTOME_PATHWAY | R-HSA-8957275: Post-translational protein phosphorylation | 2 | 108 | 0.000445033 | TIMP1|FBN1 |
| REACTOME_PATHWAY | R-HSA-381426: Regulation of IGF transport and uptake by IGFBPs | 2 | 125 | 0.000592278 | TIMP1|FBN1 |
| REACTOME_PATHWAY | R-HSA-114608: Platelet degranulation | 2 | 128 | 0.000620396 | FAM3C|TIMP1 |
| REACTOME_PATHWAY | R-HSA-109582: Hemostasis | 3 | 617 | 0.000666968 | GNG7|FAM3C|TIMP1 |
| REACTOME_PATHWAY | R-HSA-76005: Response to elevated platelet cytosolic Ca2+ | 2 | 133 | 0.000668675 | FAM3C|TIMP1 |
| REACTOME_PATHWAY | R-HSA-1474228: Degradation of the extracellular matrix | 2 | 140 | 0.000739233 | TIMP1|FBN1 |
| REACTOME_PATHWAY | R-HSA-1474244: Extracellular matrix organization | 2 | 298 | 0.003226 | TIMP1|FBN1 |
Moreover, protein interactions among the DEGs were predicted using STRING tools as presented in Figure 1C. The connectivity degree of each node was calculated. The results showed that there was a functional link between FBN1 and TIMP1.
FBN1 and TIMP1 expression in reactive lymph node hyperplasia and DLBCL tissues
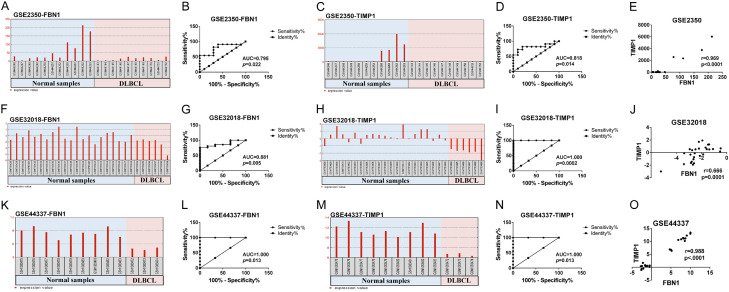
FBN1 and TIMP1 expression levels were first examined based on the expression profiles in these GEO datasets. As shown in Figure 2, DLBCL tissues showed significantly increased mRNA expression levels of FBN1 and TIMP1, compared to normal lymph node tissues (Figure 2A, 2C, 2F, 2H, 2K and 2M). Based on expression profile in these GEO datasets, the patients were divided into high expression group and low expression group. The ROC curves were used to evaluate the diagnostic accuracy of DEGs for expression level (Figure 2B, 2D, 2G, 2I, 2L and 2N). The AUC could be mapped to compare different screening genes. Based on these findings, FBN1 and TIMP1 could be considered as new biomarkers for prognosis. Furthermore, Pearson correlation analysis showed an obvious positive correlation between FBN1 and TIMP1 (Figure 2E, 2J and 2O).
Figure 2.
The expression levels and correlation of FBN1 and TIMP1 in both DLBCL and normal lymph node tissues from GSE2350, GSE32018 and GSE44337 datasets. A, C, F, H, K and M. The expression levels of FBN1 and TIMP1 in DLBCL tissues compared to normal lymph node tissues from GSE2350, GSE32018 and GSE44337 datasets. B, D, G, I, L and N. The ROC curves were analyzed to evaluate the diagnostic accuracy of DEGs for expression level. E, J and O. The correlation of mRNA levels of FBN1 and TIMP1 as analyzed by Pearson correlation analysis.
Relationship between FBN1 and TIMP1 expression levels and clinicopathological parameters of DLBCL
The association between FBN1 or TIMP1 and the clinicopathological characteristics of patients with DLBCL has been shown in Table 3. Positive expression of FBN1 in DLBCL was significantly higher in patients, in Ann Arbor stage (III-IV) group (χ2=5.223, P=0.022), with higher IPI score (3-5) (χ2=5.284, P=0.021) and in the non-GCB group (χ2=12.729, P=0.001). Comparison of the clinical characteristics of patients with high versus low expression of TIMP1 showed that patients with high TIMP1 expression were more frequently associated with B symptoms (χ2=5.297, P=0.021), Ann Arbor stage (III-IV) (χ2=12.194, P=0.001), had higher IPI scores (3-5) (χ2=5.559, P=0.018) and were in the non-GCB group (χ2=7.379, P=0.007). However, there was no significant difference between their expression levels and other clinicopathological characteristics.
Table 3.
Clinicopathological characteristics of patients with DLBCL based on the protein expression levels of FBN1 and TIMP1
| Clinicopathologic characteristics | Total | FBN1 | χ2 | p-value | TIMP1 | χ2 | p-value | ||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| high expression | low expression | high expression | low expression | ||||||
| Age | |||||||||
| ≤60 | 45 | 35 | 10 | 30 | 15 | ||||
| >60 | 65 | 50 | 15 | 0.016 | 0.899 | 50 | 15 | 0.941 | 0.332 |
| Sex | |||||||||
| Male | 63 | 51 | 12 | 49 | 14 | ||||
| Female | 47 | 34 | 13 | 0.699 | 0.403 | 31 | 16 | 1.347 | 0.246 |
| Primary site | |||||||||
| Nodal | 56 | 45 | 11 | 44 | 12 | ||||
| Extranodal | 54 | 40 | 14 | 0.312 | 0.576 | 36 | 18 | 1.409 | 0.235 |
| B sympotoms | |||||||||
| Absent | 63 | 49 | 14 | 40 | 23 | ||||
| Present | 47 | 36 | 11 | 0.007 | 0.933 | 40 | 7 | 5.297 | 0.021* |
| ECOG PS | |||||||||
| <2 | 87 | 70 | 17 | 67 | 20 | ||||
| ≥2 | 23 | 15 | 8 | 1.617 | 0.204 | 13 | 10 | 2.887 | 0.089 |
| Serum lactate dehydrogenase | |||||||||
| Normal | 41 | 30 | 11 | 30 | 11 | ||||
| Elevated | 69 | 55 | 14 | 0.309 | 0.578 | 50 | 19 | 0.019 | 0.888 |
| No. of extranodal sites | |||||||||
| <2 | 86 | 70 | 16 | 64 | 22 | ||||
| ≥2 | 24 | 15 | 9 | 2.815 | 0.093 | 16 | 8 | 0.245 | 0.621 |
| Ann Arbor stage | |||||||||
| I-II | 64 | 44 | 20 | 38 | 26 | ||||
| III-IV | 46 | 41 | 5 | 5.223 | 0.022* | 42 | 4 | 12.194 | 0.001* |
| International prognostic index | |||||||||
| Low (0-2) | 79 | 56 | 23 | 52 | 27 | ||||
| High (3-5) | 31 | 29 | 2 | 5.284 | 0.021* | 28 | 3 | 5.559 | 0.018* |
| Bone marrow involvement | |||||||||
| Absent | 87 | 68 | 19 | 65 | 22 | ||||
| Present | 23 | 17 | 6 | 0.023 | 0.879 | 15 | 8 | 0.417 | 0.518 |
| Bulky mass (cm) | |||||||||
| <10 | 101 | 80 | 21 | 74 | 27 | ||||
| ≥10 | 9 | 5 | 4 | 1.458 | 0.227 | 6 | 3 | 0.001 | 0.972 |
| Hans classification | |||||||||
| GCB | 29 | 15 | 14 | 15 | 14 | ||||
| Non-GCB | 81 | 70 | 11 | 12.729 | 0.001* | 65 | 16 | 7.379 | 0.007* |
| Treatment regimen | |||||||||
| R-CHOP | 95 | 74 | 21 | 68 | 27 | ||||
| Others | 15 | 11 | 4 | 0.004 | 0.952 | 12 | 3 | 0.136 | 0.712 |
P<0.05.
We then verified FBN1 and TIMP1 expression in TCGA database using the GEPIA web server and found that both these genes were overexpressed in tumor tissues (Figure 3A and 3C, P<0.05). However, GEPIA analysis of the two genes revealed that there was no significant difference in survival between the cancerous and normal group (Figure 3B and 3D). In addition, FBN1 and TIMP1 protein levels were examined by western blotting using 30 normal lymph node tissues and 30 DLBCL tissues. The results showed that both FBN1 and TIMP1 were highly expressed in DLBCL tissues than in normal lymph node tissues (Figure 3E and 3F).
Figure 3.
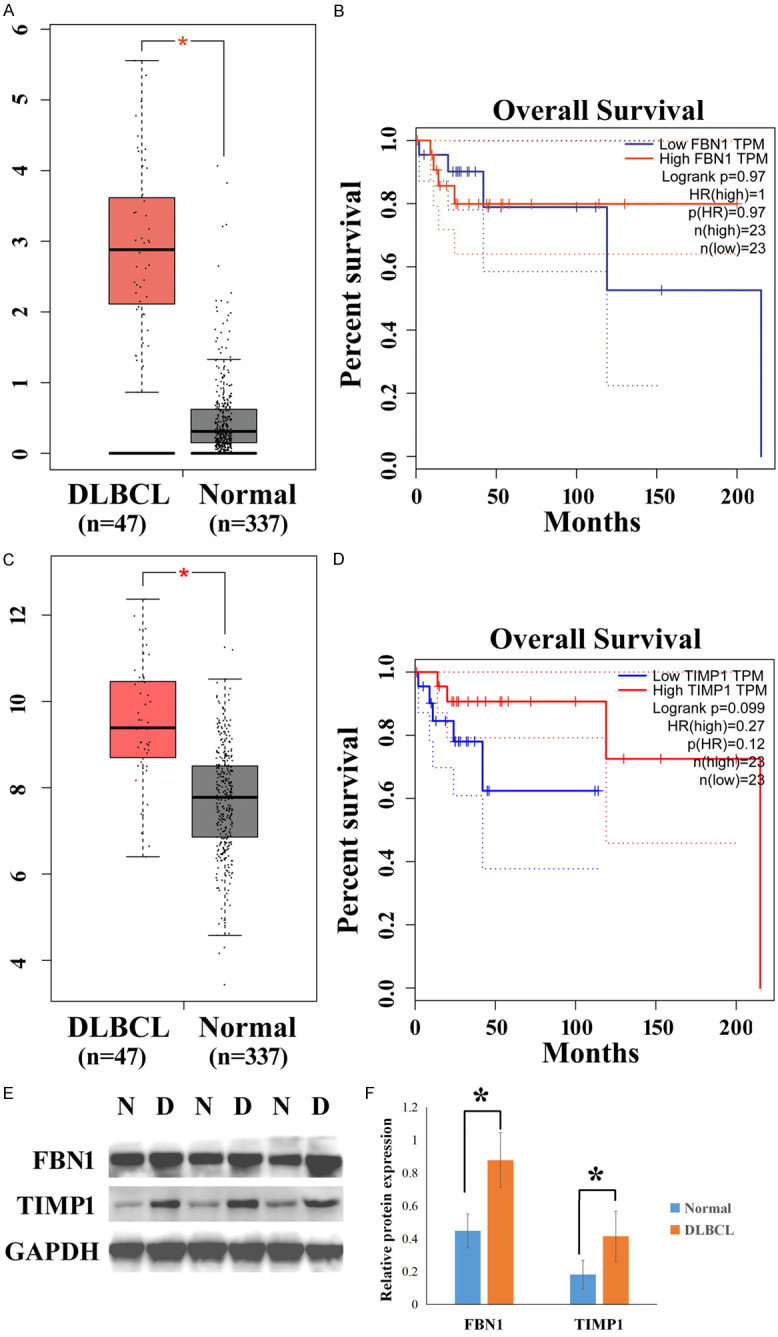
The expression levels of FBN1 and TIMP1 in TCGA database using the GEPIA web server and in clinical samples. A and C. The expression levels of FBN1 and TIMP1 in 47 DLBCL tissues and 337 normal lymph node tissues in TCGA database. B and D. The overall survival rates of patients with DLBCL and normal lymph nodes were analyzed in TCGA database using the GEPIA online tool. E. The expression levels of FBN1 and TIMP1 proteins were examined by western blotting. F. Column graphs represent the statistical results in 30 normal lymph node tissues and 30 DLBCL tissues. DLBCL or D: DLBCL samples; Normal or N: normal lymph node samples. *P<0.05.
Inhibition of FBN1 downregulated the expression of TIMP1 and the role of FBN1/TIMP1 interaction in regulating DLBCL cell migration
To detect whether there was any interaction between FBN1 and TIMP1, the two DLBCL cell lines (OCI-LY1 and SU-DHL-2) were treated with specific siRNAs and their protein levels were examined. The results showed that upon treatment with FBN1 siRNA, there was a significant decrease in the expression levels of FBN1 and TIMP1 (Figure 4A and 4B) and the downregulation of TIMP1 attenuated TIMP1 protein expression, but not FBN1 expression (Figure 4C and 4D).
Figure 4.
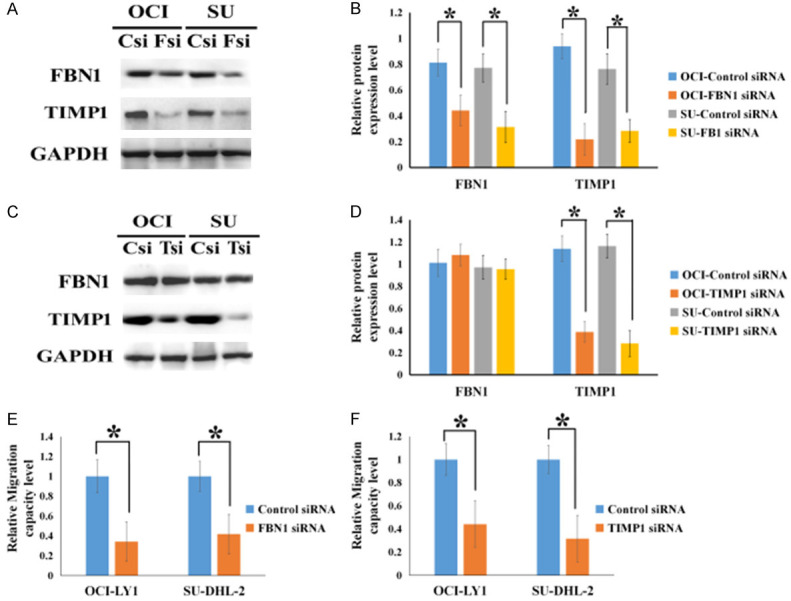
Inhibition of FBN1 downregulated the expression of TIMP1 and the role of FBN1/TIMP1 interaction in regulating DLBCL cell migration. (A) The protein expression levels of FBN1 and TIMP1 in FBN1 siRNA-treated OCI-LY1 and SU-DHL-2 cells. (B) Column graphs represent the statistical results of (A) experiment performed multiple times. (C) The protein expression levels of FBN1 and TIMP1 in TIMP1 siRNA-treated OCI-LY1 and SU-DHL-2 cells. (D) Column graphs represent the statistical results of (C) experiment performed multiple times. (E and F) The migratory ability of OCI-LY1 and SU-DHL-2 cells upon treatment with either FBN1 siRNA or TIMP1 siRNA. OCI: OCI-LY1; SU: SU-DHL-2; Csi: control siRNA; Fsi: FBN1 siRNA; Tsi: TIIMP1 siRNA; *P<0.05.
To verify whether FBN1/TIMP1interaction contributes to DLBCL cell motility, we examined DLBCL cell migration. The migratory ability of DLBCL cells was reduced upon treatment with either FBN1 or TIMP1 siRNA (Figure 4E and 4F). This indicated that FBN1/TIMP1 interaction drives DLBCL cell motility to promote dissemination.
FBN1/TIMP1 interaction is involved in DLBCL progression through the activation of Wnt/β-catenin signaling [16,17].
Increasing evidence indicates that Wnt/β-catenin promotes cancer progression. To validate the effect of FBN1/TIMP1 interaction on Wnt signaling activity, we examined the expression levels of Wnt-related molecules. The downregulated expression of either FBN1 or TIMP1 in OCI-LY1 and SU-DHL-2 cells inhibited the expression levels of Wnt target genes, including Wnt3, GSK3β, β-catenin, c-Myc and cyclin D1, suggesting the inactivation of the Wnt signaling pathway (Figure 5A-D). Interestingly, in cells treated with FBN1 siRNA, simultaneous overexpression of TIMP1 resulted in an increase of these proteins. This provided evidence that FBN1 triggered the activation of the Wnt pathway possibly through upregulating TIMP1 expression.
Figure 5.
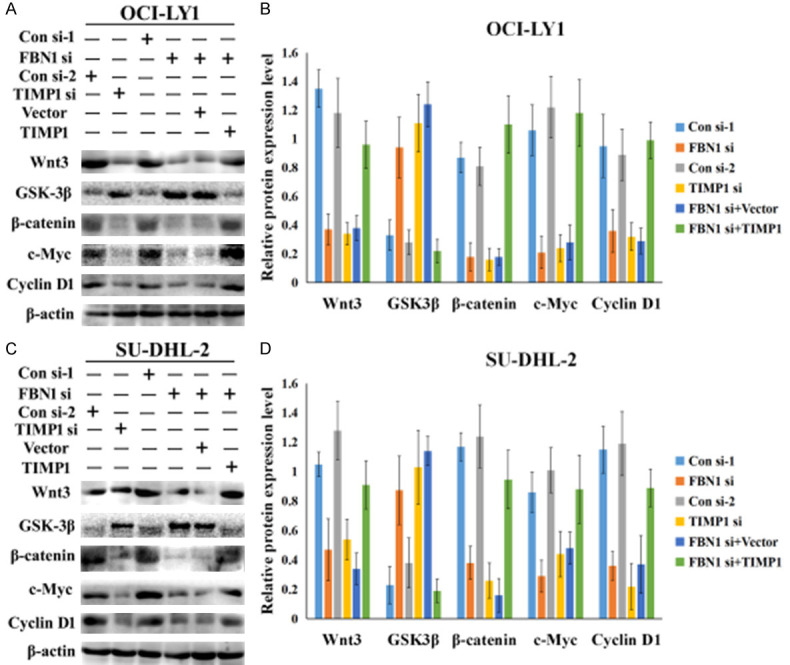
Inhibition of FBN1 downregulated the expression of TIMP1 and the role of FBN1/TIMP1 in regulating DLBCL cell migration. Western blotting was performed to detect the protein expression levels of Wnt3, GSK3β, β-catenin, c-Myc and cyclin D1 in OCI-LY1 cells (A) and SU-DHL-2 cells (C). Downregulating the expression of either FBN1 or TIMP1 in OCI-LY1 and SU-DHL-2 cells altered the expression levels of Wnt target genes, including Wnt3, GSK3β, β-catenin, c-Myc and cyclin D1. Changes in the expression levels of these proteins were examined in cells overexpressing TIMP1 and treated with FBN1 siRNA. (B and D) Column graphs represent the statistical results of (A) and (C) experiments performed multiple times. CON si: control siRNA; FBN si: FBN siRNA; TIMP1 si: TIMP1 siRNA; Vector: pCDNA3.1; TIMP1: pCDNA3.1-TIMP1-wild type plasmid; *P<0.05.
Discussion
To augment the therapeutic effect of DLBCL, it might be meaningful to validate critical chromatin modifiers and explore their tumorigenic mechanisms [18]. The different outcomes of patients with local stage and disseminated stage DLBCL demonstrated a change in the basic characteristics of tumor cells during the dissemination/progression of this disease [19]. However, the present understanding of DLBCL cell migration is rather limited compared to the plentiful knowledge regarding lymphomagenesis and tumor growth.
In this study, three gene expression profile datasets were integrated and 19 commonly altered DEGs were identified. The 19 DEGs were classified by GO term analysis into three groups, biological process (BP), cellular component (CC) and molecular function (MF) and the KEGG pathway enrichment analysis of the DEGs was performed using the functional annotation tool DAVID. Finally, PPI network was established, and the most significant modules (FBN1 and TIMP1) were filtered.
Fibrillin-1, an extracellular matrix glycoprotein and a chief component of microfibrils, is suggested to maintain cell attachment and to influence cell differentiation and migration [20]. Mutations in fibrillin-1 cause the autosomal dominant disorder Marfan syndrome and other related diseases of the connective tissue collectively named as type-1 fibrillinopathies [21]. It has been reported that FBN1 expression is significantly higher in germ cell neoplasia and gastric cancer compared to normal tissues [22,23]. In this study, FBN1 showed higher expression in DLBCL than in normal lymph nodes. The expression of FBN1 in DLBCL was significantly higher in patients of Ann Arbor stage III-IV, with higher IPI score (3-5) and in non-GCB group. These results suggested that FBN1 might act as an oncogene in DLBCL progression. However, we did not find any significant difference in survival between patients with high and low expression of FBN1.
In order to further study whether there was a correlation between FBN1 and TIMP1, specific siRNAs were used to knock down their expression respectively. The data showed that downregulation of FBN1 decreased TIMP1 protein expression. However, TIMP1 knock down had no effect on FBN1 protein expression. Therefore, FBN1 might be upstream of TIMP1. Recently, clinical studies have shown that aberrant expression of TIMP1 was associated with unfavorable prognosis in several tumors [8], [24-27]. We observed higher expression of TIMP1 in DLBCL tissues than that in normal lymph nodes. Moreover, TIMP1 expression is correlated with tumor progression, and suppression of TIMP1 enhanced tumor response to gemcitabine and radiotherapy [28]. This finding was consistent with our results as well. The results indicated that patients with high TIMP1 expression were more frequently associated with B symptoms, Ann Arbor stage (III-IV), higher IPI scores (3-5) and were in the non-GCB group. These observations validated that TIMP1 was positively correlated with the malignant progression of DLBCL.
Several reports have suggested that FBN1 and TIMP1 play important roles in certain cellular functions. FBN1 might be implicated in cell-stroma interaction with respect to signaling, attachment and migration [29]. Silencing of FBN1 could inhibit the proliferative, migratory and invasive abilities of GC cells [23]. TIMP1 could significantly increase clonogenic survival, migration and invasion [28]. We also verified that the downregulation of either FBN1 or TIMP1 could attenuate the migratory ability of DLBCL cell lines. Moreover, we demonstrated that FBN1/TIMP1 interaction was involved in DLBCL progression through the activation of Wnt/β-catenin signaling. It has also been reported that FBN1 increased the expression of β-catenin, N-cadherin and Wnt1 of the Wnt/β-catenin signal pathway, and decreased E-cadherin expression [23]. TIMP1 could also regulate the expression levels of E-cadherin, β-catenin and MMPs [30]. Taken together, these data indicate that FBN1/TIMP1 interaction could play a critical role in DLBCL progression.
By performing a series of bioinformatics analyses, we determined that two important proteins (FBN1 and TIMP1) were upregulated in DLBCL tissues. Our research confirmed that both FBN1 and TIMP1 were positively correlated with DLBCL progression. Further analysis revealed that FBN1/TIMP1 interaction could increase the cell migration of DLBCL by regulating the Wnt signaling pathway known to be involved in DLBCL cell migration. Although the underlying mechanisms regarding the interaction between FBN1 and TIMP1 require further clarification, FBN1 and TIMP1 might be potential therapeutic targets for DLBCL treatment.
Acknowledgements
This work was supported by Shenyang Plan Project of Science and Technology (Grant No. 18-014-4-20), the authors declare no competing financial interests. Funding agency did not participate in the design of the study, collection, analysis, interpretation of data and in writing the manuscript.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bauer WM, Aichelburg MC, Griss J, Skrabs C, Simonitsch-Klupp I, Schiefer AI, Kittler H, Jäger U, Zeyda M, Knobler R, Stingl G. Molecular classification of tumour cells in a patient with intravascular large B-cell lymphoma. Br J Dermatol. 2018;178:215–221. doi: 10.1111/bjd.15841. [DOI] [PubMed] [Google Scholar]
- 2.Zhang G, Wang H, Zhu K, Yang Y, Li J, Jiang H, Liu Z. Investigation of candidate molecular biomarkers for expression profile analysis of the Gene expression omnibus (GEO) in acute lymphocytic leukemia (ALL) Biomed Pharmacother. 2019;120:109530. doi: 10.1016/j.biopha.2019.109530. [DOI] [PubMed] [Google Scholar]
- 3.Del Cid JS, Reed NI, Molnar K, Liu S, Dang B, Jensen SA, DeGrado W, Handford PA, Sheppard D, Sundaram AB. A disease-associated mutation in fibrillin-1 differentially regulates integrin-mediated cell adhesion. J Biol Chem. 2019;294:18232–18243. doi: 10.1074/jbc.RA119.011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin M, Zhao S, Liu G, Huang Y, Yu C, Zhao Y, Wang L, Zhang Y, Yan Z, Wang S, Liu S, Liu J, Ye Y, Chen Y, Yang X, Tong B, Wang Z, Yang X, Niu Y, Li X, Wang Y, Su J, Yuan J, Zhao H, Zhang S, Qiu G. Identification of novel FBN1 variations implicated in congenital scoliosis. J Hum Genet. 2020;65:221–230. doi: 10.1038/s10038-019-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Urban JP, Yu J. The distribution of fibrillin-2 and LTBP-2, and their co-localisation with fibrillin-1 in adult bovine tail disc. J Anat. 2012;220:164–72. doi: 10.1111/j.1469-7580.2011.01455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue H, Wang J, Chen R, Hou X, Li J, Lu X. Gene signature characteristic of elevated stromal infiltration and activation is associated with increased risk of hematogenous and lymphatic metastasis in serous ovarian cancer. BMC Cancer. 2019;19:1266. doi: 10.1186/s12885-019-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckfeld C, Häußler D, Schoeps B, Hermann CD, KrÜger A. Functional disparities within the TIMP family in cancer: hints from molecular divergence. Cancer Metastasis Rev. 2019;38:469–481. doi: 10.1007/s10555-019-09812-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Sun W, Qin Y, Wu C, He L, Zhang T, Shao L, Zhang H, Zhang P. Knockdown of KDM1A suppresses tumour migration and invasion by epigenetically regulating the TIMP1/MMP9 pathway in papillary thyroid cancer. J Cell Mol Med. 2019;23:4933–4944. doi: 10.1111/jcmm.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song G, Xu S, Zhang H, Wang Y, Xiao C, Jiang T, Wu L, Zhang T, Sun X, Zhong L, Zhou C, Wang Z, Peng Z, Chen J, Wang X. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res. 2016;35:148. doi: 10.1186/s13046-016-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Y, Wan D, Song W. Dryofragin inhibits the migration and invasion of human osteosarcoma U2OS cells by suppressing MMP-2/9 and elevating TIMP-1/2 through PI3K/AKT and p38 MAPK signaling pathways. Anticancer Drugs. 2016;27:660–8. doi: 10.1097/CAD.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 11.Song T, Dou C, Jia Y, Tu K, Zheng X. TIMP-1 activated carcinoma-associated fibroblasts inhibit tumor apoptosis by activating SDF1/CXCR4 signaling in hepatocellular carcinoma. Oncotarget. 2015;6:12061–79. doi: 10.18632/oncotarget.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdullah ML, Hafez MM, Al-Hoshani A, Al-Shabanah O. Anti-metastatic and anti-proliferative activity of eugenol against triple negative and HER2 positive breast cancer cells. BMC Complement Altern Med. 2018;18:321. doi: 10.1186/s12906-018-2392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SY, Kim YH, Kim Y, Lee SJ. Aromatic-turmerone attenuates invasion and expression of MMP-9 and COX-2 through inhibition of NF-κB activation in TPA-induced breast cancer cells. J Cell Biochem. 2012;113:3653–62. doi: 10.1002/jcb.24238. [DOI] [PubMed] [Google Scholar]
- 14.Oliveros JC. (2007-2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index. Html.
- 15.Zhang Z, Zhang G, Kong C, Bi J, Gong D, Yu X, Shi D, Zhan B, Ye P. EIF2C, Dicer, and Drosha are up-regulated along tumor progression and associated with poor prognosis in bladder carcinoma. Tumour Biol. 2015;36:5071–9. doi: 10.1007/s13277-015-3158-z. [DOI] [PubMed] [Google Scholar]
- 16.Chiarini F, Paganelli F, Martelli AM, Evangelisti C. The role played by Wnt/β-catenin signaling pathway in acute lymphoblastic leukemia. Int J Mol Sci. 2020;21:E1098. doi: 10.3390/ijms21031098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Orozco E, Sanchez-Fernandez A, Ortiz-Parra I, Ayala-San Nicolas M. WNT signaling in tumors: the way to evade drugs and immunity. Front Immunol. 2019;10:2854. doi: 10.3389/fimmu.2019.02854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Wang X, Li Q, Chen W, Zhang N, Kong Y, Lv J, Cao L, Lin D, Wang X, Xu G, Wu X. FBXL10 contributes to the development of diffuse large B-cell lymphoma by epigenetically enhancing ERK1/2 signaling pathway. Cell Death Dis. 2018;9:46. doi: 10.1038/s41419-017-0066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SJ, Kang HJ, Kim JS, Oh SY, Choi CW, Lee SI, Won JH, Kim MK, Kwon JH, Mun YC, Kwak JY, Kwon JM, Hwang IG, Kim HJ, Park J, Oh S, Huh J, Ko YH, Suh C, Kim WS. Comparison of treatment strategies for patients with intestinal diffuse large B-cell lymphoma: surgical resection followed by chemotherapy versus chemotherapy alone. Blood. 2011;117:1958–65. doi: 10.1182/blood-2010-06-288480. [DOI] [PubMed] [Google Scholar]
- 20.Abbas Y, Carnicer-Lombarte A, Gardner L, Thomas J, Brosens JJ, Moffett A, Sharkey AM, Franze K, Burton GJ, Oyen ML. Tissue stiffness at the human maternal-fetal interface. Hum Reprod. 2019;34:1999–2008. doi: 10.1093/humrep/dez139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McInerney-Leo AM, West J, Wheeler L, Leo PJ, Summers KM, Anderson L, Brown MA, West M, Duncan EL. Compound heterozygous mutations in FBN1 in a large family with Marfan syndrome. Mol Genet Genomic Med. 2020;8:e1116. doi: 10.1002/mgg3.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cierna Z, Mego M, Jurisica I, Machalekova K, Chovanec M, Miskovska V, Svetlovska D, Kalavska K, Rejlekova K, Kajo K, Mardiak J, Babal P. Fibrillin-1 (FBN-1) a new marker of germ cell neoplasia in situ. BMC Cancer. 2016;16:597. doi: 10.1186/s12885-016-2644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang D, Zhao D, Chen X. MiR-133b inhibits proliferation and invasion of gastric cancer cells by up-regulating FBN1 expression. Cancer Biomark. 2017;19:425–436. doi: 10.3233/CBM-160421. [DOI] [PubMed] [Google Scholar]
- 24.Omar OM, Soutto M, Bhat NS, Bhat AA, Lu H, Chen Z, El-Rifai W. TFF1 antagonizes TIMP-1 mediated proliferative functions in gastric cancer. Mol Carcinog. 2018;57:1577–1587. doi: 10.1002/mc.22880. [DOI] [PubMed] [Google Scholar]
- 25.Zeng C, Chen Y. HTR1D, TIMP1, SERPINE1, MMP3 and CNR2 affect the survival of patients with colon adenocarcinoma. Oncol Lett. 2019;18:2448–2454. doi: 10.3892/ol.2019.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham V, Cao G, Parambath A, Lawal F, Handumrongkul C, Debs R, DeLisser HM. Involvement of TIMP-1 in PECAM-1-mediated tumor dissemination. Int J Oncol. 2018;53:488–502. doi: 10.3892/ijo.2018.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zajkowska M, Gacuta E, Kozłowska S, Lubowicka E, Głażewska EK, Chrostek L, Szmitkowski M, Pawłowski P, Zbucka-Krętowska M, Ławicki S. Diagnostic power of VEGF, MMP-9 and TIMP-1 in patients with breast cancer. A multivariate statistical analysis with ROC curve. Adv Med Sci. 2019;64:1–8. doi: 10.1016/j.advms.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 28.D’Costa Z, Jones K, Azad A, van Stiphout R, Lim SY, Gomes AL, Kinchesh P, Smart SC, Gillies McKenna W, Buffa FM, Sansom OJ, Muschel RJ, O’Neill E, Fokas E. Gemcitabine-induced TIMP1 attenuates therapy response and promotes tumor growth and liver metastasis in pancreatic cancer. Cancer Res. 2017;77:5952–5962. doi: 10.1158/0008-5472.CAN-16-2833. [DOI] [PubMed] [Google Scholar]
- 29.Tseleni-Balafouta S, Gakiopoulou H, Fanourakis G, Voutsinas G, Litsiou H, Sozopoulos E, Balafoutas D, Patsouris E. Fibrillin expression and localization in various types of carcinomas of the thyroid gland. Mod Pathol. 2006;19:695–700. doi: 10.1038/modpathol.3800578. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Liu Y, Lu L, Yang L, Yin S, Wang Y, Qi Z, Meng J, Zang R, Yang G. Fibrillin-1, induced by Aurora-A but inhibited by BRCA2, promotes ovarian cancer metastasis. Oncotarget. 2015;6:6670–83. doi: 10.18632/oncotarget.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.