Abstract
LncRNAs and miRNAs are emerging players in epithelial ovarian cancer (EOC). LncRNA MALAT-1 and miR-22 play vital roles in the onset and development of multiple cancers. Both of them are abnormally expressed in ovarian cancer, but the molecular basis for their involvement in EOC is unclear. In this study, we found MALAT-1 was up-regulated but miR-22 was down-regulated in EOC tissues and cell lines when compared to normal ovarian epithelial cell line IOSE80. Both of MALAT-1shRNA and miR-22 mimics inhibited ovarian cell proliferation, migration, and invasion, while simultaneously overexpressing MALAT-1 and miR-22 largely canceled out this inhibitory effect. Consistently, MALAT-1 silencing and miR-22 overexpression restrained tumor growth and metastasis to lungs in nude mice, which could be largely counteracted by co-overexpressing MALAT-1 and miR-22. Mechanistically, MALAT-1 targeted and sponged miR-22, counteracting its inhibitory effect on c-myc and c-myc-mediated epithelial-mesenchymal transition. Our findings for the first time demonstrated that MALAT-1 supports EOC progression through sponging miR-22, providing a novel insight into the role of MALAT-1 in ovarian cancer.
Keywords: Epithelial ovarian cancer, LncRNA MALAT-1, miRNA-22, c-myc
Introduction
Ovarian cancer is a type of gynecological cancer that seriously threatens woman health worldwide, with the overall 5-year survival rates below 45% [1]. Epithelial ovarian cancer (EOC), as the most common type of all ovarian carcinoma, accounts for 90% of all cases and is more aggressive than non-epithelial subtypes [1,2]. EOC is often associated with low survival rates because of late-stage diagnoses, high risk of metastasis and chemoresistance. However, precise diagnostic factors for this disease are in urgent need and molecular basis for its progression is not fully understood.
LncRNAs, regardless of its non-coding feature, could regulate expression of target genes through epigenetic, transcriptional and post-transcriptional levels [3,4], playing a vital role in the initiation and progression of ovarian cancer. Metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1), located at chromosome 11q13, was firstly found in non-small cell lung cancer and considered to be an independent prognostic factor of bronchogenic carcinoma [5]. It was abnormally up-regulated and involved in metastasis of several cancers, including breast cancer, liver carcinoma, and lung cancer [6-8]. Recently, it was reported to be highly expressed and involved in proliferation and migration in ovarian cancer [9]. Moreover, MALAT-1 was positively associated with distant metastasis of EOC [10] and could promote metastasis via activating PI3K-AKT pathway [11].
Whereas, the underlying regulatory mechanism is still not fully elucidated. miRNAs, whose lengths are 19-22 nucleotides, are key regulators of gene expression at the post-transcriptional level and emerging miRNAs have been evidenced to be involved in tumor development [12]. MiR22 is a highly conserved miRNA in mammalian species, and play dual roles during tumor onset and progression [13]. For instance, it acts as an oncogene in leukemia and prostate cancer but plays an anti-cancer role in lung and colorectal cancer [13-15]. miR-22 was also reported to be significantly down-regulated in EOC tumors and might serve as a suppressive gene through modulating VEGF and P53 [16].
In the present study, we find an interaction between miR-22 and MALAT-1 and report that MALAT-1 promotes c-myc-mediated epithelial-mesenchymal transition and supports EOC progression through acting as a sponge for miR-22.
Materials and methods
Patient samples
Patient EOC samples (n=20) were collected in Xiangya Hospital, Central South University during the period from September 2015 to September 2016. The inclusion criteria is: the patients were confirmed as serous/mucinous EOC by pathological examination and they had not received preoperative radiotherapy, chemotherapy, hormones, and other treatments. This study was approved by the ethic committee of Xiangya Hospital, Central South University and informed consents were obtained from patients.
Cell culture and transfection
The ovarian cancer cell lines (SKOV3 and CAOV3) and the normal ovarian epithelial cell line IOSE80 were purchased from Fenghui Biotechnology (Hunan, China). SKOV3 was cultured with DMEM/F12 media containing 10% FBS (Gibco, USA) and CAOV3 and IOSE80 were cultured with DMEM media containing 10% FBS. PcDNA3.1-MALAT-1 (Fenghui Biotech), MALAT-1 shRNAs (Genepharma, China), miR-22 mimics and inhibitors (Genepharma, China), and corresponding negative controls were transfected into SKOV3 cells using Lipofectamine 2000 according to the instructions.
MTT assay
Twenty-four hours after transfection, SKOV3 cells were seeded into 96-well plates at a density of 1,000 cells/well. At 0, 24, 48, 72 h, cell proliferation was determined by MTT assay. Briefly, 20 μL of 5 mg/mL MTT (Sigma-Aldrich, USA) was added and incubated for 4 h in the cell incubator at 37°C, thereafter the supernatants were removed and 100 μL DMSO was added to dissolve the formazan. Optical absorbance at 492 nm was measured on the microplate reader (Spectralmax M5, Molecular Devices, USA). The optical absorbance was used to represent cell proliferation capacity.
Quantiative PCR
Total RNAs of cell samples and tumor specimens were isolated using Trizol (Invitrogen, USA) according to the manufacturer’s instructions and then were reverse-transcribed into cDNA using reverse-transcription kit (Thermo, USA). SYBR® Green Real-time PCR Master Mix was applied for quantitative real-time PCR on a Bio-rad instrument (CFX96, USA) using the following parameters: 95°C for 5 min, followed by 35 cycles of 95°C for 20 s, and 58°C for 50 s. RNA expression levels were calculated using the 2-ΔΔCt method. The primer sequences were listed in Table 1.
Table 1.
Primer sequences
| Gene | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| MALAT-1 | CAGTGGGGAACTCTGACTCG | GTGCCTGGTGCTCTCTTACC |
| miR-22 | ACACTCCAGCGGGAAGCTGCCAGTTGAAGA | TGGTGTCGTGGAGTCG |
| c-Myc | GATTCTCTGCTCTCCTCGAC | TCCAGACTCTGACCTTTTGC |
| GAPDH | GTCAACGGATTTGGTCTGTATT | AGTCTTCTGGGTGGCAGTGAT |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTGCGT |
Note: MALAT-1: Metastasis-associated lung adenocarcinoma transcript-1.
Wound-healing assay
Twenty-four hours after transfection, SKOV3 cells were seeded into 6-well plates at a density of 5 * 105 cells/well. When the cells grew to confluence, they were wounded by a sterile 200 μL pipette tip, followed by PBS wash. Then the hungry DMEM/F12 media containing 0.05% FBS was added and cells were photographed. After incubation for another 24, 48, 72 h, the same fields were photographed. The wound widths were calculated by Image J software (v 1.49, NIH, Bethesda, MD, USA). The healed wound widths were used to represent cell migration distances.
RNA-immunoprecipitation (RIP) assay
The RIP assay was performed using RIP-Assay Kit (MBL, Japan) according to the manufacturer’s instructions. Briefly, cell lysate from SKOV3 cells were incubated with anti-Argonaute2-conjugated magnetic beads or control IgG-conjugated magnetic beads overnight at 4°C. Thereafter, the protein component was digested by proteinase K treatment to obtain the precipitated RNA, which was then quantified by quantiative PCR.
Transwell invasion assay
Cell invasion ability was assessed using a 24-well plate transwell system (Corning, USA, pore size 8 μm). Twenty-four hours after transfection, SKOV3 cells resuspended into hungry media were seeded into the top-chambers of transwells which were pre-coated with 100 μL matrigel (1:3 diluted, Corning, USA) and the bottom chambers were immersed into 600 μL complete media. The transwells were fixed with 4% PFA, stained with 0.1% crystal violet solution (Beyotime, China). The invasive cells were photographed and manually counted.
Western blots
Total protein was extracted from cell samples using RIPA Lysis Buffer (Beyotime, China) complemented with PMSF (Beyotime, China), followed by SDS-PAGE separation and electro-transfer onto PVDF membranes. After blocking the membranes using 5% skimmed milk, they were incubated with anti-c-myc (#18583, 1:1000 dilution), E-cadherin (#14472, 1:1000 dilution), Vimentin (#5741, 1:1000 dilution), and Snai1 (#3879, 1:1000 dilution) primary antibodies at 4°C overnight and then corresponding HRP-conjugated secondary antibodies (anti-rabbit, #14708, 1:2000 dilution; anti-mouse, #14708, 1:3000 dilution) at room temperature for 2 h. The protein expressions were visualized using ECL kit (Beyotime, China). The intensity of protein bands was analyzed using ImageJ software. The antibodies were all purchased from Cell Signaling Technology, USA.
Luciferase reporter assay
Dual-luciferase vectors containing wild-type or mutant LncRNA MALAT-1 3’UTR were purchased from Hanbio Biotechnology Co., Ltd. (Shanghai, China), and then co-transfected with miR-22 mimics or its control into HEK293T cells. After 48 hours, cells were collected and luciferase activities were measured using the Dual Luciferase Reporter Gene Assay Kit (Beyotime, China).
Animal experiments
Female BALB/c-nude mice (4-6 weeks old) were purchased from Hunan SJA Laboratory Animal Co., Ltd and maintained in the specific-pathogen-free environment with free access to water and food. The SKOV3 cells stably expressed MALAT-1 shRNA, scrambled control, MALAT-1 + miR-22, or miR-22 were established, respectively. For each mouse, 4 * 106 cells were resuspended in 100 μL PBS and injected subcutaneously into the left axilla of nude mice. In the following 4 weeks, tumor growth was measured every 2-3 days. Tumor volumes = tumor length * width2/2. The protocols for animal experiments were approved by the Ethics Committee of Central South University (No. 201703644).
Statistical analysis
All statistical analysis was conducted using SPSS 24.0. Quantitative results were expressed as mean ± standard deviation (x̅ ± sd). Student’s t-test was used to compare the differences between two groups. Comparisons among multiple groups were performed using one-way ANOVA followed by Tukey’s post hoc test. Two-tailed P<0.05 was considered statistically significant.
Results
MALAT-1 is up-regulated while miR-22 is down-regulated in EOC tissues and cell lines
The expression profiles of MALAT-1 and miR-22 were determined in EOC tissues and cell lines using QPCR. Compared with normal epithelial ovarian cell line IOSE80, MALAT-1 was significantly up-regulated in epithelioid ovarian cancer cell lines (SKOV3 and CAOV3) (Figure 1). On the contrary, miR-22 level was significantly higher in IOSE80 cells that in SKOV3 and CAOV3 cells (Figure 1A). Furthermore, we observed significantly increased MALAT1 expression and significantly decreased miR-22 expression in EOC tissues compared with those in tumor-adjacent normal tissues (Figure 1B). Herein, we speculated that MALAT-1 and miR-22 might be involved in EOC development and progression.
Figure 1.
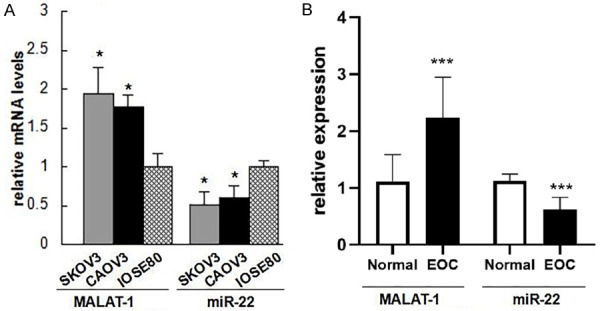
MALAT-1 is up-regulated while miR-22 is down-regulated in epithelial ovarian cancer (EOC) tissues and cell lines. A: RNA levels of MALAT-1 and miR-22 indicated cell lines; B: And EOC or tumor-adjacent tissues (n=14) were determined by qPCR. Data are expressed as mean ± standard deviation (x̅ ± sd). *P<0.05 vs. IOSE80 cells, or tumor-adjacent normal tissues. MALAT-1: Metastasis-associated lung adenocarcinoma transcript-1; EOC: epithelial ovarian cancer.
MiR-22 is a target of MALAT-1
In consideration of the reverse expression profiles of MALAT-1 and miR-22 levels in EOC and normal samples, we searched Targetscan and microrna.org to identify whether there was an interaction between MALAT-1 and miR-22. As shown in Figure 2A, there was indeed a putative direct interaction between them. To further validate this, the dual-luciferase reporter assay was conducted (Figure 2B). HEK293T cells co-expressing wild-type MALAT-1 and miR-22 mimics exhibited significantly reduced luciferase intensity, when compared with that of cells co-expressing mutant MALAT-1 and miRNA mimics, indicating MALAT-1 functions as a sponge for miR-22 in EOC cells. In order to further validate the interaction between MALAT-1 and miR-22, RNA Immunoprecipitation (RIP) assay was performed. The results showed that the levels of both MALAT-1 and miR-22 were enriched in anti-Ago2 group but not in the control IgG group (Figure 2C). These findings demonstrate the interaction between MALAT-1 and miR-22.
Figure 2.
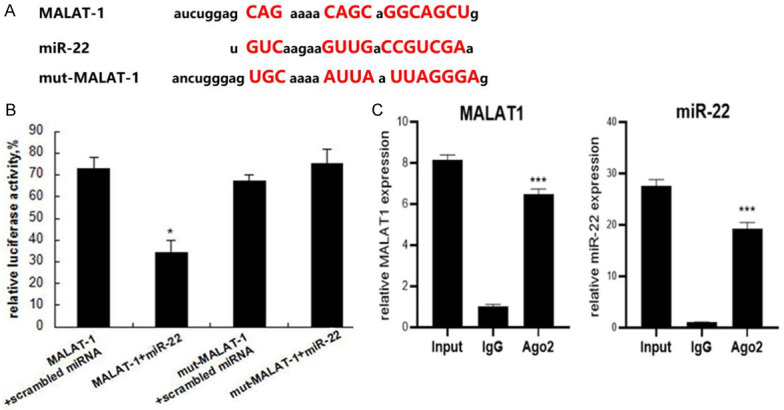
miR-22 is a target of MALAT-1. A: The sequence comparison between wide-type or mutant MALAT-1 and miR-22; B: miR-22 and luciferase plasmids cloned with wild-type or mutant MALAT-1 were transfected into HEK293T cells, the luciferase activities in cell lysates were determined; C: RIP assay was performed to validate the interaction between MALAT1 and miR-22 in SKOV3 cells. Data are expressed as mean ± standard deviation (x̅ ± sd) and represent three replicated experiments *P<0.05. MALAT-1: Metastasis-associated lung adenocarcinoma transcript-1.
MALAT-1 promotes EOC cell proliferation, migration and invasion through suppressing miR-22
To demonstrate the detailed functions of MALAT-1/miR-22 in EOC development, we altered MALAT-1 and miR-22 levels in SKOV3 cells so as to investigate their influences on cell proliferation, migration, and invasion. Figure 3A showed that MALAT-1 and miR-22 were successfully overexpressed or inhibited. Especially, MALAT1 inhibition caused markedly up-regulation of miR-22 levels while MALAT1 was not affected by miR-22 mimic and inhibitor, which further proves that MALAT-1 acts as a sponge for miR-22 in EOC cells. In MTT assay of cell proliferation capacity, MALAT-1 overexpression promoted SKOV3 cell proliferation and MALAT-1 inhibition exhibited the opposite effect (Figure 3B). On the contrary, miR-22 mimic impaired while miR-22 inhibitor enhanced SKOV3 cell proliferation (Figure 3B). Apart from the effects on proliferation, both of MALAT-1 silencing and miR-22 overexpression significantly inhibited migration of SKOV3 cells, which was largely counteracted by co-transfection of MALAT-1 and miR-22 (Figure 3C). Similar to changes in EOC cell migration, both of MALAT-1 silencing and miR-22 overexpression suppressed cell invasion, while co-overexpression of MALAT-1 and miR-22 canceled out the effect of each other mostly (Figure 3D). Collectively, these results indicate MALAT-1 promotes the proliferative, migratory, and invasive capacities of EOC cells through serving as a sponge for miR-22 and thus suppressing it.
Figure 3.
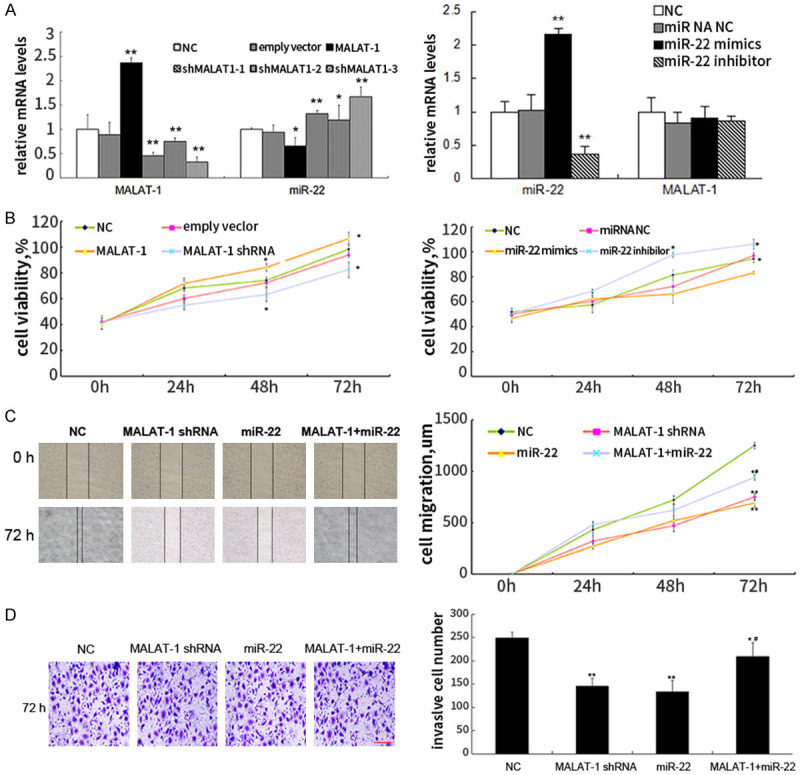
MALAT-1 promotes EOC cell proliferation, migration and invasion through suppressing miR-22. A: RNA levels of MALAT-1 and miR-22 in SKOV3 cells were examined 24 h after transfection of MALAT-1 overexpression vector, MALAT-1 shRNA, miR-22 mimics, and inhibitors using qPCR; B: The proliferative viability was assayed by MTT reagents; C: MALAT-1 shRNA, miR-22 mimics, or MALAT-1 and miR-22 mimics were introduced into SKOV3 cells, respectively. Migration capacities of these cells were assessed using scratching assay; D: Cell invasion assay was conducted using transwells and visualized using crystal violet staining. Data are expressed as mean ± standard deviation (x̅ ± sd) and represent three replicated experiments. *P<0.05 vs. the negative control (NC), **P<0.01 vs. the NC, and #P<0.05 vs. the miR-22 group. MALAT-1: Metastasis-associated lung adenocarcinoma transcript-1.
MALAT-1 enhanced epithelial-mesenchymal transition of EOC cells via MiR22/c-myc axis
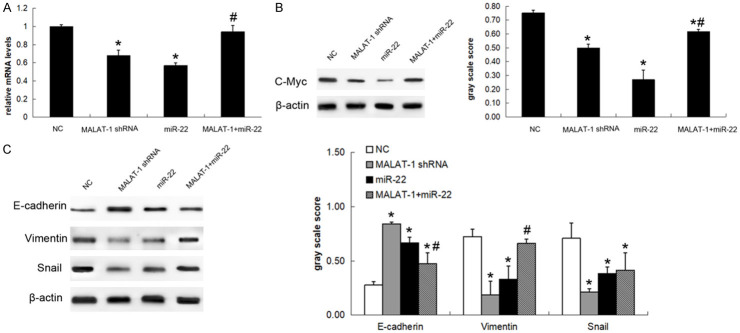
MiR-22 was previously reported to inhibit c-myc activity so as to suppress epithelial-mesenchymal transition (EMT) of breast cancer cells and it could target snail and inhibit EMT of lung cancer cells [17,18]. In order to demonstrate the possible mechanism underlying the functions of MALAT-1/miR-22, we examined the expression of c-myc and EMT markers after silencing or overexpressing MALAT-1 and miR-22. The RNA expression levels, immunoblot images, and gray-scale analysis in Figure 4 showed that shRNA-mediated MALAT-1 silencing down-regulated c-myc (Figure 4A and 4B), vimentin, and snail but up-regulated E-cadherin (Figure 4C), meanwhile miR-22 overexpression presented the similar effects (Figure 4A-C). However, the effects of miR-22 were mostly counteracted when MALAT-1 and miR-22 were co-transfected into SKOV3 cells, except that slight rescue of snail levels was observed.
Figure 4.
MALAT-1 enhanced epithelial-mesenchymal transition (EMT) of EOC cells via miR22/c-myc axis. A: MALAT-1 shRNA, miR-22 mimics, or MALAT-1 and miR-22 mimics were introduced into SKOV3 cells. The c-myc RNA levels were examined by qPCR; B: The c-myc protein was detected using immunoblots and then quantified by gray-scale analysis; C: The epithelial-mesenchymal transition markers, including E-cadherin, vimentin, snai1, were detected by immunoblots and then quantified by gray-scale analysis. Data are expressed as mean ± standard deviation (x̅ ± sd) and represent three replicated experiments. *P<0.05 vs. the negative control (NC), and #P<0.05 vs. the miR-22 group. MALAT-1: Metastasis-associated lung adenocarcinoma transcript-1.
MALAT-1 inhibition reduces in vivo ovarian cancer growth and metastasis through suppressing miR-22
Finally, we validated the modulation of MALAT-1/miR-22 on ovarian cancer growth and metastasis in nude mouse tumor models. MALAT-1-silenced, miR-22-overexpressed, MALAT-1/miR-22 co-overexpressed, or negative control SKOV3 cells were subcutaneously inoculated or injected into tail veins. Tumor growth or lung metastasis was monitored in the following 4 weeks. As shown in Figure 5A, both of MALAT-1 silencing and miR-22 overexpression markedly restrained SKOV3 xenograft growth while co-overexpression of MALAT-1 and miR-22 partly counteracted this inhibitory effect, as evidenced by the reduced tumor weight (Figure 5B) and volume (Figure 5C). Additionally, we determined the levels of MALAT-1 and miR-22 in tumor xenografts after sacrificing all mice and confirmed MALAT-1 and miR-22 maintained silenced or overexpressed as treated until the end of experiments (Figure 5D). Of special interest, co-overexpressing MALAT-1 and miR-22 resulted in mildly elevated miR-22 levels in the end (Figure 5D), indicating MALAT-1 did not exactly cancel out the effect of miR-22 overexpression, which was in agreement with the data of subcutaneous tumor xenograft volumes and weights. Consistent with the subcutaneous tumor xenograft results, mouse lung metastasis nodules were significantly reduced by MALAT-1 silencing or miR-22 overexpression, whereas this effect could be largely, though not completely, reversed by co-overexpressing MALAT-1 and miR-22 (Figure 5E). These in vivo results were in line with our in vitro data of MALAT-1 and miR-22, supporting the conclusion that MALAT-1 promotes growth and metastasis of epithelial ovarian cancer via inhibiting miR-22.
Figure 5.
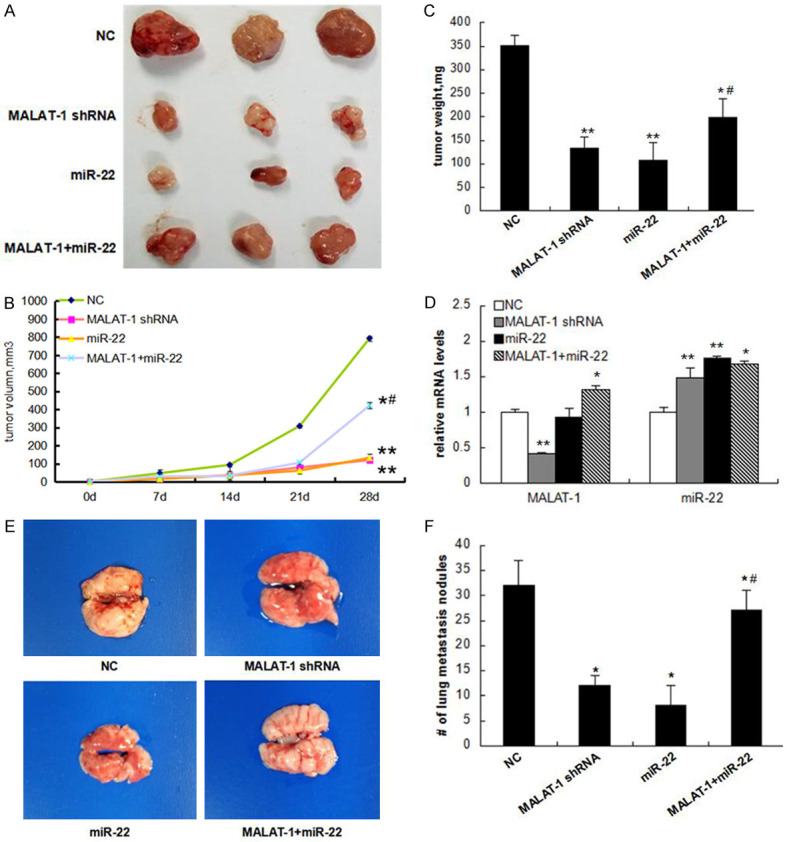
MALAT-1 restrains ovarian cancer growth and metastasis in nude mice through suppressing miR-22. SKOV3 cells stably expressed MALAT-1 shRNA, miR-22, or MALAT-1 and miR-22 were established, respectively. These cell lines were inoculated subcutaneously into nude mice or intravenously injected into nude mouse tails (4 * 106 cells/mouse). A: 4 weeks after inoculation, mice were sacrificed and the tumors were photographed; B: Tumor weights; C: Volumes in the 4 weeks were recorded. D: Expression levels of MALAT-1 and miR-22 in mouse tumor samples were determined by qPCR; F: For metastasis models, mouse lungs were photographed; E: Tumor nodules in lungs were counted. Data are mean ± standard deviation (x̅ ± sd). *P<0.05 vs. the negative control (NC), and #P<0.05 vs. the miR-22 group. MALAT-1: Metastasis-associated lung adenocarcinoma transcript-1.
Discussion
EOC is one of the major causes of mortality for women around the world, with 5-year survival rates lower than 45% [19]. Although several theories have explained its carcinogenesis, such as the incessant ovulation theory, the underlying mechanisms for its high risk of invasion and metastasis remain unclear [19,20]. LncRNAs and miRNAs are emerging contributors during EOC progression [21,22]. lncRNA MALAT-1 and miR-22 were previously reported to be abnormally expressed in multiple tumors, including ovarian cancer [9,16], however, the detailed modulation mechanisms are not fully elucidated. In this study, we found that MALAT-1 serves as a molecular sponge for miR-22, which can inhibit tumor cell proliferation and EMT-mediated invasion by targeting c-myc. By this mean, MALAT-1 acts as an onco-lncRNA contributing to the progression of ovarian cancer.
MALAT1 has been reported to be significantly up-regulated in many cancers and promote cancer progression through distinct mechanisms depending on the tumor types. For example, MALAT-1 promoted lung adenocarcinoma metastasis by enhancing transcription of motility-related genes [23], while it contributed to metastasis of kidney clear-cell tumor via acting as a sponge for miR-200, finally canceling out its inhibition on ZEB2 [24]. In our case, we initially observed up-regulated MALAT-1 both in EOC tissues and cell lines (Figure 1), which is in agreement with the results of Wu et al [9]. Then, we found MALAT-1 presented supportive role in tumor growth, invasion, and metastasis in vitro and in vivo (Figures 3 and 5). In line with this, MALAT-1 was reported to be associated with proliferation and distant metastasis of not only EOC, but also other cancers, including lung cancer, esophageal squamous cell carcinoma, and pancreatic cancer [8,25]. These collective re|sults confirmed the oncogenic role of MALAT-1 in tumors [26].
In our present study, we for the first time revealed that miR-22 was under direct modulation of MALAT-1 (Figures 2 and 3), which is well-known as a suppressor in many tumors, including breast cancer and colorectal cancer [13,27,28]. We observed miR-22 inhibited EOC cell growth and metastasis both in vivo and in vitro. Similar to our results, miR-22 positively correlated with EOC patient survival [29] and played a suppressive role in ovarian cancer [30]. Specially, we observed that the suppressive function of miR-22 on tumor progression was mostly, though not completely, counteracted by MALAT-1 (Figures 3 and 5), indicating MALAT-1 mainly functions through serving as a sponge for miR-22 in EOC.
c-Myc, located at chromosome 8q24, is a crucial oncogene which is under the regulation of Wnt signaling. It is well-established that c-myc supports tumor progression through modulating cell transformation and metabolism [31]. c-myc has been evidenced to be involved in EMT of various tumors, such as ovarian cancer [32,33]. Our results demonstrated miR-22 could inhibit c-myc expression and thus down-regulate EMT-associated vimentin and snai1 but augment E-cadherin expression markers (Figure 4). Consistently, c-myc activity was considered to be modulated by miR-22 via targeting MYCBP in breast cancer cells [17]. Taken together, MALAT-1/miR-22 participated in EOC progression through c-myc-mediated EMT.
On the other hand, our present work also has some limitations. First, the prognostic values of MALAT-1 and miR-22 were not investigated. To address this issue, we will analyze the association between MALAT-1/miR-22 expression and the overall survival/disease-free survival of ovarian cancer patients in the future. Second, whether MALAT-1 is an ovarian cancer-specific onco-lncRNA, or it can also exert oncogenic role in multiple cancer types is still unknown. Finally, besides their direct influences on tumor cells, the role of MALAT-1/miR-22 in modulating tumor immunity in ovarian cancer also deserves further exploration.
In conclusion, we revealed for the first time that MALAT-1 targeted miR-22 in EOC cells, sponging its inhibitory effect on tumor growth and metastasis in vitro and in vivo through modulating c-myc-mediated EMT. Our findings provide a novel insight into mechanisms for malignant progression of EOC.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–24. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S, Wu SY, Ivan C, Ferracin M, Dennison JB, Millward NMZ, Nagaraja AS, Gharpure KM, McGuire M, Sam N, Armaiz-Pena GN, Sadaoui NC, Rodriguez-Aguayo C, Calin GA, Drapkin RI, Kovacs J, Mills GB, Zhang W, Lopez-Berestein G, Bhattacharya PK, Sood AK. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13:2395–2402. doi: 10.1016/j.celrep.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao MY, Qiu Y, Yang B, Sun Li, Hei K, Du X, Li YM. Long non-coding RNAs involved in gynecological cancer. Int J Gynecol Cancer. 2014;24:1140–1145. doi: 10.1097/IGC.0000000000000212. [DOI] [PubMed] [Google Scholar]
- 5.Ji P, Diederichs S, Wang WB, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 6.Zhao ZY, Chen CJ, Liu Y, Wu CF. 17β-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun. 2014;445:388–393. doi: 10.1016/j.bbrc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 8.Shen LQ, Chen L, Wang YS, Jiang XC, Xia HP, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 9.Wu LQ, Wang XY, Guo Y. Long non-coding RNA MALAT1 is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp Ther Med. 2017;13:3055–3060. doi: 10.3892/etm.2017.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen QJ, Su YY, He XP, Zhao WA, Wu CX, Zhang WB, Si XM, Dong BW, Zhao LY, Gao YF, Yang XW, Chen JH, Lu J, Qiao XM, Zhang YC. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett. 2016;12:1361–1366. doi: 10.3892/ol.2016.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21:3176–3184. [PubMed] [Google Scholar]
- 12.Ji WD, Sun B, Su CQ. Targeting MicroRNAs in cancer gene therapy. Genes. 2017;8:21. doi: 10.3390/genes8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JY, Li Y, Ding MM, Zhang HH, Xu XM, Tang JL. Molecular mechanisms and clinical applications of MIR-22 in regulating malignant progression in human cancer (Review) Int J Oncol. 2017;50:345–355. doi: 10.3892/ijo.2016.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi G, Picciotto M, Russo A, Giordano A, Adamo V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol. 2014;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- 15.Zhang GJ, Xia SS, Tian HP, Liu ZL, Zhou T. Clinical significance of miR-22 expression in patients with colorectal cancer. Med Oncol. 2012;29:3108–3112. doi: 10.1007/s12032-012-0233-9. [DOI] [PubMed] [Google Scholar]
- 16.Li F, Yao L, Zhang XH, Xia YH, Cheng J, Lou XL, Jiang QH. Expression of miRNA-22 in ovarian cancer and effect of miRNA-22 over-expression on SKOV-3 ovarian cancer cell proliferation, migration and invasion. Chin J Pathophysiol. 2016;32:2251–2255. [Google Scholar]
- 17.Xiong J, Du Q, Liang Z. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene. 2010;29:4980. doi: 10.1038/onc.2010.241. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci. 2017;21:3598–3604. [PubMed] [Google Scholar]
- 19.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Dubeau L. The cell of origin of ovarian epithelial tumours. Lancet Oncol. 2008;9:1191–1197. doi: 10.1016/S1470-2045(08)70308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martini P, Paracchini L, Caratti G, Mello-Grand M, Fruscio R, Beltrame L, Romualdi C. lncRNAs as novel indicators of patients’ prognosis in stage i epithelial ovarian cancer: a retrospective and multicentric study. Clin Cancer Res. 2017;23:2356–2366. doi: 10.1158/1078-0432.CCR-16-1402. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang QH, Bützow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Xiao HB, Tang K, Liu PJ, Chen K, Hu JH, Zeng J, Xiao W, Yu G, Yao WM, Zhou H, Li H, Pan YT, Li AP, Ye ZQ, Wang J, Xu H, Huang QH. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XY, Li M, Wang ZQ, Han SC, Tang XH, Ge YX, Zhou LQ, Zhou CC, Yuan QP, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao F, Hu H, Yuan CC, Wang L, Jiang WH, Jin ZL, Guo Z, Wang LW. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 27.Kong LM, Liao CG, Zhang Y, Xu J, Li Y, Huang W, Chen ZN. A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014;74:3764. doi: 10.1158/0008-5472.CAN-13-3555. [DOI] [PubMed] [Google Scholar]
- 28.Zhang HH, Tang JL, Li C, Kong JL, Wang JY, Wu YH, Xu EP, Lai MD. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Letters. 2015;356:781–790. doi: 10.1016/j.canlet.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 29.Wan WN, Zhang YQ, Wang XM, Liu YJ, Zhang YX, Que YH, Li P. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol. 2014;9:178. doi: 10.1186/s13000-014-0178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Liang SH, Yu HL, Zhang J, Ma D, Lu X. An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecol Oncol. 2010;119:543–548. doi: 10.1016/j.ygyno.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho KB, Cho MK, Lee WY, Kang KW. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 2010;293:230–239. doi: 10.1016/j.canlet.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Vergara D, Merlot B, Lucot JP, Collinet P, Vinatier D, Fournier I, Salzet M. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett. 2010;291:59–66. doi: 10.1016/j.canlet.2009.09.017. [DOI] [PubMed] [Google Scholar]