Highlights
-
•
Two subtypes with distinct default mode network profiles exist in major depression.
-
•
Subtypes of major depression are robust in validation datasets across brain atlases.
-
•
Hyper- & hypo-connectivity DMN subgroups have comparable clinical symptom variables.
-
•
Future studies should examine whether two subtypes have differing treatment response.
Keywords: Biotypes, Default mode network, Major depressive disorder, Resting-state fMRI, Machine learning
Abstract
Background
Major depressive disorder (MDD) is heterogeneous disorder associated with aberrant functional connectivity within the default mode network (DMN). This study focused on data-driven identification and validation of potential DMN-pattern-based MDD subtypes to parse heterogeneity of the disorder.
Methods
The sample comprised 1397 participants including 690 patients with MDD and 707 healthy controls (HC) registered from multiple sites based on the REST-meta-MDD Project in China. Baseline resting-state functional magnetic resonance imaging (rs-fMRI) data was recorded for each participant. Discriminative features were selected from DMN between patients and HC. Patient subgroups were defined by K-means and principle component analysis in the multi-site datasets and validated in an independent single-site dataset. Statistical significance of resultant clustering were confirmed. Demographic and clinical variables were compared between identified patient subgroups.
Results
Two MDD subgroups with differing functional connectivity profiles of DMN were identified in the multi-site datasets, and relatively stable in different validation samples. The predominant dysfunctional connectivity profiles were detected among superior frontal cortex, ventral medial prefrontal cortex, posterior cingulate cortex and precuneus, whereas one subgroup exhibited increases of connectivity (hyperDMN MDD) and another subgroup showed decreases of connectivity (hypoDMN MDD). The hyperDMN subgroup in the discovery dataset had age-related severity of depressive symptoms. Patient subgroups had comparable demographic and clinical symptom variables.
Conclusions
Findings suggest the existence of two neural subtypes of MDD associated with different dysfunctional DMN connectivity patterns, which may provide useful evidence for parsing heterogeneity of depression and be valuable to inform the search for personalized treatment strategies.
1. Introduction
Major depressive disorder (MDD) is one of the most common mental disorders and is a heterogeneous entity characterized by depressed mood, loss of pleasurable feelings or interest, and anhedonia (Goldberg, 2011). Mounting studies suggest that patients with MDD are associated with altered functional connectivity (FC) within the default mode network (DMN) (Goya-Maldonado et al., 2016, Kaiser et al., 2015). In patients with depression, hyperconnectivity of DMN is closely related to the disordered self-referential thought and maladaptive rumination (Berman et al., 2011, Sheline et al., 2009, Yan et al., 2019). Persistent increased FC within the anterior subnetwork of the DMN may increase risk for relapse in recovered MDD patients (Li et al., 2013). In addition, baseline centrality of posterior DMN may predict early improvement following short-term antidepressant therapy for patients with MDD (Shen et al., 2015). By contrast, reduced FC of the DMN is associated with symptom severity as reported in patients with recurrent MDD (Yan et al., 2019). Inconsistencies among previous reports of DMN alterations for patients with MDD may potentially reflect the heterogeneity of this disorder (Williams, 2016). Elucidation of the relationships of specific patterns of aberrant connectivity of DMN to MDD may contribute to our understanding of the neuropathophysiological mechanisms underlying heterogeneity of symptoms in this complex disorder (Berman et al., 2011, Kaiser et al., 2015).
As focal element of the NIMH research domain criteria (RDoC), differentiation of brain functional dysconnectivity patterns to clinical diagnosis may enable further relevant description and refinement of subtypes of MDD (Beijers et al., 2019, Insel et al., 2010). It is established that clinical symptomatology and putative biological substrates for the diagnosis of depression are variable and inconsistent at the individual patient level (Drysdale et al., 2017). Preclinical experimental studies suggest that brain connectivity-based subtypes of depression correlate with an atypical connectivity pattern which may be masked by central tendency measures in traditional group-based analysis (Price et al., 2017a). Encouraging findings have been reported for connectivity-based depression biotypes in the study of Drysdale and colleagues, as four subtypes of depression associated with differing patterns of aberrant brain connectivity, and one subtype effectively responded to targeted neurostimulation therapies (Drysdale et al., 2017). The putative subtypes of neural circuit dysfunction travel across conventional diagnostic categories, and may track with clinically relevant phenotypes of depression (Price et al., 2017a, Price et al., 2017b). The ability to parse the heterogeneity of MDD may yield biologically based subgroups that will increase our ability to predict clinical outcomes and elucidate underlying disease mechanisms (Williams, 2016). This promising approach to heterogeneity at the neuroimaging level is hampered by small sample size of currently available reports. Studies using large sample sizes and exploring biotypes of MDD with differing DMN patterns are limited but much needed.
Application of multivariate modeling and machine learning algorithms to neuroimaging, provides unprecedented opportunities for large-scale imaging studies of pooled data across multiple sites, to identify brain phenotypes that more closely map for MDD the likely underpinnings of neuropsychopathology (Smith and Nichols, 2018, Woo et al., 2017). By increasing the numbers of participants across sites, data pooling may boost statistical power and probably accelerate progress in brain mapping (Costafreda, 2009). Although aggregating data across multiple sites with different scanners and acquisition protocols can increase sample heterogeneity, multi-site data could delineate a more comprehensive aberrant pattern of the disease with a more representative sample of participants (Ma et al., 2018, Suckling et al., 2010). The present study was initiated from the REST-meta-MDD Project in China (Yan et al., 2019).
We conducted a data-driven, brain-based categorization approach to resting-state FC within the DMN in patients with MDD from multi-site datasets, to parse the neural circuit dysfunctions that distinct typed of depression within traditional diagnostic boundaries. An independent single-site (unseen) dataset was used to validate the DMN-pattern-based MDD subtypes identified in multi-site discovery (training) dataset. Reproducible MDD subgroups with differing patterns of DMN were validated across different sites and diverse populations. In this way, we have applied machine learning to elucidate the depression biotypes with dysconnectivity patterns of DMN, which may eventually guide more effective personalized treatment.
2. Material and methods
2.1. Participants
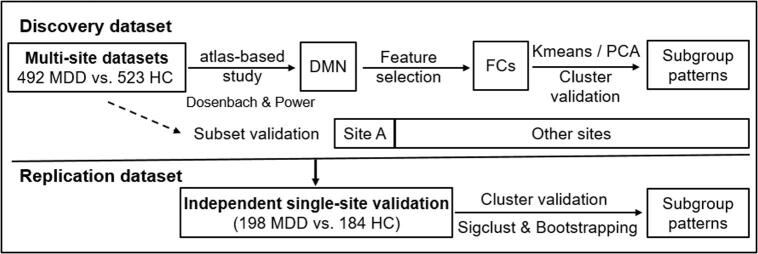
This study was based on the REST-meta-MDD Project of resting-state fMRI initiated in China. Patients diagnosed with first-episode or recurrent MDD were recruited. Symptom severity was evaluated using the Hamilton Depression Rating Scale (HAMD). All participants provided informed consent in accordance with requirement of the ethics committee of the local Institutional Review Boards. In this study, the overall dataset comprised 1397 participants including 690 patients with MDD and 707 HC registered from thirteen clinical sites. Steps for data cleansing and quality control are listed in Fig. S1. Subject demographics are displayed in Table 1. The discovery (training) dataset included 492 patients with MDD and 523 HC from twelve clinical sites to select features and identify subgroups in relation to DMN patterns in patients with MDD. The replication (unseen) dataset had 198 patients from an independent single site to further validate the MDD subtypes with differing DMN patterns.
Table 1.
Demographic characteristics of participants.
| Sites | Total | MDD (F) | HC (F) | Age MDD | Age HC | EDL MDD | EDL HC | Illness duration (months) | HAMD total scores | FED/non-FED/unknown | Medication (Y/N) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery | |||||||||||
| Site A | 318 | 149 (92) | 169 (103) | 28.05 (8.87) | 24.99 (4.92) | 13.80 (3.20) | 15.66 (2.66) | 7.00 | 22.34 (4.43) | 90 / 54 / 0 | 41 / 108 |
| Site B | 104 | 39 (32) | 65 (39) | 31.36 (10.07) | 29.37 (10.42) | 11.11 (2.89) | 13.05 (2.39) | 8.00 | 26.15 (6.22) | 36 / 1 / 2 | 11 / 28 |
| Site C | 95 | 63 (42) | 32 (17) | 30.49 (7.14) | 29.59 (5.00) | 13.70 (3.37) | 14.59 (2.82) | 6.00 | 21.29 (3.45) | 63 / 0 / 0 | 0 / 63 |
| Site D | 93 | 31 (18) | 62 (35) | 31.03 (9.79) | 34.95 (11.85) | 12.06 (2.63) | 13.13 (2.05) | 32.04 | 20.94 (4.96) | 2 / 29 / 0 | NA |
| Site E | 138 | 71 (40) | 67 (39) | 31.42 (8.09) | 30.76 (7.89) | 13.90 (2.91) | 15.16 (2.27) | 3.00 | 24.82 (4.85) | NA | NA |
| Site F | 40 | 20 (17) | 20 (13) | 38.60 (10.51) | 44.25 (11.58) | 11.75 (3.88) | 10.80 (4.83) | 11.50 | 23.25 (2.59) | 11 / 9 / 0 | 19 / 1 |
| Site G | 47 | 23 (12) | 24 (12) | 32.26 (7.57) | 28.75 (4.58) | 14.22 (3.79) | 14.79 (2.75) | 4.00 | 21.91 (3.03) | 23 / 0 / 0 | NA |
| Site H | 26 | 11 (7) | 15 (9) | 27.18 (8.50) | 28.47 (10.89) | 12.91 (4.11) | 14.67 (3.54) | 36.00 | 23.27 (6.54) | 6 / 5 / 0 | 0 / 11 |
| Site I | 65 | 28 (15) | 37 (22) | 37.68 (8.97) | 35.81 (9.52) | 11.63 (4.01) | 16.09 (3.25) | 18.00 | 22.82 (4.16) | 22 / 6 / 0 | 22 / 6 |
| Site J | 12 | 6 (4) | 6 (2) | 28.83 (13.21) | 26.83 (2.86) | 12.50 (2.43) | 17.00 (1.55) | 25.00 | 22.50 (3.62) | 2 / 4 / 0 | 0 / 6 |
| Site K | 35 | 29 (24) | 6 (1) | 34.21 (9.35) | 31.33 (2.25) | 12.72 (2.83) | 13.67 (0.82) | 12.00 | 24.28 (4.71) | 19 / 10 / 0 | 0 / 29 |
| Site L | 42 | 22 (12) | 20 (8) | 35.18 (10.30) | 24.35 (7.07) | 11.68 (2.82) | 13.30 (2.13) | 25.50 | 23.45 (5.60) | NA | 20 / 2 |
| Replication | |||||||||||
| Site M | 382 | 198 (132) | 184 (121) | 35.70 (10.11) | 32.40 (11.99) | 11.29 (3.18) | 13.95 (3.61) | 24.00 | 21.90 (4.49) | 163 / 35 / 0 | 82 / 116 |
| Total | 1397 | 690 (479) | 707 (421) | 32.45 (9.76) | 30.43 (10.22) | 12.54 (3.38) | 14.41 (3.20) | – | – | – | – |
MDD, patients with major depressive disorder. HC, healthy controls. F, female. EDL, years of educational level. FED, first-episode depression. Median duration of illness. Mean (Standard Deviation). NA, not available. The thirteen sites were respectively located in: A, the West China Hospital of Sichuan University, Chengdu. B, the First Affiliated Hospital of China Medical University, Shenyang. C, the Second Xiangya Hospital of the Central South University, Changsha. D, the Beijing Anding Hospital of Capital Medical University, Beijing. E, the Peking University Sixth Hospital, Beijing. F, the Affiliated Guangji Hospital of Soochow University, Suzhou. G, the Second Xiangya Hospital of Central South University, Changsha. H, the Shanghai Mental Health Center, Shanghai Jiao Tong University, Shanghai. I, the Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou. J, the First Affiliated Hospital of Chongqing Medical University, Chongqing. K, the First Affiliated Hospital of Chongqing Medical University, Chongqing. L, the Second Xiangya Hospital of Central South University, Changsha. M, the Southwest University, Chongqing.
2.2. Image acquisition & data preprocessing
fMRI data were preprocessed according to a standardized preprocessing protocol on Data Processing Assistant for Resting Resting-State fMRI (DPARSF) (Yan and Zang, 2010). Preprocessing steps mainly included: discard the first 10 volumes, slice-timing correction, realignment, coregistration, normalization and nuisance regression. Data preprocessing code are available on the https://github.com/Chaogan-Yan/DPARSF. Here, we used the default preprocessing parameters. Nuisance signals including the Friston-24 head motion parameters, white matter, cerebrospinal fluid and whole brain global signal were regressed out for each individual (Friston et al., 1996). To minimize head motion effects, subjects with mean framewise displacement larger than 0.3 mm were excluded from the analysis (Power et al., 2012).
2.3. Atlas-based analysis
For each participant, rs-fMRI data, brain regions of DMN were defined as 5-mm radius spheres placed at coordinates based on the Dosenbach 160 atlas (Dosenbach et al., 2010). The time series of thirty-four brain regions were extracted for the DMN, and temporal correlation coefficients of the network were converted to z-values by using Fisher’s r-to-z transformation. Multiple linear regression was used to control for age-, sex- and site-related effects on FC (34*35/2 = 595 connections) by regressing the z-values on the subjects' ages and dummy variables for sex and sites (Drysdale et al., 2017). We also computed the FC of fifty-eight nodes within the DMN (58*59/2 = 1711 connections) based on the Power atlas (Power et al., 2011). Fig. 1 illustrates the flowchart for the data analysis strategy. Analyses were computed in Python.
Fig. 1.
Data analysis flowchart.
2.4. Data analyses for biotypes
2.4.1. Feature selection
In the discovery dataset, FC with low variance were considered as irrelevant features. VarianceThreshold (as a baseline dimensionality reduction algorithm) was used to remove low-variance FC (here threshold = 0.01), and then SelectFdr (f_classif) was applied to identify significant FC to differentiate patients with MDD from HC with false discovery rate (FDR) correction (PFDR < 0.05). Based on the Dosenbach atlas, twenty-eight FC within the DMN were selected as brain features. The selected FC are shown in Table S1 & Fig. S2.A-B.
2.4.2. Cluster analysis and cluster validation
Using the identified FC of DMN, K-means clustering was conducted for class formation in patients with MDD. To evaluate clustering validity, Calinski-Harabasz (CH) score and silhouette score were used to determine the optimal cluster number with the largest numerical values. Finally, principal component analysis (PCA) was applied to visualize the clustering of patient subgroups. See details in the Supplementary material.
2.4.3. Independent replication dataset validation
To validate DMN-pattern-based MDD subtypes defined in the discovery dataset, the selected FC within the DMN were extracted from the replication dataset - an independent cohort. Cluster analysis and cluster validation, as described above, were then applied to the replication dataset to explore DMN-pattern-based patient subgroups. Using the mean FC (averaged across the selected connections), Pearson correlation and multiple comparisons were applied between patient subgroups in the discovery and replication datasets.
2.4.4. Subset dataset validation
Here we conducted further analysis in subsets of the discovery dataset, to confirm the patient subgroups based on the dataset of site A and a subset dataset comprising the remaining discovery sites. Patients with MDD in site A were either first-episode drug-naive, or exhibited recurrent episodes but had not taken antidepressants for at least three months.
2.4.5. Statistical significance of clustering
To assess significance of clustering, a SigClust approach was conducted to test whether a cluster followed a single Gaussian distribution (Liu et al., 2008). Here, an R package SigClust was used. We also tested the significance of clustering with bootstrapping method. The average of CH scores with the optimal cluster based on the original dataset was plotted in the bootstrapping distribution. See details in the Supplementary material.
2.4.6. Statistical analysis
Demographic characteristics (age and education level) and severity of clinical symptoms (HAMD total score and subscale scores) were compared between patient subgroups using two-sample t tests. Categorical variables were compared between patient subgroups using χ2 test. Illness durations were compared using Mann–Whitney U tests. Pearson correlation was used to measure the association between HAMD total score, illness duration, age and the selected FC, and effect size (ES) was calculated between patient subgroups. All statistical test results were considered significant with PFDR < 0.05.
3. Results
3.1. MDD subgroups with differing DMN patterns identified in the discovery dataset
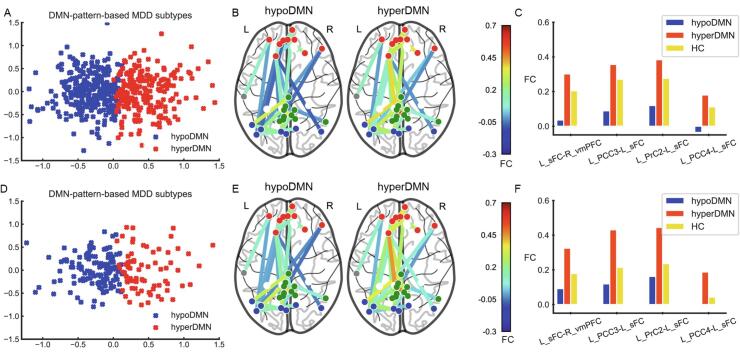
In the discovery dataset, based on the selected FC within the DMN, the cluster result achieved the maximum for the CH score and silhouette score, when the cluster number was equal to two (see Fig. S3.A & Fig. S4.A). Results of the SigClust and bootstrapping also suggested that two clusters best represented the underlying data structure (see Fig. S5.A & Fig. S6). Clustering subgroups were visualized by PCA (see Fig. 2.A). Subsequent analyses mainly focused on these two patient subgroups with differing DMN patterns. In the discovery dataset, subgroup 1 had 288 patients (58.54%), and subgroup 2 had 204 patients (41.46%). Relative to HC, subgroup 1 was characterized by decreased FC in the DMN (hypoDMN), while for subgroup 2 hyperconnectivity was predominant (hyperDMN). Compared to the hypoDMN, the hyperDMN subgroup showed relatively increased FC within the DMN, especially between the left superior frontal cortex (sFC) and left precuneus cortex (PrC), left sFC and left posterior cingulate cortex (PCC), and left sFC and right ventral medial prefrontal cortex (vmPFC) (see Fig. 2.C & Table S2).
Fig. 2.
DMN-Pattern-Based MDD subtypes in discovery and replication datasets. (A). Clusters are plotted using PCA in discovery dataset. Two-dimensional principal subspace for patient subgroups. The X axis represents the value of first principal component that accounts for the largest possible variance in the dataset. The Y axis represents the value of second principal component. (B). Patient subgroups in the discovery dataset. hypoDMN, subgroup with hypoconnectivity within the DMN. hyperDMN, subgroup with hyperconnectivity within the DMN. (C). FDR-corrected FC between each pair of groups (hypoDMN, hyperDMN and HC) in the discovery dataset. Blue bar, hypoDMN. Red bar, hyperDMN. Yellow bar, HC. (D). Clusters are plotted using PCA in the replication dataset. (E). Patient subgroups in the replication dataset. (F). FDR-corrected FC between each pair of groups (hypoDMN, hyperDMN and HC) in the replication dataset. In (B) & (E), red dots represent frontal regions. Grey dots represent temporal regions. Green dots represent PCC and PrC. Blue dots represent occipital and parietal regions. In (C) & (F), sFC, superior frontal cortex. PCC, posterior cingulate cortex. PrC, precuneus. vmPFC, ventral medial prefrontal cortex. L, left. R, right. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
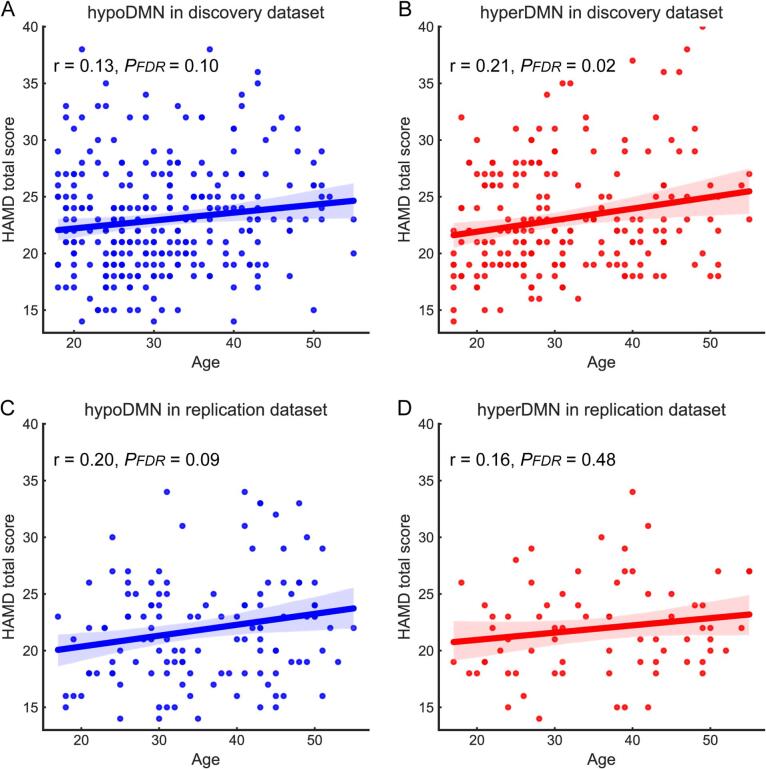
In the correlation analysis, age was positively correlated with HAMD total score (r = 0.21, PFDR = 0.02) in the hyperDMN subgroup, but not in the hypoDMN (see Fig. 3. A&B). However, there was no significant linear relationship between the FC and clinical variables in each patient subgroup. No statistically significant differences were found between patient subgroups in age, sex, years of educational level, illness duration, and first-episode or not, medicated or not, HAMD total scores or subscale scores (see Table S3).
Fig. 3.
Correlation between age and HAMD total score for each patient subgroup. (A). hypoDMN subgroup in the discovery dataset (B). hyperDMN in the discovery dataset (C). hypoDMN in the replication dataset (D). hyperDMN in the replication dataset. r, correlation coefficient. FDR, false discovery rate. HAMD, Hamilton Depression Rating Scale.
3.2. DMN-pattern-based MDD subtypes validated in the replication dataset
In the replication dataset, using the selected FC of the DMN based on the Dosenbach atlas, a cluster number of two achieved the maximum for the CH score and silhouette score (see Fig. S3.B & Fig. S4.B). Results of the SigClust and bootstrapping also suggested that two clusters were the best fit for the data structure (see Fig. S5.B & Fig. S7). The DMN-pattern-based patient subgroups were also visualized by PCA (see Fig. 2.D). In the replication dataset, 127 patients (64.14%) were placed in the hypoDMN subgroup and 71 patients (35.86%) were placed in the hyperDMN subgroup. Using the FC within the DMN identified in the discovery dataset, patient subgroups were validated in the replication dataset – the hypoDMN characterized with hypoconnectivity, the hyperDMN marked by increased FC (see Fig. 2.E & Table S4).
In the correlation analysis, age was positively correlated with illness duration in the hypoDMN subgroup (r = 0.29, PFDR = 0.01), but not in the hyperDMN. However, no significant linear correlation was found between age and other variables in each patient subgroup (see Fig. 3 C&D). There were no significant differences between these patient subgroups with respect to demographic and clinical symptom variables in the replication dataset (see Table S5).
3.3. Patient subgroups between discovery and replication datasets
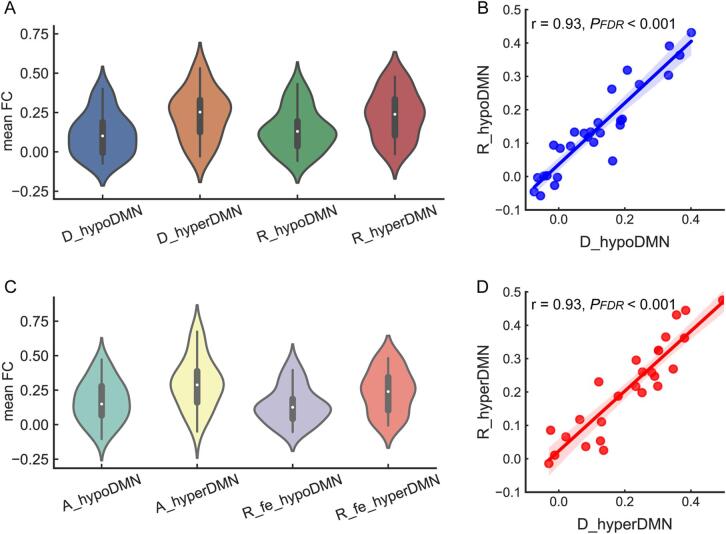
Based on the mean of selected twenty-eight connections, significant differences were observed between the two subgroups in the discovery dataset (ES = 0.82, PFDR = 0.01) and the replication dataset (ES = 0.65, PFDR = 0.03), respectively. Compared to the hypoDMN subgroup in the discovery dataset: there were no differences in the mean FC between the hypoDMN subgroup in the replication dataset (ES = 0.20, PFDR = 0.55), but significant differences between the hyperDMN in the replication dataset (ES = 0.84, PFDR = 0.01). Relative to hyperDMN subgroup in the discovery dataset, no significant differences in the mean FC were detected in the hyperDMN subgroup in the replication dataset (ES = 0.01, PFDR = 0.99), but there were differences in the mean FC between the hypoDMN subgroup in the replication dataset (ES = 0.64, PFDR = 0.03). The mean FC for each respective subgroup is displayed using a violin plot in Fig. 4.A.
Fig. 4.
Comparisons of mean FC within the DMN for patient subgroups (based on Dosenbach atlas). A. The violin plot for patient subgroups in the discovery and replication datasets. The X axis represents subgroups in these two datasets. D_hypoDMN and D_hyperDMN for subgroups in the discovery dataset. R_hypoDMN and R_hyperDMN for subgroups in the replication dataset. The Y axis represents mean FC values. B. correlation of mean FC between patient subgroups with hypoconnectivity of the DMN (hypoDMN) in the discovery and replication datasets. C. correlation of mean FC between patient subgroups with hyperconnectivity of the DMN (hyperDMN) in these two datasets. D. The violin plot for patient subgroups in the site A and first-episode MDD subset of replication dataset. The X axis represents subgroups in these two datasets. A_hypoDMN and A_hyperDMN for subgroups in the dataset of site A. R_fe_hypoDMN and R_fe_hyperDMN for subgroups in the first-episode MDD subset of replication dataset. The Y axis represents the mean FC.
In relation to the mean FC, the hypoDMN patient subgroups in the discovery and replication datasets were highly correlated (r = 0.93, PFDR < 0.001). The hyperDMN subgroup in the discovery dataset was highly correlated with the hyperDMN subgroup in the replication dataset (r = 0.93, PFDR < 0.001). See Fig. 4.B & D.
3.4. Repeatability of DMN-pattern-based MDD subtypes in subset analysis
Based on the selected FC, an optimal cluster number of two best represented the data structure for the dataset of site A - a subset of discovery dataset (see Fig. S3.C & Fig. S4.C). The DMN-pattern-based patient subgroups were also validated in the dataset of site A (see Fig. S8.A-C & Table S6). A violin plot was used to display the distribution for patient subgroups of site A and first-episode MDD subset of replication dataset (See Fig. 4C). In the dataset of site A, patients of hypoDMN tended to have higher anxiety scores in the HAMD subscale than those of hyperDMN subgroup (ES = 0.39, PFDR = 0.07). However, no statistically significant differences in demographic and clinical variables were detected between patient subgroups (see Table S7).
3.5. Repeatability of DMN-pattern-based MDD subtypes based on power atlas
Based on the DMN of the Power atlas, thirty-seven FC within the DMN were selected as features for subtyping patients with MDD. The selected FC are shown in Fig. S2.C-D. The cluster analysis for the CH score and silhouette score are displayed in Fig. S3.D-F & Fig. S4.D-F. Results of the SigClust and bootstrapping also suggested that two clusters were optimal to represent the data structure both in the discovery and replication datasets (see Fig. S5.C-D, Fig. S9 & Fig. S10). Results of subtyping based on the Power atlas were further validated the existence of DMN-pattern-based patient subgroups (see Fig. S11-12).
4. Discussion
In the present study, we applied data-driven machine learning and multivariable statistics to multi-site datasets and identified two MDD subtypes with distinct connectivity patterns of DMN at rest. Patient subgroups with differing DMN patterns were validated in an independent single-site replication dataset across different brain atlases. Compared to HC, one patient subgroup was characterized by hyperconnectivity within the DMN, especially the connectivity between the prefrontal areas and PCC/PrC, and one subgroup had decreased connectivity. The hyperDMN subgroup had age-related severity of depressive symptoms in the discovery dataset, but lacking of the correlation in the replication dataset. In the subset analysis of site A – almost included first-episode drug-naive MDD patients, the hypoDMN subgroup tended to have more severe anxiety symptoms than the hyperDMN subgroup. However, in this study, there was no significant diverse regarding demographic and clinical variables between patient subgroups.
In the current study, the DMN-pattern-based patient subgroups displayed distinguished connectivity between prefrontal regions and PCC/PrC. The prefrontal areas and PCC/PrC are critical element regions of the DMN, which are involved in monitoring one’s own internal state and emotions, self-referential processing and memory consolidation (Buckner et al., 2008). Additionally, altered structural connectivity between these nodes of the DMN are also considered as the prominent neural features related to MDD (Korgaonkar et al., 2014). Furthermore, prior studies have suggested that antidepressants could selectively normalize hyperconnectivity of the posterior rather than anterior DMN, which suggests the DMN may be a potential therapeutic target and predictive biomarker in patients with MDD (Brakowski et al., 2017, Li et al., 2013). Patients with MDD could have increased connectivity within the DMN at the group level both at rest and during task performance (Berman et al., 2011, Goya-Maldonado et al., 2016, Kaiser et al., 2015, Sheline et al., 2009). Hyperconnectivity within the DMN could be a significant issue in the pathophysiology of both acute and chronic clinical manifestations of depression (Posner et al., 2013). Moreover, patients with MDD characterized by hyperconnectivity of the DMN at baseline were likely to benefit from short-term antidepressant treatment and achieve acute remission (Korgaonkar et al., 2019). By contrast, hypoconnectivity of the DMN was associated with severity of clinical symptoms and non-response to first-line antidepressants in patients with MDD (Korgaonkar et al., 2019, Price et al., 2017a, Price et al., 2017b, Yan et al., 2019). Prior research may provide a hint that subgroup of MDD with DMN hyperconnectivity are more likely to benefit acutely from antidepressant treatments than those with DMN hypoconnectivity. Future longitudinal studies are needed to examine this hypothesis and determine whether non-remitters of MDD would be clustered to the subtype with hypoconnectivity of the DMN. Parsing heterogeneity of the disorder and identifying treatments capable of addressing the subtypes of MDD with differing brain patterns may be particularly clinically impactful. Future longitudinal studies could accelerate the pace of biotypes guided treatment in depression.
Mounting studies attempt to parse the heterogeneity of depression and categorize subtypes to predict external measures of functioning, clinical outcomes and neurobiological underpinnings (Drysdale et al., 2017, Feder et al., 2017, Price et al., 2017a, Price et al., 2017b, Williams, 2016). Two subtypes of MDD with differing profiles of brain circuits were identified in a prior study, and the subgroup characterized by lacking DMN connectivity and increased dorsal anterior cingulate connectivity was associated with high rates of comorbid anxiety and recurrence (Price et al., 2017a). Subgroup of MDD with hyperconnectivity of brain pattern defined during the positive mood had decreased resting-state connectivity of the DMN and experienced higher self-reported symptoms (Price et al., 2017b). Two potential subgroups of MDD with distinct connectivity of depression-related brain regions were differed in symptom severity and disease duration (Feder et al., 2017). Results of the present study also suggest that patients with MDD mainly exhibit two subtypes with dysfunctional connectivity of the DMN, which are identified in a large multi-site sample and replicated in an independent single-site dataset. Our prior study also reported that subtype of MDD with widespread white-matter abnormalities had more neurocognitive deficit (Liang et al., 2019). Findings of subtypes with distinct profiles of brain abnormalities may reveal the potential neurobiological substrates of key sources of heterogeneity in depression (Price et al., 2017a, Williams, 2016). Additionally, inflammation and oxidative stress could also be implicated in differentiating the distinct subtypes of depression (Gonda et al., 2019). Moreover, subgroups of individuals with MDD characterized by differing brain abnormalities may reflect qualitatively distinct genetic factors influence in the illness trajectory. The latent genetic subtype of MDD defined by the whole-exome genotyping data had increased common genetic substrates associated with the disorder, and affected by paranoid symptoms and more anxiety (Yu et al., 2017).
Results of this study found that the hyperDMN subgroup had age-related severity of depressive symptoms in the discovery dataset, but not in the replication dataset. Previous studies have reported that hypoconnectivity within the DMN is correlated with age-related structural and cognitive changes in healthy individuals (Geerligs et al., 2015, Vidal-Piñeiro et al., 2014). Age and taking medications can be used to predict severity of depressive symptoms in midlife and older women with depression (Gathuru et al., 2015). Youth with posttraumatic stress disorder exhibited age-related increased connectivity within the DMN, which probably indicated that trauma exposure during childhood could have effects on the DMN (Patriat et al., 2016). Whether the childhood trauma or life events could mediate the relationship between age and depressive symptom severity in the hyperDMN subgroup would clearly require further testing in future studies. In this study, relative to the hyperDMN, subgroup of hypoDMN was likely to have more severe anxiety symptoms. Consistent with previous studies, decreased functioning of the DMN was associated with anxiety disorders and high trait anxiety (Price et al., 2017a, Sylvester et al., 2012). However, no statistical significances were detected in demographic and clinical variables between patient subgroups in the discovery and replication datasets. Given limited information, subgroups of MDD with differing DMN patterns appeared to have comparable demographic and clinical features. This might coincide with the prior perspective that biotypes with distinct brain-circuit-based patterns could be independent of specific clinical features, and not be clinically deterministic (Price et al., 2017b). Future studies could focus on a more fine-grained set of clinical features and neurocognitive assessments to better characterize the differences between the subgroups of MDD with differing DMN profiles.
Certain limitations should be addressed. First, the clinical information, such as comorbid conditions (disturbances in chronobiologic rhythms, obsessional traits and psychotic symptoms), early life stress, treatment details and the number of prior depressive episodes, was not fully recorded for each patient due to variations in data management practices across different research sites. This issue limited our power to analyze the effects of clinical variables on DMN-pattern-based MDD subtypes. We were unable to account for potential moderators such as medication. Second, the current study is based on cross-sectional analysis. It will be fruitful to conduct the longitudinal follow-up study that may help to develop reliable biomarkers for predicting treatment effects and enable prediction of prognosis of the subgroups. Third, the present study is based on the analyses of FC within the DMN. Whether the FC of other resting-state networks, or a consideration of inter-network analysis could be used to define the subtypes of MDD is unclear, but worthy of examination.
5. Conclusions
Using a data-driven approach with multi-site datasets, the present study provides robust evidence for MDD biotypes with distinct DMN patterns. The current results represent strong evidence for the identification and validation of two MDD subtypes with dysfunctional connectivity patterns of DMN, supporting a robust sub-categorization of depression. It is significant that the biotype classification is reproducible using data from different clinical recruitment sites and different MRI scanners. Taken together, our findings highlight the accelerating promise of parsing biological heterogeneity in major neuropsychiatric illnesses such as MDD based on neurological biomarkers. Our data-driven approach has the capacity to reveal heterogeneity within large-scale brain networks in depression that is overlooked in conventional group comparisons and eventually could inform the development of more tailored diagnoses and treatments.
6. Author statement
Chao-Gan Yan, Tao Li and Sugai Liang conceived of the presented idea. Sugai Liang and Chao-Gan Yan performed the computations. Tao Li and Deng Wei verified the analytical methods. Sugai Liang and Chao-Gan Yan designed the figures. Sugai Liang wrote the draft. Andrew J. Greenshaw, Qiang Wang and Xi-Nian Zuo reviewed and revised the manuscript. The REST-meta-MDD Project has been done at multiple research sites in China. Each author has contributed significantly to the manuscript, and the submitted work has been reviewed and approved by all of the authors. None of the authors have conflicts of interest to report regarding this manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors would like to thank all those who assisted with this project. The authors also recognize, with great appreciation, all the study participants who contributed to this project.
Role of Funding Source
This work was supported by the National Natural Science Foundation of China (81801326 to S.G.L., 81630030 to T.L., 81671774 to C.G.Y., 81630031 to C.G.Y., 81461168029 to T.L., and 81471740 to X.N.Z.), the National Key R&D Program of China (2017YFC1309902 to C.G.Y. & 2016YFC0904300 to T.L.), the National High-tech R&D Program of China (863 Program) (2015AA020513 to M.H.X.), the Hundred Talents Program and the 13th Five-year Informatization Plan (XXH13505) of Chinese Academy of Sciences, Beijing Municipal Science & Technology Commission (Z161100000216152 to C.G.Y. and Z171100000117016 to T.M.S.), Science and Technology Program of Zhejiang Province (2015C03037 to Y.F.Z.), Brain Research Special Program of Guangdong Science and Technology Department (2018B030334001 to T.L.), and 1.3.5 Project for disciplines of excellence, West China Hospital, Sichuan University (ZY2016103 and ZY2016203 to T.L. and X.H.M.). Dr. Xi-Nian Zuo was partly supported by the National R&D Infrastructure and Facility Development Program of China, “Fundamental Science Data Sharing Platform” (DKA2019-12-02-21), and the Guangdong Key Area R&D Program (2019B030335001). Funders of the study had no role in study design, data collection, data analysis, data interpretation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2020.102514.
Contributor Information
Chao-Gan Yan, Email: yancg@psych.ac.cn.
Tao Li, Email: litaohx@scu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Beijers L., Wardenaar K.J., van Loo H.M., Schoevers R.A. Data-driven biological subtypes of depression: systematic review of biological approaches to depression subtyping. Mol. Psychiatry. 2019;24:888–900. doi: 10.1038/s41380-019-0385-5. [DOI] [PubMed] [Google Scholar]
- Berman M.G., Peltier S., Nee D.E., Kross E., Deldin P.J., Jonides J. Depression, rumination and the default network. Soc. Cogn. Affect. Neurosci. 2011;6:548–555. doi: 10.1093/scan/nsq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakowski J., Spinelli S., Dorig N., Bosch O.G., Manoliu A., Holtforth M.G., Seifritz E. Resting state brain network function in major depression – depression symptomatology, antidepressant treatment effects, future research. J. Psychiatr. Res. 2017;92:147–159. doi: 10.1016/j.jpsychires.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Buckner R.L., Andrews-Hanna J.R., Schacter D.L. The brain's default network: anatomy, function, and relevance to disease. Ann. N. Y. Acad. Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Costafreda S.G. Pooling FMRI data: meta-analysis, mega-analysis and multi-center studies. Front. Neuroinform. 2009;3:33. doi: 10.3389/neuro.11.033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U., Nardos B., Cohen A.L., Fair D.A., Power J.D., Church J.A., Nelson S.M., Wig G.S., Vogel A.C., Lessov-Schlaggar C.N., Barnes K.A., Dubis J.W., Feczko E., Coalson R.S., Pruett J.R., Jr., Barch D.M., Petersen S.E., Schlaggar B.L. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale A.T., Grosenick L., Downar J., Dunlop K., Mansouri F., Meng Y., Fetcho R.N., Zebley B., Oathes D.J., Etkin A., Schatzberg A.F., Sudheimer K., Keller J., Mayberg H.S., Gunning F.M., Alexopoulos G.S., Fox M.D., Pascual-Leone A., Voss H.U., Casey B.J., Dubin M.J., Liston C. Erratum: Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat. Med. 2017;23:264. doi: 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder S., Sundermann B., Wersching H., Teuber A., Kugel H., Teismann H., Heindel W., Berger K., Pfleiderer B. Sample heterogeneity in unipolar depression as assessed by functional connectivity analyses is dominated by general disease effects. J. Affect. Disord. 2017;222:79–87. doi: 10.1016/j.jad.2017.06.055. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Williams S., Howard R., Frackowiak R.S., Turner R. Movement-related effects in fMRI time-series. Magn. Reson. Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Gathuru I.M., Odukoya O.K., Thorpe J.M. Under treatment of depression among midlife and older adults in the United States. Soc Pharm J. 2015;1(1) doi: 10.17795/spj-2343. [DOI] [Google Scholar]
- Geerligs L., Renken R.J., Saliasi E., Maurits N.M., Lorist M.M. A brain-wide study of age-related changes in functional connectivity. Cereb. Cortex. 2015;25:1987–1999. doi: 10.1093/cercor/bhu012. [DOI] [PubMed] [Google Scholar]
- Goldberg D. The heterogeneity of “major depression”. World Psychiatry. 2011;10:226–228. doi: 10.1002/j.2051-5545.2011.tb00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda X., Petschner P., Eszlari N., Baksa D., Edes A., Antal P., Juhasz G., Bagdy G. Genetic variants in major depressive disorder: from pathophysiology to therapy. Pharmacol. Ther. 2019;194:22–43. doi: 10.1016/j.pharmthera.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Goya-Maldonado R., Brodmann K., Keil M., Trost S., Dechent P., Gruber O. Differentiating unipolar and bipolar depression by alterations in large-scale brain networks. Hum. Brain Mapp. 2016;37:808–818. doi: 10.1002/hbm.23070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T., Cuthbert B., Garvey M., Heinssen R., Pine D.S., Quinn K., Sanislow C., Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Kaiser R.H., Andrews-Hanna J.R., Wager T.D., Pizzagalli D.A. Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiatry. 2015;72:603–611. doi: 10.1001/jamapsychiatry.2015.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar M.S., Fornito A., Williams L.M., Grieve S.M. Abnormal structural networks characterize major depressive disorder: a connectome analysis. Biol. Psychiatry. 2014;76:567–574. doi: 10.1016/j.biopsych.2014.02.018. [DOI] [PubMed] [Google Scholar]
- Korgaonkar M.S., Goldstein-Piekarski A.N., Fornito A., Williams L.M. Intrinsic connectomes are a predictive biomarker of remission in major depressive disorder. Mol. Psychiatry. 2019;25(7):1537–1549. doi: 10.1038/s41380-019-0574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Liu L., Friston K.J., Shen H., Wang L., Zeng L.L., Hu D. A treatment-resistant default mode subnetwork in major depression. Biol. Psychiatry. 2013;74:48–54. doi: 10.1016/j.biopsych.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Liang S., Wang Q., Kong X., Deng W., Yang X., Li X., Zhang Z., Zhang J., Zhang C., Li X.M., Ma X., Shao J., Greenshaw A.J., Li T. White matter abnormalities in major depression biotypes identified by diffusion tensor imaging. Neurosci. Bull. 2019;35:867–876. doi: 10.1007/s12264-019-00381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hayes D.N., Nobel A., Marron J.S. Statistical significance of clustering for high-dimension, low–sample size data. J. Am. Stat. Assoc. 2008;103:1281–1293. doi: 10.1198/016214508000000454. [DOI] [Google Scholar]
- Ma Q., Zhang T., Zanetti M.V., Shen H., Satterthwaite T.D., Wolf D.H., Gur R.E., Fan Y., Hu D., Busatto G.F., Davatzikos C. Classification of multi-site MR images in the presence of heterogeneity using multi-task learning. Neuroimage Clin. 2018;19:476–486. doi: 10.1016/j.nicl.2018.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriat R., Birn R.M., Keding T.J., Herringa R.J. Default-mode network abnormalities in pediatric posttraumatic stress disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2016;55:319–327. doi: 10.1016/j.jaac.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner J., Hellerstein D.J., Gat I., Mechling A., Klahr K., Wang Z., McGrath P.J., Stewart J.W., Peterson B.S. Antidepressants normalize the default mode network in patients with dysthymia. JAMA Psychiatry. 2013;70:373–382. doi: 10.1001/jamapsychiatry.2013.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.B., Gates K., Kraynak T.E., Thase M.E., Siegle G.J. Data-driven subgroups in depression derived from directed functional connectivity paths at rest. Neuropsychopharmacology. 2017;42:2623–2632. doi: 10.1038/npp.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.B., Lane S., Gates K., Kraynak T.E., Horner M.S., Thase M.E., Siegle G.J. Parsing heterogeneity in the brain connectivity of depressed and healthy adults during positive mood. Biol. Psychiatry. 2017;81:347–357. doi: 10.1016/j.biopsych.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline Y.I., Barch D.M., Price J.L., Rundle M.M., Vaishnavi S.N., Snyder A.Z., Mintun M.A., Wang S., Coalson R.S., Raichle M.E. The default mode network and self-referential processes in depression. Proc. Natl. Acad .Sci. U.S.A. 2009;106:1942–1947. doi: 10.1073/pnas.0812686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yao J., Jiang X., Zhang L., Xu L., Feng R., Cai L., Liu J., Wang J., Chen W. Sub-hubs of baseline functional brain networks are related to early improvement following two-week pharmacological therapy for major depressive disorder. Hum. Brain Mapp. 2015;36:2915–2927. doi: 10.1002/hbm.22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Nichols T.E. Statistical challenges in “big data” human neuroimaging. Neuron. 2018;97:263–268. doi: 10.1016/j.neuron.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Suckling J., Barnes A., Job D., Brenan D., Lymer K., Dazzan P., Marques T.R., MacKay C., McKie S., Williams S.R., Williams S.C., Lawrie S., Deakin B. Power calculations for multicenter imaging studies controlled by the false discovery rate. Hum. Brain Mapp. 2010;31:1183–1195. doi: 10.1002/hbm.20927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester C.M., Corbetta M., Raichle M.E., Rodebaugh T.L., Schlaggar B.L., Sheline Y.I., Zorumski C.F., Lenze E.J. Functional network dysfunction in anxiety and anxiety disorders. Trends Neurosci. 2012;35:527–535. doi: 10.1016/j.tins.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Piñeiro D., Valls-Pedret C., Fernández-Cabello S., Arenaza-Urquijo E.M., Sala-Llonch R., Solana E., Bargalló N., Junqué C., Ros E., Bartrés-Faz D. Decreased Default Mode Network connectivity correlates with age-associated structural and cognitive changes. Front. Aging Neurosci. 2014;6:256. doi: 10.3389/fnagi.2014.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.M. Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry. 2016;3:472–480. doi: 10.1016/S2215-0366(15)00579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.W., Chang L.J., Lindquist M.A., Wager T.D. Building better biomarkers: brain models in translational neuroimaging. Nat. Neurosci. 2017;20:365–377. doi: 10.1038/nn.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Zang Y. DPARSF: a MATLAB toolbox for“ pipeline” data analysis of resting-state fMRI. Front. Syst. Neurosci. 2010;4:13. doi: 10.1038/nn.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.G., Chen X., Li L., Castellanos F.X., Bai T.J., Bo Q.J., Cao J., Chen G.M., Chen N.X., Chen W., Cheng C., Cheng Y.Q., Cui X.L., Duan J., Fang Y.R., Gong Q.Y., Guo W.B., Hou Z.H., Hu L., Kuang L., Li F., Li K.M., Li T., Liu Y.S., Liu Z.N., Long Y.C., Luo Q.H., Meng H.Q., Peng D.H., Qiu H.T., Qiu J., Shen Y.D., Shi Y.S., Wang C.Y., Wang F., Wang K., Wang L., Wang X., Wang Y., Wu X.P., Wu X.R., Xie C.M., Xie G.R., Xie H.Y., Xie P., Xu X.F., Yang H., Yang J., Yao J.S., Yao S.Q., Yin Y.Y., Yuan Y.G., Zhang A.X., Zhang H., Zhang K.R., Zhang L., Zhang Z.J., Zhou R.B., Zhou Y.T., Zhu J.J., Zou C.J., Si T.M., Zuo X.N., Zhao J.P., Zang Y.F. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 2019;116:9078–9083. doi: 10.1073/pnas.1900390116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Arcos-Burgos M., Licinio J., Wong M.L. A latent genetic subtype of major depression identified by whole-exome genotyping data in a Mexican-American cohort. Transl. Psychiatry. 2017;7 doi: 10.1038/tp.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.