Abstract
Objective:
In recent years, research on microRNAs (miRNAs) associated with coronary artery disease (CAD) has attracted considerable attention. However, findings of these studies on the validity of circulating miRNAs in CAD diagnosis are controversial. A meta-analysis was therefore conducted to determine the potential value of miRNAs as biomarkers in CAD diagnosis.
Methods:
Relevant documents on miRNAs expression levels in the diagnosis of CAD were searched and collected from Pubmed, Embase, and Web of Science. They were collected from the time of inception of the database till January 31, 2020. A meta-analysis was conducted using Stata14.0 software. Forest maps were studied and a comprehensive evaluation of the diagnostic value of the expression levels of mRNAs in CAD was conducted using statistical indicators such as the summary receiver operating characteristic curve.
Results:
Overall, 14 studies were included, with 38 data sets, involving 29 miRNAs with 846 cases and 898 controls. The meta-analysis revealed that the average sensitivity and specificity of miRNAs for CAD diagnosis were 0.80 (0.75–0.84) and 0.78 (0.75–0.81), respectively. The positive likelihood, negative likelihood, and diagnostic odds ratios were 3.7 (3.1–4.4), 0.26 (0.21–0.33), and 14 (10–21), respectively, and the area under the curve was 0.85 (0.82–0.88). Subgroup analysis revealed that the accuracy in the Asian population was higher than that in the non-Asian population. Multiple miRNAs may be more diagnostically accurate than single miRNAs. MiRNAs in whole blood were more accurate than those in plasma, serum, and peripheral blood mononuclear cells. The diagnostic performance of the quantitative real-time polymerase chain reaction group was better than that of the qPCR group.
Conclusion:
According to our study, miRNAs may be a new, non-invasive diagnostic tool for the diagnosis of CAD. As a screening tool in clinical practice, it has potential diagnostic value and is worthy of clinical promotion. Considering the number and quality of the studies included in this meta-analysis, the above conclusion requires more quality research to verify it.
Keywords: diagnostic, microRNAs, biomarker, coronary artery disease, meta-analysis
Introduction
Coronary artery disease (CAD) is one of the diseases with the highest morbidity and mortality in the world and can seriously threaten human health. It is the most common type of organ disease caused by atherosclerosis. Many atherosclerotic processes such as inflammatory response, oxidative stress, hemodynamic changes, and endothelial cell damage are associated with CAD (1). At present, although there are some biomarkers of CAD such as interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), and interleukin-18 (IL-18) (2, 3), these often lack specificity. Thus, novel biomarkers are still required to achieve earlier, more accurate and sensitive detection, and provide clinicians with a basis for the early diagnosis and treatment of CAD.
MicroRNAs (miRNAs) are a class of small non-coding RNAs having a length of 18–22 nt. They can bind to the 3’ untranslated region of target mRNA for inhibiting translation of the mRNA at the posttranscriptional level and regulate the expression of corresponding target genes (4). New research also shows that miRNAs can bind to the 5’ end of target genes (5) and activate their expression (6, 7). MicroRNAs are involved in cell differentiation, growth, and apoptosis (8-8). In recent years, much research has shown that miRNAs have an important regulatory role in the occurrence and development of CAD (12-12). Coronary atherosclerosis mainly causes CAD. Once the unstable plaque ruptures, miRNAs in the cellular components of the plaque are released into the circulating blood. These cellular components may simultaneously also adsorb miRNAs in the circulating blood. The release of miRNAs from the myocardial tissue of the upper infarct into the blood causes abnormal expression of miRNAs in the circulating blood of patients with coronary heart disease (15-17). Therefore, the increase or decrease in miRNAs is very closely associated with CAD. To date, many abnormal expression levels of miRNAs have been found in the blood of CAD patients, particularly significantly increased miRNAs, such as miRNA-208a, miRNA-499, and miRNA-149, which may become diagnostic biomarkers of CAD (18, 19).
Aside from this, miRNAs can stably exist and be detected in the circulation, and blood samples can be conveniently collected. Due to their tissue specificity and different expression profiles in different diseases, circulating miRNAs are expected to become new non-invasive biomarkers for the diagnosis of CAD.
However, whether miRNAs can be used for the diagnosis of CAD is not completely clear. This topic is controversial at present. To further explore the relationship between miRNAs and CAD, this study quantitatively evaluated the relationship between miRNAs and CAD diagnosis through a meta-analysis so as to provide additional evidence to improve clinical treatment decisions.
Methods
This review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (20) and the recommendations of the Cochrane Collaboration (21).
Search strategy
Studies were retrieved from Pubmed, Embase, and Web of Science, from the time of inception of the database until January 31, 2020. The search terms included “coronary heart disease,” “coronary artery disease,” “miRNA,” “miR,” “miRNAs,” “microRNA,” “diagnos*,” “sensitivity and specificity,” “ROC curve,” etc. These were searched as keywords to comprehensively and systematically collect relevant literature. Simultaneously, published literature related to miRNAs diagnosis CAD was collected retrospectively.
Inclusion and exclusion criteria
Inclusion criteria: (1) Research subjects: Patients diagnosed with CAD by coronary angiography, including stable CAD (with or without angina), regardless of age, gender, and ethnicity; (2) CAD population as the observation group, and non-CAD disease or healthy population as the control group; (3) Research information is reliable; (4) Ability to extract the original four-cell table data, true positive (TP), false positive (FP), false negative (FN), true negative (TN); (5) Study types include diagnostic trials, case-control, retrospective, or prospective studies. Exclusion criteria: (1) The research object is non-human research; (2) Sensitivity and specificity analysis was not performed in the original text; (3) Studies without data such as reviews and meeting records; (4) Repeated publications.
Data extraction and quality assessment
Two independent reviewers screened the literature based on the inclusion and exclusion criteria and then further searched and read the full-text review. Cross-checking was completed, and group discussions were resolved when the opinions were inconsistent. The extracted content included: the first author, publication year, country, specimen, sample size, detection method, miRNA type, sensitivity, specificity, TPs, FPs, FNs, TNs, and AUC.
This study used QUADAS2 (22). Quality evaluation was performed on the included studies, and each item was assessed using “Yes,” ;”No”, and “Unclear”.
Statistical methods
Stata 14.0 (StataCorp, College Station, TX, USA) software was used for statistical analysis. TP, FP, FN, TN, combined sensitivity, and specificity of each study were summarized. A comprehensive summary receiver operating characteristic curve (SROC) was drawn and the area under the curve (AUC) was calculated. The Spearman correlation coefficient was used to evaluate heterogeneity caused by the threshold effect. A p value of <0.05 indicated that the heterogeneity was caused by the threshold effect and a p value of >0.05 indicated that the heterogeneity was caused by other reasons, which were further analyzed. Chi-squared (χ2) and I2 tests were used for statistical heterogeneity analysis: χ2 test for heterogeneity of each study and I2 test for qualitative heterogeneity. When I2>50% or p<0.1, it indicated that there was heterogeneity between the research results, and a random effect model was used. When I2<50% or p>0.1, it indicated that there was no heterogeneity among the studies, and a fixed effect model was used. The test level was α=0.05. Meta-regression and subgroup analysis were used to explore potential heterogeneity. A subgroup analysis was conducted based on ethnicity (Asian vs. non-Asian), detection methods for miRNAs (qPCR vs. qRT-PCR), sample sources (blood vs. plasma vs. serum vs. Peripheral blood mononuclear cells (PBMCs), miRNAs profiling (single miRNA vs. multiple miRNAs), and aberrant expression (upregulation vs. downregulation). The stability of the sensitivity analysis test results was evaluated. Deek’s funnel chart was used to assess for publication bias.
Results
Literature search
Figure 1 is a flow chart of study selection. A total of 283 articles were obtained. Endnote was used to delete 86 duplicate articles. After reading the title and abstract, 151 articles were found to be inconsistent with the topic and were excluded. The remaining articles were then read and 28 of them were excluded. Ultimately, 14 articles were eligible for inclusion (23-23).
Figure 1.
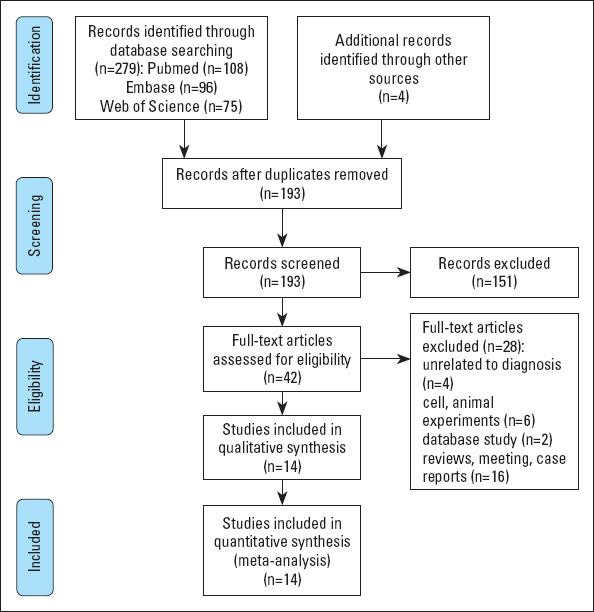
Flow diagram of the study selection for the present meta-analysis
Study characteristics
Fourteen studies were included(23-23), which contained 38 datasets, of which contained 38 datasets, of which 4 datasets analyzed multiple miRNAs for the joint diagnosis of CAD, and 34 data sets analyzed single miRNAs for the diagnosis of CAD. Different literatures contain the same miRNAs. Therefore, there are only 29 kinds of miRNAs (27). One study was conducted in Europe (24), 12 in Africa (23, 25, 26, 28-36), and one in Asia. The meta-analysis included 846 cases and 898 controls. These are listed in Table 1.
Table 1.
Characteristics of studies included in the present meta-analysis
| Study | Year | Country | Sample size (Patients/controls) | Mean age (Patients/controls) | Detected sample | miRNA | Detection method |
|---|---|---|---|---|---|---|---|
| Wu and Zhang (32) | 2018 | China | 119/96 | 59±11/57±10 | PBMCs | miRNA-126(down) | qPCR |
| Dong et al. (25) | 2017 | China | 161/149 | 61.35±7.10/61.08±7.51 | PBMCs | miRNA-24(up), miRNA-33a(up), miRNA-103a(up), miRNA-122(up) | qPCR |
| Quan et al. (29) | 2018 | China | 73/59 | 65.67±11.59/59.67±9.86 | Plasma | miRNA-146a | qRT-PCR |
| Amr et al. (24) | 2018 | Egypt | 46/20 | 57.0±6.2/58.1±1.1 | Blood | miRNA-126(down) | qRT-PCR |
| Faccini et al. (27) | 2017 | France | 69/32 | 58.4±9.0/57.3±11.6 | Plasma | let-7c(down), miRNA-145 (down), miRNA-155(down), | qRT-PCR |
| Guo et al. (28) | 2018 | China | 300/100 | 56.2 | Blood | miRNA-223 | qRT-PCR |
| Sayed et al. (30) | 2015 | China | 65/32 | 53 | Plasma | miRNA-149(down), miRNA-424,(down), miRNA-765(up) | qRT-PCR |
| Wang et al. (31) | 2014 | China | 92/34 | 65.2±10.5/59.4±13.1 | Serum | miRNA-487a(up), miRNA-29b(up), miRNA-502(up), | qRT-PCR |
| miRNA-208(up), miRNA-215(up) | |||||||
| Zhang et al. (35) | 2017 | China | 290/110 | 59.2 | Blood | miRNA-208a(up) | qRT-PCR |
| Zhang et al. (33) | 2018 | China | 102/92 | 59.6±9.7/57.2±8.5 | Plasma | miRNA-126(down), miRNA-210(down), miRNA-378(down), | qPCR |
| Zhou et al. (36) | 2015 | China | 67/67 | NA | Plasma | miRNA-260(up), miRNA-574-5p(up) | qRT-PCR |
| Zhang et al. (39) | 2020 | China | 88/67 | NA | Blood | miRNA-29a-3p(up), miRNA-574-3p(up), miRNA-574-5p (up) | qRT-PCR |
| Du et al. (26) | 2016 | China | 40/40 | 34.20+5.93/36.58+3.96 | Serum | miRNA-196b-5p (down), miRNA-3613-3p(down), | qRT-PCR |
| miRNA-145-3p (down), miRNA-190a-5p(down) | |||||||
| Ali Sheikh et al. (23) | 2015 | China | 69/20 | 72.53±4.31/71.7±5.2 | Plasma | miRNA-765(up), miRNA-149 | qRT-PCR |
qPCR - quantitative real-time PCR; qRT-PCR - quantitative reverse transcription–PC; PBMCs - peripheral blood mononuclear cells; NA - data not available
Quality assessment
The evaluators independently evaluated the quality of eligible studies according to the QUADAS-2 standard assessment, as shown in Table 2.
Table 2.
Methodological quality evaluation of the included literature
| Study | Risk of bias | Applicability | |||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Patient selection | Index test | Reference standard | |
| Sayed et al. (30) 2015 | L | U | U | L | L | U | U |
| Faccini et al. (27) 2017 | H | L | L | L | L | L | L |
| Zhang et al. (33) 2018 | H | U | L | L | U | U | L |
| Zhou et al. (36) 2015 | U | U | U | L | L | L | L |
| Dong et al. (25) 2017 | H | U | L | L | H | U | L |
| Amr et al. (24) 2018 | H | U | L | U | L | U | L |
| Guo et al. (28) 2018 | L | L | U | L | L | L | L |
| Du et al. (26) 2016 | H | L | L | L | U | L | L |
| Quan et al. (29) 2018 | H | U | L | L | L | L | L |
| Wang et al. (31) 2014 | L | U | U | L | L | U | L |
| Wu and Zhang (32) 2018 | H | H | U | L | L | L | L |
| Zhang et al. (35) 2017 | L | U | L | L | L | L | L |
| Zhang et al. (39) 2020 | L | U | L | L | L | L | L |
| Ali Sheikh et al. (23) 2015 | U | U | U | L | L | L | L |
U - unclear risk of bias; L - low risk of bias; H - high risk of bias
Diagnostic accuracy of circulating miRNAs in CAD
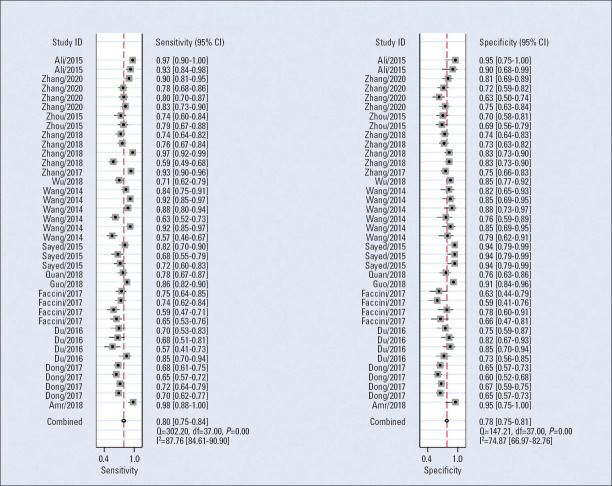
The Spearman correlation coefficient used for diagnostic results was 0.51 (p=0.26), and although the ROC plan did not show a “shoulder-arm” distribution, there was no threshold effect between studies. Heterogeneity test results found that there was significant heterogeneity between studies. The random effects model was used for meta-analysis. The average sensitivity and specificity of miRNAs for CAD diagnosis were 0.80 (0.75–0.84) and 0.78 (0.75–0.81), respectively. The positive likelihood, negative likelihood, and diagnostic odds ratios were 3.7 (3.1–4.4), 0.26 (0.21–0.33), and 14 (10–21), respectively, and the AUC was 0.85 (0.82–0.88), indicating that miRNAs have a reference value for CAD diagnosis. The corresponding sensitivity, specificity, AUC, and 95% CI are shown in Figures 2 and 3.
Figure 2.
Forest plots for studies on overall miRNAs used in the diagnosis of coronary artery disease among the 38 studies included in the meta-analysis
Figure 3.
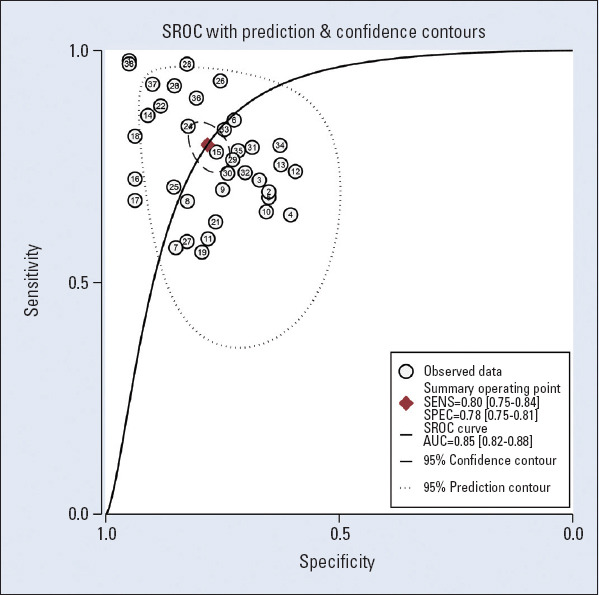
The summary receiver operating characteristic curves (SROCs) of circulating miRNAs for the diagnosis of coronary artery disease
Subgroup analysis and heterogeneity analysis
Subgroup analysis was based on ethnicity (Asian vs. non-Asian), detection methods for miRNAs (qPCR vs. qRT-PCR), sample sources (blood vs. plasma vs. serum vs. PBMCs), miRNAs profiling (single miRNA vs. multiple miRNAs), and aberrant expression (upregulation vs. downregulation). All results are shown in Table 3. Comparing circulating miRNAs in patients with different ethnicities, the accuracy in the Asian population (sensitivity 0.79, average specificity 0.75, diagnostic odds ratio 14.79, and AUC 0.86) was higher than that in the non-Asian population (sensitivity 0.73, average specificity 0.70, diagnostic odds ratio 6.61, and AUC 0.74).
Table 3.
Summary estimates of diagnostic criteria and their 95% confidence intervals
| Subgroup | n | SEN (95% CI) | SPE (95% CI) | DOR (95% CI) | AUC |
|---|---|---|---|---|---|
| Ethnicity | |||||
| Asian | 33 | 0.79 (0.77-0.80) | 0.75 (0.74-0.79) | 14.79 (10.32-21.20) | 0.86 |
| Non-Asian | 5 | 0.73 (0.67-0.77) | 0.70 (0.62-0.77) | 6.61 (2.71-16.12) | 0.74 |
| miRNA profiling | |||||
| Single miRNA | 34 | 0.78 (0.76-0.79) | 0.75 (0.73-0.77) | 13.6 (9.44-19.60) | 0.86 |
| Multiple miRNA | 4 | 0.81 (0.76-0.85) | 0.76 (0.69-0.81) | 13.26 (5.51-31.79) | 0.84 |
| Specimen | |||||
| Blood | 7 | 0.88 (0.85-0.90) | 0.78 (0.74-0.81) | 25.64 (11.61-56.61) | 0.91 |
| Plasma | 16 | 0.76 (0.74-0.79) | 0.77 (0.74-0.80) | 13.21 (7.98-21.86) | 0.86 |
| Serum | 10 | 0.77 (0.74-0.80) | 0.81 (0.77-0.85) | 15.53 (8.26-29.19) | 0.88 |
| PBMCs | 5 | 069 (0.66-0.72) | 0.67 (0.64-0.71) | 4.89 (3.14-7.63) | 0.75 |
| Altered miRNA | |||||
| Upregulation | 19 | 0.79 (0.77-0.81) | 0.72 (0.70-0.74) | 13.23 (8.09-21.65) | 0.85 |
| Downregulation | 15 | 0.75 (0.72-0.77) | 0.81 (0.78-0.83) | 14.11 (8.12-24.53) | 0.85 |
| Method | |||||
| qPCR | 9 | 0.72 (0.69-0.74) | 0.71 (0.68_0.74) | 7.39 (4.54-12.03) | 0.80 |
| qRT-PCR | 29 | 0.81 (0.79-0.83) | 0.78 (0.76-0.80) | 16.63 (11.16-24.78) | 0.87 |
| Overall | 0.80 (0.75-0.84) | 0.78 (0.75-0.81) | 14 (10-21) | 0.85 |
n - number ; Cl - confidence intervals; DOR - diagnostic odds ratio; SEN - sensitivity; SPE - specificity; AUC - area under curve
Compared with a single miRNA, the diagnosis of multiple miRNAs had a higher sensitivity and specificity. The sensitivity increased from 0.78 to 0.81, and the specificity increased from 0.75 to 0.76.
Based on the analysis of different sample sources, the miRNAs in blood was more accurate than the miRNAs in plasma, serum, and PBMC. The average specificity, diagnostic odds ratio, and AUC were 0.78, 25.64, and 0.91, respectively.
For the detection methods of miRNAs, the diagnostic performance of the qRT-PCR group (sensitivity 0.81, average specificity 0.78, diagnostic odds ratio 16.63, and AUC 0.87) was better than that of the qPCR group (sensitivity 0.72, average specificity 0.71, diagnostic odds ratio 7.39, and AUC 0.80).
In addition, we conducted further studies to determine if upregulated miRNAs had better diagnostic accuracy than downregulated miRNAs. The results showed that although the sensitivity (0.79 and 0.75, respectively) and specificity (0.72 and 0.81, respectively) were slightly different, the AUC of both groups was 0.85, and the upregulated and downregulated miRNAs may have similar diagnostic performance.
Due to the heterogeneity of this meta-analysis, regression analysis was conducted on factors such as ethnicity, miRNA profiling, altered miRNA, detection methods fo r miRNAs, and sample size. The results show that the sensitivity was affected by the detection method and aberrant expression, whereas specificity was affected by the test method, aberrant expression, and sample size. This is shown in Figure 4.
Figure 4.
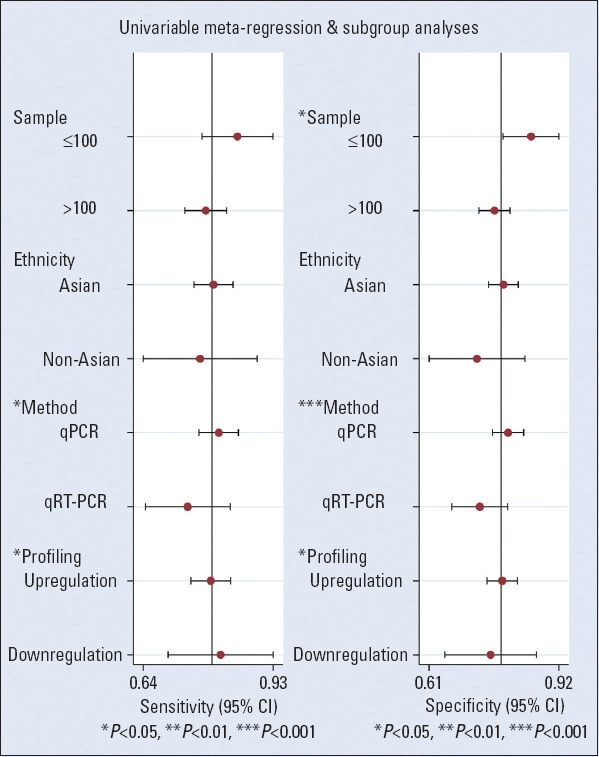
Univariate meta-regression and subgroup analyses for sensitivity and specificity of miRNAs for the diagnosis of coronary artery disease
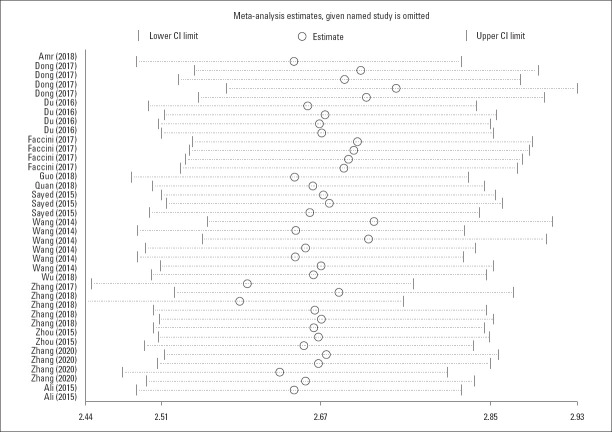
Sensitivity analysis
Sensitivity analysis tests the stability of the results of this meta-analysis. The results show that our results were stable and not affected by a single study. This has been shown in Figure 5.
Figure 5.
Sensitivity analysis for all eligible studies
Publication bias
Deek’s funnel plot asymmetry test explored the publication bias in diagnostic accuracy, and the results showed no publication bias (p=0.07). This is shown in Figure 6.
Figure 6.
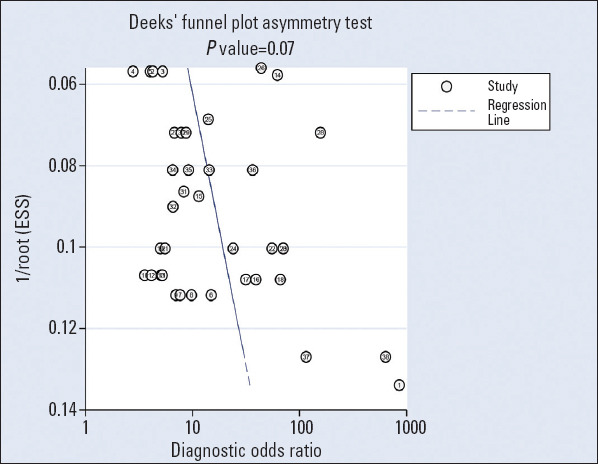
Deek’s funnel plots used to estimate publication bias for discrimination of miRNAs in patients with coronary artery disease
Discussion
CAD is a global health problem and the leading cause of death. How to improve the early diagnosis of CAD has been an important direction for cardiovascular disease research. Therefore, specific and sensitive CAD diagnostic markers are urgently required.
Many studies have found miRNAs in human peripheral blood and over 100 of these have been confirmed in serum (37). Subsequent research tended to use miRNAs as biomarkers for tumor diagnosis. Zheng et al. (38) found that miRNA-106b and miRNA-195 are good classifiers of hepatocellular carcinoma (HCC) and may have key roles in the progression of HCC. Studies have also shown that the AUC of miRNAs in diagnosing HCC is 0.991 (39). Some studies have shown that serum miRNA-21 is upregulated in patients with colorectal cancer (40, 41). Other studies have shown that miRNA-101 is downregulated in colorectal cancer (42). As the detection of miRNAs becomes increasingly sensitive and convenient, research on miRNAs has gradually turned to the field of clinical application. At present, research on new biomarkers indicates that miRNAs may play a key role in CAD because of the differences in miRNA expression levels observed between CAD patients and healthy controls. miRNAs are the main regulators of the functions of cardiomyocytes, endothelial cells, vascular smooth muscle cells (VSMCs), and platelets, which refer to the initiation and progression of atherosclerosis as the key cause of CAD (1). In the process of atherosclerosis, miR-33a can inhibit cell cycle progression and proliferation of VSMCs by downregulating the expression of p53 and cmyc genes (43). Besides, miRNA-21 can promote the proliferation of VSMCs and reduce apoptosis, which plays a role in regulating the formation of vascular neointima (44). In human myocardial progenitor cells, miRNA-1 can directly act on the target gene Spred1 and inhibit its expression, thus promoting angiogenesis and differentiation (45). Nitric oxide (NO) is a vascular protective substance regulated by endothelial nitric oxide synthase (eNOS) and has anti-atherosclerotic effects (46). miRNA-214 is associated with eNOS activity, which can decrease the expression of the eNOS gene (47). Inhibiting the expression of miRNA-214 has an anti-atherosclerotic effect, thus reducing the incidence of coronary heart disease (48). Inflammation is known to be a major contributor to atherogenesis. miRNA-155 can also inhibit the mitogen-activated protein kinase 10 gene, thereby reducing the production of inflammatory cytokines and slowing down the progression of atherosclerosis (49).
However, for different studies (24, 26, 27, 31, 32, 34, 35), the results of miRNAs in CAD are controversial. Previous systematic reviews (50-50) have evaluated whether miRNAs can be used as biomarkers for the diagnosis and prognosis of CAD, but these included only those evaluated by bibliometrics and descriptive analyses. However, whether miRNAs are potential diagnostic biomarkers and how their diagnostic ability is mediated is not yet clear, and the inconsistencies are not yet resolved. Our meta-analysis includes specific types of common miRNAs and diagnostic meta-analysis was used for analysis. Therefore, the conclusions of our study are more stable and significant than those of previous studies.
In this meta-analysis, we included 14 studies (23-23), containing 38 data sets of miRNA expression profiles to systematically and comprehensively evaluate the potential diagnostic value of circulating miRNAs as diagnostic markers of CAD. Results of the meta-analysis showed that the average sensitivity was 0.80 (0.75–0.84), the average specificity was 0.78 (0.75–0.81), the positive likelihood ratio was 3.7 (3.1–4.4), and the negative likelihood ratio was 0.26 (0.21–0.33). The diagnostic odds ratio and the AUC were 14 (10–21) and 0.85 (0.82–0.88), respectively. This shows that miRNAs have a certain reference value for CAD diagnosis. One study showed that the AUC of miRNA-126 in diagnosing CAD was 0.98 (24). The predictive value of miR-126 for CAD may be explained by its essential roles in the mediation of endothelial cell activities and inflammatory responses (32). Moreover, miRNA-223 and miRNA-149 have relatively good potential as biomarkers for the diagnosis and prognosis of CAD patients, with diagnostic AUCs of 0.933 and 0.938, respectively (28, 30). miRNA-181b exerts its anti-atherosclerotic effect by inhibiting the proliferation and migration of vascular smooth muscle cells (53). miRNA-130a can regulate angiogenesis by inhibiting the expression of target genes (54). These findings may lead to the development of novel therapeutic modalities for the prevention and treatment of this disease. From the results of this study, the combined sensitivity and specificity of miRNAs in the diagnosis of CAD are high. The SROC (AUC) was approximately 0.85. In addition to this, the diagnostic odds ratio, as a combined measure of the sensitivity and specificity of miRNAs for CAD diagnosis, was 14, thus reflecting higher diagnostic performance. These results indicate that miRNAs have a high diagnostic value in the diagnosis of CAD. However, due to the heterogeneity of the combined effect amount, in this case, the combined analysis and interpretation of the results may not be accurate. Therefore, to address heterogeneity, subgroup and meta-regression analyses were performed to explore the source of heterogeneity.
In the subgroup analysis, the results of the present study showed that the accuracy in the Asian population was higher than that in the non-Asian population. Because most of the meta-analyses included in this study were conducted in Asia, the results may be biased and need to be supported by research in other regions. The results of this study may be more suitable for the diagnosis of Asian populations. Compared with a single miRNA, multiple miRNAs may be more diagnostic. The occurrence and development of CAD are diverse, and single miRNAs may lack specificity in the diagnosis of CAD, whereas multiple miRNAs with complex molecular mechanisms may be more valuable. Diagnostic accuracy of miRNAs in blood was better than those in plasma, serum, and PBMCs. Because miRNAs in blood can resist digestion by ribonuclease, they are not susceptible to changes in temperature, pH, and storage time, and thus have great stability (55). The detection method of miRNAs is also an important factor that affects sensitivity and specificity. Our subgroup analysis found that the diagnostic performance of the qRT-PCR group was better than that of the qPCR group, of which the AUC was as high as 0.87. One of the studies that were included (24) reported that the AUC of miRNAs detected by qRT-PCR for diagnosing CAD was as high as 98%. Therefore, qRT-PCR may be the tool of choice for miRNA detection.
There was some heterogeneity in the combined results of this meta-analysis. First, whether the heterogeneity was caused by non-threshold effects was analyzed. The Spearman correlation coefficient for diagnostic results was 0.51 (p=0.26), indicating that there was no threshold effect. For ethnicity, miRNA profiling, and altered regression analysis were performed on miRNAs, miRNA detection methods, and sample size. It was found that sensitivity was affected by the detection method and aberrant expression, and specificity was affected by the measurement method, aberrant expression, and sample size. The above factors are the main sources of heterogeneity.
A sensitivity analysis was performed to evaluate the stability of the study, which suggested that the results of the study are stable. Besides, Deek’s funnel plot asymmetry test showed no publication bias.
The following are the limitations of this study. (1) There were differences between the populations of the studies included in the observation group, which may have affected the results. (2) This study had some heterogeneity, and while some sources of heterogeneity were explained, different cut-off values were used in the included studies, which may also be a cause of heterogeneity. (3) There was a certain bias in the methodology of the included studies, and there may be design and implementation bias in the experimental results. (4) The sample size of some of the included studies was small. (5) Most of the included studies were retrospective case-control studies, which may have increased the selection bias. These limitations may have biased the meta-analysis results and affected the reliability of the results.
Conclusion
In summary, miRNAs, as novel biomarkers for the diagnosis of CAD, present a better diagnostic value, which can help clinicians diagnose CAD in a more timely and accurate manner. With the advent of technology and the development of new high-throughput miRNA detection methods, the sensitivity and specificity of detection will increase, and costs will decrease. The study of circulating miRNAs will eventually provide new methods for the diagnosis and treatment of CAD.
Footnotes
Conflict of interest: None declared.
Peer-review: Externally peer-reviewed.
Authorship contributions: Concept – Q.F., Y.L.; Design – Q.F., Y.L.; Supervision – Y.L.; Fundings – Y.W.; Materials – Z.X., J.L.; Data collection &/or processing – X.Z.; Analysis &/or interpretation – X.Z.; Literature search – Q.F.; Writing – Q.F., Y.W.; Critical review – Q.F., Y.W.
References
- 1.Ziegler T, Abdel Rahman F, Jurisch V, Kupatt C. Atherosclerosis and the Capillary Network; Pathophysiology and Potential Therapeutic Strategies Cells. 2019;9:50. doi: 10.3390/cells9010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lubrano V, Balzan S. Consolidated and emerging inflammatory markers in coronary artery disease. World J Exp Med. 2015;5:21–32. doi: 10.5493/wjem.v5.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yayan J. Emerging families of biomarkers for coronary artery disease: inflammatory mediators. Vasc Health Risk Manag. 2013;9:435–56. doi: 10.2147/VHRM.S45704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Panda AC, Sahu I, Kulkarni SD, Martindale JL, Abdelmohsen K, Vindu A, et al. miR-196b-mediated translation regulation of mouse insulin2 via the 5'UTR. PLoS One. 2014;9:e101084. doi: 10.1371/journal.pone.0101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859–68. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 9.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–20. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 11.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 12.Michell DL, Vickers KC. HDL and microRNA therapeutics in cardiovascular disease. Pharmacol Ther. 2016;168:43–52. doi: 10.1016/j.pharmthera.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li HY, Zhao X, Liu YZ, Meng Z, Wang D, Yang F, et al. Plasma MicroRNA-126-5p is Associated with the Complexity and Severity of Coronary Artery Disease in Patients with Stable Angina Pectoris. Cell Physiol Biochem. 2016;39:837–46. doi: 10.1159/000447794. [DOI] [PubMed] [Google Scholar]
- 14.Satoh M, Takahashi Y, Tabuchi T, Tamada M, Takahashi K, Itoh T, et al. Circulating Toll-like receptor 4-responsive microRNA panel in patients with coronary artery disease: results from prospective and randomized study of treatment with renin-angiotensin system blockade. Clin Sci (Lond) 2015;128:483–91. doi: 10.1042/CS20140417. [DOI] [PubMed] [Google Scholar]
- 15.Wang KJ, Zhao X, Liu YZ, Zeng QT, Mao XB, Li SN, et al. Circulating MiR-19b-3p, MiR-134-5p and MiR-186-5p are Promising Novel Biomarkers for Early Diagnosis of Acute Myocardial Infarction. Cell Physiol Biochem. 2016;38:1015–29. doi: 10.1159/000443053. [DOI] [PubMed] [Google Scholar]
- 16.Shan Z, Qin S, Li W, Wu W, Yang J, Chu M, et al. An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells:Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. J Am Coll Cardiol. 2015;65:2526–37. doi: 10.1016/j.jacc.2015.03.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taurino C, Miller WH, McBride MW, McClure JD, Khanin R, Moreno MU, et al. Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci (Lond) 2010;119:335–43. doi: 10.1042/CS20100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karakas M, Schulte C, Appelbaum S, Ojeda F, Lackner KJ, Münzel T, et al. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur Heart J. 2017;38:516–23. doi: 10.1093/eurheartj/ehw250. [DOI] [PubMed] [Google Scholar]
- 19.Schulte C, Karakas M, Zeller T. microRNAs in cardiovascular disease - clinical application. Clin Chem Lab Med. 2017;55:687–704. doi: 10.1515/cclm-2016-0576. [DOI] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses:the PRISMA statement. Ann Intern Med. 2009;151:264–9. doi: 10.7326/0003-4819-151-4-200908180-00135. W64. [DOI] [PubMed] [Google Scholar]
- 21.Higgins JP, Green S. Cochrane Handbook for Systematic Reviews of Interventions 2008. URL;https: //handbook-5-1.cochrane.org/ [Google Scholar]
- 22.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 23.Ali Sheikh MS, Xia K, Li F, Deng X, Salma U, Deng H, et al. Circulating miR-765 and miR-149:potential noninvasive diagnostic biomarkers for geriatric coronary artery disease patients. Biomed Res Int. 2015;2015:740301. doi: 10.1155/2015/740301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amr KS, Abdelmawgoud H, Ali ZY, Shehata S, Raslan HM. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br J Biomed Sci. 2018;75:82–7. doi: 10.1080/09674845.2017.1402404. [DOI] [PubMed] [Google Scholar]
- 25.Dong J, Liang YZ, Zhang J, Wu LJ, Wang S, Hua Q, et al. Potential Role of Lipometabolism-Related MicroRNAs in Peripheral Blood Mononuclear Cells as Biomarkers for Coronary Artery Disease. J Atheroscler Thromb. 2017;24:430–41. doi: 10.5551/jat.35923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du Y, Yang SH, Li S, Cui CJ, Zhang Y, Zhu CG, et al. Circulating MicroRNAs as Novel Diagnostic Biomarkers for Very Early-onset (</=40 years) Coronary Artery Disease. Biomed Environ Sci. 2016;29:545–54. doi: 10.3967/bes2016.073. [DOI] [PubMed] [Google Scholar]
- 27.Faccini J, Ruidavets JB, Cordelier P, Martins F, Maoret JJ, Bongard V, et al. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci Rep. 2017;7:42916. doi: 10.1038/srep42916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo JF, Zhang Y, Zheng QX, Zhang Y, Zhou HH, Cui LM. Association between elevated plasma microRNA-223 content and severity of coronary heart disease. Scand J Clin Lab Invest. 2018;78:373–8. doi: 10.1080/00365513.2018.1480059. [DOI] [PubMed] [Google Scholar]
- 29.Quan X, Ji Y, Zhang C, Guo X, Zhang Y, Jia S, et al. Circulating MiR-146a May be a Potential Biomarker of Coronary Heart Disease in Patients with Subclinical Hypothyroidism. Cell Physiol Biochem. 2018;45:226–36. doi: 10.1159/000486769. [DOI] [PubMed] [Google Scholar]
- 30.Sayed AS, Xia K, Li F, Deng X, Salma U, Li T, et al. The diagnostic value of circulating microRNAs for middle-aged (40-60-year-old) coronary artery disease patients. Clinics (Sao Paulo) 2015;70:257–63. doi: 10.6061/clinics/2015(04)07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Pei Y, Zhong Y, Jiang S, Shao J, Gong J. Altered serum microRNAs as novel diagnostic biomarkers for atypical coronary artery disease. PLoS One. 2014;9:e107012. doi: 10.1371/journal.pone.0107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Zhang J. miR-126 in Peripheral Blood Mononuclear Cells Negatively Correlates with Risk and Severity and is Associated with Inflammatory Cytokines as well as Intercellular Adhesion Molecule-1 in Patients with Coronary Artery Disease. Cardiology. 2018;139:110–8. doi: 10.1159/000484236. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Hao J, Sun X, Zhang Y, Wei Q. Circulating pro-angiogenic micro-ribonucleic acid in patients with coronary heart disease. Interact Cardiovasc Thorac Surg. 2018;27:336–42. doi: 10.1093/icvts/ivy058. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Zhang Y, Xue S, Ding H, Wang Y, Qi H, et al. Clinical significance of circulating microRNAs as diagnostic biomarkers for coronary artery disease. J Cell Mol Med. 2020;24:1146–50. doi: 10.1111/jcmm.14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Li HH, Yang R, Yang BJ, Gao ZY. Association between circulating microRNA-208a and severity of coronary heart disease. Scand J Clin Lab Invest. 2017;77:379–84. doi: 10.1080/00365513.2017.1328740. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Shao G, Chen X, Yang X, Huang X, Peng P, et al. miRNA 206 and miRNA 574-5p are highly expression in coronary artery disease. Biosci Rep. 2015;36:e00295. doi: 10.1042/BSR20150206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Q, Wei X, Rao J, Zhou C. Identification of key miRNAs in the progression of hepatocellular carcinoma using an integrated bioinformatics approach. PeerJ. 2020;8:e9000. doi: 10.7717/peerj.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Xu H, Song W, Zhang P, Song Y. Potential microRNA panel for the diagnosis and prediction of overall survival of hepatocellular carcinoma with hepatitis B virus infection. World J Gastrointest Oncol. 2020;12:383–93. doi: 10.4251/wjgo.v12.i4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Miguel Pérez D, Rodriguez Martínez A, Ortigosa Palomo A, Delgado Ureña M, Garcia Puche JL, Robles Remacho A, et al. Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci Rep. 2020;10:3974. doi: 10.1038/s41598-020-60212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabry D, El-Deek SEM, Maher M, El-Baz MAH, El-Bader HM, Amer E, et al. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1α-VEGF signaling pathway. Mol Cell Biochem. 2019;454:177–89. doi: 10.1007/s11010-018-3462-1. [DOI] [PubMed] [Google Scholar]
- 42.Chen MB, Yang L, Lu PH, Fu XL, Zhang Y, Zhu YQ, et al. MicroRNA-101 down-regulates sphingosine kinase 1 in colorectal cancer cells. Biochem Biophys Res Commun. 2015;463:954–60. doi: 10.1016/j.bbrc.2015.06.041. [DOI] [PubMed] [Google Scholar]
- 43.Blandino G, Valerio M, Cioce M, Mori F, Casadei L, Pulito C, et al. Metformin elicits anticancer effects through the sequential modulation of DICER and c-MYC. Nat Commun. 2012;3:865. doi: 10.1038/ncomms1859. [DOI] [PubMed] [Google Scholar]
- 44.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–43. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Mil A, Vrijsen KR, Goumans MJ, Metz CH, Doevendans PA, Sluijter JP. MicroRNA-1 enhances the angiogenic differentiation of human cardiomyocyte progenitor cells. J Mol Med (Berl) 2013;91:1001–12. doi: 10.1007/s00109-013-1017-1. [DOI] [PubMed] [Google Scholar]
- 46.Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis:new actors for an old movie. Nutr Metab Cardiovasc Dis. 2012;22:937–43. doi: 10.1016/j.numecd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Jamaluddin MS, Weakley SM, Zhang L, Kougias P, Lin PH, Yao Q, et al. miRNAs: roles and clinical applications in vascular disease. Expert Rev Mol Diagn. 2011;11:79–89. doi: 10.1586/erm.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Mao H, Chen JY, Wen S, Li D, Ye M, et al. Increased expression of microRNA-221 inhibits PAK1 in endothelial progenitor cells and impairs its function via c-Raf/MEK/ERK pathway. Biochem Biophys Res Commun. 2013;431:404–8. doi: 10.1016/j.bbrc.2012.12.157. [DOI] [PubMed] [Google Scholar]
- 49.Zhu J, Chen T, Yang L, Li Z, Wong MM, Zheng X, et al. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One. 2012;7:e46551. doi: 10.1371/journal.pone.0046551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barraclough JY, Joan M, Joglekar MV, Hardikar AA, Patel S. MicroRNAs as Prognostic Markers in Acute Coronary Syndrome Patients-A Systematic Review. Cells. 2019;8:1572. doi: 10.3390/cells8121572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur A, Mackin ST, Schlosser K, Wong FL, Elharram M, Delles C, et al. Systematic review of microRNA biomarkers in acute coronary syndrome and stable coronary artery disease. Cardiovasc Res. 2020;116:1113–24. doi: 10.1093/cvr/cvz302. [DOI] [PubMed] [Google Scholar]
- 52.Navickas R, Gal D, Laucevičius A, Taparauskaitė A, Zdanytė M, Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease:a systematic review. Cardiovasc Res. 2016;111:322–37. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee SH, Jung YD, Choi YS, Lee YM. Targeting of RUNX3 by miR-130a and miR-495 cooperatively increases cell proliferation and tumor angiogenesis in gastric cancer cells. Oncotarget. 2015;6:33269–78. doi: 10.18632/oncotarget.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Gonzalo-Calvo D, Cenarro A, Garlaschelli K, Pellegatta F, Vilades D, Nasarre L, et al. Translating the microRNA signature of microvesicles derived from human coronary artery smooth muscle cells in patients with familial hypercholesterolemia and coronary artery disease. J Mol Cell Cardiol. 2017;106:55–67. doi: 10.1016/j.yjmcc.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]