To the Editor:
We read with interest the letter by Roeker et al., on SARS-CoV-2 antibody responses in twenty-one patients with chronic lymphocytic leukemia (CLL), reporting that 67% generated antibodies to the SARS-CoV-2 Nucleoprotein antigen [1]. Their report highlighted the need to characterise the serological response to SAR-CoV-2 in immune compromised patients, particularly those with haematological malignancies, to help inform patient management and public health strategies. Patients with acute leukemia also represent a particularly high-risk group, who are reported to have some of the poorest outcomes of COVID-19 [2] and are hypothesised to have impaired immune SARS-CoV-2 responses due to either disease- or treatment-associated immune dysfunction. This has resulted in recommendations to reduce the risk of COVID-19 in these patients, including restructuring of clinical services, shielding and alterations in therapy to minimise their immune suppressive consequence [3]. Here we report SARS-CoV-2 antibody responses in a cohort of patients with acute leukemia and COVID-19 receiving systemic anti-cancer therapy (SACT) at University College London Hospital during the first wave of the pandemic.
Longitudinal serum samples were collected from nine patients with acute leukaemia, of whom eight had PCR-confirmed SARS-CoV-2 infection and one had a clinical diagnosis of COVD-19. Five patients had AML, three B-ALL and one T-ALL. Four patients commenced SACT prior to developing COVID-19 and five presented with leukemia and COVID-19. All patients received SACT within 28 days of developing COVID-19. Four patients received less myelosuppressive regimens (venetoclax azacitdine or gilteritinib) in accordance with NICE/NCRI COVID-19 guidance for acute leukaemia. COVID-19 symptoms were assigned from mild to severe [4], with two patients requiring ITU and mechanical ventilation. The median time between symptom onset and PCR diagnosis was 2.5 days (IQR 8.25), median duration of PCR positivity was 18.5 days (IQR 22) (Supplementary Fig. 1) and four patients received a potential COVID-19 modifying agent (tocilizumab, anakinra, remdesivir or dexamethasone). All patients survived and were discharged from hospital, with a median duration of illness of 30 days (IQR 30). Further patient demographics are described in Table 1.
Table 1.
Demographics, patient and disease characteristics, treatment and outcomes in patients with acute leukemia and COVID-19.
| Patients (n = 9) | A | B | C | D | E | F | H | J | K |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 45–49 | 25–29 | 35–39 | 20–24 | 60–64 | 50–54 | 35–39 | 55–59 | 75–79 |
| Sex | F | M | M | M | F | M | M | M | F |
| Ethnicity | Caucasian | Caucasian | Caucasian | South Asian | Caucasian | Caucasian | Caucasian | Caucasian | Caucasian |
| Haematological diagnosis | AML | AML | B ALL | AML | B ALL | T ALL | AML | B ALL relapsed | AML |
| Haematological disease features | IDH2 mt | FLT3 mt; NPM1 WT | Normal CGN | Bi-allelic CEBPA mt, GATA2 mt | t(9;22), mono 7 | None | NPM1 mt, MECOM +1 | t(9;22) | Complex karyotype (del 5q, TP53 loss, mono 16, amplification KMT2A) |
| Haemtological chemo-/immuno-therapy | Ven/Aza | AraC, Gilteritinib | Blinatumumab (prev UKALL14) | DA |
UKALL 60+ Ph+ induction |
UKALL 14 Consolidation 1 | Ven/Aza | Mini FLA-Ida + imatinib, (prev UKALL14) | Ven/Aza |
| Days from haematological diagnosis to COVID-19 | 1 | 0 | 246 | 61 | 1 | 272 | 9 | 52 | 0 |
| Comorbidities | None | None | None | None |
HTN T2DM |
None | None | None | COPD |
| Smoking history | Ex-smoker | None | None | None | None | None | Ex-smoker | Smoker | Ex-smoker |
| Presenting symptoms of COVID-19 | Fever, tooth abscess, myalgia, fatigue | Fever | Fever, collapse | Neutropaenic fever | Cough, diarrhoea | Neutropaenic fever | Fever, cough | Fever | Fever, shortness of breath, palpitations |
| Days from symptom onset to COVID-19 | 35 | 3 | 2 | 0 | 28 | 1 | 3 | 1 | 27 |
| CXR/CT findings | None | Ground glass | Ground glass | Multiple areas of consolidation | Ground glass | None | Bilateral consolidation | Mild atelectasis | Ground glass |
| ITU admission | No | Yes | No | No | No | No | Yes | No | No |
| Max FiO2 | 21 | 100 | 85 | 21 | 21 | 21 | 100 | 24 | 60 |
| Max fever | 39.5 | 40.5 | 39.7 | 38.6 | 37.7 | 37.8 | 40.7 | 38 | 39.9 |
| COVID severity scorea | 0 | 4 | 3 | 0 | 0 | 0 | 4 | 1 | 3 |
| COVID-19 modifying treatment | None | Dex | Anakinra | None | Remdesivir | None | Tocilizumab | None | None |
| Duration of PCR positivity (days)b | 8d | 59 | 33 | 11 | 8 | 12 | 32 | 25 | NA |
| Days from symptom onset to negative PCR | 43 | 62 | 35 | 11 | 36 | 13 | 35 | 26 | NA |
| Duration of illness (days)c | 15 | 50 | 30 | 14 | 45 | 15 | 43 | 9 | 64 |
| Outcome | Alive, OP | Alive, OP | Alive, OP | Alive, OP | Alive, OP | Alive, OP | Alive, OP | Alive, OP | Alive, OP |
All patients consented for excess serum to be stored and used as part of the “UCL Biobank for Studying Health and Disease—Haematology Project”, reference no NC10.13.
AML acute myeloid leukaemia, B-ALL B-lymphoblastic leukaemia, T-ALL T lymphoblastic leukaemia, Ven/aza venetoclax/azacytidine, DA daunorubicin, AraC; Dex dexamethasone, HTN hypertension, COPD chronic obstructive pulmonary disease, T2DM type 2 diabetes mellitus, OP outpatient.
aCOVID-19 severity score as previously defined [4]: 0—asymptomatic OR no requirement for supplemental oxygen; 1—supplemental oxygen (Fi02 < 0.4) for ≥12 h; 2—supplemental oxygen (Fi02≥0.4) for ≥12 h, 3—requirement for NIV/CPAP OR proning OR supplemental oxygen (Fi02 > 0.6) for ≥12 h; 4—intubation and ventilation OR supplemental oxygen (Fi02 > 0.8) AND peripheral Sp02 < 90% (no known T2RF or <85% if known T2RF) for ≥12.
bRT PCR for SARS-CoV-2 was performed on samples from a combined nose and throat swab specimen.
cDuration of illness defined as the period between diagnosis and cessation of treatment for COVID-19 that would mandate inpatient treatment (step down from ITU or discharge from the COVID ward).
dThis patient subsequently tested positive one day after initial negative (for four days) again 21 days after second negative test (for eight days) (Supplementary Fig. 2).
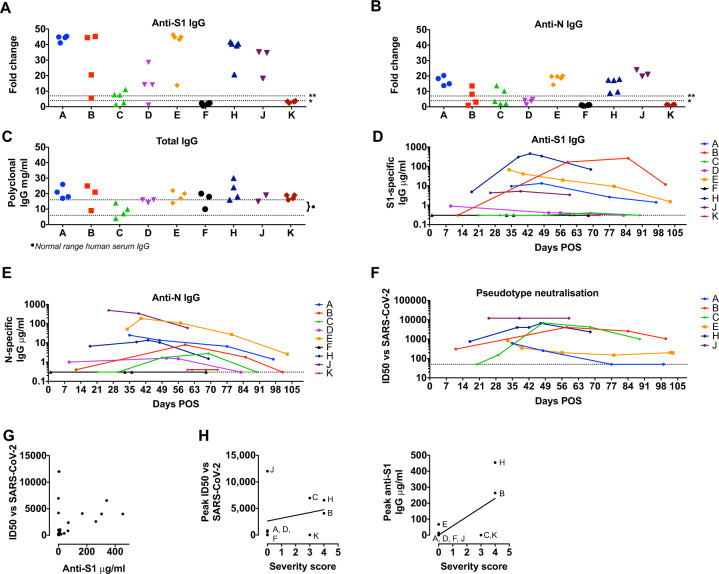
Serum samples were taken a median of 9.5 days after positive PCR test for SARS-CoV-2 (range 1–25 days) and subsequent longitudinal serum samples taken between 2 and 103 days post onset of symptoms (POS). These were screened for anti-SARS-CoV-2 antibodies using ELISA to the external Spike glycoprotein (S1 subunit) and internal Nucleoprotein (N) [4–6]. Total serum IgG was within in the normal range in each case, excluding hypogammaglobinaemia. Seven of eight patients (88%) with PCR-confirmed SARS-CoV-2 had IgG responses to S1 and N (Fig. 1a, b and appendix). Classifying patient samples into 7-day intervals POS (Supplementary Fig. 2) showed that seroconversion to S appeared to precede N, with only two patients seroconverting to both by day 30 (Supplementary Fig. 2 and Supplementary Tables 1–3). Overall seroconversion rates of 88% were similar to the general population [4, 6, 7] and higher than that reported by Roeker et al. for CLL [1]. Seroconversion appeared delayed in our cohort, with 50% seroconverting by day 28, compared to 90% of healthy individuals [7], although this requries confirmation in a larger study. The temporal dynamics of SARS-CoV-2 responses in patients were measured using a semi-quantitative assay of S1 and N antibodies. Again, the overall magnitude and persistence of SARS-CoV-2 IgG (up to >100 days POS) were similar to the general population. Furthermore, the patterns of antibody responses seen were as expected during an acute infection, namely; increasing titre reflective of seroconversion, declining titre suggestive of waning responses, and maintained titres (Fig. 1d, e). Similar kinetics of anti-N and anti-S1 antibody were seen, albeit with differences in overall antibody titre.
Fig. 1. Temporal dynamics and anti-viral function of SARS-CoV-2 antibody responses in acute leukaemia.
Serum samples from patients A–K were assayed on ELISA plates pre-coated with S1 (a) or N (b). Antigen production and assay conditions were as previously described [4–6] except that all samples were treated with 0.5% NP40 before dilution in ELISA buffer. Absorbance was measured at 405 nm and data expressed as fold-change above blank background. The limit of detection (seropositivity) was determined as fold change >4 and is indicated by the dotted line marked *. The dotted line marked ** indicates the limit of quantification in the assay depicted in d–e, determined by the linear range of the standard curve. c ELISA plates were coated overnight at 4 °C with goat anti-human F(ab)’2. Serum samples from patients A–K, and commercial polyclonal IgG standard, were titrated in ELISA buffer and added to the ELISA plate. Binding was detected with goat anti-human IgG conjugated to peroxidase and absorbance read at 450 nm. IgG concentrations in serum were calculated based on interpolation from the IgG standard results using a four-parameter logistic (4PL) regression curve fitting model. Dotted lines marked with }* indicate the normal average range for IgG in human serum. d, e Sera supplemented with 0.5% NP40 from patients A–K were diluted in ELISA buffer and then added to a blocked ELISA plates pre-coated with the indicated antigen and three lanes of goat anti-human F(ab)’2 as per (c). Binding was detected with anti-IgG conjugated to alkaline phosphatase and absorbance measured at 405 nm. Antigen-specific IgG concentrations in serum were calculated based on interpolation from the IgG standard results using a four-parameter logistic (4PL) regression curve fitting model. The dotted line indicates the limit of quantification, which is determined by the linear range of the standard curve and higher than the limit of detection in a and b. f. Sequential serum samples from seropositive patients were titrated in duplicate and pre-incubated with luciferase-encoding HIV pseudotyped with the SARS-CoV-2 Spike for 1 h prior to the addition of HeLa cells expressing human ACE2 as previously described [7]. ID50 titres were only calculated in GraphPad Prism where at least two data points exhibited >50% neutralisation. ID50 values for each patient are plotted on the y-axis against days POS on the x-axis. g. ID50 values plotted against semi-quantitative S1 titres (μg/ml) for each sample. There is a trend to increasing ID50 with higher S1 titres but linear regression does not show a significant correlation (r = 0.06915, p = 0.0925). All data are from at least three independent experiments. h ID50 values and anti-S1 (μg/ml) values plotted against COVID-19 severity score for each patient as cited in Table 1. A trend line is shown but there were insufficient sample numbers to assess correlation and significance.
To address whether the antibodies generated by patients with acute leukemia were functional and able to inhibit SARS-CoV-2 infection, the capacity of serum to block viral infection in vitro was measured using an established pseudotyped SARS-CoV-2 neutralisation assay [7, 8] (Fig. 1f). Six of the seven (86%) patients who seroconverted to SARS-CoV-2 generated antibodies able to inhibit virus infection, measured by determining the concentration of antibody capable of inhibiting SARS-CoV-2 infection by 50% (ID50%). In general, neutralisation correlated with anti-S1 IgG levels (Fig. 1g); however, some patients showed strong neutralisation despite low anti-S1 titres, suggesting the presence of particularly potent antibodies similar to previous reports in individuals without cancer [7]. Importantly anti-S1 IgG titres and neutralising responses broadly correlated with clinical COVID-19 disease severity (Fig. 1h), although this correlation was not absolute with variation between individuals’ responses observed. Again, this is in keeping with studies the general population and patients with haematological disease [1, 4, 7].
Our finding that the majority of patients with acute leukemia on SACT are able to make antibody responses to SARS-CoV-2 has important implications for patient management, population-based serological monitoring and clinical decision making for this high-risk group. While our analysis of the kinetics, magnitude and function of SARS-CoV2 antibody responses in individuals with acute leukaemia align with similar SARS-CoV-2 serology studies on non-haematology patients [7], longitudinal sampling suggested a potential delay to SARS-CoV-2 seroconversion in patients with acute leukemia. This must be confirmed in a larger cohorts and in different subgroups of patients to determine if this is a feature of patients with haematological malignancy. Future studies should not only confirm but extend analysis of seroconversion rates, incorporating regular PCR testing to determine the precise onset and duration of SARS-CoV-2 infection, and frequent, longitudinal serum sampling to capture the emerging antibody response in greater depth. Notably, the ability to correlate antibody responses with function highlights the importance of measuring anti-S antibody responses alongside anti-N. Whether serological responses vary by leukaemia type or SACT regimen, and the influence this has on patient outcomes are essential next steps for investigation.
Supplementary information
Acknowledgements
We are extremely grateful to all the patients who participated in this study and the NHS staff that provided their clinical care. We would like to thank, Leo James and Jakub Luptak (LMB) for the provision of the plasmid encoding the N protein, and James E Voss (TSRI) for providing the Hela-ACE2. LEM and LM are supported by an MRC Career Development Award (MR/R008698/1) to LEM. CJ is supported by a Wellcome Trust Investigator award (108079/Z/15/Z). RG acknowledges funding from Cure Cancer@ UCL.
Author contributions
Designed the study: JO, LEM, CJ; Sample curation: JO, JZ, RG; Performed experiments: LM, LEM, CR-S; Generated reagents: AR, CR, CE, PC; Analysed data: JO, JZ, LM, LEM; Wrote/edited the manuscript: JO, LEM, CJ, LM, JZ, AK, RG.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
All clinical information was recorded and blood samples taken as routine standard of care. Patients were consented to allow any excess serum to be stored and used as part of the UCL Biobank for Studying Health and Disease, Haematology Project, reference no NC10.13, approved by the Leeds (East) Research Ethics Committee, UK.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
J. O’Nions, Email: jenny.onions@nhs.net
L. E. McCoy, Email: l.mccoy@ucl.ac.uk
Supplementary information
The online version of this article (10.1038/s41375-020-01103-2) contains supplementary material, which is available to authorized users.
References
- 1.Roeker LE, Knorr DA, Pessin MS, Ramanathan LV, Thompson MC, Leslie LA, et al. Anti-SARS-CoV-2 antibody response in patients with chronic lymphocytic leukemia. Leukemia. 2020;34:3047–9. doi: 10.1038/s41375-020-01030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Passamonti F, Cattaneo C, Arcaini L, Bruna R, Cavo M, Merli F, et al. Clinical characteristics and risk factors associated with COVID-19 severity in patients with haematological malignancies in Italy: a retrospective, multicentre, cohort study. Lancet Haematol. 2020;10:e737–45. doi: 10.1016/S2352-3026(20)30251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeidan AM, Boddu PC, Patnaik MM, Bewersdorf JP, Stahl M, Rampal RK, et al. Special considerations in the management of adult patients with acute leukaemias and myeloid neoplasms in the COVID-19 era: recommendations from a panel of international experts. Lancet Haematol. 2020;7:e601–12. doi: 10.1016/S2352-3026(20)30205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickering S, Betancor G, Galao RP, Merrick B, Signell AW, Wilson HD, et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog. 2020;16:e1008817. doi: 10.1371/journal.ppat.1008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houlihan CF, Vora N, Byrne T, Lewer D, Kelly G, Heaney J, et al. Pandemic peak SARS-CoV-2 infection and seroconversion rates in London frontline health-care workers. Lancet. 2020;396:e6–7. doi: 10.1016/S0140-6736(20)31484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng, KW, Faulkner N, Cornish G, Rosa A, Hussain S, Harvey R, et al. Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020. 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed]
- 7.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 human infection. Nat Micro. 2020 doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–50. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.