Abstract
Human stem cells bear a great potential for multiple therapeutic applications but at the same time constitute a major threat to human health in the form of cancer stem cells. The molecular processes that govern stem cell maintenance or differentiation have been extensively studied in model organisms or cell culture, but it has been difficult to extrapolate these insights to therapeutic applications. Recent advances in the field suggest that local and global changes in histone modifications that affect chromatin structure could influence the capability of cells to either maintain their stem cell identity or differentiate into specialized cell types. The enzymes that regulate these modifications are therefore among the prime targets for potential drugs that can influence and potentially improve the therapeutic application of stem cells. In this review, we discuss recent findings on the role of histone modifications in stem cell regulation and their potential implications for clinical applications.
Keywords: chromatin, epigenetics, histone modifications, cell cycle, metabolism, stem cells
In this article, Axel Imhof and colleagues discuss recent findings on the role of histone modifications in stem cell regulation. In particular, this article focuses on the effects of global processes such as the cell cycle or the metabolic state on chromatin maturation and links them to targeted therapeutic interventions aiming to reset the epigenetic landscape.
Main Text
Introduction
In pluripotent stem cells of most multicellular eukaryotes, the epigenome undergoes global remodeling during commitment to a specific lineage (Atlasi and Stunnenberg 2017). Growing evidence suggests that maintenance of the pluripotent state as well as lineage commitment is controlled by global epigenetic mechanisms such as DNA methylation, histone modifications, and regulation of ATP-dependent remodeling of chromatin structure (Zhou et al., 2011). As the roles of DNA methylation, histone variants, and ATP-dependent remodelers on stem cell biology have been extensively reviewed (Turinetto and Giachino 2015; Wu and Sun 2006), we will focus on the role of histone modifications and histone-modifying enzymes.
The chromatin landscape of pluripotent cells has been intensely investigated in mouse embryonic stem cells (mESCs), which have higher acetylation and lower methylation levels than differentiated cells (Bhanu et al., 2016). These signatures of a more “active” chromatin conformation are consistent with the findings of an increased transcriptional activity (Efroni et al., 2008) and a hyperdynamic behavior of chromatin-associated factors in ESCs (Meshorer et al., 2006). In addition to the surprisingly high dynamics of stem cell chromatin, detailed chromatin immunoprecipitation (ChIP)-ChIP and ChIP-PCR studies revealed a co-localization of active and repressive chromatin marks at promoters and enhancers of developmentally regulated genes in mESCs (Azuara et al., 2006; Bernstein et al., 2006). Such a “bivalent’’” chromatin signature of H3K4me3 and H3K27me3 is thought to mark genes that are repressed in ESCs but poised to allow for alternative fates. H3K4me3 and H3K27me3 also overlap at thousands of genes in human ESCs (hESCs), but few genes exhibit H3K27me3 alone. This indicates that bivalency is the default chromatin state at key developmental control genes marked by H3K27me3 in hESCs (Harikumar and Meshorer 2015).
Deposition of H3K27me3 is mediated by Polycomb repressive complex 2 (PRC2) via its catalytic subunit EZH1/2. Trimethylation of K4 within the H3 tail is mediated by the SETD1 (COMPASS) and MLL-containing complexes. Mutations in either H3K27 or H3K4 methyltransferase result in severe defects in ESC growth. The genetic ablation of various enzymes belonging to PRC2 causes the removal of repressive H3K27me3 marks, reduced self-renewal, and upregulation of mesendoderm genes in hESCs (Collinson et al., 2016; Shan et al., 2017), whereas the removal of the H3K4 methyltransferase SET1A results in a lack of proliferation and an increase in apoptosis (Bledau et al., 2014; Fang et al., 2016; Sze et al., 2017).
Recruitment of Modifiers
Recruitment of the H3K4 methyltransferase-containing complexes to active promoters is thought to be mediated by different complex subunits that interact with the C-terminal domain of Pol II, unmethylated CpG islands, or site-specific DNA-binding transcription factors and long non-coding RNAs (Voigt et al., 2013). The mechanisms governing the recruitment of PRC2 to specific genomic targets are not yet fully understood (Holoch and Margueron 2017). It appears that recruitment of PRC2 is, similar to Set1A and MLL-containing complexes, likely not (only) based on the recognition of specific sequences but rather on composition-modulated physical properties of DNA, such as the shape of the DNA, nucleosome density, or overall chromatin conformation (Laugesen et al., 2019).
Despite similar modes of recruitment, the two modifications show a markedly different distribution within the genome. Whereas H3K4me3 is often found within promoter regions, where it is frequently found at a few nucleosomes covering this domain (Stewart-Morgan et al., 2019), H3K27me2 and me3 usually cover larger genomic loci. In contrast to the rather localized H3K4me3 mark, H3K27me3 is thought to spread from an initial site of recruitment, which is mediated by an allosteric activation of the PRC through the product of its methylation (Ragazzini et al., 2019) or by an autoactivation through methylation of the JARID subunit (Sanulli et al., 2015). This difference in distribution is also reflected by the much smaller amount of histone H3 methylated at K4 compared with K27-methylated H3 (Alabert et al., 2015). So why is there such a difference in the localization and amount of those modifications in eukaryotic cells?
Modulation of Histone Modification States
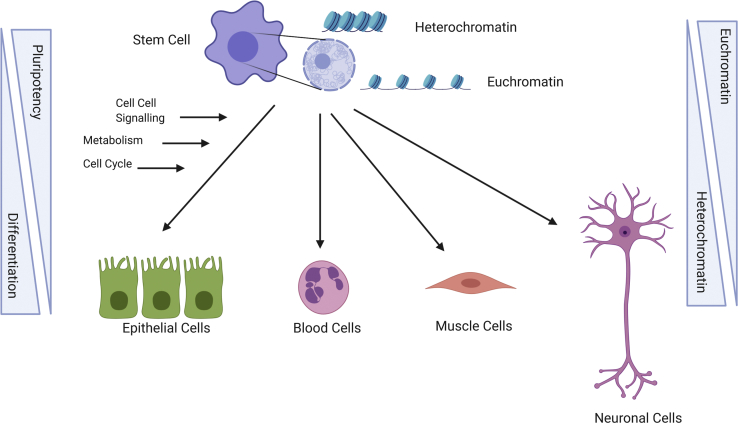
There is increasing evidence that the number of specific histone modifications, such as the bivalent marks in ESCs, as well as their position within the genome depends not only on the recruitment of the enzymes that catalyze the modification but also on the level of demethylases, the metabolic state of the cell, and the speed of the cell cycle (Figure 1).
Figure 1.
General Concept of Stem Cell Differentiation and Its Influences by Epigenetics
Pluripotent stem cells can differentiate into lineage-restricted progenies. Intrinsic properties of stem cells as well as extrinsic factors such as cell-cell signaling and metabolism influence stem cell self-renewal and differentiation. Progression from stem cell to differentiated progeny is accompanied by remarkable epigenetic changes from euchromatin to heterochromatin. Figure was created with BioRender.com.
Metabolic Effects on Histone Modifications
Histone-modifying enzymes as well as enzymes that remove certain modifications rely on the presence of key metabolites such as acetyl-CoA, S-adenosyl methionine (SAM), NAD, or 2 oxoglutarate (Katada et al., 2012). The intracellular concentrations of these metabolites highly depend on the physiological status of the cell and nutrient availability. Most stem cells have a very specialized metabolism and are more dependent on glycolysis and less on oxidative phosphorylation for energy production (Facucho-Oliveira and St John, 2009). Moreover, mESCs depend on Thr and hESCs on Met to maintain their pluripotency (Shiraki et al., 2014). The removal of these amino acids results in a drop in SAM levels and concomitantly of particular histone methylation sites such as H3K4 (Shyh-Chang et al., 2013). Importantly, not only histone methylation but also histone acetylation is influenced by stem cell-specific metabolic pathways. Stem cells produce cytosolic acetyl-CoA via glycolysis and the subsequent pyruvate-derived citrate flux by action of the ATP citrate lyase. Blockage of this pathway results in a decrease in histone acetylation and early differentiation of hESCs (Moussaieff et al., 2015). From these studies it became clear that global changes such as differences in metabolism can have very specific effects during cell differentiation.
Histone Post-translational Modification Kinetics across the Cell Cycle
Another global effect that influences the entire chromatin within the nucleus is the cell cycle. With each cycle, newly formed, largely unmodified histones are incorporated into the DNA, leading to an overall dilution of most histone modifications (Jasencakova et al., 2010). Within newly replicated chromatin, half of the histone H3 molecules carry H3K14ac, H3K18/K23ac, and H3K9me1 and half of the H4 molecules carry H4K5ac and H4K12ac at levels closely mirroring the modification pattern of soluble histones in pre-deposition complexes (Jasencakova et al., 2010; Loyola et al., 2006), suggesting that these post-translational modifications are maintained within the first minutes of incorporation (Alabert et al., 2015). During chromatin maturation, K27me1, K36me1, and K27me2 are imposed on new histones (Alabert et al., 2015). This marking occurs soon after deposition onto newly replicated DNA, arguing for a quick deposition. However, further methylations on heterochromatic marks are imposed on the newly incorporated histones with a much slower kinetics that is similar to the cell cycle (Alabert et al., 2015), making the length of the cell cycle an important regulator of global chromatin methylation. Consistent with this hypothesis, resting and slowly dividing cells tend to accumulate histone methylations, whereas cells with a short cell cycle carry more acetylations (Alabert et al., 2015; Bhanu et al., 2016; Leroy et al., 2013). Assuming that methylated histones are more refractory to reprogramming due to the slower turnover of this modification, the length of the cell cycle can therefore have a major effect on cellular plasticity. In fact, studies investigating the effect of DNA synthesis on cellular reprogramming efficiency revealed a positive correlation between these two processes (Tsubouchi et al., 2013).
In recent publications, computational modeling approaches have been used to investigate and model the role of the modification kinetics in setting up specific chromosomal domains and conferring epigenetic plasticity (Alabert et al., 2020; Schuh et al., 2020). Two aspects common to the modeling approaches are: (1) the existence of potentially competing modifications, such as H3K4me (active) and H3K9me (inactive) or H3K27me (inactive) and H3K36me (active), and (2) a low contribution of cell-cycle-mediated modification dilution to the total modification profile. Although there are clear exceptions, like the above-mentioned bivalent mark, a widely accepted model of chromatin shaping is the fixation of histones in specific states by antagonistic modifications. This provides a barrier to changes in cell identity that can be overridden globally by cell proliferation when modification patterns are diluted genome wide through the incorporation of new naive histones during replication. Recently, profiling of histone modifications in tumor samples has revealed pervasive K27me3 loss and K36me2 gain across different cancers (Noberini et al., 2019). In addition to mutations in histones and chromatin regulators that directly affect the K27/K36 methylation balance, a more general reduction in K27me3 might thus be driven by changes in proliferation rates during cellular transformation. These strong effects of global and rather unspecific processes, such as the cell cycle or the metabolic state, on chromatin maturation therefore need to be considered in the context of targeted therapeutic intervention aimed at resetting the epigenetic landscape.
Current Implications and Treatments according to Epigenetics and Histone Modifications in Stem Cells
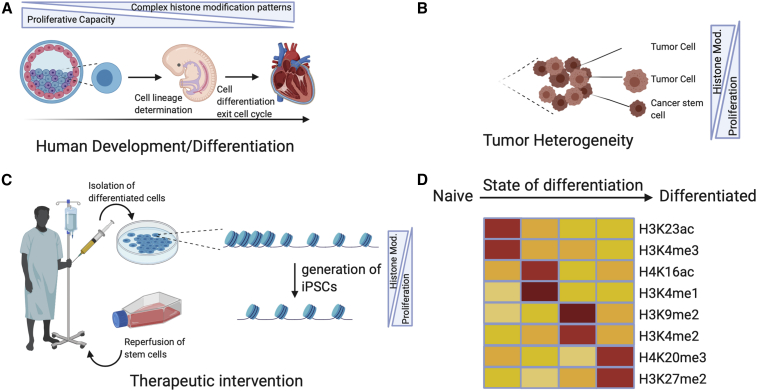
Stem cells represent a promising tool for new clinical concepts in support of cellular therapy for a variety of human diseases and injuries. At the same time many human cancers are fed by a small population of cancer stem cells (CSCs), which represents a major challenge for tumor treatment (Figure 2). A better understanding of the mechanisms that are associated with stemness and self-renewal but also cell differentiation is therefore urgently needed to translate our knowledge into clinical applications.
Figure 2.
Histone Modifications in Development and Therapeutic Intervention
(A) Schematic illustration of stem cell differentiation into tissue with decreasing proliferative capacity and increasing histone modification decoration.
(B) Schematic illustration of colorectal tumor heterogeneity with changes in histone modifications fostering high proliferation rates in distinct cancer stem cells.
(C) Concept of stem cell reprogramming for therapeutic intervention by isolation of differentiated cells and inducing them into a pluripotent state to bring them back into a diseased body. iPSCs, induced pluripotent stem cells.
(D) Schematic representation of the correlation of the stem cells’’ developmental hierarchy with their epigenetic status according to Schneider et al. (2011). Figure was created with BioRender.com.
Cancer Stem Cells
CSCs represent a small subpopulation of cells often found in dedicated niches of various tumor types. Unlike normal stem cells, whose cell-cycle transitions are strictly regulated, CSCs carry various pro-oncogenic mutations that enable uncontrolled proliferation, which facilitates the capacity for (1) asymmetric division, (2) reconstituting a differentiated tumor upon transplantation, (3) participating in the epithelial-mesenchymal transition, and (4) resistance to conventional therapies (Batlle and Clevers 2017). By virtue of these properties, CSCs can promote, enhance, and sustain malignant phenotypes and have been causally linked to tumor aggressiveness, heterogeneity, disease stage, the onset of relapse and metastasis, and ultimately poor clinical outcome (Batlle and Clevers 2017). The importance of improved strategies for targeting, suppressing, and eradicating CSCs can therefore not be overstated.
Histone-Linked Chromatin Dysregulation as a Player in CSC Formation and Maintenance
In recent years it has become clear that epigenetic dysregulation of chromatin plays a major role in the formation of CSCs and is often vital for CSC self-renewal during tumor growth. Studies in pediatric glioblastoma (GBM) have provided key insights. Initial studies demonstrated the presence of two recurrent gain-of-function mutations within histone variant H3.3, each affecting the K27M and G34R/V amino acids (Wu et al., 2012). Subsequent studies showed that the K27M mutant H3.3 results in aberrant recruitment of PRC2 to H3.3 and a subsequent inhibition of its enzymatic activity. This inhibition leads to a genome-wide reduction in H3K27me3 (Bender et al., 2013; Lewis et al., 2013) and a reestablishment of self-renewal (Funato et al., 2014). In addition to pediatric GBMs, there are several examples in which disruptions of the histone code have been linked to the formation of CSCs in other cancer types (reviewed in Liu et al., 2017; Vincent and Van Seuningen, 2012). For instance, malfunction of the KMT2A/MLL gene, which encodes a histone methyltransferase (HMT) that plays a key role in the regulation of enhancer accessibility through chromatin modifications, has been linked to the formation of leukemia stem cells (LSCs) in both acute myeloid leukemia (AML) and acute lymphoblastic leukemia (Shilatifard 2012). Similarly, studies have shown that the p300/CBP coactivator family (Zhao et al., 2011), as well as the transcriptional coactivators MOZ and MORF (Yang and Ullah 2007), which have intrinsic histone acyltransferase activity, is crucial for the establishment and maintenance of LSCs. Concerning histone deacetylases (HDACs), HDAC1 and HDAC7 were, for example, found to be overexpressed in CSCs originating from both breast and ovarian tissues, contributing not only to the CSC phenotype but also to the maintenance of stem cell properties (Witt et al., 2017). In addition to histone modifiers, induction of CSCs can be affected by mutations in structural proteins involved in higher-order chromatin conformation, chromatin-remodeling complexes, and proteins involved in DNA methylation, including DNA methyltransferases (DNMTs) and methylcytosine dioxygenases (TET1 and TET2). However, consideration of these factors is beyond the focus of this paper. For a thorough review, refer to Wainwright and Scaffidi (2017).
Cancer Stem Cell Epigenetics as a Therapeutic Target
From the studies discussed thus far, it is clear that (1) CSCs often contribute greatly to the initiation and propagation of cancer (Batlle and Clevers 2017) and (2) the formation and maintenance of CSCs during tumor growth appear to be highly dependent on their chromatin state (Liu et al., 2017; Makena et al., 2020; Vincent and Van Seuningen 2012). In keeping with this, the intrinsic reversibility of epigenetic modifications provides a window through which basic knowledge can be leveraged toward therapeutic targeting of CSCs, thereby reducing cancer morbidity and mortality. Unsurprisingly, pharmacological inhibition of many chromatin-interacting proteins, and subsequent interference with the upstream regulators of oncoproteins, has in recent years been shown to target CSCs and inhibit the growth of various cancer types in cell cultures, animal models, and preclinical human studies. As reviewed elsewhere (Makena et al., 2020), there are many possible therapeutic strategies against CSCs. However, for the purposes of this paper the discussion will be limited mostly to epigenetic therapies that target CSCs via direct modulation of the histone code. Below, we will discuss different strategies as they relate to various drug types.
HDAC inhibitors (HDACis) are drugs that interfere with HDACs and lead to increased levels of histone acetylation and subsequent de-repression of gene expression. In both preclinical models and landmark clinical trials, various HDACis have shown efficacy in arresting the cell cycle and inducing apoptosis in differentiated cancer cells, resulting in inhibition of tumor growth (Toh et al., 2017). In addition to killing normal cancer cells, a broad spectrum of HDACis have demonstrated efficacy in the specific suppression or elimination of CSCs. However, the suppressive effects of HDACis on CSCs are not yet fully understood (reviewed in Lin et al., 2019) and have been shown to be achieved through different pathways and different modes of action (Table 1). First, HDACis can reprogram chemotherapy-resistant CSCs back into differentiated cells that are sensitive to chemotherapy. Through the use of different types of HDACis, differentiation of CSCs has been triggered in a wide range of cancer cell types, including pancreatic (Meidhof et al., 2015), endometrial adenocarcinoma (Uchida et al., 2005), and breast cancer (Munster et al., 2001; Salvador et al., 2013). Second, HDACis can reverse the differentiated state of cancer cells. For example, HDACis have been used to reprogram breast cancer cells into quiescent stem cell-like cancer cells that were subsequently characterized by a shift of energy metabolism into the pentose phosphate pathway, which could then be targeted by inhibition of glucose-6-phosphate dehydrogenase (Debeb et al., 2016), suggesting a potential dual-therapy approach. In addition to interfering with differentiation protocols, HDACis have been shown to induce selective death of CSCs in various cancer models, such as AML (Guzman et al., 2014; Yan et al., 2019), pancreatic cancer (Nalls et al., 2011), sarcoma (Di Pompo, Salerno et al., 2015), and chronic myelogenous leukemia (Zhang et al., 2010).
Table 1.
Functions of Different HDAC Inhibitors in CSC Targeting
| HDAC Inhibitor | Cancer Type | Effect on CSCs | Mechanism | Reference |
|---|---|---|---|---|
| SAHA | pancreatic | impairs self-renewal capacity | inhibition of miR-34a-Notch signaling and epithelial-mesenchymal transition | (Nalls et al., 2011) |
| head and neck | reverses cisplatin resistance | downregulation of Nanog expression | (Kumar et al., 2015) | |
| Mocetinostat | pancreatic | represses stemness/resensitization to chemotherapy | interference with ZEB1 and restoration of miR-203 expression | (Meidhof et al., 2015) |
| TSA and SAHA | endometrial adenocarcinoma | induction of differentiation | upregulation of glycodelin | (Uchida et al., 2005) |
| Abexinostat | breast | induction of differentiation | involvement of Xist and potentially BRCA1 | (Salvador et al., 2013) |
| AR-42 | leukemia | Apoptosis | inhibition of NF-κB and Hsp90 functions | (Guzman et al., 2014) |
| Romidepsin | leukemia | Apoptosis | upregulation of genes involved in the inflammatory response and apoptosis pathways | (Yan et al., 2019) |
Although there are still only a few studies that have directly evaluated the effects of the inhibition of HMTs and histone demethylases (HDMs) in CSCs, they show promise of revealing a potential function of these drugs in therapeutic applications. Several studies have shown that drugs targeting different histone lysine methyltransferases (HKMTs) demonstrate efficacy at reducing CSC stemness in different cancer types, which is mirrored by significant anti-tumor activity. Some examples of effectively targeted HKMTs include (1) EZH2 (KMT6) (Yu et al., 2017), (2) DOT1L (KMT4) (Daigle et al., 2013), (3) G9a/EHMT2 (Kim et al., 2013), and (4) SUV39H1 and SUV39H2 (Lai et al., 2015). Some examples of effectively targeted HDMs include the histone lysine demethylases (1) LSD1 (Schenk et al., 2012) and (2) the Jumonji domain-containing (JmjC) enzyme UTX/KDM6A (Lee et al., 2007). Inhibition of UTX/KDM6A leads to the suppression of CSCs in breast cancer (Yan et al., 2017) and ovarian cancer (Sakaki et al., 2015), while enhancing radiosensitivity in various cell lines (Rath et al., 2018).
In addition to HDACs, HMTs, and HDMs, pharmacological targeting of DNMTs and epigenetic readers such as bromodomain and extra-terminal (BET) proteins has also been shown to ablate CSCs and represent additional therapeutic approaches (Toh et al., 2017; Wainwright and Scaffidi 2017). For example, exposure of ovarian cancer stem-like cells to the DNMT inhibitor SGI-110 resulted in a reduction of stem cell properties (Wang et al., 2014). BET inhibitors, which hinder reactions between BET proteins and acetylated histones and transcription factors, have been shown to inhibit the targeting of BRD4 to and subsequent transcriptional repression of several oncogenes in a wide range of CSC types, including c-MYC, which is essential to the survival of many CSCs (Wainwright and Scaffidi 2017).
In the above reports, epigenetic drugs were usually tested as the sole agent. However, numerous studies have demonstrated that the efficacy of these drugs can often be enhanced significantly by co-treatments with similar drug types, such as simultaneous inhibition of EZH2 and G9a, or by combining with other drug types, such as the DNMT inhibitors azacitidine and decitabine, or conventional chemotherapies such as platinum, cytarabine, and irinotecan therapy (Toh et al., 2017; Wainwright and Scaffidi 2017).
When exploring the landscape of the diverse epigenetic factors that are responsible for regulating and maintaining the CSC phenotype, there are potential caveats and experimental/technical challenges that need to be considered. First, CSCs are relatively rare in solid tumors and often occur in relatively low numbers, which complicates the isolation procedure (Witt et al., 2017). Furthermore, once they are successfully isolated, it is challenging to expand CSCs in the in vitro setting, primarily because they rapidly differentiate into non-stem tumor cells under standard culture conditions (Fillmore and Kuperwasser 2008). Second, there is a notable variation and overlap of CSC surface markers among different tumor types, while many of the same surface markers are also expressed on normal cells and cells that are in a transitionary state (Makena et al., 2020). This not only further complicates the isolation of specific CSCs for study, but also complicates the targeting of specific CSCs for therapeutic purposes. Taken together, these results highlight the importance of identifying more and increasingly specific markers for CSCs. Last, as they often target whole enzyme families, the epigenetic drugs described in this paper have a broad spectrum of inhibition and often affect a variety of non-histone effector molecules (Makena et al., 2020). On one hand, this makes it exceedingly difficult to elucidate the true mechanisms that underlie the mode of action of a specific drug in general and in specific cases. Second, such unspecific targeting may not be the most suitable treatment option for many solid tumors, especially in cases where there is a big difference in the epigenetic landscape and expression of specific epigenetic regulators between CSCs and the rest of the tumor (Vincent and Van Seuningen 2012). This highlights the importance of (1) exercising caution when developing and testing such drugs, (2) identifying and clearly mapping out the epigenetic reprogramming that transpires during the formation of CSCs, and (3) an improved understanding of the differences between normal stem cells and CSCs. This will aid in the development of new drugs or approaches with enhanced specificity, enabling the targeting of specific CSCs and limiting aberrant disruptions of tissue homeostasis. As indicated earlier, this is substantiated by the improvements in patient outcomes that are achieved by the use of epigenetic drugs that have greater specificity (e.g., that target an individual epigenetic modifier protein) and/or by combinatory drug approaches.
Stem Cell Epigenetics and Therapeutics in Regenerative Medicine
Beyond the domain of oncology, epigenetic principles can also be leveraged in stem cell-based regeneration of damaged tissues. First, drugs affecting epigenetic modifiers can be used to improve differentiation protocols in stem cells and their derivatives. An effective stem cell reprogramming should guarantee a complete remodeling of epigenetic memory of the initial somatic cell, followed by the establishment of the epigenetic signature of the new type of cell to be differentiated (Figure 2). In line with this, the epigenome is frequently used as a biomarker of efficiency and safety in stem cell differentiation. An alternative strategy involves an active stimulation of the natural niche of adult stem cells after tissue injury, which has enormous translational potential because it does not involve cellular transplantation. Such epigenetic-based stimulation of stem cells has shown some initial positive effects in the field of cardiovascular and neurological disorders (Ganai et al., 2016; Gilsbach et al., 2018). For example, HDACi treatment has been shown to induce neuronal differentiation in adult progenitor cells (Hsieh et al., 2004). Similarly, HDACi treatment improves cardiac progenitor cell function and has been shown to positively affect the repair of ischemic myocardium upon cellular transplantation (Guo et al., 2018), as well as suppressing cardiac fibrosis (Williams et al., 2014). In the latter case, selective inhibition of class I HDACs results in the suppression of angiotensin II-mediated cardiac fibrosis, mainly by blocking the progression of cardiac fibroblasts through the cell cycle (blocking them in the G0/G1 phase), which is achieved by inhibition of Rb phosphorylation through de-repression of the genes encoding the p15 and p57 cyclin-dependent kinase inhibitors (Williams et al., 2014).
In summary, recent studies on the role of histone modifications for stem cell maintenance and differentiation have revealed a surprising role of global effects like the cell cycle or metabolic pathways on what so far has been thought to be constituted by specific signaling pathways. Although these effects are very likely modulatory rather than deterministic, they can be easily targeted and therefore bear potential for supplementary treatments. In the future such global treatment could potentially be used to improve stem cell-based regenerative therapies or to selectively target CSCs, thereby facilitating the development of improved, and perhaps more personalized, therapeutic possibilities.
Author Contributions
M.V.-A., A.B., S.H., and A.I. wrote the manuscript with input from all authors. A.I. conceptualized and created the figures.
Acknowledgments
A.B. is supported by the Georg Förster Research Fellowship Program of the Alexander von Humboldt Foundation. The financial assistance of the Alexander von Humboldt Foundation is hereby acknowledged.
Conflicts of Interests
A.I. and M.V.-A. are co-founders of EpiQMAx GmbH. A.I. and S.H. are SAB members of Volition RX.
References
- Alabert C., Barth T.K., Reveron-Gomez N., Sidoli S., Schmidt A., Jensen O.N., Imhof A., Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C., Loos C., Voelker-Albert M., Graziano S., Forne I., Reveron-Gomez N., Schuh L., Hasenauer J., Marr C., Imhof A., Groth A. Domain model explains propagation dynamics and stability of histone H3K27 and H3K36 methylation landscapes. Cell Rep. 2020;30:1223–1234.e8. doi: 10.1016/j.celrep.2019.12.060. [DOI] [PubMed] [Google Scholar]
- Atlasi Y., Stunnenberg H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017;18:643–658. doi: 10.1038/nrg.2017.57. [DOI] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A.G. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Batlle E., Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- Bender S., Tang Y., Lindroth A.M., Hovestadt V., Jones D.T., Kool M., Zapatka M., Northcott P.A., Sturm D., Wang W. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhanu N.V., Sidoli S., Garcia B.A. Histone modification profiling reveals differential signatures associated with human embryonic stem cell self-renewal and differentiation. Proteomics. 2016;16:448–458. doi: 10.1002/pmic.201500231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledau A.S., Schmidt K., Neumann K., Hill U., Ciotta G., Gupta A., Torres D.C., Fu J., Kranz A., Stewart A.F., Anastassiadis K. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141:1022–1035. doi: 10.1242/dev.098152. [DOI] [PubMed] [Google Scholar]
- Collinson A., Collier A.J., Morgan N.P., Sienerth A.R., Chandra T., Andrews S., Rugg-Gunn P.J. Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 2016;17:2700–2714. doi: 10.1016/j.celrep.2016.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle S.R., Olhava E.J., Therkelsen C.A., Basavapathruni A., Jin L., Boriack-Sjodin P.A., Allain C.J., Klaus C.R., Raimondi A., Scott M.P. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeb B.G., Lacerda L., Larson R., Wolfe A.R., Krishnamurthy S., Reuben J.M., Ueno N.T., Gilcrease M., Woodward W.A. Histone deacetylase inhibitor-induced cancer stem cells exhibit high pentose phosphate pathway metabolism. Oncotarget. 2016;7:28329–28339. doi: 10.18632/oncotarget.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pompo G., Salerno M., Rotili D., Valente S., Zwergel C., Avnet S., Lattanzi G., Baldini N., Mai A. Novel histone deacetylase inhibitors induce growth arrest, apoptosis, and differentiation in sarcoma cancer stem cells. J. Med. Chem. 2015;58:4073–4079. doi: 10.1021/acs.jmedchem.5b00126. [DOI] [PubMed] [Google Scholar]
- Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D., Buetow K.H. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facucho-Oliveira J.M., St John J.C. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. Rep. 2009;5:140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- Fang L., Zhang J., Zhang H., Yang X., Jin X., Zhang L., Skalnik D.G., Jin Y., Zhang Y., Huang X. H3K4 methyltransferase Set1a is A key Oct4 coactivator essential for generation of Oct4 positive inner cell mass. Stem Cells. 2016;34:565–580. doi: 10.1002/stem.2250. [DOI] [PubMed] [Google Scholar]
- Fillmore C.M., Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K., Major T., Lewis P.W., Allis C.D., Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai S.A., Ramadoss M., Mahadevan V. Histone Deacetylase (HDAC) Inhibitors - emerging roles in neuronal memory, learning, synaptic plasticity and neural regeneration. Curr. Neuropharmacol. 2016;14:55–71. doi: 10.2174/1570159X13666151021111609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach R., Schwaderer M., Preissl S., Grüning B.A., Kranzhöfer D., Schneider P., Nührenberg T.G., Mulero-Navarro S., Weichenhan D., Braun C. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun. 2018;9:391. doi: 10.1038/s41467-017-02762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Bai Y., Zhang L., Zhang B., Zagidullin N., Carvalho K., Du Z., Cai B. Cardiomyocyte differentiation of mesenchymal stem cells from bone marrow: new regulators and its implications. Stem Cell Res. Ther. 2018;9:44. doi: 10.1186/s13287-018-0773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M.L., Yang N., Sharma K.K., Balys M., Corbett C.A., Jordan C.T., Becker M.W., Steidl U., Abdel-Wahab O., Levine R.L. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol. Cancer Ther. 2014;13:1979–1990. doi: 10.1158/1535-7163.MCT-13-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A., Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015;16:1609–1619. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Margueron R. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 2017;42:531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Hsieh J., Nakashima K., Kuwabara T., Mejia E., Gage F.H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z., Scharf A.N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Katada S., Imhof A., Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Kim Y., Kim Y.S., Kim D.E., Lee J.S., Song J.H., Kim H.G., Cho D.H., Jeong S.Y., Jin D.H., Jang S.J. BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production. Autophagy. 2013;9:2126–2139. doi: 10.4161/auto.26308. [DOI] [PubMed] [Google Scholar]
- Kumar B., Yadav A., Lang J.C., Teknos T.N., Kumar P. Suberoylanilide hydroxamic acid (SAHA) reverses chemoresistance in head and neck cancer cells by targeting cancer stem cells via the downregulation of nanog. Genes Cancer. 2015;6:169–181. doi: 10.18632/genesandcancer.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.S., Chen J.Y., Tsai H.J., Chen T.Y., Hung W.C. The SUV39H1 inhibitor chaetocin induces differentiation and shows synergistic cytotoxicity with other epigenetic drugs in acute myeloid leukemia cells. Blood Cancer J. 2015;5:e313. doi: 10.1038/bcj.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A., Højfeldt J.W., Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell. 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Di Croce L., Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Leroy G., Dimaggio P.A., Chan E.Y., Zee B.M., Blanco M.A., Bryant B., Flaniken I.Z., Liu S., Kang Y., Trojer P., Garcia B.A. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin. 2013;6:20. doi: 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.W., Müller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., Sausville E.L., Lukow D.A., Liu L., Chait A.R. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl Med. 2019;11:eaaw8412. doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Li S., Wu N., Cho K.S. Acetylation and deacetylation in cancer stem-like cells. Oncotarget. 2017;8:89315–89325. doi: 10.18632/oncotarget.19167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Makena M.R., Ranjan A., Thirumala V., Reddy A.P. Cancer stem cells: road to therapeutic resistance and strategies to overcome resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165339. doi: 10.1016/j.bbadis.2018.11.015. [DOI] [PubMed] [Google Scholar]
- Meidhof S., Brabletz S., Lehmann W., Preca B.T., Mock K., Ruh M., Schüler J., Berthold M., Weber A., Burk U. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol. Med. 2015;7:831–847. doi: 10.15252/emmm.201404396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Munster P.N., Troso-Sandoval T., Rosen N., Rifkind R., Marks P.A., Richon V.M. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001;61:8492–8497. [PubMed] [Google Scholar]
- Nalls D., Tang S.N., Rodova M., Srivastava R.K., Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noberini R., Restellini C., Savoia E.O., Raimondi F., Ghiani L., Jodice M.G., Bertalot G., Bonizzi G., Capra M., Maffini F.A. Profiling of epigenetic features in clinical samples reveals novel widespread changes in cancer. Cancers (Basel) 2019;11:723. doi: 10.3390/cancers11050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragazzini R., Pérez-Palacios R., Baymaz I.H., Diop S., Ancelin K., Zielinski D., Michaud A., Givelet M., Borsos M., Aflaki S. EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nat. Commun. 2019;10:3858. doi: 10.1038/s41467-019-11800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath B.H., Waung I., Camphausen K., Tofilon P.J. Inhibition of the histone H3K27 demethylase UTX enhances tumor cell radiosensitivity. Mol. Cancer Ther. 2018;17:1070–1078. doi: 10.1158/1535-7163.MCT-17-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki H., Okada M., Kuramoto K., Takeda H., Watarai H., Suzuki S., Seino S., Seino M., Ohta T., Nagase S. GSKJ4, A selective Jumonji H3K27 demethylase inhibitor, effectively targets ovarian cancer stem cells. Anticancer Res. 2015;35:6607–6614. [PubMed] [Google Scholar]
- Salvador M.A., Wicinski J., Cabaud O., Toiron Y., Finetti P., Josselin E., Lelièvre H., Kraus-Berthier L., Depil S., Bertucci F. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res. 2013;19:6520–6531. doi: 10.1158/1078-0432.CCR-13-0877. [DOI] [PubMed] [Google Scholar]
- Sanulli S., Justin N., Teissandier A., Ancelin K., Portoso M., Caron M., Michaud A., Lombard B., da Rocha S.T., Offer J. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk T., Chen W.C., Göllner S., Howell L., Jin L., Hebestreit K., Klein H.U., Popescu A.C., Burnett A., Mills K. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.D., Arteaga-Salas J.M., Mentele E., David R., Nicetto D., Imhof A., Rupp R.A. Stage-specific histone modification profiles reveal global transitions in the Xenopus embryonic epigenome. PLoS One. 2011;6:e22548. doi: 10.1371/journal.pone.0022548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh L., Loos C., Pokrovsky D., Imhof A., Rupp R., Marr C. Computational modeling reveals cell-cycle dependent kinetics of H4K20 methylation states during Xenopus embryogenesis. bioRxiv. 2020 doi: 10.1101/2020.05.28.110684. [DOI] [Google Scholar]
- Shan Y., Liang Z., Xing Q., Zhang T., Wang B., Tian S., Huang W., Zhang Y., Yao J., Zhu Y. PRC2 specifies ectoderm es and maintains pluripotency in primed but not naïve ESCs. Nat. Commun. 2017;8:672. doi: 10.1038/s41467-017-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F., Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J.J., Zhu H. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Morgan K.R., Reverón-Gómez N., Groth A. Transcription restart establishes chromatin accessibility after DNA replication. Mol. Cell. 2019;75:284–297.e6. doi: 10.1016/j.molcel.2019.04.033. [DOI] [PubMed] [Google Scholar]
- Sze C.C., Cao K., Collings C.K., Marshall S.A., Rendleman E.J., Ozark P.A., Chen F.X., Morgan M.A., Wang L., Shilatifard A. Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 2017;31:1732–1737. doi: 10.1101/gad.303768.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh T.B., Lim J.J., Chow E.K. Epigenetics in cancer stem cells. Mol. Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T., Soza-Ried J., Brown K., Piccolo F.M., Cantone I., Landeira D., Bagci H., Hochegger H., Merkenschlager M., Fisher A.G. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell. 2013;152:873–883. doi: 10.1016/j.cell.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V., Giachino C. Histone variants as emerging regulators of embryonic stem cell identity. Epigenetics. 2015;10:563–573. doi: 10.1080/15592294.2015.1053682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H., Maruyama T., Nagashima T., Asada H., Yoshimura Y. Histone deacetylase inhibitors induce differentiation of human endometrial adenocarcinoma cells through up-regulation of glycodelin. Endocrinology. 2005;146:5365–5373. doi: 10.1210/en.2005-0359. [DOI] [PubMed] [Google Scholar]
- Vincent A., Van Seuningen I. On the epigenetic origin of cancer stem cells. Biochim. Biophys. Acta. 2012;1826:83–88. doi: 10.1016/j.bbcan.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright E.N., Scaffidi P. Epigenetics and cancer stem cells: unleashing, Hijacking, and restricting cellular plasticity. Trends Cancer. 2017;3:372–386. doi: 10.1016/j.trecan.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cardenas H., Fang F., Condello S., Taverna P., Segar M., Liu Y., Nephew K.P., Matei D. Epigenetic targeting of ovarian cancer stem cells. Cancer Res. 2014;74:4922–4936. doi: 10.1158/0008-5472.CAN-14-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.M., Golden-Mason L., Ferguson B.S., Schuetze K.B., Cavasin M.A., Demos-Davies K., Yeager M.E., Stenmark K.R., McKinsey T.A. Class I HDACs regulate angiotensin II-dependent cardiac fibrosis via fibroblasts and circulating fibrocytes. J. Mol. Cell Cardiol. 2014;67:112–125. doi: 10.1016/j.yjmcc.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt A.E., Lee C.W., Lee T.I., Azzam D.J., Wang B., Caslini C., Petrocca F., Grosso J., Jones M., Cohick E.B. Identification of a cancer stem cell-specific function for the histone deacetylases, HDAC1 and HDAC7, in breast and ovarian cancer. Oncogene. 2017;36:1707–1720. doi: 10.1038/onc.2016.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Sun Y.E. Epigenetic regulation of stem cell differentiation. Pediatr. Res. 2006;59:21r–25r. doi: 10.1203/01.pdr.0000203565.76028.2a. [DOI] [PubMed] [Google Scholar]
- Yan B., Chen Q., Shimada K., Tang M., Li H., Gurumurthy A., Khoury J.D., Xu B., Huang S., Qiu Y. Histone deacetylase inhibitor targets CD123/CD47-positive cells and reverse chemoresistance phenotype in acute myeloid leukemia. Leukemia. 2019;33:931–944. doi: 10.1038/s41375-018-0279-6. [DOI] [PubMed] [Google Scholar]
- Yan N., Xu L., Wu X., Zhang L., Fei X., Cao Y., Zhang F. GSKJ4, an H3K27me3 demethylase inhibitor, effectively suppresses the breast cancer stem cells. Exp. Cell Res. 2017;359:405–414. doi: 10.1016/j.yexcr.2017.08.024. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Ullah M. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26:5408–5419. doi: 10.1038/sj.onc.1210609. [DOI] [PubMed] [Google Scholar]
- Yu Y., Deng P., Yu B., Szymanski J.M., Aghaloo T., Hong C., Wang C.Y. Inhibition of EZH2 promotes human embryonic stem cell differentiation into mesoderm by reducing H3K27me3. Stem Cell Reports. 2017;9:752–761. doi: 10.1016/j.stemcr.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Strauss A.C., Chu S., Li M., Ho Y., Shiang K.D., Snyder D.S., Huettner C.S., Shultz L., Holyoake T., Bhatia R. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Glazov E.A., Pattabiraman D.R., Al-Owaidi F., Zhang P., Brown M.A., Leo P.J., Gonda T.J. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]