Summary
The ten-eleven translocation factor TET1 and its conferred epigenetic modification 5-hydroxymethylcytosine (5hmC) have important roles in maintaining the pluripotent state of embryonic stem cells (ESCs). We previously showed that TET1 is also essential to maintain the stem cell state of trophoblast stem cells (TSCs). Here, we establish an integrated panel of absolute 5hmC levels, genome-wide DNA methylation and hydroxymethylation patterns, transcriptomes, and TET1 chromatin occupancy in TSCs and differentiated trophoblast cells. We show that the combined presence of 5-methylcytosine (5mC) and 5hmC correlates with transcriptional activity of associated genes. Hypoxia can slow down the global loss of 5hmC that occurs upon differentiation of TSCs. Notably, unlike in ESCs and epiblast cells, most TET1-bound regions overlap with active chromatin marks and TFAP2C binding sites and demarcate putative trophoblast enhancer regions. These chromatin modification and occupancy patterns are highly informative to identify novel candidate regulators of the TSC state.
Keywords: trophoblast stem cells, epigenetics, DNA methylation, TET1, DNA hydroxymethylation, enhancers, chromatin interactions, embryonic stem cells, gene regulation
Highlights
-
•
5hmC to 5mC ratios correlate with gene activity in TS cells
-
•
TS cell differentiation-associated loss of 5hmC is slowed down in hypoxia
-
•
TET1 binding in TS cells forms long-range interactions with key trophoblast genes
-
•
Intergenic TET1 binding sites in TS cells demarcate putative trophoblast enhancers
TET1 and its conferred epigenetic modification 5-hydroxymethylation are critical for maintaining pluripotency of embryonic stem cells. In this article, Senner and colleagues describe in-depth the role of this modification, in conjunction with DNA methylation, in trophoblast stem cells, and determine a distinct role for TET1 in demarcating putative trophoblast enhancers and thereby establishing trophoblast-specific gene networks.
Introduction
DNA methylation is crucial for proper embryonic development. Its involvement in key developmental processes, such as the silencing of transposable elements, imprinted gene expression, and transcriptional repression of the inactive X chromosome has been well established (Greenberg and Bourc'his, 2019). Hypomethylation of the trophectoderm (TE) compared with the inner cell mass at the blastocyst stage (Santos et al., 2002) may suggest that DNA methylation is less important in the TE-derived trophoblast lineage (Sakaue et al., 2010). However, it is clear that while DNA methylation is not required for establishment of the TE, placental development in de novo DNA methyltransferase (Dnmt) knockouts (KOs) is severely compromised (Arima et al., 2006, Branco et al., 2016, Sakaue et al., 2010), with particular defects in cell adhesion and syncytiotrophoblast formation that highlight an important role of DNA methylation at non-imprinted loci specifically for trophoblast development (Branco et al., 2016).
The importance of DNA methylation is also manifest in trophoblast stem cells (TSCs). TSCs can be derived from the blastocyst or early post-implantation extraembryonic ectoderm. They can be maintained in culture as stem cells or differentiated into various placental cell types, and contribute exclusively to the placenta in chimeras (Tanaka et al., 1998). While the trophoblast compartment in vivo is globally hypomethylated, cultured TSCs are not. We previously showed that TSCs in culture have DNA methylation levels equivalent to those observed in embryonic stem cells (ESCs) (Senner et al., 2012). This increase in methylation compared with the in vivo tissue is not stochastic, however, but rather reinforces trophoblast-specific patterns of gene expression and repression. In particular, we found a set of methylated CpG islands (CGIs) that regulate genes specific to embryonic differentiation, and described a lineage-identifying “extraembryonic signature” based on CGI methylation profiles. In addition to genes required for embryonic development, pluripotency genes Pou5f1 (Oct4) and Nanog are also repressed by DNA methylation in TSCs (Hattori et al., 2004, Hattori et al., 2007). Moreover, Dnmt triple KO (Dnmt1-, Dnmt3a-, and Dnmt3b-null) TSCs display cell adhesion defects (Branco et al., 2016) and misregulate the expression of many genes, including those of the prolactin-like cluster, Rhox genes, and several imprinted genes (Sakaue et al., 2010). Together these data suggest that DNA methylation functions as a transcriptional regulator in the trophoblast lineage in vitro as well as in vivo.
The ten-eleven translocation (TET) family of Fe(II) and 2-oxoglutatarate-dependent DNA dioxygenases catalyzes the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethycytosine (5hmC) as the first step in an active demethylation process (He et al., 2011, Ito et al., 2011, Tahiliani et al., 2009). TET proteins (TET1-3) have been extensively studied in ESCs and embryonic lineages where they are associated with the pluripotency program as well as the regulation of key developmental genes (Wu et al., 2018). Although their expression levels are lower in extraembryonic stem cells compared with ESCs, TET proteins also play an important role in supporting the stem cell character of TSCs. Similar to the situation in ESCs, Tet1 is the most highly expressed member of the Tet genes in TSCs (Chrysanthou et al., 2018). Tet1 KO TSCs undergo an epithelial-to-mesenchymal transition and exhibit adhesion as well as cell-cycle defects, suggesting diverse roles for TET1 within the trophoblast lineage. Further evidence that Tet1 is important for trophoblast development comes from Tet1 KO mice (Dawlaty et al., 2011). These mice are viable and have only a mild phenotype with developmental delay apparent at embryonic day 12.5 and mutant pups appearing smaller. Interestingly, tetraploid complementation was found to rescue this phenotype, indicating a role for Tet1 in placental development.
In this study we aimed to specifically characterize the role of Tet1 in global methylation dynamics in the trophoblast lineage. We generated genome-wide 5hmC and 5mC profiles using methylated and hydroxymethylated DNA immunoprecipitation sequencing (meDIP-seq and hmeDIP-seq, respectively), which we analyzed alongside TET1 chromatin immunoprecipitation sequencing (ChIP-seq) (Chrysanthou et al., 2018), promoter capture Hi-C (high-throughput chromosome conformation capture) (Schoenfelder et al., 2018), and RNA sequencing (RNA-seq) data from Tet1 KO TSCs (Chrysanthou et al., 2018). We found absolute 5hmC levels to be correlated with the stem cell state and to exhibit a genomic distribution pattern similar to 5mC, with enrichment within genes and regulatory elements. The balance of 5mC and 5hmC, particularly at CGIs, is important for gene expression. TET1 specifically is involved in maintaining higher levels of 5hmC in a low-oxygen environment, associates with transcriptionally active genes both directly and through enhancer-promoter interactions, and keeps a subset of key promoters and CGIs free of 5mC. Overall, these data show an important epigenetic modulatory function of TET1 and 5hmC in preserving the stem cell state of the trophoblast compartment.
Results
Global Distribution Patterns of Methylation and Hydroxymethylation Are Similar in TSCs
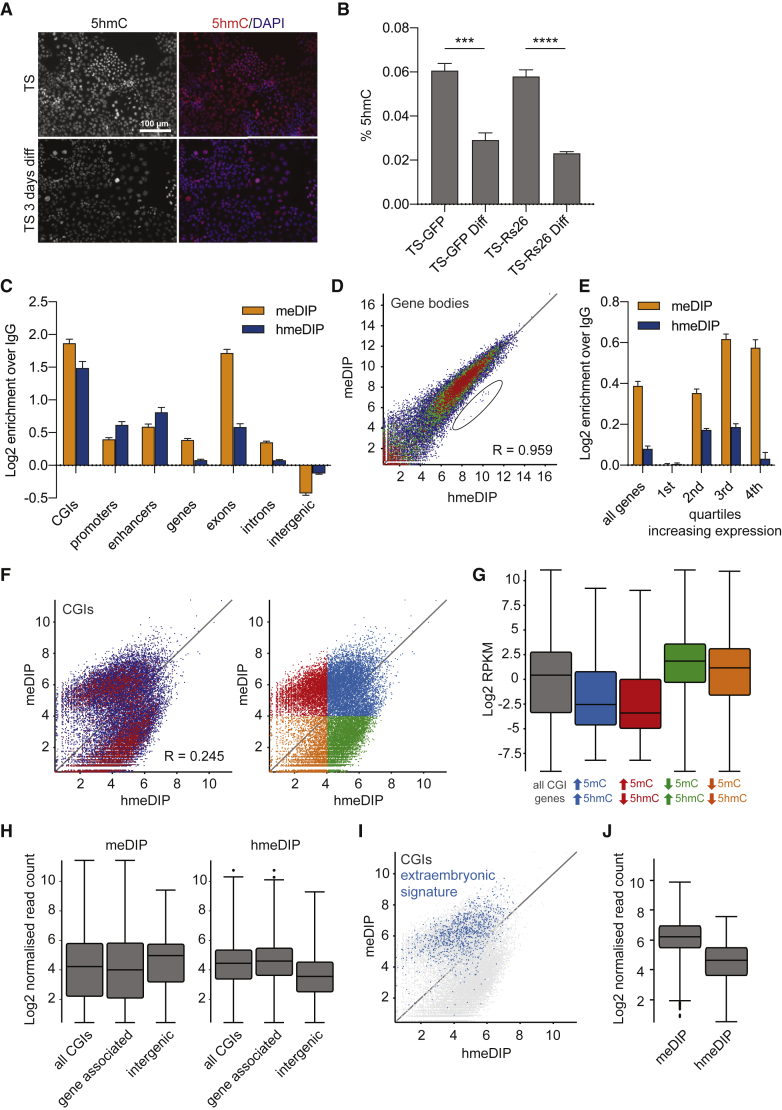
We previously showed that mRNA and protein expression of TET1 and TET2 is correlated with the TSC state (Chrysanthou et al., 2018). To determine if 5hmC has the same correlation we carried out immunofluorescence and mass spectrometry on TSCs cultured in stem cell conditions and upon differentiation for 3 days. We detected robust staining intensity for 5hmC in undifferentiated TSCs, which was reduced upon differentiation (Figure 1A). Mass spectrometry revealed around 0.06% of cytosines to harbor the 5hmC modification (Figure 1B). This is in contrast to around 0.2% 5hmC/C in ESCs (Figure S1A) and around 4% 5mC/C in both ESCs and TSCs (Senner et al., 2012). The mass spectrometric analysis also showed that the level of 5hmC decreased in differentiated TSCs (Figure 1B), thus confirming the association of 5hmC with the stem cell state in trophoblast.
Figure 1.
Global Comparison of 5mC and 5hmC in Trophoblast Stem Cells
(A) 5hmC immunofluorescence of TSCs in stem cell conditions ("TS", top) and after 3 days of differentiation ("TS 3 days diff", bottom). Red, 5hmC; blue, DAPI.
(B) Global 5hmC levels in TS-GFP and TS-R26 lines measured by mass spectrometry, expressed as a percentage of total cytosines (n = 3). ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001 (unpaired t test).
(C) Relative enrichments of meDIP (orange) and hmeDIP (blue) sequencing reads over IgG control across different genomic features (n = 3 each). Enrichments were converted to a log2 scale.
(D) Scatterplot showing log2 normalized read counts mapping to gene bodies from meDIP and hmeDIP-seq (n = 3 each). R, Pearson's correlation. Outliers corresponding to mitochondrial genes are circled.
(E) Relative enrichments of meDIP (orange) and hmeDIP (blue) sequencing reads over IgG control mapping to all genes and genes divided into quartiles of increasing expression levels (n = 3 each).
(F) Scatterplot showing log2 normalized meDIP and hmeDIP-seq (n = 3 each) read counts mapping to CpG islands (CGIs). R, Pearson's correlation. Divisions into 5mC-high and 5hmc-high (blue), 5mC-high and 5hmC-low (red), 5mC-low and 5hmC-high (green), and 5mC-low and 5hmC-low (orange) are indicated on the right-hand scatterplot. A normalized log2 read count value of 4 was used as cutoff to define “high” and “low” populations.
(G) Box-whisker plots showing expression levels (log2 reads per kilobase per million mapped reads [RPKM]) of all CGI-regulated genes (gray) and those with CGIs falling into the four CGI categories indicated in (F). RNA-seq data (n = 3) are from Chrysanthou et al. (2018).
(H) Box-whisker plots showing log2 normalized read counts at all CGIs, gene-associated CGIs and intergenic CGIs from meDIP-seq (left) and hmeDIP-seq (right) (n = 3 each).
(I) Scatterplot showing log2 normalized read counts mapping to CGIs from meDIP and hmeDIP-seq (n = 3 each). Extraembryonic methylated (“extraembryonic signature”) CGIs are highlighted in blue.
(J) Box-whisker plots showing log2 normalized read counts at extraembryonic methylated CGIs from meDIP-seq (left) and hmeDIP-seq (right). n = 3 each.
See also Figure S1.
To compare the global distribution of both 5mC and 5hmC, we carried out meDIP-seq and hmeDIP-seq in TSCs. While absolute levels of 5mC and 5hmC are quite different, the overall distribution patterns were similar (Figure S1B), consistent with 5mC being the substrate for 5hmC. When investigating individual genomic features, we found both modifications to be enriched in regulatory elements, such as CGIs (Illingworth et al., 2010), promoters (−1 kb to +100 bp around transcriptional start sites) and TSC enhancers (Schoenfelder et al., 2018), and depleted in intergenic regions (Figures 1C, S1C, and S1D). This is consistent with previous data for 5mC in TSCs (Senner et al., 2012) and for 5hmC in other cell types (Ficz et al., 2011, Pastor et al., 2011, Williams et al., 2011, Wu et al., 2011a).
We then investigated features that were differentially enriched for 5mC or 5hmC. We found a higher level of enrichment of 5mC than 5hmC within genes, especially exons (Figure 1C). Despite the enrichment difference, however, the correlation of both modifications was still high: where one modification was abundant, the other one was also enriched. This was evident when normalized read counts of 5mC and 5hmC over genes were compared (Figure 1D). We noted only a small group of outlier genes with a higher 5hmC:5mC ratio that corresponded to mitochondrial genes. Both 5mC and 5hmC have been reported in both mouse and human mitochondrial DNA (Shock et al., 2011). The relatively high abundance of 5hmC suggests dynamic regulation of mitochondrial genes in TSCs.
When genes were divided into quartiles depending on their level of transcription and these groups assessed for relative enrichment of 5mC and 5hmC, we found that genes with no or very low expression were largely devoid of either modification (Figure 1E). The enrichment of both 5mC and 5hmC increased through the second and third quartiles. Interestingly, in the fourth quartile, representing the most highly expressed genes, 5mC enrichment remained high, whereas 5hmC enrichment decreased. While 5mC-enrichment over gene bodies has been previously shown to be correlated with active transcription (Varley et al., 2013), this finding is in contrast to reports of 5hmC over gene bodies correlating with gene expression (Wu et al., 2011a). Indeed, we did not detect appreciable differences in expression levels of genes whose exons were either consistently enriched in 5mC or 5hmC (Figure S1E).
DNA Methylation to Hydroxymethylation Ratio at CGIs Correlates with Gene Expression
Since CGI methylation patterns are a particularly distinctive feature of stem cells (Senner et al., 2012), we examined how 5mC and 5hmC compare at CGIs in TSCs (Figure 1F). Plotting enrichment levels of 5hmC against 5mC identified two distinct populations of CGIs, one that exhibited high levels of 5mC with varying levels of 5hmC, and another with high levels of 5hmC with varying levels of 5mC. We separated the CGIs into four groups according to their 5mC:5hmC ratio, using a log2 normalized read count of 4 to divide the data into the “high” and “low” groups indicated on the scatterplot (Figure 1F). Genes associated with low 5mC and high 5hmC were most highly expressed while those with high 5mC and low 5hmC displayed lower levels of transcription (Figure 1G). This correlation had previously been described for promoters in ESCs (Ficz et al., 2011), and reflects a rapid turnover of methylation at actively transcribed genes. To further confirm this, we assessed the relative levels of 5mC and 5hmC at gene-associated (within a gene body ±2 kb) versus intergenic CGIs where more stable silencing may be expected. Indeed, we found intergenic CGIs to have higher 5mC and lower 5hmC levels than gene-associated CGIs (Figures 1H and S1F). Similarly, our previously described extraembryonic signature CGIs, which are commonly associated with embryonic genes that are stably silenced in extraembryonic stem cell types, displayed relatively high enrichment of 5mC and relatively low enrichment of 5hmC (Figures 1I and 1J), consistent with their repressive function in TSCs.
Hydroxymethylation Is Reduced in Differentiating TSCs
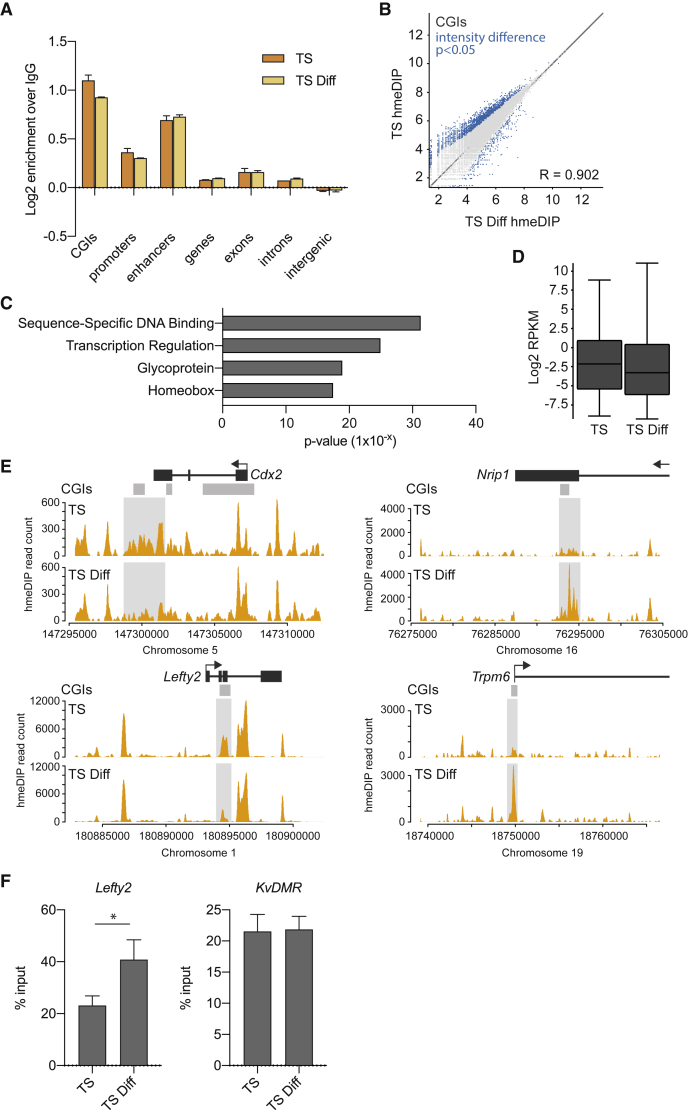
TSC differentiation is associated with a loss in 5hmC (Figures 1A and 1B). To determine where in the genome this depletion occurred, we carried out hmeDIP-seq on TSCs in stem cell conditions (TS) and upon 3 days of differentiation (TS Diff). The resulting profiles were very similar to those generated in the previous experiment described in Figure 1 (Figure S2). While the distribution of 5hmC remained broadly similar across the genome, the loss of 5hmC appeared to specifically occur at CGIs (Figures 2A and 2B). Genes associated with CGIs that lost the most 5hmC (Table S1) were significantly enriched for gene ontology (GO) terms such as DNA-binding, homeobox, and transcriptional regulation, as well as glycoprotein (Figure 2C; Huang et al., 2009). These genes also had modestly lower levels of transcription in differentiated TSCs (Figure 2D). Together, these epigenetic changes may underpin the shifting transcriptional circuitry as TSCs exit the stem cell state and differentiate.
Figure 2.
Comparison of 5hmC Levels and Distribution in Undifferentiated and Differentiated TSCs
(A) Relative enrichments of hmeDIP sequencing reads from TSCs ("TS", orange) and 3-day differentiated TSCs ("TS Diff", light orange) over IgG pull-down controls across different genomic features (n = 2 each). Enrichments were converted to a log2 scale.
(B) Scatterplot showing log2 normalized hmeDIP-seq read counts mapping to CGIs in TSCs and 3-day differentiated TSCs (n = 2 each). An intensity difference filter was applied using SeqMonk software to identify significant differences (blue). R, Pearson's correlation.
(C) Gene ontology analysis of genes associated with CGIs that lose 5hmC in differentiated TSCs. Gene-associated CGIs were defined as being within 1 kb upstream of the transcription start site or falling within the gene body. Analysis was carried out using DAVID (Huang et al., 2009
(D) Box-whisker plots showing expression levels (log2 RPKM) determined by RNA-seq (n = 1) of genes associated with CGIs that lose 5hmC in differentiated TSCs.
(E) Examples of hmeDIP-seq enrichment (n = 2) across genes that are downregulated upon loss of 5hmC (Cdx2, Lefty2) or conversely that are upregulated with a gain of 5hmC (Nrip1, Trpm6) upon TS differentiation.
(F) meDIP-qPCR experiment showing gain of methylation at the Lefty2 CGI in differentiated TSCs. Data are mean ± SEM (n = 3). ∗p < 0.05 (unpaired t test). KvDMR was used as a control region.
See also Figures S2 and S3.
While globally, 5hmC enrichment levels at CGIs were not strongly correlated with transcriptional activity of associated genes, there were a few notable exceptions (Figures 2E and S3A). Cdx2, a core TSC transcription factor known to be downregulated early in TSC differentiation (Tanaka et al., 1998, Latos et al., 2015a, Lee et al., 2019), lost 5hmC from a CGI associated with a putative 3′-enhancer in differentiated TSCs (Watts et al., 2011). Interestingly, depletion of 5hmC was also observed at Lefty2, a transcription factor that is similarly downregulated upon TSC differentiation (Latos et al., 2015a) and indeed is one of the most strongly downregulated genes in Tet1 KO TSCs (Chrysanthou et al., 2018) (Figure 2E). Conversely, Nrip1, a gene involved in placental maturation (Sõber et al., 2015), and Trpm6 expressed in syncytiotrophoblast (Suzuki et al., 2017), both gained 5hmC at CGIs in line with an increase in their transcription levels upon differentiation. To further investigate potential changes in 5mC at these loci, we carried out meDIP-qPCR in TSCs and those differentiated in culture for 3 days. As expected, Lefty2 significantly gained 5mC (Figure 2F) upon differentiation, in line with a shifting 5hmC:5mC ratio in favor of methylation, associated with gene repression. The other three loci exhibited no statistically significant changes in 5mC (Figure S3B). This observation is consistent with CGI hydroxymethylation being more sensitively regulated with transcriptional activity than methylation. Even in the absence of notable 5mC changes, alterations in 5hmC still alter the ratio of the two marks and hence are, in combination, an important modulator of gene activity.
Low-Oxygen Conditions Maintain Higher Levels of Hydroxymethylation in TSCs
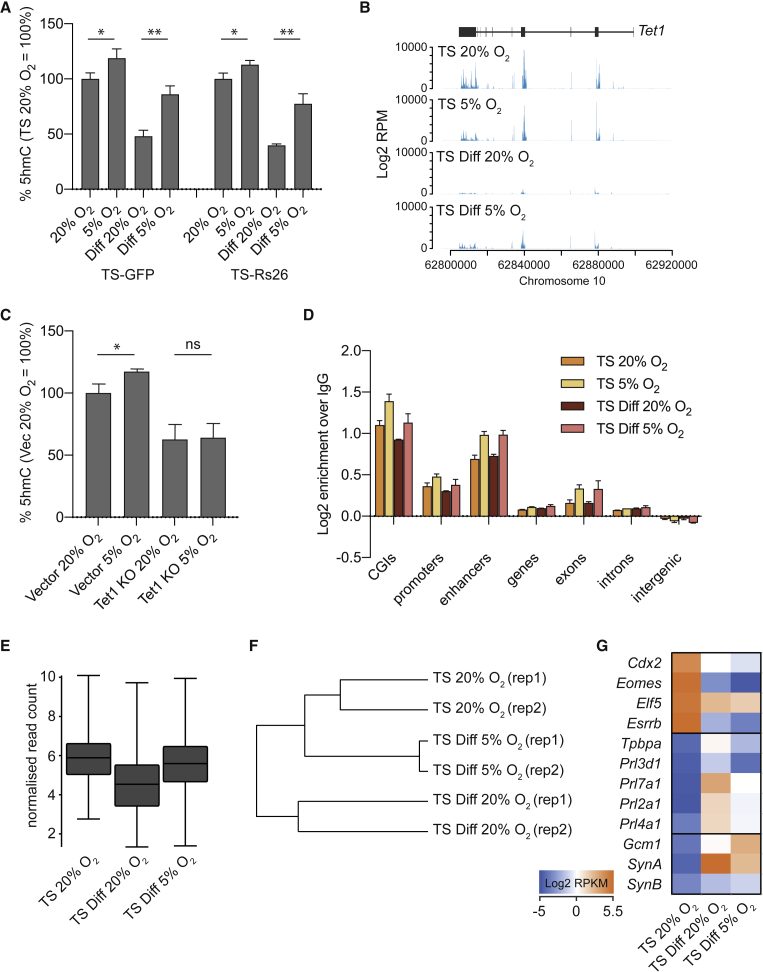
We previously reported that trophoblast tissue from embryonic day 7.5 had higher levels of 5hmC than TSCs in culture (Senner et al., 2012). We hypothesized that this may be due to the lower-oxygen environment of the developing trophoblast before the establishment of placental circulation (around 3%). Since TET catalytic activity is dependent on Fe(II) and hence a reducing environment (Ponnaluri et al., 2013), we tested the effect of oxygen tension on 5hmC levels and distribution specifically in TSCs. TSCs exposed to low-oxygen (5%) conditions exhibited significantly higher amounts of 5hmC both in the TSC state and upon differentiation (Figure 3A). Indeed, 5% O2 preserved 5hmC levels in differentiating conditions at levels more similar to those of undifferentiated TSCs grown in standard 20% O2 conditions. The increased levels of 5hmC were correlated with higher expression of Tet1 and Tet2 at both the RNA and protein levels (Figures 3B and S4A–S4C). This low-oxygen-dependent increase in 5hmC was not observed in Tet1 KO TSCs, suggesting that TET1 was the main activity conferring the increased 5hmC levels in 5% O2 (Figure 3C). Further to this, an increase in the nuclear:cytoplasmic ratio of TET1 protein was observed in low-oxygen conditions, indicative of a greater nuclear availability of enzyme to catalyze the conversion of 5mC to 5hmC in 5% O2 (Figure S4D).
Figure 3.
Changes in 5hmC Levels and Distribution at 5% Oxygen
(A) Global 5hmC levels as measured by mass spectrometry in two TSC lines (TS-GFP and TS-R26) in the stem cell state and upon differentiation for 3 days (Diff) at 20% and 5% O2 (n = 3 each). 5hmC levels are expressed as a percentage of 5hmC detected in undifferentiated TSCs at 20% O2. ∗p < 0.05, ∗∗p < 0.005 (unpaired t test).
(B) RNA-seq data showing differences in Tet1 expression levels in TSCs ("TS") at 20% and 5% O2, and TSCs differentiated for 3 days ("TS Diff") at 20% O2 and 5% O2 (n = 1 each).
(C) Global 5hmC levels, measured by mass spectrometry, in empty vector control TSCs and Tet1 KO TSCs generated by CRISPR (Chrysanthou et al., 2018) cultured at 20% and 5% O2 (n = 3 each). 5hmC levels are expressed as a percentage of the 5hmC amount detected in vector controls at 20% O2. ∗p < 0.05 (unpaired t test).
(D) Relative enrichment of 5hmC across different genomic features in TSCs grown at 20% (orange) and 5% O2 (light orange), and after 3 days differentiation at 20% (red) and 5% O2 (pink). hmeDIP-seq: n = 2 each. Enrichments were converted to a log2 scale.
(E) Box-whisker plot showing log2 normalized RNA-seq read counts over CGIs that lose 5hmC in TSCs differentiated for 3 days at 20% O2, as described in Figure 2. The box-whisker plot shows data from TSCs at 20% O2, and from TSCs differentiated for 3 days at 20% and 5% O2 (n = 1 each).
(F) Data store tree diagram based on a Pearson's correlation distance matrix of hmeDIP-seq reads mapping to CGIs in TSCs at 20% O2 and after 3 days differentiation in 20% and 5% O2.
(G) Heatmap of RNA-seq data showing expression levels (log2 RPKM) of TSC markers (Cdx2, Eomes, Elf5, Esrrb), giant cell markers (Tpbpa, prolactin-like genes) and syncytiotrophoblast markers (Gcm1, SynA, SynB) in TSCs at 20% O2, TSCs differentiated for 3 days at 20% O2, and TSCs differentiated for 3 days at 5% O2 (n = 1 each).
See also Figure S4.
We then carried out hmeDIP-seq to assess where in the genome 5hmC levels were increased in 5% O2. CGIs, promoters, enhancers, and exons all became more enriched for 5hmC (Figure 3D). This increase seemed to be global (Figure S4E) without many significant outliers. However, those CGIs that lost the most 5hmC upon TSC differentiation in 20% O2 (Figure 2B) retained 5hmC levels in 5% O2 that were more similar to those in undifferentiated TSCs (Figure 3E). Indeed, hierarchical clustering of CGI 5hmC enrichment indicated that TS Diff at 5% O2 were more similar to undifferentiated TSCs than to TS Diff at 20% O2 (Figure 3F). Although TSC transcription factors Cdx2, Eomes, Elf5, and Esrrb were downregulated appropriately in 5% O2, terminal differentiation markers Tpbpa, Prl3d1, Prl7a1, Prl4a1, Prl2a1, SynA, and SynB were not upregulated to the same extent in 5% O2 compared with normoxic conditions (Figure 3G). While these differentiation markers are not direct targets of 5hmC, their attenuated upregulation is likely a secondary consequence of the persistent 5hmC marks at stem cell state-associated transcription factors (Figures 2C and 2D). Gcm1, an early syncytiotrophoblast marker that usually peaks after 1–2 days of differentiation (Anson-Cartwright et al., 2000), remained more highly expressed in TS Diff at 5% versus TS Diff at 20% (Figure 3G), as expected if differentiation was delayed. Overall, these data suggest that the process of differentiation may be slowed down in 5% O2 due to increased TET1 and 5hmC levels.
TET1 Associates with Active Genes
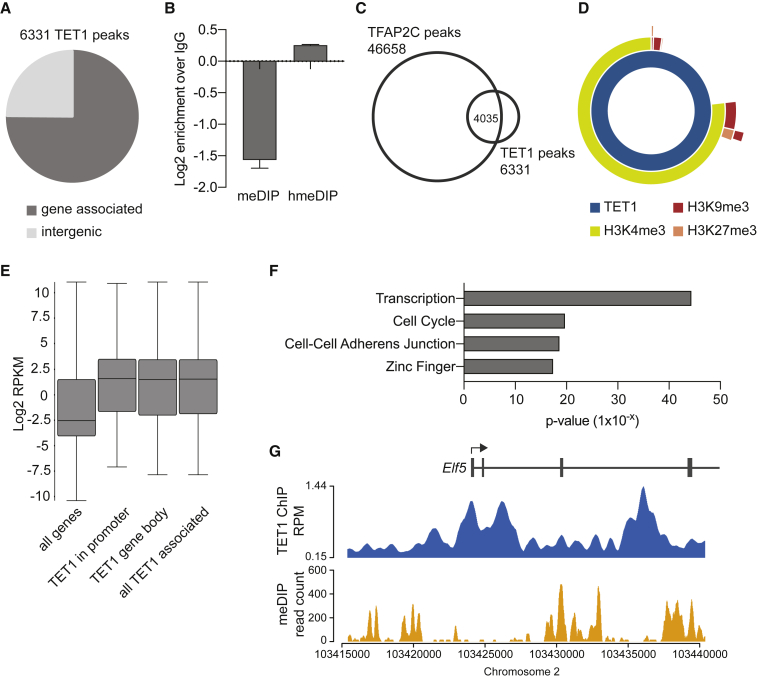
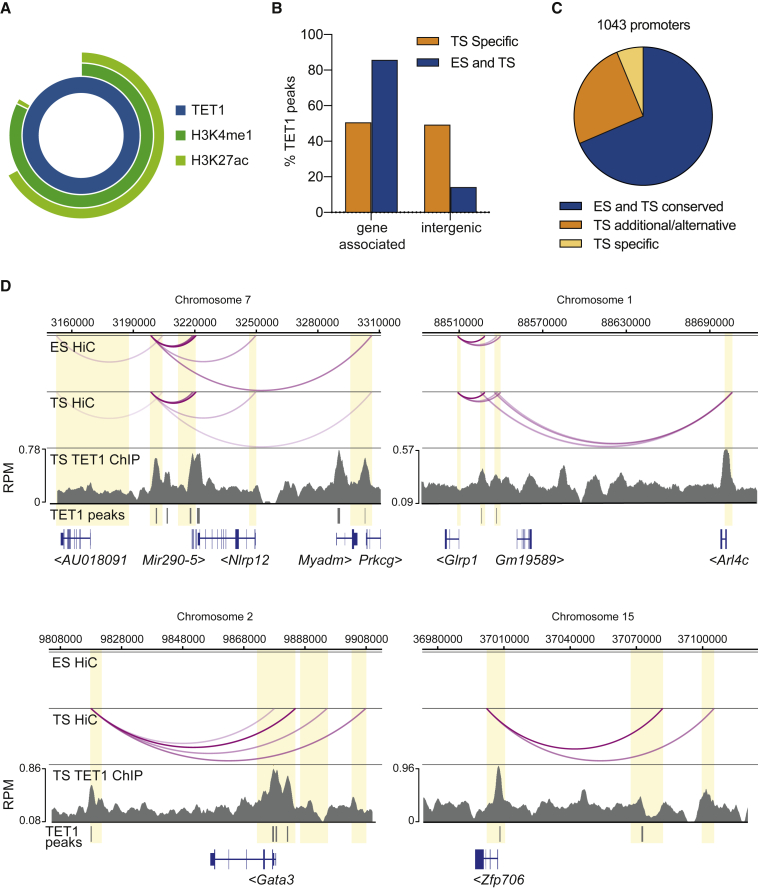
To better understand how TET1 regulates the TSC state, we assessed TET1 binding patterns (ChIP-seq) in TSCs and identified 6,331 TET1 peaks, three-quarters of which were associated with a gene (±2 kb) (Figure 4A). As expected, TET1 peaks were enriched for 5hmC and depleted in 5mC (Figure 4B). Intriguingly, almost two-thirds of all TET1 peaks overlapped with TFAP2C binding sites (Figure 4C). The gene-associated TET1 peaks also extensively overlapped with the active histone modification H3K4me3 but showed very little overlap with repressive marks H3K27me3 and H3K9me3 (Figure 4D) (Chuong et al., 2013). Consistent with this, TET1-associated genes (i.e., genes with a TET1 peak within the gene body ±2 kb) were more highly expressed than average (Figure 4E). Thus, it appears that TET1 is predominantly associated with active genes in TSCs. TET1-bound genes in TSCs were enriched for GO terms, such as transcription, cell cycle, cell-cell adherens junction, and zinc finger (Figure 4F), consistent with the recognized role of TET1 in the regulation of the TSC cycle, EMT, and the trophoblast stem cell state (Chrysanthou et al., 2018). We also specifically noted that all of our previously described “gatekeeper” genes, whose differential promoter methylation constitutes an epigenetic barrier between the trophoblast and embryonic lineages (Ng et al., 2008, Cambuli et al., 2014), harbored TET1 peaks in their hypomethylated regions, indicating that TET1 plays a role in maintaining their expression in the trophoblast lineage (Figures 4G, S5A, and S5B). This role is likely to be more modulatory than essential as only Elf5 is differentially expressed in Tet1 KO TSCs (Chrysanthou et al., 2018). In fact, of more than 4,000 TET1-bound genes, only 56 were differentially expressed in Tet1 KO TSCs (Figure S5C). This may in part be due to our high stringency peak calling, but is also consistent with a previous study reporting minimal gene expression changes upon deletion of Tet1 in extraembryonic ectoderm (Khoueiry et al., 2017). Among the TET1-bound genes that were differentially expressed was Cdh1 as described above and the key trophoblast transcription factor Gata3.
Figure 4.
Analysis of TET1 ChIP-Seq Data
(A) Pie chart showing 6331 TET1 ChIP-seq (n = 3) peaks in TSCs (Chrysanthou et al., 2018), of which 4,745 were gene-associated (± 2 kb of a gene) and 1,574 intergenic.
(B) Relative enrichments of meDIP and hmeDIP sequencing reads over IgG controls mapping to TET1 peaks (n = 3 each). Enrichments were converted to a log2 scale.
(C) Venn diagram showing overlap of TET1 and TFAP2C peaks.
(D) Sunburst diagram showing overlap of gene-associated TET1 peaks with active histone modification H3K4me3, and repressive modifications H3K9me3 and H3K27me3.
(E) Box-whisker plot showing expression (log2 RPKM, n = 3) of all genes, genes with TET1 peaks in the promoter, in the gene body, and all TET1-associated genes combined.
(F) Gene ontology analysis of TET1-associated genes. Analysis was carried out using DAVID (Huang et al., 2009).
(G) TET1 peaks at the hypomethylated Elf5 locus as an example of TET1 binding to “gatekeeper” genes.
See also Figure S5.
Intergenic TET1 Is Associated with Extensive Promoter Interactions
One-quarter of TET1 peaks were located in intergenic regions (Figure 4A). These intergenic peaks overlapped extensively with enhancer marks H3K4me1 and H3K27ac (Chuong et al., 2013) (Figure 5A). To determine if these intergenic TET1 peaks may be TSC-specific enhancers, we incorporated previously published TET1 ChIP-seq data from ESCs (Williams et al., 2011) into our analysis. Using the same peak calling parameters as for the TSC data, we identified 89,074 TET1 peaks in ESCs of which a similar proportion, just over a quarter, mapped to intergenic regions (Figure S5D). Two-thirds of the TET1 peaks found in TSCs were shared with those in ESCs, whereas the remaining third (1,921/6,331) was only found in TSCs. This collection of TSC-specific TET1 peaks was disproportionately enriched in intergenic regions (Figure 5B). We then looked for interactions between these TSC-specific intergenic sites and promoters in our recently published promoter capture Hi-C dataset (Schoenfelder et al., 2018), and identified 1,041 interacting promoters (Table S2). These fell into three categories (Figure 5C): (1) those conserved between ESCs and TSCs, such as the Mir290-5 cluster, (2) those where the intergenic site is involved in promoter interactions in both ESCs and TSCs but additional or alternative promoter interactions are present in TSCs, such as the Arl4c promoter, and (3) those that were entirely unique to TSCs, such as the Gata3 and Zfp706 promoters (Figure 5D). Interestingly Arl4c is downregulated in Tet1 KO TSCs, indicating a direct regulation through this promoter-enhancer interaction. Gata3 is also downregulated in Tet1 KO TSCs. Gata3, therefore, appears to be regulated by TET1 directly both by TET1 binding to its promoter and through this enhancer-promoter interaction.
Figure 5.
Analysis of Intergenic TET1 Peaks
(A) Sunburst diagram showing overlap of intergenic TET1 peaks with enhancer-associated histone modifications H3K4me1 and H3K27Ac.
(B) Graph showing the percentage of TSC-specific TET1 peaks, and TET1 peaks that are common to ESCs ("ES") and TSCs ("TS"), that are gene-associated or intergenic. TET1 ChIP-seq data from ESCs was from Williams et al. (2011) (n = 2).
(C) Pie chart showing proportions of different types of intergenic TET1 peak interactions derived from promoter capture Hi-C data (Schoenfelder et al., 2018): those conserved between ESCs and TSCs, those where the intergenic site is involved in promoter interactions in both ESCs and TSCs but additional or alternative promoter interactions are present in TSCs, and those that were entirely unique to TSCs.
(D) Examples of intergenic TET1 peak interactions. Interactions at the Mir290-5 cluster (top left) are conserved between ESCs and TSCs. Interactions at the Arl4c locus (top right) include some that are present in ESCs and TSCs but additional promoter interactions are present in TSCs. Interactions shown at the Gata3 (bottom left) and Zfp706 (bottom right) promoters are unique to TSCs. TET1 peaks are indicated by the gray bars; yellow shaded boxes indicate interacting restriction fragments of the Hi-C analysis.
See also Figure S5.
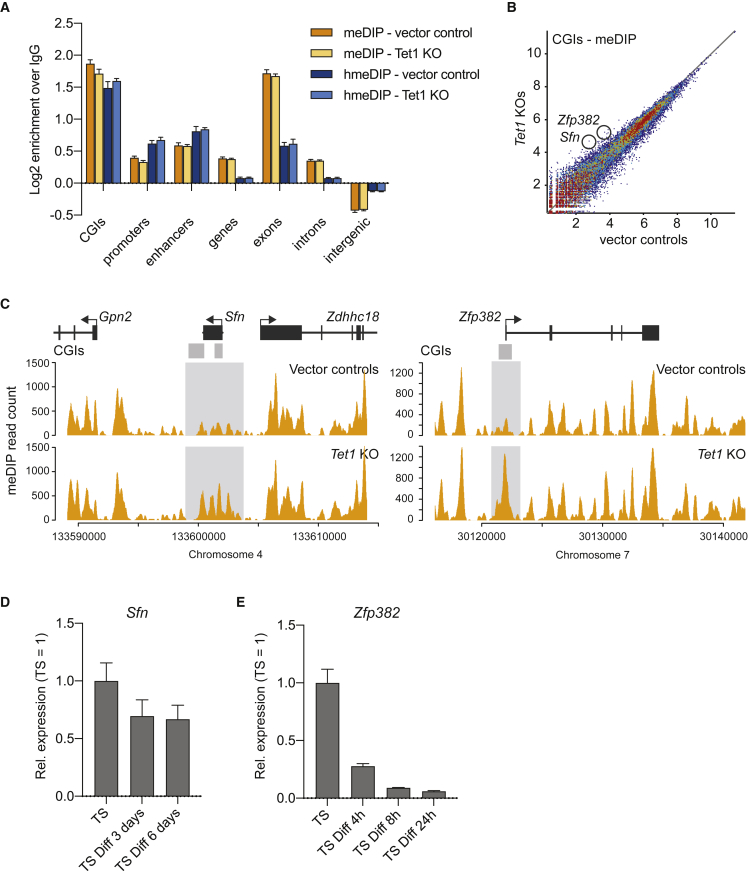
Gain of 5mC in Tet1-Deficient TSCs Identifies Novel Candidate Trophoblast Regulators
To further understand how TET1 binding may contribute to the regulation of gene expression in the trophoblast lineage, we carried out meDIP-seq and hmeDIP-seq in Tet1 KO TSCs. The distribution of 5hmC was largely unchanged in Tet1 KOs (Figures 6A and S6A). We reasoned that if a gene was regulated by TET1, its regulatory regions may exhibit increased 5mC levels in the Tet1-null situation. We therefore screened for CGIs and promoters that gained 5mC in the Tet1 KOs (Figures 6B and S6B). Among these, two genes, Sfn and Zfp382, stood out because they both gained 5mC at specific CGIs (Figure 6C), and because of their expression pattern in KO TSCs: Sfn, also known as 14-3-3 sigma, was significantly downregulated in Tet1 KO TSCs (Chrysanthou et al., 2018 and Figure S4C). Zfp382 contains a TET1 peak and was also downregulated to some extent in Tet1 KO TSCs, albeit not significantly (Figure S6C). It is likely, therefore, that TET1 directly regulates both Sfn and Zfp382 in TSCs, and suggests previously undiscovered roles for these two genes in trophoblast biology. It should be noted that the gain of methylation at Zfp382 was observed in only two out of three of our replicates (Figure S6D), indicating that the generation of KO clones resulted in some epigenetic variability and/or that other factors are involved in keeping this CGI free of methylation. Sfn and Zfp382 exhibited distinct expression patterns (Figure 6D). Sfn expression was maintained during TSC differentiation, pointing to a role in both the stem cell state and in differentiating trophoblast. In contrast, Zfp382 expression was rapidly downregulated after only 4 h in differentiating conditions, implicating this zinc-finger protein in the maintenance of the undifferentiated TSC state. Overall, these examples highlight the power of our approaches to identify putative novel regulators of mouse TSC biology.
Figure 6.
5mC and 5hmC Distribution in Tet1 KO Trophoblast Stem Cells
(A) Relative enrichments of meDIP vector control (orange), meDIP Tet1 KO (light orange), hmeDIP vector control (dark blue), and hmeDIP Tet1 KO (light blue) sequencing reads, normalized to IgG control and expressed as log2 transformed values, across different genomic features (n = 3 each).
(B) Scatterplot showing log2 normalized meDIP-seq read counts mapping to CGIs in vector control and Tet1 KO TSCs (n = 3 each). CGIs associated with Sfn and Zfp382 are circled.
(C) meDIP-seq reads mapping to the Sfn and Zfp382 loci in vector controls and Tet1 KO TSCs (n = 3 each).
(D) RT-qPCR of Sfn in undifferentiated TSCs and TSCs differentiated for 3 and 6 days. Expression is normalized to housekeeping genes Sdha and Dynein with the undifferentiated time point set to 1. Data are mean ± S.E.M. (n = 3).
(E) RT-qPCR of Zfp382 in undifferentiated TSCs and TSCs differentiated for 4, 8, and 24 h. Expression is normalized to housekeeping genes Sdha and Gapdh with the undifferentiated time point set to 1. Data are mean ± SEM (n = 3).
See also Figure S6.
Discussion
In this study, we describe the distribution of 5mC and 5hmC in TSCs. Much like in ESCs, both modifications were enriched in regulatory elements and within gene bodies. Enrichment of 5mC was greater within gene bodies than 5hmC. Gene body methylation has been extensively described in ESCs (Lister et al., 2009) where it is believed to facilitate splicing or to prevent aberrant initiation of transcription (Greenberg and Bourc'his, 2019). The comparatively lower levels of 5hmC imply that gene body methylation is relatively stable. Conversely, hydroxymethylation was more highly enriched in CGIs, promoters, and enhancers, suggesting a more dynamic regulation of DNA methylation at these regulatory genomic features with frequent turnover of the modification by TET enzymes. At CGIs, the balance of 5mC and 5hmC correlated with transcription levels of the associated genes, where low 5mC and high 5hmC correlated with higher expression levels, whereas high 5mC and low 5hmC was indicative of silent or lowly expressed genes. Furthermore, CGIs not associated with active transcription, such as intergenic CGIs and CGIs known to be highly methylated in extraembryonic lineages (Senner et al., 2012), exhibited a higher enrichment of 5mC and a lower abundance 5hmC.
Like Tet1 expression (Chrysanthou et al., 2018), we found that the absolute abundance of 5hmC was associated with the TSC state, with levels dropping to about 40%–50% upon differentiation for only 3 days in culture. This is perhaps to be expected as dynamic regulation of the methylation landscape would be necessary in a multipotent cell type, but then becomes less crucial as cells differentiate and gene expression patterns become fixed.
Interestingly, culturing cells in 5% O2 during differentiation maintained 5hmC levels and seemed to delay the differentiation process. The higher levels of 5hmC levels in 5% O2 may be due to an increased pool of Fe(II), higher levels of TET1 mRNA and protein levels, or indeed the higher nuclear:cytoplasmic ratio of TET1 in TSCs cultured in 5% versus 20% O2. Although the low-oxygen environment did not prevent the downregulation of critical stem cell transcription factors, it seemed to delay the upregulation in particular of trophoblast giant cell differentiation markers. Culture in low-oxygen conditions may indeed better reflect the situation in vivo where O2 levels in the developing trophoblast compartment are around 3% before the establishment of feto-maternal blood flow in the placenta. The slower differentiation rate in the lower O2 environment may allow the TSC pool to sufficiently expand before differentiation.
We further found that genes associated with a TET1 peak were on average more highly expressed and that these TET1 peaks overlapped extensively with the active histone modification H3K4me3, while there was almost no overlap with repressive marks H3K9me3 or H3K27me3. This is in stark contrast to the results of the TET1 ChIP-seq in ESCs (Williams et al., 2011, Wu et al., 2011b) where TET1 peaks were detected at both active and repressed genes. In epiblast cells, TET1 was even shown to repress a majority of target genes (Khoueiry et al., 2017). Furthermore, in ESCs TET1 associates with Polycomb group proteins and is found at bivalent genes marked by both H3K27me3 and H3K4me3. TSCs have little H3K27me3 and hence bivalently marked regions are not a frequent occurrence (Alder et al., 2010, Rugg-Gunn et al., 2010). Together, these results suggest that the TET1 protein does not have the same repressive functions in the trophoblast lineage perhaps due to differing interacting partners in this context. We found extensive overlap of TET1 peaks with previously identified TFAP2C peaks (Latos et al., 2015b). It has been suggested that pioneer transcription factors can recruit TET proteins to particular loci (Lio et al., 2016, Yang et al., 2016). It is tempting to speculate that TFAP2C may recruit TET1 to maintain the hypomethylated state of key regulatory elements in TSCs.
Perhaps most importantly, we found that intergenic TET1 peaks had extensive overlap with enhancer marks H3K4me1 and H3K27ac, many of which were only observed in TSCs and not in ESCs, leading us to postulate that these may constitute trophoblast enhancers. We found that while some of these putative enhancer-promoter interactions are shared with ESCs, others are unique to TSCs and may reveal genes with important roles in the trophoblast lineage, as exemplified by Gata3. We previously found that enhancers bound by TET1 were strongly associated with a higher TSC:ESC expression ratio from interacting promoters. Further there was an association of TET1 bound enhancers with genes that are expressed in TSCs and are then downregulated upon differentiation (Schoenfelder et al., 2018).
Together, these data suggest that TET1 is involved in modulating gene expression in TSCs both directly at the promoter and through longer-range enhancer-promoter interactions in a TSC-specific manner. Indeed, Gata3 is an example of a single gene that appears to be regulated in this way. Moreover, we identify two genes, Sfn and Zfp382, as examples where TET1 seems to be directly required to maintain a low methylation state at their regulatory CGIs, to ensure high expression levels of both genes in the TSC state.
Taken together, here we describe the regulatory functions of 5hmC and TET1 in TSCs. Although 5hmC is less abundant globally in TSCs than in ESCs, we show that it introduces an important layer of gene modulation that is critical to maintain the stem cell state in the trophoblast compartment, in particular through its role in demarcating trophoblast-specific promoter and enhancer sites.
Experimental Procedures
Materials Availability
All reagents used throughout this study are detailed in the Experimental Procedures. Detailed protocols are available upon request.
Resource Availability
Data and Code Availability
All high-throughput sequencing data have been deposited with the GEO under accession number GEO: GSE139049. Additional datasets used in this study are detailed in Table S3.
TSC Culture
TSC lines used were blastocyst-derived TS-Rs26 and TS-GFP (a kind gift of the Rossant lab, Toronto, Canada), as well as Tet1 CRISPR KO and corresponding vector control cell lines (Chrysanthou et al., 2018). TSCs were cultured in routine conditions (Senner et al., 2012, Tanaka et al., 1998). Throughout, “n” denotes independent samples/experiments.
Immunofluorescence
TSCs were grown on coverslips, fixed with 4% paraformaldehyde for 10 min and permeabilized with PBS/0.1% Triton X-100 for 15 min. Further details on staining procedure are provided in the Supplemental Information. Antibodies used were anti-5hmC (Active Motif, 39769) at 1:2,000 dilution, and anti-TET1 (GeneTex, GTX125888) diluted 1:750. Primary antibodies were detected with anti-rabbit Alexa Fluor 568 (Thermo Fisher Scientific, diluted 1:500). Nuclei were counter-stained with DAPI.
Mass Spectrometry
Genomic DNA was digested using DNA Degradase Plus (Zymo Research) according to the manufacturer's instructions and analyzed on a Q-Exactive mass spectrometer (Thermo Scientific) fitted with a nanoelectrospray ion-source (Proxeo). Mass spectral data for C, 5mC, and 5hmC were acquired in high-resolution full-scan mode, and also in selected reaction monitoring (SRM) mode. SRM data, monitoring the transitions 228 → 112.0505 (C), 242 → 126.0662 (5mC), and 258 → 142.0611 (5hmC), were generated by higher-energy collisional dissociation fragmentation using a 1-mass unit parent ion isolation window, a relative collision energy of 10%. Peak areas for the fragment ions were obtained from extracted ion chromatograms of the relevant scans and quantified by external calibration relative to synthetic standards 2′-deoxycytidine (Sigma), and 5-methyl- and 5-hydroxymethyl-2′-deoxycytidine (Berry & Associates). Samples were analyzed in triplicate.
meDIP-Seq and hmeDIP-Seq
MeDIP-seq and hmeDIP-seq was carried out as described previously (Ficz et al., 2011, Senner et al., 2012). In brief, purified genomic DNA was sonicated to yield 150- to 600-bp fragments, and adaptors for paired-end sequencing (Illumina) were ligated using NEBNext DNA Sample Prep Reagent Set 1 (New England Biolabs). Immunoprecipitations were carried out using 500 ng DNA per sample, 1.25 μg anti-5mC antibody (Eurogentec BI-MECY-0100) or anti-5hmC antibody (Active Motif, 39769), and 10 μL Dynabeads coupled with M-280 sheep anti-mouse immunoglobulin G or Protein G (Invitrogen). Pulled down DNA was amplified for 12 (meDIP) or 15 (hmeDIP) cycles with adapter-specific indexed primers. Final clean-up and size selection was carried out with AMPure-XP SPRI beads (Beckman Coulter). Libraries were quantified and assessed using the Kapa Library Quantification Kit (Kapa Biosystems) and Bioanalyzer 2100 System (Agilent). Indexed libraries were sequenced (50-bp paired-end) on an Illumina HiSeq 2500 sequencer. Raw FASTq data were trimmed with TrimGalore, using default parameters, and mapped to the Mus musculus GRCm38 genome assembly using Bowtie2 v.2.2.6, allowing only a single hit per read and guided by gene models from Ensembl v.61. Data analysis was carried out using SeqMonk software (www.bioinformatics.babraham.ac.uk).
RNA-Seq
Total RNA was extracted using TRI Reagent (Sigma), followed by DNase treatment using the TURBO-DNA-free Kit (Life Technologies). mRNA was isolated using the Dynabeads mRNA Purification Kit (Life Technologies) and processed into indexed, strand-specific libraries using the ScriptSeq v.2 RNA-Seq Library Preparation Kit (Epicentre). Libraries were quantified and assessed using the Kapa Library Quantification Kit (Kapa Biosystems) and Bioanalyzer 2100 System (Agilent). Indexed libraries were sequenced with a 100-bp single-end protocol on an Illumina HiSeq 2500 sequencer. Raw FASTq data were trimmed with TrimGalore, using default parameters, and mapped to the Mus musculus GRCm38 genome assembly using TopHat v.2.0.12. Data were quantitated using the RNA-seq quantitation pipeline in SeqMonk software (www.bioinformatics.babraham.ac.uk). For all analyses we excluded some extreme outliers which we believe to be mapping artifacts and a small region on chromosome 11, which is deleted in the TS-Rs26 cell line.
Real-Time qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen). cDNA synthesis was typically performed on 2 μg RNA with RevertAid M-MuLV Reverse Transcriptase (Fermentas) according to the manufacturer's instructions. Real-time qPCR was carried out using SYBR Green JumpStart ReadyMix (Sigma) on a Bio-Rad CFX96 Real-Time PCR Detection System. Data were normalized against housekeeping reference genes Sdha, Dynein, or Gapdh. Primer information is provided in the Supplemental Experimental Procedures.
Author Contributions
C.E.S., S.C., and H.-Y. L. carried out the experiments. C.E.S., S.B., and M.R.B. performed bioinformatics analysis. C.E.S. and M.H. designed the experiments, performed data interpretation, and wrote the manuscript.
Acknowledgments
We thank the Babraham Institute's Bioinformatics, Sequencing, Mass Spectrometry, and Imaging Facilities for their contributions toward this work. We also thank Georgia Lea for helpful discussions. This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC, UK), by a Medical Research Council, UK DTP studentship to S.C., by a Next-Generation Fellowship from the Center for Trophoblast Research, University of Cambridge, to C.E.S., and by a Tier I Canada Research Chair to M.H.
Published: May 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.04.009.
Contributor Information
Claire E. Senner, Email: ces207@cam.ac.uk.
Myriam Hemberger, Email: myriam.hemberger@ucalgary.ca.
Supplemental Information
References
- Alder O., Lavial F., Helness A., Brookes E., Pinho S., Chandrashekran A., Arnaud P., Pombo A., O'Neill L., Azuara V. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137:2483–2492. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson-Cartwright L., Dawson K., Holmyard D., Fisher S.J., Lazzarini R.A., Cross J.C. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 2000;25:311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- Arima T., Hata K., Tanaka S., Kusumi M., Li E., Kato K., Shiota K., Sasaki H., Wake N. Loss of the maternal imprint in Dnmt3Lmat-/- mice leads to a differentiation defect in the extraembryonic tissue. Dev. Biol. 2006;297:361–373. doi: 10.1016/j.ydbio.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Branco M.R., King M., Perez-Garcia V., Bogutz A.B., Caley M., Fineberg E., Lefebvre L., Cook S.J., Dean W., Hemberger M., Reik W. Maternal DNA methylation regulates early trophoblast development. Dev. Cell. 2016;36:152–163. doi: 10.1016/j.devcel.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambuli F., Murray A., Dean W., Dudzinska D., Krueger F., Andrews S., Senner C.E., Cook S.J., Hemberger M. Epigenetic memory of the first cell fate decision prevents complete ES cell reprogramming into trophoblast. Nat. Commun. 2014;5:5538. doi: 10.1038/ncomms6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysanthou S., Senner C.E., Woods L., Fineberg E., Okkenhaug H., Burge S., Perez-Garcia V., Hemberger M. A critical role of TET1/2 proteins in cell-cycle progression of trophoblast stem cells. Stem Cell Rep. 2018;10:1355–1368. doi: 10.1016/j.stemcr.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E.B., Rumi M.A., Soares M.J., Baker J.C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M.M., Ganz K., Powell B.E., Hu Y.C., Markoulaki S., Cheng A.W., Gao Q., Kim J., Choi S.W., Page D.C., Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Greenberg M.V.C., Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- Hattori N., Nishino K., Ko Y.G., Hattori N., Ohgane J., Tanaka S., Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Hattori N., Imao Y., Nishino K., Hattori N., Ohgane J., Yagi S., Tanaka S., Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Illingworth R.S., Gruenewald-Schneider U., Webb S., Kerr A.R., James K.D., Turner D.J., Smith C., Harrison D.J., Andrews R., Bird A.P. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoueiry R., Sohni A., Thienpont B., Luo X., Velde J.V., Bartoccetti M., Boeckx B., Zwijsen A., Rao A., Lambrechts D., Koh K.P. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat. Genet. 2017;49:1061–1072. doi: 10.1038/ng.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos P.A., Goncalves A., Oxley D., Mohammed H., Turro E., Hemberger M. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat. Commun. 2015;6:7776. doi: 10.1038/ncomms8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos P.A., Sienerth A.R., Murray A., Senner C.E., Muto M., Ikawa M., Oxley D., Burge S., Cox B.J., Hemberger M. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 2015;29:2435–2448. doi: 10.1101/gad.268821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Q.E., Bailey A., Lopez-Tello J., Sferruzzi-Perri A.N., Okkenhaug K., Moffett A., Rossant J., Hemberger M. Inhibition of phosphoinositide-3-kinase signaling promotes the stem cell state of trophoblast. Stem Cells. 2019;37:1307–1318. doi: 10.1002/stem.3052. [DOI] [PubMed] [Google Scholar]
- Lio C.W., Zhang J., González-Avalos E., Hogan P.G., Chang X., Rao A. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife. 2016;5:e18290. doi: 10.7554/eLife.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R.K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M., McLoughlin E.M., Brudno Y., Mahapatra S., Kapranov P. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnaluri V.K., Maciejewski J.P., Mukherji M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem. Biophys. Res. Commun. 2013;436:115–120. doi: 10.1016/j.bbrc.2013.05.077. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Cox B.J., Ralston A., Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl. Acad. Sci. U S A. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Mifsud B., Senner C.E., Todd C.D., Chrysanthou S., Darbo E., Hemberger M., Branco M.R. Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages. Nat. Commun. 2018;9:4189. doi: 10.1038/s41467-018-06666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F., Hendrich B., Reik W., Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Sakaue M., Ohta H., Kumaki Y., Oda M., Sakaide Y., Matsuoka C., Yamagiwa A., Niwa H., Wakayama T., Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr. Biol. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Senner C.E., Krueger F., Oxley D., Andrews S., Hemberger M. DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells. 2012;30:2732–2745. doi: 10.1002/stem.1249. [DOI] [PubMed] [Google Scholar]
- Shock L.S., Thakkar P.V., Peterson E.J., Moran R.G., Taylor S.M. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U S A. 2011;108:3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sõber S., Reiman M., Kikas T., Rull K., Inno R., Vaas P., Teesalu P., Marti J.M.L., Mattila P., Laan M. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci. Rep. 2015;5:13336. doi: 10.1038/srep13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Watanabe M., Saito C.T., Tominaga M. Expression of the TRPM6 in mouse placental trophoblasts; potential role in maternal-fetal calcium transport. J. Physiol. Sci. 2017;67:151–162. doi: 10.1007/s12576-016-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A.K., Nagy A., Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.A., Zhang C., Klein-Szanto A.J., Kormish J.D., Fu J., Zhang M.Q., Zaret K.S. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 2011;7:e1002277. doi: 10.1371/journal.pgen.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., D'Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., D'Alessio A.C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li G., Xie R. Decoding the role of TET family dioxygenases in lineage specification. Epigenetics Chromatin. 2018;11:58. doi: 10.1186/s13072-018-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.A., Zhao J.C., Fong K.W., Kim J., Li S., Song C., Song B., Zheng B., He C., Yu J. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 2016;44:8153–8164. doi: 10.1093/nar/gkw498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All high-throughput sequencing data have been deposited with the GEO under accession number GEO: GSE139049. Additional datasets used in this study are detailed in Table S3.