Abstract
Membrane-free intracellular biocondensates are enclosures of proteins and nucleic acids that form by phase separation. Extensive ensembles of nuclear “membraneless organelles” indicate their involvement in genome regulation. Indeed, nuclear bodies have been linked to regulation of gene expression by formation of condensates made of chromatin and RNA processing factors. Important questions pertain to the involvement of membraneless organelles in determining cell identity through their cell-type-specific composition and function. Paraspeckles provide a prism to these questions because they exhibit striking cell-type-specific patterns and since they are crucial in embryogenesis. Here, we outline known interactions between paraspeckles and chromatin, and postulate how such interactions may be important in regulation of cell fate transitions. Moreover, we propose long non-coding RNAs (lncRNAs) as candidates for similar regulation because many form foci that resemble biocondensates and exhibit dynamic patterns during differentiation. Finally, we outline approaches that could ascertain how chromatin-associated membraneless organelles regulate cellular differentiation.
Keywords: phase separation, paraspeckles, NEAT1, chromatin, pluripotent stem cells, differentiation, RNA-binding protein, lncRNA, biocondensates, membraneless organelles
In this review, Drukker and colleagues describe emerging roles of nuclear membraneless organelles in chromatin and how they affect development. Based on studies of stem cells, they describe primarily the roles of a key form of membraneless organelles, named paraspeckles, and the roles that lncRNAs might play in the phase separation of membraneless organelles in the chromatin during development.
Main Text
Introduction
Eukaryotic cells are scattered with micrometer-scale intracellular bodies that are held together by multivalent interactions of nucleic acids and proteins (Shin and Brangwynne, 2017). As their name implies, “membraneless organelles” differ from classical organelles by forming macromolecular assemblies that are not enclosed by lipid bilayers (Figure 1A). The function of membraneless organelles is thought to be connected to the regulation of biochemical processes by spatiotemporal cellular control of their components. The peculiar behavior of membraneless organelles relies on phase separation; namely, entropy-driven “demixing” of components, which produces distinct liquid droplets coined “condensates” (Shin and Brangwynne, 2017). Evidence of the stable liquid-like state of membraneless organelles include their round shape, deformability, and the ability to exchange components with the surrounding environment (Boeynaems et al., 2018; Hyman et al., 2014). Some membraneless organelles can also adopt viscous gel- or solid-like state depending on the concentration of components and their intermolecular connections (Boeynaems et al., 2018).
Figure 1.
Hypothesized Modes of Regulation That Could Affect the Functions of Nuclear Membraneless Organelles
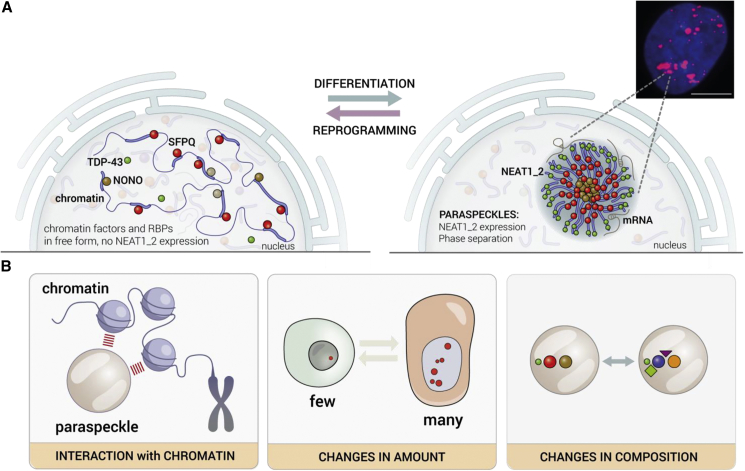
(A) A schematic illustration of paraspeckle assembly and disassembly as an example of a nuclear membraneless organelle that forms by nuclear liquid-liquid phase separation (LLPS). Paraspeckle assembly relies on lncRNA NEAT1_2, chromatin factors, and RBPs, such as SFPQ, and NONO. The figure illustrates that the RNA-binding protein TDP-43 undergoes LLPS in paraspeckles during the differentiation of PSCs, which thereby affects its function. Nucleus of human fibroblast on top to illustrate phase-separated lncRNA complexes with RBPs and chromatin factors. NEAT1_2 probe is depicted in red and DAPI DNA stain in blue. Scale bar: 10 μm.
(B) Principal modes of regulation that affect nuclear membraneless organelles with possible functional cell-type-specific outcomes: left, the interactions of membraneless organelles with chromatin factors expressed and positioned in the genome in a cell-specific manner; middle, the number of condensates formed in a cell-type-specific manner; and right, the cell-type-specific composition of membraneless organelles.
In recent years, a growing list of cellular structures has been associated with phase separation behavior, including P granules in Caenorhabditis elegans (Brangwynne et al., 2009), germ granules in Drosophila melanogaster (Kistler et al., 2018), the nucleoli (Brangwynne et al., 2011; Feric et al., 2016), stress granules (Mateju et al., 2017), and the centrosome (Raff, 2019). Furthermore, an ensemble of nuclear membraneless organelles has been identified through biochemical analyses, including OPT domains, histone body locus, polycomb group (PcG) bodies, splicing speckles, paraspeckles, and nuclear stress bodies (Table 1). Advancements in fluorescent microscopy and photobleaching techniques have expanded the list with an additional set of condensed domains, including heterochromatin, the nuclear pore complex, transcriptional puffs, super enhancers, and clusters of transmembrane receptors (Gomes and Shorter, 2019). It is therefore apparent that a multitude of vastly different membraneless organelles reside in the nucleus, which raises intriguing questions about their relationship with the chromatin, nuclear architecture, and gene expression. Of particular interest are questions pertaining to the amount and composition of nuclear membraneless organelles in different types of cells, and how their cell-type-specific functions are thereby determined.
Table 1.
Properties of Nuclear Membraneless Condensates
| Macromolecular Complex | lncRNA | Protein Interactions | Developmental Process | Chromatin Interactions | Literature Describing Phase Separation |
|---|---|---|---|---|---|
| Nucleolus | pRNA, PAPAS, IGS16 RNA, IGS22 RNA, IGS28 RNA, PNCTR (perinucleolar compartment), LoNA, SLERT | fibrillarin, nucleolin, dyskerin | chromatin organization in embryonic gene activation | chromatin organization of rRNA genes | Brangwynne et al. (2011); Feric et al. (2016); Weber and Brangwynne, (2015) |
| OPT domain/DNA repair compartments | none reported | Oct1, PTF, 53BP1 | none reported | contain gamma-H2AX, a marker of DNA damage | Kilic et al. (2019) |
| PcG body | TUG1 | RING1, BMI1, CBX2 | organization of genome architecture in ESCs | repressive function on gene clusters | Plys et al. (2019); Tatavosian et al. (2019) |
| PML body | none reported | PML, Sp100, p53 | telomeric chromatin integrity and self-renewal in ESCs | associated with gene-rich, transcriptionally active genomic regions | Banani et al. (2016) |
| Paraspeckle | NEAT1, lincRNA-p21 | SFPQ, PSPC1, NONO TDP43, FUS, BRG1 | regulation of PSC differentiation | NEAT1 binding to actively transcribed genes; co-localization with H3K4me3 | Yamazaki et al. (2018) |
| Splicing speckle | MALAT1, 7SK RNA | SC35, SRSF1, SRSF3 | none reported | MALAT1 binding to actively transcribed genes; co-localization with H3K36me3 | Greig et al. (2020) |
| Nuclear stress body | HSATIII | SAFB, SRSF1, SRSF7, HSF1 | none reported | assembled on specific pericentric heterochromatic regions | none reported |
| Histone locus body | Y3/Y3∗∗ RNA | FLASH, NPAT | none reported | assembled at replication-dependent histone genes | Hur et al. (2020) |
| Cajal body | TERC RNA | Coilin, SMN1 | small nuclear ribonucleoprotein particle assembly during embryogenesis | dynamic shuttling between chromatin and interchromatin space; contribute to genome organization of CB-associated genes | none reported |
| Gem | none reported | SMN1, GEMIN2-8 | none reported | none reported | none reported |
| XIST foci | XIST | Spen | X-chromosome inactivation during female development | Spread along condensed X-chromosome | Cerase et al. (2019) (hypothesis) |
The regulation of the chromatin is highly dynamic in development as it determines cellular identity in multicellular organisms. Zygotic genome activation marks the first event that is accompanied by chromatin rearrangements, including the exchange of paternal protamine proteins by maternal nucleosomes that contain the histone variant H3.3 (Perino and Veenstra, 2016). The first cell fate decisions (namely, of trophectoderm progenitors and pluripotent cells of the inner cell mass [ICM]) are accompanied by de novo DNA methylation (Santos et al., 2002) and histone H3 arginine and lysine methylation (Paul and Knott, 2014). Important rearrangements of the chromatin also take place in the differentiation of pluripotent stem cells (PSCs). In the undifferentiated state, PSCs exhibit transcriptionally active “open” chromatin that is maintained by ATP-dependent chromatin-remodeling complexes of the SWI/SNF, CHD, ISWI, and INO80 family. Perturbations of these complexes generally lead to severe differentiation defects (Gaspar-Maia et al., 2011). PSCs also harbor bivalent histone domains, which serve as a priming mechanism for developmental genes during differentiation (Blanco et al., 2020). Acting together, patterns of histone modifications that promote and repress transcription, de novo DNA methylation, and chromatin remodeling, configure the chromatin landscapes to govern the specification of numerous types of differentiated cells (Argelaguet et al., 2019; Xiang et al., 2020). Furthermore, extensive topological restructuring of chromatin interactions that takes place at megabase scale is important for the productive differentiation (Dixon et al., 2015). Considering the intricate restructuring of the chromatin and the vast expanses of intranuclear domains, the association of the chromatin with membraneless organelles is therefore likely to play a key role in the regulation of cellular differentiation (Figure 1B).
Phase separation has been shown to take place extensively in the chromatin. Histone tails, the cohesin complex, and insulator CTCC-binding factor (CTCF) complexes (Gibson et al., 2019; Ryu et al., 2020; Zirkel et al., 2018) exhibit the characteristic attributes. Importantly, the heterochromatin also exhibits features of phase separation mediated by the aggregation of heterochromatin-binding protein HP1 (Strom et al., 2017), although the latter is not instructive for formation of heterochromatin foci in living cells (Erdel et al., 2020). Moreover, distinct nuclear membraneless organelles, including nucleoli (Birch and Zomerdijk, 2008), paraspeckles, splicing speckles (West et al., 2014), and promyelocytic leukemia protein (PML) bodies (Wang et al., 2004) have been found to associate with the chromatin, either by interactions mediated by long non-coding RNAs (lncRNAs) in the case of splicing speckles and paraspeckles, or by chromatin-binding proteins in the case of nucleoli and PML bodies. Another common feature of these nuclear domains, namely paraspeckles, splicing speckles, nucleoli, and Cajal bodies, is their association with RNA-binding proteins (RBPs) that regulate the assembly of ribonucleoprotein complexes (Baβler and Hurt, 2019; Staněk, 2016) and post-transcriptional processes such as splicing and polyadenylation (Modic et al., 2019; Spector and Lamond, 2011). This raises the intriguing possibility that membraneless organelles could be involved in cell-type-specific functions through association with chromatin factors and RBPs.
In this review, we discuss nuclear membraneless organelles in development and cell-type-specific context, and their interactions with chromatin. We focus on paraspeckles as a leading example of a membraneless organelle that has been implicated in the regulation of differentiation states and that has a multifaceted interplay with chromatin. Biochemical modes of membraneless organelle assembly, paraspeckle-associated RBPs, and their disease associations have been covered in detail in other reviews (Alberti and Dormann, 2019; Fox et al., 2018; Hyman et al., 2014; Knott et al., 2016). We emphasize the relevance of lncRNAs for deepening the understanding of nuclear membraneless organelles and chromatin because many nuclear lncRNAs can produce foci that resemble condensates, and they associate with RBPs with phase separation properties. RBPs and lncRNAs also exhibit striking lineage and differentiation stage patterns, which further indicate important connections to the chromatin in the context of phase separation. Finally, we outline directions for future research of novel connections between membraneless organelles, chromatin, and cell fate.
Nuclear Membraneless Organelles in Developmentally Regulated Chromatin Interactions
Current understanding of nuclear membraneless organelles indicates that some are ubiquitous across different types of cells. This includes the nucleolus (Brangwynne et al., 2011; Feric et al., 2016; Weber and Brangwynne, 2015), splicing speckles (Greig et al., 2020), PcG bodies (Plys et al., 2019; Tatavosian et al., 2019), OPT domains (Kilic et al., 2019), histone locus bodies (Hur et al., 2020), and PML bodies (Banani et al., 2016) (Table 1). Despite their constitutive appearance, considerable phenotypic variability has been documented between different types of cells, and between cells within populations. For example, light microscopy revealed that tumor cells tend to exhibit large nucleoli (Derenzini et al., 1998), and the rate of proliferation of tumor cells has been shown to rely on the abundance and the size of nucleoli to enhance overall protein synthesis (Farley et al., 2015). Similarly, hepatocytes exhibit large nucleoli (Boyer et al., 2012) that support their proliferation and high protein synthesis rates (Sayegh and Lajtha, 1989). Further indications include changes of nucleolar size, which are associated with cellular stress taking place during viral infection and DNA damage (Tiku et al., 2018; Weeks et al., 2019). Moreover, the function of PcG bodies seems to be tightly connected to cell fate as these domains exhibit transitions from large to small foci by restructuring the chromatin; for example, during neuronal differentiation (Ren et al., 2008). This indicates that despite being ubiquitous, membraneless organelles can have cell-type-specific functions.
Additional membraneless organelles exhibit an even higher degree of cell-type-specific features. For example, Cajal bodies are apparent in embryonic and fetal tissues and neuronal cells, but not in many other types of somatic cells (Sawyer et al., 2016). In the case of paraspeckles, this is very pronounced. Formerly, the appearance of paraspeckles was thought to be random, because their abundance in cancer cells is highly variable. However, recent studies implicated paraspeckles in embryonic development, PSC differentiation, and cell-type-specific regulation (Grosch et al., 2020; Hupalowska et al., 2018; Modic et al., 2019; Wang et al., 2018). Paraspeckles were found to disappear prior to the formation of the pluripotent ICM of embryos and reappear upon the differentiation of PSCs. It was shown that paraspeckles cluster the histone methyltransferase CARM1 in the chromatin, an association that is required for embryonic development (Hupalowska et al., 2018). Germ layer differentiation is accompanied by paraspeckle formation, which is regulated by the RBP TDP-43, and depletion of paraspeckles impairs human PSC differentiation and mouse embryonic development (Grosch et al., 2020; Modic et al., 2019). Moreover, different cell types exhibit a vast range in the number of paraspeckles (Grosch et al., 2020). It is therefore apparent that investigating mechanisms by which the cell-type-specific appearance of paraspeckles is regulated, and the interactions of paraspeckles with chromatin in different types of embryonic cells, can serve to dramatically expand the understanding of the connections between developmental processes, chromatin, and attributes of membraneless organelles.
Paraspeckles in Chromatin Interactions
The functions of membraneless organelles are often difficult to discern from their components in free form. In this context, paraspeckles provide a unique opportunity to interrogate the biology of biocondensates, because their scaffold is encoded by a lncRNA gene called Nuclear Paraspeckle Assembly Transcript 1 (NEAT1), which can be manipulated with relative ease (Yamazaki et al., 2018). NEAT1_2 is the long isoform of the gene, and it serves as an anchor of over 40 proteins that have been shown in tumor cell lines to co-localize with the lncRNA (Figure 2). Importantly, most of the interacting proteins have annotated functions that are independent of paraspeckles. For example, SFPQ (PSF), NONO (p54nrb), and PSPC1 belong to the multifunctional Drosophila behavior/human splicing (DBHS) factors that produce homo- and heterodimer complexes that can directly bind DNA and RNA and regulate transcription and post-transcriptional processes (Knott et al., 2016). The fact that two of these proteins, namely SFPQ and NONO, are also required for the assembly of paraspeckles (Sasaki et al., 2009) means that they have functions that are related and some that are unrelated to paraspeckles. For example, NONO has been shown to promote pluripotency by enhancing Erk signaling in mouse PSCs (Ma et al., 2016), a role that is clearly unrelated to paraspeckles owing to the fact that undifferentiated PSCs do not express Neat1_2 (Chen and Carmichael, 2009). Also, PSPC1 exhibits a paraspeckle-independent function in PSCs by recruiting the TET2 DNA methyltransferase to transcriptionally active loci (Guallar et al., 2018). Moreover, SFPQ mediates cytoplasmic mRNA transport in axons of motor neurons (Cosker et al., 2016), which exhibit only very few paraspeckles (Shelkovnikova et al., 2018). Similarly, additional proteins that are essential for paraspeckle formation, namely HNRNPK, BRG1, and RBM14, have functions that are unrelated to paraspeckles in the self-renewal of mouse PSCs (Chen et al., 2018a; Lin et al., 2014; Zhang et al., 2014). Because essential paraspeckle proteins are engaged in multifaceted regulation across the genome, it is apparent that primary functions of paraspeckles are connected to spatiotemporal regulation of the chromatin.
Figure 2.
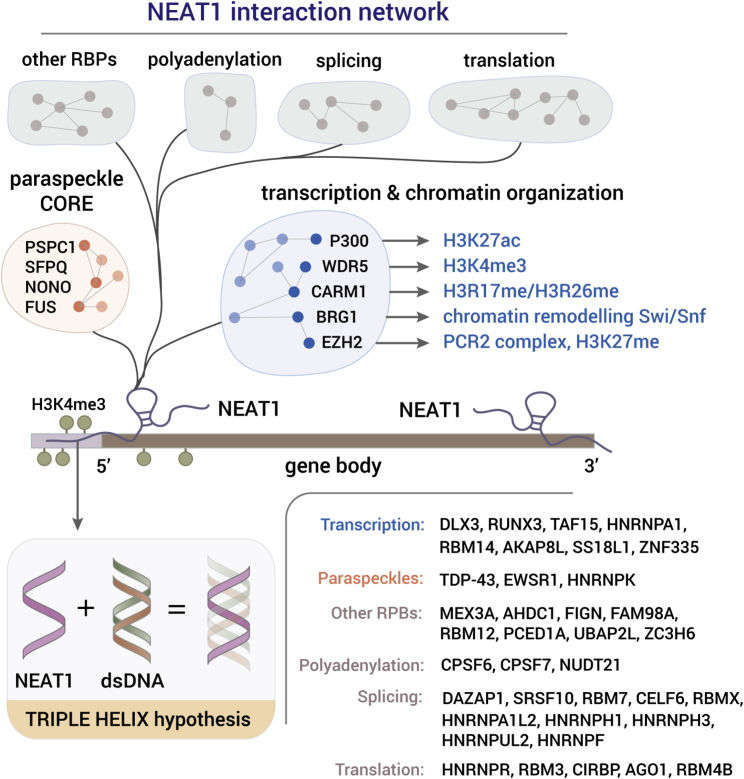
The Interactome of Paraspeckles and NEAT1_2
NEAT1 is predominantly associated with transcription start and termination sites (West et al., 2014), and it interacts with a network of RBPs and chromatin factors. NEAT1 binding to dsDNA is likely mediated via the formation of sequence-specific RNA:DNA triple-helix structures (Sentürk Cetin et al., 2019). Proteins that are known to localize in paraspeckles (Naganuma et al., 2012) are classified in functional categories as indicated at the bottom right corner of the figure. Hypothesized connections between chromatin factors that are known to localize to paraspeckles, known effects of NEAT1/paraspeckles on the chromatin, and the functions of the respective chromatin factors are shown.
Many studies have linked paraspeckles to modulation of histones and nucleosomes, although the question of whether paraspeckles actually determine the genomic locations of histone-modifying complexes and in what cell types is still largely open. Genome-wide crosslinking and pull-down assays using complementary oligonucleotides showed that hundreds of genomic regions are associated with NEAT1 RNA in a tumor cell line. NEAT1 RNA was enriched primarily in actively transcribed genes with highest peaks close to the transcription start and termination sites (Figure 2). Many of these loci also exhibited H3K4me3 enrichment, a marker of actively transcribed genes, and interestingly many were co-occupied by another lncRNA named MALAT1 (West et al., 2014). Further evidence for the connection between paraspeckles and histone modifications is the knock-down of NEAT1 in murine neuroblastoma cells, which led to a significant reduction of the H3K4me2 mark (Butler et al., 2019). The question therefore is whether enzymes that catalyze di- and trimethylation of H3K4 are localized by paraspeckles in the first place, or whether paraspeckles are recruited to actively transcribed regions. Several studies indicate that NEAT1 RNA regulates the positioning of histone marks that promote transcription. For example, the acetylase transferase complex P300/CBP, which catalyzes H3K27 acetylation (Wang et al., 2019b), and WDR5, a subunit of an H3K4 methyltransferase complex (Ahmed et al., 2018), have been shown to interact with NEAT1, and BRG1, BRM, and BAF155, all subunits of the SWI/SNF complex chromatin-remodeling complex, form foci that co-localize with paraspeckles (Kawaguchi et al., 2015).
Contrarily, additional studies indicate that paraspeckles can regulate gene repression. It was shown in glioblastoma (Chen et al., 2018b; Wang et al., 2020) and murine myoblasts (Wang et al., 2019a), that NEAT1 RNA interacts with EZH2, a member of the polycomb repressive complex 2 (PRC2). Moreover, a link has been established in glioblastoma between NEAT1, EZH2, and H3K27me3, the latter being a marker of transcriptional silencing (Chen et al., 2018a; 2018b; Wang et al., 2020). Whether interaction with EZH2 leads to similar outcomes in different cell types remains an open question. Also, it is self-evident that the activity of EZH2 in undifferentiated PSCs is not mediated by paraspeckles because NEAT1 is not expressed in this state. Collectively, these data indicate the possibility that paraspeckles are involved in positioning of repressive histone marks.
Evidence of interactions and positioning of histone-modifying enzymes by paraspeckles raise the question of the mode(s) of interaction between DNA and paraspeckles. Mounting evidence points to direct interaction by binding to double-stranded DNA (dsDNA). First, many DNA-binding domains were identified in NEAT1_2 (Kuo et al., 2019) and in vitro mobility shift assays showed direct binding of NEAT1 RNA to target DNA by formation of dsDNA:RNA triple-helix structures (Sentürk Cetin et al., 2019). Second, as we showed, DNA intercalating small molecules rapidly dissolve paraspeckles in vivo, indicating that the molecules perturb the tethering of NEAT1 RNA in the dsDNA (Grosch et al., 2020). Third, the putative locations of the interactions were shown to be sequence-dependent and cell-type-specific (Sentürk Cetin et al., 2019; Katsel et al., 2019; Zhang et al., 2019). Taken together, this evidence indicates that paraspeckles participate in the regulation of chromatin in a manner that determines cell-type-specific functions. Moreover, it is evident that paraspeckles have complex modes of interaction with the chromatin, as well as a dynamic composition in different types of cells, which as a result can determine their cell-type-specific functions. To further substantiate the roles of paraspeckles in chromatin regulation, it is important to analyze the composition, genomic locations, and functions of paraspeckles in a cell-type-specific manner.
Paraspeckle Regulation in Stem Cell Differentiation and Development
To overcome limitations of orthogonal cell line models in paraspeckle research, we recently established an atlas of differentiated cells from human PSCs for the purpose of characterizing developmental and general modes of paraspeckle regulation. By analyzing over 20 human differentiated cell types, we observed highly dynamic developmental patterns of paraspeckles in the multipotent progenitors of the three germ layers and their differentiated progeny (Figure 3). For example, we found that motor neurons and astrocytes, which are derived from a common neural progenitor population, exhibit vastly different numbers of paraspeckles, namely about 15 on average per cell in astrocytes and ∼2 in motor neurons (Grosch et al., 2020). Intriguingly, it seemed that developmental trajectories and duration of differentiation did not explain the dynamic patterns of paraspeckles, but rather the size of the nucleus correlated with the amount. We noted that the size of the nucleus varies considerably between different types of cells and that the number of paraspeckles is scaled accordingly, although some exceptions exist; e.g., of low numbers of paraspeckles in hepatocytes. Moreover, we noted that the number of paraspeckles is correlated to the nucleus size distribution in individual cells (Figure 3). Since depletion of NEAT1 did not lead to overt changes in nuclei size, we concluded that it is the size of the nucleus that determines the number of paraspeckles and not vice versa.
Figure 3.
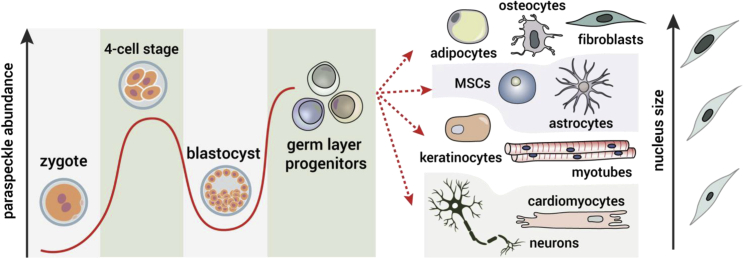
The Number of Paraspeckles Relies on Nucleus Size and Stage-Specific Regulation
Paraspeckles are highly abundant at the four-cell stage before being downregulated upon blastocyst differentiation. Germ layer differentiation is accompanied by an increase in the number of paraspeckles and mature cell types exhibit paraspeckle numbers that often correspond to the size of their nuclei.
How the nucleus regulates the number of paraspeckles according to its size is an open question. The answer may lie in chromatin factors. Since the overall transcriptional activity of cells is regulated by the size of the nucleus in many cases (Marguerat and Bähler, 2012), one explanation might be that the transcription of NEAT1 relies on the global upregulation of RNA polymerase II activity, a connection that was demonstrated between nucleoli and nucleus size (Weber and Brangwynne, 2015). However, in the case of paraspeckles, this mechanism is less plausible, at least for certain types of cells, because RNA polymerase II in PSCs is very active (Efroni et al., 2008) and these cells do not exhibit paraspeckles. However, it is important to consider that PSCs might be a special case for downregulation of paraspeckles in transcriptionally active cells, since alternative polyadenylation (APA) of NEAT1 by the RBP TDP-43 in undifferentiated cells maintains the low number of paraspeckles in these cells (Modic et al., 2019). Despite that, we could show that the number of paraspeckles in mouse cells is smaller and proportional to a nucleus size difference of approximately 40% between mouse and human cells (Figure 3) but not to the difference in genome size, which amounts to ∼14% (Chinwalla et al., 2002). Collectively, these observations provide an enticing indication that paraspeckles are scaled by the volume of the nucleus. It is therefore important to consider the possibility that the associations of paraspeckles with chromatin are being determined by and have functions connected to the size of the nucleus, such as global protein synthesis.
The complex functions associated with paraspeckles in development support a notion that their interactions in the chromatin are involved in the regulation of cellular plasticity and differentiation. Analysis of morula-stage embryos in the mouse has shown that paraspeckles accumulated the arginine methyltransferase CARM1, which drives the development of the embryo proper but not extra-embryonic tissues. In this context, downregulation of Neat1, or a different gene essential for paraspeckles, namely Nono, has led to the abnormal upregulation of Cdx2, and formation of trophoblasts as a result (Hupalowska et al., 2018). Paraspeckles have also been implicated in the differentiation of PSCs; downregulation of Neat1 in murine PSCs slowed the exit from pluripotency, and embryos from a 2n-4n aggregated complementation assay using Neat1 knock-down embryonic stem cells (ESCs) exhibited defects in primitive streak formation (Modic et al., 2019). Contrarily, human ESCs lacking NEAT1 exhibited accelerated differentiation. The phenotypic difference of mouse and human PSCs lacking NEAT1 is intriguing, but it is not unusual that the functions of mouse and human gene orthologues diverged in evolution (Liao and Zhang, 2008). Perhaps the different phenotypes are connected to the substantially larger nuclei in human cells and the higher number of paraspeckles in human cells in general (Grosch et al., 2020). Furthermore, unexpectedly, the knockout of Neat1 did not uncover overt developmental aberrations in newborn mice, but fewer parturitions of Neat1−/− females with smaller litter size were observed (Nakagawa et al., 2014). Collectively, these results encompass significant advancements that were recently made using stem cell systems and embryological studies with respect to understanding the molecular basis of the developmental phenotypes associated with paraspeckles, a research direction that should expand to resolve gaps in understanding the diverse and species-specific functions of paraspeckles.
Developmentally Regulated lncRNA Foci and Their Connection to Phase Separation in the Chromatin
A key question about condensates of chromatin factors is whether their tethering to the genome is mediated by lncRNAs. Despite little being known about phase separation of lncRNAs other than NEAT1, several lines of evidence raise the possibility that chromatin factors, lncRNAs, and phase separation could be regulated in concert and in a cell-type-specific manner. The development of several genome-wide techniques in recent years has helped to map the locations of RNA-DNA interactions, although few experiments have dealt with cell-type-specific aspects. Mapping RNA-Genome Interactions (MARGI) (Sridhar et al., 2017), Capture Hybridization Analysis of RNA Targets (CHART) (West et al., 2014), Chromatin Isolation by RNA Purification (ChIRP) (Chu et al., 2011), RNA Antisense Purification (RAP) (Engreitz et al., 2015), and Global RNA Interactions with DNA by Deep Sequencing (GRID-Seq) (Li et al., 2017) have produced extensive evidence for direct interplay between lncRNAs and the chromatin. These studies revealed the existence of hundreds of chromatin-bound lncRNAs that were mapped to numerous sites in the genome, but they left open the question of whether lncRNAs build macromolecular structures. Nevertheless, if considered together with a screening study showing that dozens of lncRNAs form nuclear foci (Cabili et al., 2015), and evidence that nuclear lncRNAs are associated with chromatin factors and RBPs (Jonas et al., 2020; Saxena and Carninci, 2011), one could conclude that the phenomenon of membraneless foci is quite likely very broad. Indeed, evidence that lncRNAs might be involved in phase separation of membraneless organelles was recently provided by Pitchiaya et al. (2019), who showed that RNAs localize in distinct domains of mammalian cytoplasmic P-bodies where they interact with P-body proteins.
The cell-type-specific expression of lncRNAs has been demonstrated in comprehensive studies of mouse tissues (Ravasi et al., 2006) and human tissues (Werner et al., 2017), and high cell-to-cell variation was demonstrated by single cell sequencing (Gawronski and Kim, 2017). Furthermore, we showed recently that many lncRNAs that form nuclear foci in addition exhibit highly dynamic and diverse expression patterns during the differentiation of hPSCs toward multiple cell types (Grosch et al., 2020). This could mean that membraneless lncRNA granules and their interactions with chromatin play key roles in developmental regulation. In line with this possibility, other studies have shown that PSCs express thousands of lncRNAs (Guttman et al., 2009), and dozens have been shown to affect pluripotency (Guttman et al., 2011). The transcription of some of these lncRNAs is regulated by OCT4 and NANOG, including, for example, MIAT and AK141205, which participate in maintenance of pluripotency (Sheik Mohamed et al., 2010). Moreover, in mESCs, the lncRNA TUNA maintains pluripotency by interacting with RBPs that bind to the promoters of pluripotency factors Nanog, Sox2, and Fgf4 (Lin et al., 2014). In human PSCs, lncRNA lncPRESS1 was shown to interact with the histone H3 deacetylase SIRT6 to prevent its access to chromatin, thus maintaining histone acetylation at promoters of pluripotency genes (Jain et al., 2016). Other lncRNAs, namely lncRNA-ES1 and lncRNA-ES2, are highly expressed in hPSCs, where they interact with the repressive Polycomb protein SUZ12 and the pluripotency factor SOX2 to block neural differentiation (Ng et al., 2012). Additional developmentally regulated lncRNAs such as DIGIT and TERRA are up- and downregulated during differentiation, respectively (Daneshvar et al., 2016; Xu et al., 2018), however their functions in this process have not been fully elucidated. Taken together, these studies indicate the possibility that lncRNAs could form nuclear membraneless organelles that regulate development in mammals by determining the localization of chromatin factors (Figure 1 bottom).
The roles of RBPs in connection to the chromatin and lncRNAs might also be important. A notion with increasing credence is that RBPs can directly interact with the chromatin and interact with transcription factors. For instance, the RBP RBM25 associates with the transcription factor YY1 to regulate its chromatin association (Xiao et al., 2019). Other examples of RBPs that take part in regulating transcription include HNRNPL (Kuninger et al., 2002), SRSF2 (Ji et al., 2013), and the paraspeckle core protein NONO (Knott et al., 2016). Moreover, the aggregation of RBPs is often driven by intrinsically disordered domains (IDDs) of RBPs, such as SR splicing factors (Haynes and Iakoucheva, 2006). Therefore, lncRNAs could serve to facilitate associations of RBPs with chromatin to induce phase separation at specific sites of the genome, and thereby to regulate gene expression. Validation of lncRNAs in conjunction to RBPs partaking in nuclear phase separation is therefore important in the context of chromatin-mediated regulation of development.
Determining the Cell-Type-Specific Phase Separation Properties of lncRNAs
The formation of nuclear lncRNA foci and the dynamic expression patterns of lncRNAs are strong indicators for widespread occurrence of nuclear liquid-liquid phase separation (LLPS) with site-specific developmental functions. However, it has not been shown yet that lncRNA aggregates indeed exhibit liquid-like behavior in living cells. Investigating LLPS of numerous lncRNAs represents a dual challenge; namely, the systematic application of structural and biochemical techniques to numerous lncRNAs, and to a diversity of cell types. Common techniques used in the characterization of LLPS droplets include fluorescence recovery after photo bleaching (FRAP), time-lapse imaging, and digital holographic microscopy (Yoshizawa et al., 2020). Furthermore, analysis of IDDs of RBPs involves nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR), and high-speed atomic force microscopy (AFM). Because these workflows are complex and laborious, they are best suited for in-depth analysis of prime candidates, rather than the high-throughput characterization of cell-type-specific lncRNAs that can give rise to LLPS droplets.
In this respect, an opportunity may lie in evidence that lncRNA tethering to the genome via triple-helix DNA:RNA structure can be perturbed by small molecules that intercalate into dsDNA. We have shown recently that the treatment of live cells by small DNA-binding molecules such as Hoechst, Actinomycin D (ActD), and Mithramycin (Grosch et al., 2020) induce the genome-wide disintegration of paraspeckles in minutes. According to earlier x-ray crystallography and NMR of B-DNA and these molecules (Kamitori and Takusagawa, 1992; Sastry et al., 1995; Sriram et al., 1992), we speculate that the molecules disrupt the conformation of B-DNA leading to dissolution of DNA:RNA contacts mediated by the lncRNA NEAT1 (Figure 3). Alternative explanations, such as that ActD and other intercalators promote disintegration of lncRNAs by stalling RNA polymerase II or by inducing the translocation of paraspeckle proteins to perinucleolar caps, are less likely because (1) a similarly rapid disintegration effect was also observed with lncRNA MALAT1 foci, and (2) alpha-Amanitin, a direct inhibitor of RNA polymerase II, did not induce a disintegration of NEAT1 and MALAT1 lncRNA foci. Taken together, it is therefore plausible that analysis by next-generation sequencing of RNAs released from the chromatin following treatment of cells by ActD or other intercalators could help to systematically identify those lncRNAs that are tethered to the chromatin. It is also plausible that rearrangements of chromatin factors and RBPs that undergo LLPS by their association with lncRNAs, as is the case with paraspeckles (Figure 2), could be analyzed systematically by proteome techniques after the depletion of scaffold lncRNAs. Pull-down studies of chromatin-associated proteins in cells with and without candidate lncRNA of interest could be a way to address this question.
Open Questions About the Cell-Type-Specific Functions of Membraneless Organelles in the Chromatin
The nucleolus and paraspeckles are forerunners in the research of membraneless organelles because they are involved in embryonic development (Hupalowska et al., 2018; Ogushi et al., 2008). A chief question is how these condensates, which are associated with the chromatin and influence gene expression, participate in the organization of the genome. The fact that nuclear membraneless organelles have a diameter approximately one or two orders of magnitude larger than nucleosomes and dsDNA (500 nm compared with 11 and 2 nm respectively) indicates that individual membraneless organelles associate topologically with a multitude of genes. In line with this idea are observations that hundreds of loci are bound to NEAT1 and PcG bodies (Pirrotta and Li, 2012; West et al., 2014). Despite our limited knowledge about these structures, it is reasonable to assume that biocondensates could produce macromolecular expanses of chromatin factors in a droplet form. Whether this serves to create reservoirs of chromatin factors or to sequester them, and how dynamic is the exchange of the condensed factors with their environment, are open questions. Advances in live-cell super-resolution imaging recently revealed the first indications that liquid droplets of the transcriptional regulator protein YAP1 could serve as depositories in open chromatin regions (Cai et al., 2019). Based on the indications that paraspeckles associate with nucleosome remodeling and histone-modifying enzymes (Figure 2), a reasonable assumption is that paraspeckles and possibly other membraneless organelles are important in the establishment of repressive and active transcriptional environments. Evidence that SWI/SNF enzymes and Carm1 are integral components of paraspeckles, and that both in fact are essential for the formation of paraspeckles (Hupalowska et al., 2018; Kawaguchi et al., 2015), strongly support this notion. On the other hand, other histone-modifying proteins, such as EZH2, do not exhibit overtly granular distribution patterns, indicating that NEAT1 and/or paraspeckles generally associate with the predetermined locations of some chromatin factors. Taken together, to better understand the connections between membraneless organelles, topological organization of the chromatin, and cell identity, it is important to resolve the order of events in the association of nucleosome remodeling and histone-modifying complexes with paraspeckles. The fact that up to 20 foci of different species of membraneless organelles are found in cells (Grosch et al., 2020; Pirrotta and Li, 2012) indicates how intricate and important this form of regulation is.
In view that the genomic locations and numbers of nuclear condensates often seem to vary within populations of cells, and between different cell types, it is important to systematically analyze the genomic locations of membraneless organelles in cells representing different tissues and stages of the cell cycle. The use of PSC-derived cell types could be particularly useful for this purpose since all cell types can be produced from a single source. The generic editing of hPSCs is also highly useful for creating molecular tracking tools and dissecting mechanisms in this regard with a common genetic background. Ultimately, methods that detect the locations of DNA:RNA interactions on the single cell level could greatly advance the understanding of mechanisms that determine the locations of membraneless organelles by analyzing the genomic locations of NEAT1 in a variety of cell types.
Finally, a key question is how the regulation of chromatin factors is connected to RBPs and to what end. RBPs often harbor IDDs with liquid-like properties that have been implicated in neurodegenerative diseases, such as Parkinson disease, Huntington disease, and amyotrophic lateral sclerosis (ALS) (Elbaum-Garfinkle, 2019). This points to connections with mechanisms of condensed nuclear structures with RNAs and RBPs. The formation of nuclear granules is often associated with cellular stress factors such as hypoxia and acidosis, and nuclear stress bodies, paraspeckles, and the nucleolus are leading examples (Biamonti and Vourc'h, 2010; Fox et al., 2018; Weeks et al., 2019). The purpose of their formation in stress conditions, and whether exaggerated accumulation of canonical nuclear condensates could promote disease, are therefore key questions. Importantly, as we review here, the discovery of basic mechanisms pertaining to membraneless organelles could be facilitated by analysis of developmental systems where the number and functions of membraneless aggregates seem to be tightly regulated, as in the case of paraspeckles. The research of developmental membraneless organelles, chromatin factors, and lncRNAs, in particular paraspeckles, which can be readily manipulated and have been connected to neurodegeneration, should therefore serve to advance understanding disease mechanisms and therapy.
Author Contributions
M.G. and S.I. researched data for the article and wrote the text under supervision of M.D. D.S. designed the figures. M.D. provided the conceptual structure for this review.
Acknowledgments
Research reported in this publication was supported by the Volkswagen Foundation grant OntoTime.
References
- Ahmed A.S.I., Dong K., Liu J., Wen T., Yu L., Xu F., Kang X., Osman I., Hu G., Bunting K.M. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc. Natl. Acad. Sci. U S A. 2018;115:E8660–E8667. doi: 10.1073/pnas.1803725115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S., Dormann D. Liquid–liquid phase separation in disease. Annu. Rev. Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- Argelaguet R., Clark S.J., Mohammed H., Stapel L.C., Krueger C., Kapourani C.-A., Imaz-Rosshandler I., Lohoff T., Xiang Y., Hanna C.W. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019;576:487–491. doi: 10.1038/s41586-019-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J., Hurt E. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019;88:281–306. doi: 10.1146/annurev-biochem-013118-110817. [DOI] [PubMed] [Google Scholar]
- Biamonti G., Vourc’h C. Nuclear stress bodies. Cold Spring Harb Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch J.L., Zomerdijk J.C.B.M. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 2008;36 doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E., González-Ramírez M., Alcaine-Colet A., Aranda S., Croce L.D. The bivalent genome: characterization, structure, and regulation. Trends Genet. 2020;36:118–131. doi: 10.1016/j.tig.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Bosch L.V.D. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer T.D., Manns M.P., Sanyal A.J., Zakim D. Elsevier Health Sciences; 2012. Zakim and Boyer’s Hepatology: A Textbook of Liver Disease. [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A.A., Johnston D.R., Kaur S., Lubin F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aaw9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Feliciano D., Dong P., Flores E., Gruebele M., Porat-Shliom N., Sukenik S., Liu Z., Lippincott-Schwartz J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019;21:1578–1589. doi: 10.1038/s41556-019-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerase A., Armaos A., Neumayer C., Avner P., Guttman M., Tartaglia G.G. Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 2019;26:331–334. doi: 10.1038/s41594-019-0223-0. [DOI] [PubMed] [Google Scholar]
- Chen G., Zhang D., Zhang L., Feng G., Zhang B., Wu Y., Li W., Zhang Y., Hu B. RBM14 is indispensable for pluripotency maintenance and mesoderm development of mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2018;501:259–265. doi: 10.1016/j.bbrc.2018.04.231. [DOI] [PubMed] [Google Scholar]
- Chen Q., Cai J., Wang Q., Wang Y., Liu M., Yang J., Zhou J., Kang C., Li M., Jiang C. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-Catenin pathway by scaffolding EZH2. Clin. Cancer Res. 2018;24:684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- Chen L.-L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Chu C., Qu K., Zhong F., Artandi S.E., Chang H.Y. Genomic maps of lincRNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K.E., Fenstermacher S.J., Pazyra-Murphy M.F., Elliott H.L., Segal R.A. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat. Neurosci. 2016;19:690–696. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K., Pondick J.V., Kim B.-M., Zhou C., York S.R., Macklin J.A., Abualteen A., Tan B., Sigova A.A., Marcho C. DIGIT is a conserved long noncoding RNA that regulates GSC expression to control definitive endoderm differentiation of embryonic stem cells. Cell Rep. 2016;17:353–365. doi: 10.1016/j.celrep.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M., Trerè D., Pession A., Montanaro L., Sirri V., Ochs R.L. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D.G., Buetow K.H. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S. Matter over mind: liquid phase separation and neurodegeneration. J. Biol. Chem. 2019;294:7160–7168. doi: 10.1074/jbc.REV118.001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J., Lander E.S., Guttman M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol. Biol. 2015;1262:183–197. doi: 10.1007/978-1-4939-2253-6_11. [DOI] [PubMed] [Google Scholar]
- Erdel F., Rademacher A., Vlijm R., Tünnermann J., Frank L., Weinmann R., Schweigert E., Yserentant K., Hummert J., Bauer C. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell. 2020;78:236–249.e7. doi: 10.1016/j.molcel.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley K.I., Surovtseva Y., Merkel J., Baserga S.J. Determinants of mammalian nucleolar architecture. Chromosoma. 2015;124:323–331. doi: 10.1007/s00412-015-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar sub-compartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.H., Nakagawa S., Hirose T., Bond C.S. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski K.A., Kim J. Single cell transcriptomics of non-coding RNAs and their cell-specificity. Wiley Interdiscip. Rev. RNA. 2017;8 doi: 10.1002/wrna.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B.A., Doolittle L.K., Schneider M.W.G., Jensen L.E., Gamarra N., Henry L., Gerlich D.W., Redding S., Rosen M.K. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.e21. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E., Shorter J. The molecular language of membraneless organelles. J. Biol. Chem. 2019;294:7115–7127. doi: 10.1074/jbc.TM118.001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig J.A., Nguyen T.A., Lee M., Holehouse A.S., Posey A.E., Pappu R.V., Jedd G. Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol. Cell. 2020;77:1237–1250.e4. doi: 10.1016/j.molcel.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosch M., Ittermann S., Rusha E., Greisle T., Ori C., Truong D.-J.J., O’Neill A.C., Pertek A., Westmeyer G.G., Drukker M. Nucleus size and DNA accessibility are linked to the regulation of paraspeckle formation in cellular differentiation. BMC Biol. 2020;18:42. doi: 10.1186/s12915-020-00770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D., Bi X., Pardavila J.A., Huang X., Saenz C., Shi X., Zhou H., Faiola F., Ding J., Haruehanroengra P. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018;50:443–451. doi: 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C., Iakoucheva L.M. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalowska A., Jedrusik A., Zhu M., Bedford M.T., Glover D.M., Zernicka-Goetz M. CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell. 2018;175:1902–1916.e13. doi: 10.1016/j.cell.2018.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur W., Kemp J.P., Tarzia M., Deneke V.E., Marzluff W.F., Duronio R.J., Di Talia S. CDK-regulated phase separation seeded by histone genes ensures precise growth and function of histone locus bodies. Dev. Cell. 2020 doi: 10.1016/j.devcel.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Jain A.K., Xi Y., McCarthy R., Allton K., Akdemir K.C., Patel L.R., Aronow B., Lin C., Li W., Yang L. LncPRESS1 is a p53-regulated lncRNA that safeguards pluripotency by disrupting SIRT6 mediated de-acetylation of histone H3K56. Mol. Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C.Y., Xiao R., Burge C.B., Fu X.-D. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K., Calin G.A., Pichler M. RNA-binding proteins as important regulators of long non-coding RNAs in cancer. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21082969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitori S., Takusagawa F. Crystal structure of the 2:1 complex between d(GAAGCTTC) and the anticancer drug actinomycin D. J. Mol. Biol. 1992;225:445–456. doi: 10.1016/0022-2836(92)90931-9. [DOI] [PubMed] [Google Scholar]
- Katsel P., Roussos P., Fam P., Khan S., Tan W., Hirose T., Nakagawa S., Pletnikov M.V., Haroutunian V. The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophrenia. 2019;5:1–9. doi: 10.1038/s41537-019-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Tanigawa A., Naganuma T., Ohkawa Y., Souquere S., Pierron G., Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. U S A. 2015;112:4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler K.E., Trcek T., Hurd T.R., Chen R., Liang F.-X., Sall J., Kato M., Lehmann R. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. Elife. 2018;7:e37949. doi: 10.7554/eLife.37949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G.J., Bond C.S., Fox A.H. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 2016;44:3989–4004. doi: 10.1093/nar/gkw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger D.T., Izumi T., Papaconstantinou J., Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-C., Hänzelmann S., Sentürk Cetin N., Frank S., Zajzon B., Derks J.-P., Akhade V.S., Ahuja G., Kanduri C., Grummt I. Detection of RNA–DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019;47:e32. doi: 10.1093/nar/gkz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhou B., Chen L., Gou L.-T., Li H., Fu X.-D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017;35:940–950. doi: 10.1038/nbt.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B.-Y., Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc. Natl. Acad. Sci. U S A. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N., Chang K.-Y., Li Z., Gates K., Rana Z.A., Dang J., Zhang D., Han T., Yang C.-S., Cunningham T.J. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Karwacki-Neisius V., Tang H., Li W., Shi Z., Hu H., Xu W., Wang Z., Kong L., Lv R. Nono, a bivalent domain factor, regulates Erk signaling and mouse embryonic stem cell pluripotency. Cell Rep. 2016;17:997–1007. doi: 10.1016/j.celrep.2016.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S., Bähler J. Coordinating genome expression with cell size. Trends Genet. 2012;28:560–565. doi: 10.1016/j.tig.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Mateju D., Franzmann T.M., Patel A., Kopach A., Boczek E.E., Maharana S., Lee H.O., Carra S., Hyman A.A., Alberti S. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modic M., Grosch M., Rot G., Schirge S., Lepko T., Yamazaki T., Lee F.C.Y., Rusha E., Shaposhnikov D., Palo M. Cross-regulation between TDP-43 and paraspeckles promotes pluripotency-differentiation transition. Mol. Cell. 2019;74:951–965.e13. doi: 10.1016/j.molcel.2019.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T. Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., Fujita Y., Fujimori T., Standaert L., Marine J.-C. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.-Y., Johnson R., Stanton L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogushi S., Palmieri C., Fulka H., Saitou M., Miyano T., Fulka J. The maternal nucleolus is essential for early embryonic development in mammals. Science. 2008;319:613–616. doi: 10.1126/science.1151276. [DOI] [PubMed] [Google Scholar]
- Paul S., Knott J.G. Epigenetic control of cell fate in mouse blastocysts: the role of covalent histone modifications and chromatin remodeling. Mol. Reprod. Dev. 2014;81:171–182. doi: 10.1002/mrd.22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino M., Veenstra G.J.C. Chromatin control of developmental dynamics and plasticity. Dev. Cell. 2016;38:610–620. doi: 10.1016/j.devcel.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Li H.-B. A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchiaya S., Mourao M.D.A., Jalihal A.P., Xiao L., Jiang X., Chinnaiyan A.M., Schnell S., Walter N.G. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol. Cell. 2019;74:521–533.e6. doi: 10.1016/j.molcel.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plys A.J., Davis C.P., Kim J., Rizki G., Keenen M.M., Marr S.K., Kingston R.E. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33:799–813. doi: 10.1101/gad.326488.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff J.W. Phase separation and the centrosome: a fait accompli? Trends Cell Biol. 2019;29:612–622. doi: 10.1016/j.tcb.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Vincenz C., Kerppola T.K. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol. Cell. Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.-K., Bouchoux C., Liu H.W., Kim E., Minamino M., de Groot R., Katan A.J., Bonato A., Marenduzzo D., Michieletto D. Phase separation induced by cohesin SMC protein complexes. bioRxiv. 2020 doi: 10.1101/2020.06.13.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F., Hendrich B., Reik W., Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Sasaki Y.T.F., Ideue T., Sano M., Mituyama T., Hirose T. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry M., Fiala R., Patel D. Solution structure of Mithramycin dimers bound to partially overlapping sites on DNA. J. Mol. Biol. 1995;251:674–689. doi: 10.1006/jmbi.1995.0464. [DOI] [PubMed] [Google Scholar]
- Sawyer I.A., Hager G.L., Dundr M. Specific genomic cues regulate Cajal body assembly. RNA Biol. 2016;14:791–803. doi: 10.1080/15476286.2016.1243648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A., Carninci P. Long non-coding RNA modifies chromatin. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh J.F., Lajtha A. In vivo rates of protein synthesis in brain, muscle, and liver of five vertebrate species. Neurochem. Res. 1989;14:1165–1168. doi: 10.1007/BF00965625. [DOI] [PubMed] [Google Scholar]
- Sentürk Cetin N., Kuo C.-C., Ribarska T., Li R., Costa I.G., Grummt I. Isolation and genome-wide characterization of cellular DNA:RNA triplex structures. Nucleic Acids Res. 2019;47:2306–2321. doi: 10.1093/nar/gky1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Mohamed J., Gaughwin P.M., Lim B., Robson P., Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T.A., Kukharsky M.S., An H., Dimasi P., Alexeeva S., Shabir O., Heath P.R., Buchman V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018;13:30. doi: 10.1186/s13024-018-0263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Spector D.L., Lamond A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar B., Rivas-Astroza M., Nguyen T.C., Chen W., Yan Z., Cao X., Hebert L., Zhong S. Systematic mapping of RNA-chromatin interactions in vivo. Curr. Biol. 2017;27:602–609. doi: 10.1016/j.cub.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram M., van der Marel G.A., Roelen H.L., van Boom J.H., Wang A.H. Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342. EMBO J. 1992;11:225–232. doi: 10.1002/j.1460-2075.1992.tb05045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staněk D. Cajal bodies and snRNPs - friends with benefits. RNA Biol. 2016;14:671–679. doi: 10.1080/15476286.2016.1231359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavosian R., Kent S., Brown K., Yao T., Duc H.N., Huynh T.N., Zhen C.Y., Ma B., Wang H., Ren X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019;294:1451–1463. doi: 10.1074/jbc.RA118.006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku V., Kew C., Mehrotra P., Ganesan R., Robinson N., Antebi A. Nucleolar fibrillarin is an evolutionarily conserved regulator of bacterial pathogen resistance. Nat. Commun. 2018;9:3607. doi: 10.1038/s41467-018-06051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P.J., Islam S.A., Freemont P.S., Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu L., Zhang S., Ming Y., Liu S., Cheng K., Zhao Y. Long noncoding RNA NEAT1 suppresses hepatocyte proliferation in fulminant hepatic failure through increased recruitment of EZH2 to the LATS2 promoter region and promotion of H3K27me3 methylation. Exp. Mol. Med. 2020;52:461–472. doi: 10.1038/s12276-020-0387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zuo H., Jin J., Lv W., Xu Z., Fan Y., Zhang J., Zuo B. Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 2019;10:505. doi: 10.1038/s41419-019-1742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhao Y., Xu N., Zhang S., Wang S., Mao Y., Zhu Y., Li B., Jiang Y., Tan Y. NEAT1 regulates neuroglial cell mediating Aβ clearance via the epigenetic regulation of endocytosis-related genes expression. Cell Mol. Life Sci. 2019;76:3005–3018. doi: 10.1007/s00018-019-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hu S.-B., Wang M.-R., Yao R.-W., Wu D., Yang L., Chen L.-L. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018;20:1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- Weber S.C., Brangwynne C.P. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks S.E., Metge B.J., Samant R.S. The nucleolus: a central response hub for the stressors that drive cancer progression. Cell. Mol. Life Sci. 2019;76:4511–4524. doi: 10.1007/s00018-019-03231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.S., Sullivan M.A., Shah R.N., Nadadur R.D., Grzybowski A.T., Galat V., Moskowitz I.P., Ruthenburg A.J. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 2017;24:596–603. doi: 10.1038/nsmb.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Zhang Y., Xu Q., Zhou C., Liu B., Du Z., Zhang K., Zhang B., Wang X., Gayen S. Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat. Genet. 2020;52:95–105. doi: 10.1038/s41588-019-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R., Chen J.-Y., Liang Z., Luo D., Chen G., Lu Z.J., Chen Y., Zhou B., Li H., Du X. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell. 2019;178:107–121.e18. doi: 10.1016/j.cell.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Guo M., Zhang N., Ye S. Telomeric noncoding RNA promotes mouse embryonic stem cell self-renewal through inhibition of TCF3 activity. Am. J. Physiol. Cell Physiol. 2018;314:C712–C720. doi: 10.1152/ajpcell.00292.2017. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H., Bond C.S., Nakagawa S., Pierron G., Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70:1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- Yoshizawa T., Nozawa R.-S., Jia T.Z., Saio T., Mori E. Biological phase separation: cell biology meets biophysics. Biophys. Rev. 2020;12:519–539. doi: 10.1007/s12551-020-00680-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Lan Y., Xie A., Shi J., Zhao H., Xu L., Zhu S., Luo T., Zhao T., Xiao Y. Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements. J. Biol. Chem. 2019;294:15613–15622. doi: 10.1074/jbc.RA119.008732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li B., Li W., Ma L., Zheng D., Li L., Yang W., Chu M., Chen W., Mailman R.B. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports. 2014;3:460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel A., Nikolic M., Sofiadis K., Mallm J.-P., Brackley C.A., Gothe H., Drechsel O., Becker C., Altmüller J., Josipovic N. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell. 2018;70:730–744.e6. doi: 10.1016/j.molcel.2018.03.030. [DOI] [PubMed] [Google Scholar]