Abstract
The nucleolus is the largest compartment of the eukaryotic cell's nucleus. It acts as a ribosome factory, thereby sustaining the translation machinery. The nucleolus is also the subnuclear compartment with the highest transcriptional activity in the cell, where hundreds of ribosomal RNA (rRNA) genes transcribe the overwhelming majority of RNAs. The structure and composition of the nucleolus change according to the developmental state. For instance, in embryonic stem cells (ESCs), rRNA genes display a hyperactive transcriptional state and open chromatin structure compared with differentiated cells. Increasing evidence indicates that the role of the nucleolus and rRNA genes might go beyond the control of ribosome biogenesis. One such role is linked to the genome architecture, since repressive domains are often located close to the nucleolus. This review highlights recent findings describing how the nucleolus is regulated in ESCs and its role in regulating ribosome biogenesis and genome organization for the maintenance of stem cell identity.
Keywords: ESCs, nucleolus, hypertranscription, rRNA genes, NAD, genome organization
Highlights
-
•
The nucleolus of ESC is hyperactive but protein translation rate in ESC is low
-
•
ESC have transcriptionally hyperactive rRNA genes with open and active chromatin
-
•
The hyperactive state of rRNA genes is required to maintain pluripotency
-
•
Genome organization in and around the nucleolus changes during ESC differentiation
In this review article, Santoro and colleague describe the regulation and role of the largest subnuclear compartment of the cell, the nucleolus, in embryonic stem cells (ESCs). Specifically, the authors highlight recent findings describing how the hyperactive transcriptional state and open chromatin structure of ribosomal RNA genes, which are located in the nucleolus, are regulated and affect ribosome biogenesis activities and genome organization in ESCs and during differentiation.
Main Text
Nucleolus and Ribosomal rRNA Genes
The nucleolus is the largest subnuclear compartment in the cell's nucleus and the place where ribosome biogenesis occurs. Ribosomes are essential for protein production and their biosynthesis in the nucleolus is a complex process that requires the interplay of many factors. Ribosome biogenesis is initiated in the nucleolus by the RNA polymerase I (Pol I)-driven transcription of hundreds of ribosomal RNA (rRNA) genes that generates 45S/47S pre-rRNA. This transcript is then modified and processed to form 28S, 18S, and 5.8S rRNAs, which in turn are assembled with ribosomal proteins in the nucleolus. This process is mediated by small nucleolar RNAs, endonucleases, and exonucleases, as well as ribose-modifying enzymes that mediate the proper folding and processing of the pre-rRNAs to give rise to the pre-40S and pre-60S subunits. Both subunits are then exported to the cytoplasm, where they become competent for translation after the final maturation steps (Pelletier et al., 2018).
The nucleolus is a membrane-less compartment generally surrounded by a shell of heterochromatin. Since rRNA genes produce the overwhelming majority of RNAs, the nucleolus is the subnuclear compartment with the highest transcriptional activity in the cell. The nucleolus displays a tripartite organization that consists of the fibrillar center, the dense fibrillar component, and the granular component. These subcompartments within the nucleolus represent distinct, coexisting liquid phases (Feric et al., 2016). Importantly, the formation of the nucleolus depends on the availability of pre-rRNAs. Accordingly, the morphology and size of the nucleolus are linked to rRNA gene transcriptional activity, which in turn depends on cell growth, metabolism, and developmental state. For example, structural changes in the nucleolus are often observed in cancer (Hein et al., 2013). Furthermore, the structure and composition of the nucleolus are remarkably different between somatic cells and germ cells and during early development (reviewed in Fulka and Langerova, 2019; Kresoja-Rakic and Santoro, 2019). As discussed later, these structural changes mirror ribosome biogenesis activities as well as the transcriptional and chromatin states of rRNA genes, and might have implications in genome organization.
The detailed structure and regulation of rRNA genes have been recently reviewed (Bersaglieri and Santoro, 2019; Moss et al., 2019). Here, we briefly describe the chromatin and epigenetic regulation of rRNA genes, highlighting critical factors in the control of nucleolar activities in embryonic stem cells (ESCs). Mammalian cells bear ~200 rRNA genes per haploid genome, which are arranged in arrays of tandem repeats among different chromosomes at regions called nucleolar organizer regions. In differentiated cells, not all rRNA genes are competent for transcription. In mammalian cells, rRNA genes can be subdivided into three major classes—silent, inactive (or pseudogenes), and active—and this classification is mainly based on their transcriptional state and chromatin and epigenetic features. Silent rRNA genes belong to the class of the non-transcribing and nucleosome-packed rRNA gene fraction, which replicates in mid-late S phase and is inherited during cell division. This class of rRNA genes displays heterochromatic features such as DNA methylation, repressive histone marks like H3K9me2 and H3K9me3, and deacetylated histones (Li et al., 2005; Santoro et al., 2002; Santoro and Grummt, 2001; Stancheva et al., 1997; Zhou et al., 2002). CpG methylation at the rRNA gene promoter is responsible for transcriptional silencing since it impairs the binding of the upstream binding factor UBF, thereby abrogating the formation of the Pol I pre-initiation complex (Sanij et al., 2008; Santoro and Grummt, 2001). The establishment of rRNA gene silencing is mediated by the nucleolar remodeling complex NoRC, which is constituted of TIP5 (BAZ2A, TTF1-interacting protein 5) and SNF2H. NoRC associates with repressive factors such as DNA methyltransferases, which methylate rRNA gene promoter sequences (Guetg et al., 2010, 2012; Santoro et al., 2002; Zhou et al., 2002). The recruitment of NoRC to the rRNA gene promoter occurs through a long non-coding RNA (lncRNA)-mediated mechanism. TIP5 associates with an lncRNA called promoter-associated RNA (pRNA). This lncRNA is generated by the processing of the intergenic spacer (IGS) rRNA, which is transcribed by Pol I and originates from the rRNA spacer promoter (Mayer et al., 2006; Santoro et al., 2010). The association of TIP5 with pRNA is required for the interaction with the transcription terminator factor I (TTF1), which in turn is bound at the rRNA gene promoter (Mayer et al., 2006; Savić et al., 2014).
The rest of the rRNA genes that do not belong to the silent and CpG-methylated class represent active and inactive genes. UBF is implicated in the establishment of these two rRNA gene classes (Hamdane et al., 2014; Herdman et al., 2017; Sanij et al., 2008). Active genes associate with UBF and are nucleosome free in the coding region. UBF binds to the active rRNA genes at the spacer and main promoters, enhancer repeats, and gene bodies and allows the formation of the pre-initiation complex (Herdman et al., 2017). In contrast, inactive genes do not interact with UBF and belong to the nucleosome-packed rRNA gene chromatin as in the case of silent rRNA genes. Data have also suggested that the repressive chromatin structure of inactive rRNA genes can be mediated by the energy-dependent nucleolar silencing complex and the nucleosome remodeling and deacetylase complex in mammalian cells (Murayama et al., 2008; Xie et al., 2012). The lack of DNA methylation at inactive genes suggests that their transcriptional state can be potentially reversed. Accordingly, in the absence of UBF, active genes switch into inactive genes, a process that is accompanied by a histone H1-induced assembly of transcriptionally inactive chromatin. Importantly, the re-expression of UBF was shown to restore the active gene number, indicating that the switch from the active to the inactive state is indeed a reversible process (Sanij et al., 2008).
Accumulating evidence in several systems suggests that, even under conditions of high metabolic activity, the number of genes competent for transcription remains the same (Conconi et al., 1989; French et al., 2003; Muscarella et al., 1985). Accordingly, changes in DNA methylation state of rRNA genes (i.e., silent rRNA genes) were reported only in specific cases, such as the comparison between ESCs and differentiated cells or between some cancer types and healthy cells and during aging (reviewed in Bersaglieri and Santoro, 2019). Thus, it appears that the epigenetic regulation of silent rRNA genes is not always directly implicated in the control of ribosome biogenesis.
Increasing evidence indicates that the role of nucleolus and rRNA genes might go beyond the production of ribosomes to sustain protein synthesis. This particular aspect of the nucleolus is starting to emerge in the study of ESCs. In this review, we will discuss recent results on the regulation of nucleolar activities in ESCs and highlight their structural and functional consequences in stem cell properties that are not necessarily linked to protein synthesis.
Regulation of the Nucleolus in ESCs
ESCs derive from the pre-implantation epiblast cells and have the unrestricted potential to develop into all cells of the adult body (Nichols and Smith, 2009). ESCs are characterized by a state of hypertranscription (i.e., global elevation in nascent transcription relative to more differentiated cells) and an open chromatin conformation (Gaspar-Maia et al., 2011; Percharde et al., 2017). The majority of chromatin in ESCs is homogeneously spread and largely devoid of compact heterochromatin blocks. In contrast, chromatin in differentiated cells appears heterogeneous, with clustering of highly condensed heterochromatin at the nuclear periphery and nucleolus (Efroni et al., 2008; Savić et al., 2014) (Figure 1). Whereas the open chromatin state of ESCs has been proposed to facilitate transcriptional programs to switch rapidly upon induction of differentiation (Meshorer and Misteli, 2006), the role of the hypertranscription in ESCs is less clear. Interestingly, however, recent data pointed to a dynamic positive feedback loop between chromatin and translation by showing that acute inhibition of translation rapidly depletes euchromatic marks in mouse ESCs (mESCs) and blastocysts (Bulut-Karslioglu et al., 2018).
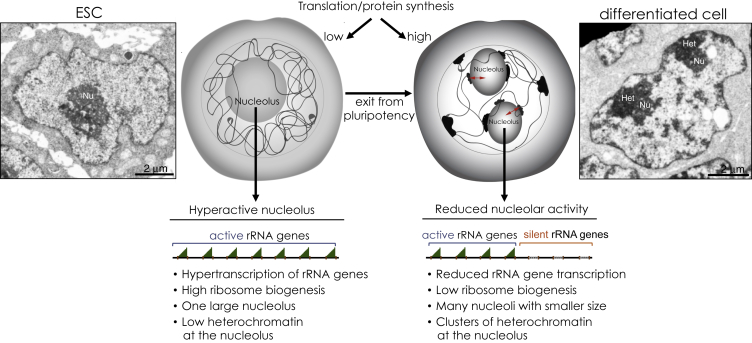
Figure 1.
Chromatin States of rRNA Genes in ESCs and Differentiated Cells
The hyperactive state of the nucleolus in ESCs is characterized by the lack of silent rRNA genes and elevated ribosome biogenesis. In contrast, upon exit from pluripotency, a fraction of rRNA genes acquire epigenetic silencing, ribosome biogenesis is reduced, and clusters of heterochromatin blocks surround the nucleolus and the nuclear periphery. The red arrows indicate the link between the nucleolar chromatin state and the rest of the genome. Representative electron microscopy images show the distinct chromatin organization between an mESC and a neural progenitor cell (NPC) 8 days after differentiation. Generally, ESCs display a single, large nucleolus (Nu), whereas differentiated cells have more nucleoli of smaller size compared with ESCs. The contrast procedure reveals in dark large and condensed heterochromatic structures (Het) particularly evident close to the nucleolus of the NPC.
The hypertranscription and active chromatin state of the ESC genome are mirrored in the nucleolus of ESCs due to the lack of heterochromatic and silent rRNA genes and the higher rRNA transcription levels relative to differentiated cells (Savić et al., 2014; Schlesinger et al., 2009) (Figure 1). DNA hypomethylation of rRNA genes and their lack of silencing was reported in both ground-state pluripotent and developmentally primed mESCs (Dalcher et al., 2019; Savić et al., 2014; Schlesinger et al., 2009). These ESC types are known to have distinct epigenetic signatures, such as a low DNA methylation content in ground-state pluripotent cells and high CpG methylation in primed mESCs (Ficz et al., 2013; Habibi et al., 2013; Leitch et al., 2013; Marks et al., 2012). Thus the lack of rRNA gene silencing seems to be a general feature of pluripotency, at least in mESCs.
It was shown that in mESCs, de novo establishment of heterochromatin at a fraction of rRNA genes (ca. 30%–40%) occurs only upon exit from pluripotency, with the acquisition of CpG methylation and an increase in repressive histone marks such as H3K9me2 and H3K9me3 (Savić et al., 2014). This process is concomitant with the downregulation of rRNA transcription upon differentiation of mESCs and human ESCs (hESCs), indicating that the reduction in nucleolar transcription is an early event once cells exit pluripotency (Savić et al., 2014; Watanabe-Susaki et al., 2014; Woolnough et al., 2016). The elevated rRNA transcriptional activity in ESCs is also evidenced by the structure of the nucleolus, since ESCs generally contain one large nucleolus, whereas differentiated cells have more nucleoli with reduced size (Meshorer and Misteli, 2006) (Figure 1). Studies in mESCs showed that the lack of rRNA gene silencing is due to the impairment of NoRC recruitment to rRNA genes (Savić et al., 2014). The abrogation of TIP5 binding to rRNA genes in ESCs was caused by the impairment of IGS-rRNA processing into mature pRNA, a reaction that is critical for TIP5 recruitment. In differentiated cells, the RNA helicase DHX9 efficiently processes IGS-rRNA into the mature pRNA, which promotes TIP5 interaction with TTF1, leading to the formation of rRNA gene silencing (Leone et al., 2017). In contrast, IGS-rRNA processing is inhibited in ESCs and abrogates TIP5 interaction with TTF1, thereby impairing the establishment of silent rRNA genes (Figure 2). Importantly, targeting of heterochromatin in the nucleolus of mESCs was shown to affect pluripotency (Savić et al., 2014). On the other hand, the impairment of heterochromatin formation at rRNA genes during ESC differentiation abolished the exit from pluripotency (Leone et al., 2017). Therefore, these results suggest that the balance between euchromatin and heterochromatin at rRNA genes has a role in cell fate specification.
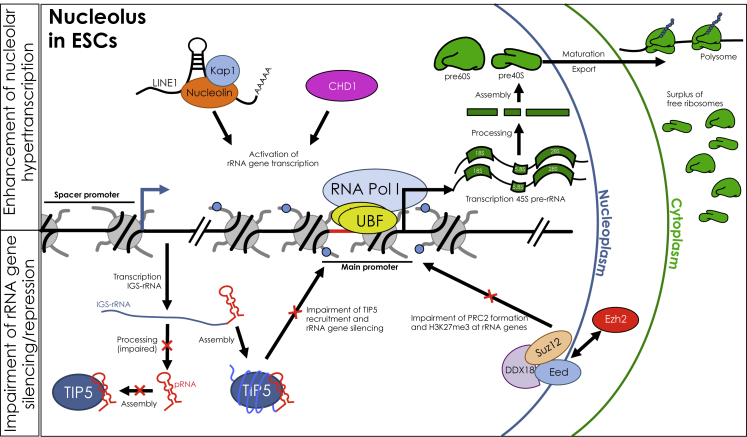
Figure 2.
Mechanisms Contributing to the Establishment of the Hyperactive State of the Nucleolus in ESCs
Hypertranscription of rRNA genes was shown to be favored by the binding of CHD1 (chromodomain helicase DNA-binding protein 1) (Guzman-Ayala et al., 2015) and LINE1-nucleolin RNA complex (Percharde et al., 2018) with rRNA genes. On the other hand, mechanisms for rRNA gene repression are impaired. The recruitment of TIP5 (TTF1-interacting protein 5) to rRNA genes is abrogated by the impairment of IGS-rRNA processing and consequent lack of formation of mature pRNA (promoter-associated RNA). The association of TIP5 with IGS-rRNA impairs TIP5 recruitment to rRNA genes and their epigenetic silencing (Leone et al., 2017; Savić et al., 2014). DDX18 (DEAD-box helicase 18) was shown to sequester PRC2 (polycomb repressive complex 2) in the outer layer of the nucleolus and impairs its formation (Zhang et al., 2020). This mechanism prevents the deposition of the repressive H3K27me3 mark onto rRNA genes. Hyperactivation of rRNA genes promotes ribosome biogenesis. However, ESCs require a low global protein synthesis rate. The enhanced ribosome biogenesis in ESCs might result in a surplus of free ribosomes, which can be used to allow rapid elevation of translation rate in response to differentiation signals (Golob et al., 2008; Sampath et al., 2008).
Data indicated that rRNA gene hypertranscription supports self-renewal. For example, stable expression of fibrillarin, which has an important role in pre-rRNA processing, prolongs the pluripotency of (m)ESCs cultured in the absence of leukemia inhibitory factor. On the other hand, fibrillarin knockdown or treatment with the Pol I inhibitor actinomycin D promotes stem cell differentiation (Watanabe-Susaki et al., 2014). Similarly, the reduction of rRNA synthesis in (h)ESCs by the Pol I inhibitor CX-5461 induces the expression of markers for all three germ layers, reduces the expression of pluripotency markers, and displaces UBF from rRNA genes (Woolnough et al., 2016). Another example linking rRNA hypertranscription and elevated ribosome biogenesis activities is provided by Nucleophosmin (NPM1), a regulator of rRNA gene transcription and ribosome processing. Indeed, NPM1 downregulation was shown to induce the expression of genes involved in mesoderm and ectoderm differentiation pathways (Johansson and Simonsson, 2010; Murano et al., 2008) (Figure 2). A role of NPM1 in pluripotency is also supported by its interaction with Mki67ip, a nucleolar phosphoprotein that is required for mESC maintenance (Abujarour et al., 2010). However, the mechanism of this process remains unclear. A role for the establishment of rRNA hypertranscription in ESCs has also been proposed for chromodomain helicase DNA-binding protein 1, a key mediator of the open chromatin state in ESCs and required for transcriptional output and development of the mouse epiblast (Gaspar-Maia et al., 2009). Chd1 inactivation in both epiblast cells and mESCs was shown to cause downregulation of rRNA gene transcription and alterations in nucleoli structure, so that they become smaller and more elongated (Guzman-Ayala et al., 2015) (Figure 2). rRNA hypertranscription state in ESCs has also been recently linked to the retrotransposon long interspersed element 1 (LINE1), which constitutes 19% and 17% of the genome in mouse and human, respectively, and is highly expressed during early development (Bodak et al., 2014; Fadloun et al., 2013). In mESCs, LINE1 transcript was shown to act as a nuclear RNA scaffold for the interaction with nucleolin, a regulator of rRNA transcription and processing, and the co-repressor tripartite motif-containing protein 28 (TRIM28/Kap1) (Rowe et al., 2010). The assembly of this RNA-protein complex was shown to play an important role in repressing a transcriptional program specific to the two-cell stage and promoting rRNA synthesis and self-renewal in ESCs (Percharde et al., 2018) (Figure 2). All these results indicated that the hypertranscription state of rRNA genes in ESCs supports the maintenance of the pluripotency state.
Several studies have also suggested that ESCs require rRNA hypertranscription to sustain elevated protein synthesis due to their high proliferative state. The RNA binding protein HTATSF1 was shown to control differentiation of both mESCs and hESCs by regulating multiple aspects of ribosome biogenesis, including rRNA transcription and processing and splicing of ribosomal protein transcripts, thereby controlling the 60S ribosomal abundance and promoting protein synthesis (Corsini et al., 2018). Furthermore, it was reported that HTATSF1 and protein synthesis levels decreased upon ESC differentiation, whereas HTATSF1 overexpression prevented differentiation. These observations led to the proposal that reduced rates of rRNA transcription and processing might cause a reduction in protein synthesis to facilitate the transition toward the post-implantation epiblast stage of mammalian development. Along the same line, recent results highlighted a role for the DEAD-box RNA helicase 18 (DDX18) in safeguarding the chromatin and transcriptional integrity of rRNA genes by counteracting the epigenetic silencing machinery to promote pluripotency (Zhang et al., 2020) (Figure 2). Specifically, it was shown that in mESCs DDX18 binds and sequesters polycomb repressive complex 2 (PRC2) in the outer layer of the nucleolus. This action prevents the deposition of the repressive H3K27me3 mark onto rRNA genes, thereby promoting hyperactive rRNA transcription, ribosome biogenesis, and global protein synthesis. However, the link between rRNA hypertranscription and protein synthesis in ESCs appears more complex than predicted (Tahmasebi et al., 2018). Several studies indicated that ESCs require a low global protein synthesis rate to maintain their overall homeostasis (Li and Wang, 2020). Accordingly, it was demonstrated that the global translation in ESCs is lower than in early differentiated cells (Ingolia et al., 2011; Sampath et al., 2008). Similar results were also observed in adult stem cells such as hair follicle stem cells, hematopoietic stem cells, and muscle stem cells (Blanco et al., 2016; Signer et al., 2014; Zismanov et al., 2016). Furthermore, upregulation in translation through inactivation of pseudouridine synthetase PUS7 in hESCs was shown to cause ESC differentiation defects (Guzzi et al., 2018). Thus, it appears that the enhanced ribosome biogenesis does not mirror an increase in protein synthesis in ESCs compared with differentiated cells. Instead, this might result in a surplus of free ribosomes, which can be used to allow rapid elevation of translational rate in response to differentiation (Golob et al., 2008; Sampath et al., 2008) (Figure 2). These results suggest that the translational control is a key modulator of stem cell maintenance and differentiation. Furthermore, they indicated that rRNA hypertranscription and high ribosome biogenesis in ESCs do not necessarily drive an increase in protein synthesis.
All the results described so far indicated that rRNA hypertranscription and the active chromatin state of rRNA genes are features of ESCs and required to maintain pluripotency. However, the hyperactive state of rRNA genes in ESCs does not correlate with protein synthesis levels, suggesting a role that goes beyond ribosome biogenesis and protein synthesis in ESCs. In recent years, increasing evidence has started to highlight additional roles of the nucleolus in several biological processes, including genome stability, regulation of cell cycle in response to stress, and nuclear architecture (Bersaglieri and Santoro, 2019; Boisvert et al., 2007). In the next section, we will discuss recent evidence pointing to the role of the nucleolus in the context of genome architecture and its role in ESCs.
Nucleolus and Genome Organization of ESCs
The recent advancements in chromatin conformation capture and high-resolution microscopy have started to unravel important details of the genome architecture in many cell types, including ESCs. Chromosomes are folded into hierarchical domains at different genomic scales, which likely enable the organization of the genome into functional compartments (Kempfer and Pombo, 2019). These structures comprise multiple levels of spatial features, including the distinct occupancy of chromosomes within the nucleus, called chromosome territories, active and repressive chromosomal compartments, topologically associating domains, and chromatin loops. An important observation initially emerging from Hi-C studies is that, at the megabase scale, chromosomes are largely segregated into two compartments, called A and B (Lieberman-Aiden et al., 2009). Compartment A is highly enriched in open and active chromatin, whereas compartment B is enriched in closed and repressed chromatin. This genome compartmentalization can change between cell types. For example, mESCs undergo a global reorganization in chromatin interactions during differentiation (Bonev et al., 2017). In particular, the interaction strength between A-type domains decreased in differentiated cells compared with mESCs, whereas contacts within the B compartment became stronger. These results are consistent with previous studies showing that the chromatin in ESCs is in a more plastic and open state than in differentiated cells (Meshorer et al., 2006).
Subnuclear compartments, including the nucleolus, have also emerged as important regulators of large-scale organization of chromosomes and function during interphase. Studies have started to suggest that the location of defined genes in specific subnuclear compartments might allow the concentration of factors (e.g., repressor or activators) and thereby facilitate functions that rely on proteins found in limiting concentrations (Gonzalez-Sandoval and Gasser, 2016).
In mammalian cells, genomic regions in close contact with the nuclear lamina at the nuclear periphery are termed lamina-associated domains (LADs) (van Steensel and Belmont, 2017). LADs mainly display features typical of heterochromatin since they mainly contain genes in a transcriptionally silent state or with low expression levels. Furthermore, LADs have a low overall gene density, correspond to late-replicating DNA, and are typically enriched for repressive histone marks (Guelen et al., 2008; Peric-Hupkes et al., 2010; Pope et al., 2014). Changes in LAD composition have also been observed during mESC differentiation (Peric-Hupkes et al., 2010). This reorganization of genomic interactions with the nuclear lamina was shown to affect many genes implicated in cellular identity, suggesting an important role in the control of gene expression programs during lineage commitment and terminal differentiation.
In recent years, the nucleolus has also started to emerge as a subnuclear compartment with an important role in the organization of genome architecture. Genomic regions positioned in close proximity to the nucleolus are known as nucleolar-associated domains (NADs) (Figure 3). Considering that the size and the number of nucleoli change according to cell state and developmental stage, it is expected that NAD composition might also change. Although the nucleolus is the subnuclear compartment with the highest transcriptional activity in the cell, increasing evidence indicates that it also attracts repressive chromatin regions (Bersaglieri and Santoro, 2019; Guetg and Santoro, 2012; Padeken and Heun, 2014). For example, centromeres and telomeres often associate with nucleoli in many cell types (Carvalho et al., 2001; Weierich et al., 2003; Zhang et al., 2004). Similarly, the repressed Kcnq1-imprinted domain on the paternal allele is frequently localized to the nucleolar periphery in mouse trophoblast stem cells and placenta tissue (Pandey et al., 2008). However, its spatial proximity to the nucleolus was shown not to be sufficient to preclude transcriptional reactivation (Fedoriw et al., 2012a). Another example of repressive domains localizing close to the nucleolus is the inactive X chromosome (Xi) of female cells. During X-chromosome inactivation (XCI), Xi forms a compact heterochromatic structure that is frequently localized close to the perinucleolar region during the S phase (Zhang et al., 2007). However, how Xi is tethered to nucleoli remains to be elucidated. A role in this process has been proposed for the lncRNA Firre, which is one of the genes found to escape XCI in humans and mice (Hacisuleyman et al., 2014). The Firre locus on the Xi was found to be located adjacent to the nucleolus. Importantly, knockdown of Firre in mouse fibroblasts disrupted its perinucleolar location and led to a decrease in H3K27me3 on the X chromosome, suggesting a role in the maintenance of this repressive chromatin feature of Xi (Yang et al., 2015). However, similar to the Kcnq1-imprinted domain, Firre knockdown was not sufficient to reactivate X-linked genes, suggesting that perinucleolar localization is not sufficient to establish transcriptional silencing. Thus, at least in these particular cases, the location of genes close to the nucleolar compartment does not lead to transcriptional silencing. A similar observation has also been reported for LADs, since targeting of a gene at the nuclear periphery does not necessarily lead to transcriptional inactivation (Finlan et al., 2008; Kumaran and Spector, 2008; Wang et al., 2018). However, examples also exist where targeting of genetic loci to the nuclear periphery can induce transcriptional silencing. MiCEE is a multicomponent ribonucleoprotein complex composed of the microRNA mir-let-7d and factors of the exosome and PRC2. It was shown that in several human cell lines MiCEE tethers loci of bidirectionally active genes to the perinucleolar region, leading to transcriptional silencing (Singh et al., 2018). Another example can be found in mESCs, where LINE1 RNA was suggested to tether LINE1 DNA loci close to the nucleolus and be required for the transcriptional silencing of genes containing LINE1 (Lu et al., 2020). Another example showing a functional relationship between nucleolar localization and gene expression was the integration of a 5S rDNA gene into a new genomic location that could significantly influence the association of the host region with the nucleolus and cause transcriptional inhibition of neighboring genes transcribed by RNA polymerase II (Fedoriw et al., 2012b).
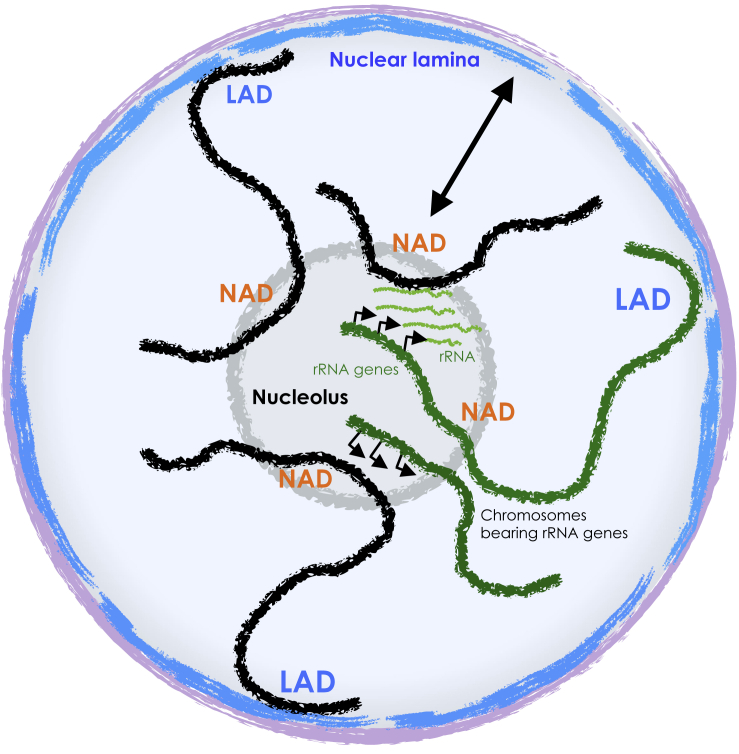
Figure 3.
Types of Genomic Contacts with the Nucleolus
Schema representing nucleolar-associated domains (NADs). Genomic contacts with the nucleolus have been identified by biochemical purification of the nucleoli and contacts with rRNA genes (HIC, 4C-seq) or rRNA transcripts (SPRITE). Some NADs were described to overlap with lamina-associated domains (LADs). The double arrow indicates the relocation of some NADs to the nuclear lamina after mitosis. Green lanes represent chromosomes bearing rRNA genes. Interchromosomal DNA contacts between chromosomes containing rRNA genes have been reported, mirroring the coalescence of rRNA genes from different chromosomes in the same nucleolus.
All the results described above suggest that the interaction of chromosomes with the nuclear periphery and nucleolus should contribute to a basal chromosome architecture and genome function. However, while the role of the nuclear periphery in the genome organization has been well documented, the function of the nucleolus remains yet underinvestigated. A major obstacle in studying NADs is that the nucleolus is a membrane-less compartment, thereby challenging the establishment of technologies for the identification of NADs as has been done for LADs (Feric et al., 2016). Attempts in the identification of NADs at the genome-wide level were mainly based on the biochemical purification of nucleoli, a method that relies on sonication of nuclei, adjusting the power so that nucleoli remain intact while the rest of the nuclei is fragmented (Muramatsu et al., 1963). To date, using this method, NADs have been characterized in mESCs (Bizhanova et al., 2020; Lu et al., 2020), mouse embryonic fibroblasts (MEFs) (Vertii et al., 2019), a few human somatic cell lines (Dillinger et al., 2017; Nemeth et al., 2010; van Koningsbruggen et al., 2010), and the plant Arabidopsis thaliana (Pontvianne et al., 2016) (Table 1). Although this is the only method that until now can be used for the mapping of NADs at the genome-wide level, some limitations should be taken into consideration. First, the biochemical purification of nucleoli can vary between cell types, making a direct comparison between NADs from different cells problematic. Furthermore, the identification of NADs upon sequencing of purified nucleoli can be biased toward the selection of repressive chromatin domains, since heterochromatin is generally more resistant to sonication than euchromatin (Becker et al., 2017). The difficulty in a precise mapping of NADs from biochemically purified nucleoli is also evident from two recent analyses of NADs in mESCs that showed several discrepancies, including a substantial difference in NAD genome coverage (7.5% in Lu et al., 2020, and 31% in Bizhanova et al., 2020). This discrepancy could also be due to the different methods applied for the purification of nucleoli of mESCs. Indeed, while in one work nucleoli were purified from cross-linked mESCs (Bizhanova et al., 2020), in other study this step was omitted (Lu et al., 2020). The two initial independent parallel studies that mapped NADs from biochemically purified nucleoli of HeLa, IMR90, and HT1080 human cell lines revealed that NADs contain repressive histone modifications and are enriched in olfactory receptor genes, zinc-finger genes, immunoglobulin gene families, and 5S RNA genes (Nemeth et al., 2010; van Koningsbruggen et al., 2010). Furthermore, centromeric and pericentromeric satellites and subtelomeric regions were also identified as NADs, confirming previous microscopy studies. However, the conclusions of these two studies were not always overlapping. While one work highlighted that NADs were enriched in tRNA genes and differed markedly from LADs (Nemeth et al., 2010), the other showed that NADs have a high density of AT-rich sequence elements and low gene density, and there was a clear overlap between NADs and LADs (van Koningsbruggen et al., 2010). A positive correlation between NADs and LADs has also been reported by imaging analyses. Tracing of LADs in the human fibrosarcoma cell line HT1080 during the cell cycle revealed that LADs from the mother cell after completion of cell division can be positioned to the nucleoli of the daughter cells (Kind et al., 2013). Similarly, NADs can relocate from nucleoli in close proximity to the nuclear envelope after mitosis (van Koningsbruggen et al., 2010) (Figure 3). Interestingly, both LADs and NADs contain significantly higher levels of LINE1 and they are depleted of short interspersed nuclear elements, which are instead enriched in euchromatic regions (i.e., compartment A) (Lu et al., 2020). Recent studies using the biochemical purification of nucleoli to map NADs revealed that the genome coverage of NADs in mESCs (31% in Bizhanova et al., 2020 or 7.5% in Lu et al., 2020) is lower than in MEFs (41%) (Vertii et al., 2019), a result that might reflect the low heterochromatic content of the ESC genome compared with differentiated cells (Gaspar-Maia et al., 2011).
Table 1.
Features of NADs
| Material | Organism | Method | Genome Coverage | Representative Sequences in NAD |
|---|---|---|---|---|
| HeLa, IMR901 | Homo sapiens | biochemical purification | 4% | ZNF, olfactory receptor, defensin, immunoglobulin, 5S RNA, tRNA genes, alpha-,beta-, (GAATG)n/(CATTC)n satellite repeats |
| HT1802 | H. sapiens | biochemical purification | NA | ZNF, olfactory receptors, immunoglobulin, 5S RNA, satellite repeats, LADs |
| Arabidopsis thaliana3 | Plant | biochemical purification and FANoS | 4.2% | subtelomeric regions; short arm of chromosome 4; transposable elements Mariner, Pogo, and Tc1; pseudo-LINE elements; tRNA; and pseudogenes. |
| IMR904 | H. sapiens | biochemical purification | 38% | satellite repeats, LTR elements, ZNF, 5S RNA, defensin, olfactory receptor genes, LADs |
| MEFs5 | Mus musculus | biochemical purification | 30% XL: 41% |
NA |
| ESCs6 | M. musculus | biochemical purification | 7.5% | LINE1-enriched genes |
| ESCs7 | M. musculus | biochemical purification | XL: 31% | NA |
| ESCs8 | M. musculus | genomic contacts with rRNA transcripts (SPRITE) | NA | centromere-proximal regions, DNA in linear proximity to rRNA genes, inactive chromatin |
| K562 and LCL9 | H. sapiens | recovery of reads containing rRNA gene contacts from Hi-C | NA | repressed and late-replicating chromatin, CTCF binding sites, ribosomal- and mitochondrial-related genes |
Methodologies, genome coverage relative to the annotated genome, and representative sequences identified as NADs are listed. Numbers refer to the NAD published works: 1Nemeth et al., 2010, 2van Koningsbruggen et al., 2010, 3Pontvianne et al., 2016, 4Dillinger et al., 2017, 5Vertii et al., 2019, 6Lu et al., 2020, 7Bizhanova et al., 2020, 8Quinodoz et al., 2018, 9Yu and Lemos, 2018. FANoS, fluorescence-activated nucleolar sorting; ZNF, zinc-finger proteins; XL, nucleoli purified from cross-linked cells.
Genomic analyses revealed that NADs can be divided into two main categories: one can associate with the nuclear periphery or nucleolus (type I NAD), the other can interact exclusively with nucleoli (type II NAD) (Bizhanova et al., 2020; Vertii et al., 2019). Type I NADs in MEFs were shown to generally display characteristics of constitutive heterochromatin, including late DNA replication and low gene expression levels as found in LADs (Vertii et al., 2019). Furthermore, type I NADs were enriched in the heterochromatic mark H3K9me3, a result that is consistent with a previous report (Dillinger et al., 2017). In contrast, type II NADs in MEFs were found to share many features with facultative heterochromatin (Vertii et al., 2019). Indeed, this class of NAD displays greater gene expression, is enriched in H3K27me3, and contains genes implicated in differentiation and development processes. Interestingly, it was also reported that type II NADs were less abundant in mESCs than in MEFs (Bizhanova et al., 2020), suggesting a distinct genome organization around the nucleolus between ESC and differentiated cells.
The recent development of ligation-capture Hi-C sequencing technology has provided alternative method for the identification of NADs. Specifically, these technologies allowed the identification of genomic contacts with the rRNA genes or rRNA transcripts (Figure 3). However, this class of genomic region has to be considered as a subclass of NADs, since not all genomic domains associating with the nucleolus must necessarily interact with the rRNA genes and their corresponding transcripts. A new technique, known as split-pool recognition of interactions by tag extension (SPRITE), identified genomic contacts in close proximity to the nucleolus by taking into account the interaction with rRNA transcripts (Quinodoz et al., 2018). Genomic sequences in contact with rRNA in mESCs are frequently found close to nucleoli and linearly close to the centromeres, a result that is consistent with previous observations showing that centromeres often co-localize on the periphery of the nucleolus. These results also provided further evidence that genomic DNA regions that are closer to the nucleolus tend to correspond to inactive chromatin. Furthermore, this method identified a class of interchromosomal DNA contacts between chromosomes containing rRNA genes, mirroring the coalescence of rRNA genes from different chromosomes in the same nucleolus (Figure 3). In contrast, however, LAD interactions generally occur between regions that are linearly close to each other rather than between chromosomes, highlighting a distinct 3D organization of NADs and LADs. Recently, the recovery of reads containing rRNA gene contacts from a published Hi-C dataset from human lymphoblastoid (LCL) and erythroleukemia (K562) cells revealed enrichment in genomic regions belonging to the chromosomal compartment B with features of repressed and late-replicating chromatin as well as CTCF binding sites (Yu and Lemos, 2018). Circularized chromosome conformation capture sequencing (4C-seq) has also been used to identify genomic contacts with defined rRNA gene sequences. A 4C-seq analysis in HEK293T cells revealed that IGS-rRNA sequences often form contacts with specific regions of different chromosomes, including the pericentromeric regions as well as regions that are characterized by H3K27ac and H3K4me3 marks and CTCF binding sites (Tchurikov et al., 2015). A recent work employed 4C-seq to identify genomic contacts with rRNA genes using an Eμ-Myc mouse model of spontaneous MYC-driven B cell lymphoma (Diesch et al., 2019). The analysis of the nucleolus in cancer is of particular interest since the increased number and/or size of nucleoli has historically been used by pathologists as a prognostic indicator of cancerous lesions (Weeks et al., 2019). The results revealed that during the progression from premalignancy to malignancy, UBF associates with a fraction of inactive genes and remodels their chromatin into an active state (Diesch et al., 2019). This process co-occurs with the establishment of contacts between rRNA genes and defined genomic loci and correlates with changes in the expression of genes that belong to pathways involved in B cell differentiation. Remarkably, the establishment of some contacts between rRNA genes and genomic domains during malignant progression depended on the formation of active rRNA genes but not on rRNA transcription, suggesting a role of the chromatin structure of rRNA genes in shaping genome organization. Consistent with these results, targeting of heterochromatin at rRNA genes in mESCs was found to induce the remodeling of the open and euchromatic genome into a condensed heterochromatic form (Savić et al., 2014). These changes included a global increase in the repressive histone mark H3K9me2 and the appearance of highly condensed heterochromatic blocks outside the nucleolus, a structure resembling the genome organization found in differentiated cells (Figure 1). The link between the rRNA gene chromatin state and the genome architecture was also found in MEF NIH3T3 cells, which display a large fraction (ca. 50%) of silent rRNA genes (Santoro and Grummt, 2001). The knockdown of TIP5 induced not only a decrease in the number of silent rRNA genes but also the reduction of silent histone marks at pericentric heterochromatin, which is often located close to the nucleolus (Guetg et al., 2010; Postepska-Igielska et al., 2013). These results suggest that the nucleolus not only is the compartment where repressive chromatin tends to be located, but also is implicated in the establishment of repressive chromatin states, an action that is luckily mediated by the chromatin state of rRNA genes.
The results using different technologies for the measurement of genomic regions localizing within the nucleolus or interacting with rRNA genes or rRNA transcripts agree in describing the nucleolus as a compartment where repressive domains are located. However, precise identification of all NADs for each cell state is still a technical limitation to fully understanding how the nucleolus regulates genome architecture and function. Furthermore, it is not yet clear how these genomic regions are tethered to the nucleolus and whether tethering to the nucleolus is the cause or a consequence of repressive chromatin states. Recent studies implicated p150, a subunit of chromatin assembly factor 1 (CAF-1), and the proliferation antigen Ki-67 in nucleolar targeting. Ki-67 depletion decreased the nucleolar association of a LacO array proximal to the rRNA repeats on chromosome 13 (Booth et al., 2014) and α-satellite DNA from chromosome 17 in HeLa cells (Matheson and Kaufman, 2017). Similarly, the p150 subunit of CAF-1 was shown to regulate the association of the 10q-telomere, 5S rDNA array, and α satellites with the nucleolus (Smith et al., 2014). The recent implication of phase separation in the formation of nucleoli (Feric et al., 2016) and heterochromatin (Larson et al., 2017; Strom et al., 2017) might also represent an attractive model to explain the assembly of repressive domains at the nucleolus. An important example is provided by studies in Drosophila showing that deletion of NPM1, which forms liquid-like droplets in the presence of RNA (Mitrea et al., 2016), is required for centromere clustering and anchoring to the nucleolus (Padeken and Heun, 2014). Recently, treatment of MEFs with 1,6-hexanediol, which is thought to perturb phase separation by abrogating liquid-like condensates through the disruption of weak hydrophobic interactions, was shown to reduce the nucleolar associations of two different type II NADs (Vertii et al., 2019). However, further studies are required to dissect how phase separation might contribute to targeting of genomic domains to the nucleolus and how these interactions are modulated during development and in disease.
Conclusions
The nucleolus is now recognized to serve essential functions in several processes in addition to ribosome synthesis. In this review, we summarized recent results showing how the regulation of nucleolar activities can influence cell fate determination. We provided examples of nucleolar regulation in ESCs, where the hyperactive state of the nucleolus was shown to be required for the maintenance of pluripotency. The fact that ESCs require low translation rates suggests that nucleolar hyperactivation is probably not required for an immediate supply of ribosomes for protein synthesis and, consequently, might serve other functions. One such important, "non-conventional" function of the nucleolus that has emerged in recent years is its connection with the regulation of genome architecture. Since the size of nucleoli increases in proportion to the amount of synthesized rRNA (Hernandez-Verdun et al., 2010), it can be assumed that the enlarged nucleolus of ESCs should reduce the nuclear volume occupied by the chromosomes and probably affect some features of the genome organization. Furthermore, the typical clustering of heterochromatin blocks at nucleoli observed in the large majority of differentiated cells is absent in ESCs. The impact of the chromatin state of rRNA genes, the genetic component of the nucleolus, on the rest of the genome and its role in the maintenance of pluripotency suggest an active role of the nucleolus in the regulation of chromatin states at a genome-wide level. The recent development of sophisticated methods for the analysis of genome organization has contributed to providing first insights into the identification of genomic regions surrounding the nucleolus. However, the full understanding of this layer of genome compartmentalization is still in its infancy due to the technical difficulties arising from the lack of a membrane that separates the nucleolus from the rest of the nucleus. Although there is a certain agreement that some repressive domains can switch in their localization between the nuclear periphery and the nucleolus, it remains uncertain whether only repressive chromatin domains associate with the nucleolus. Furthermore, we still ignore how NADs are tethered to the nucleolus and whether proximity to the nucleolus affects chromatin state and gene expression. Finally, the establishment of novel methodologies that preserve and catch most of the genomic contacts with the nucleolus and their adaptation for the analysis of genome-nucleolus interactions in single cells, as done in the case of LADs (Kind et al., 2013), would be essential to understand nucleolar dynamics during development and disease. This will provide novel insights into basic principles of genome organization and its role in gene expression and cellular function that yet remain elusive.
Author Contributions
S.G. and R.S. performed literature research and wrote the manuscript. R.S. conceived and supervised the project. Both authors reviewed and approved the final version of the manuscript.
Acknowledgments
This work was supported by the Swiss National Science Foundation, Switzerland (31003A_173056), and an ERC grant (ERC-AdG-787074-NucleolusChromatin).
References
- Abujarour R., Efe J., Ding S. Genome-wide gain-of-function screen identifies novel regulators of pluripotency. Stem Cells. 2010;28:1487–1497. doi: 10.1002/stem.472. [DOI] [PubMed] [Google Scholar]
- Becker J.S., McCarthy R.L., Sidoli S., Donahue G., Kaeding K.E., He Z., Lin S., Garcia B.A., Zaret K.S. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell. 2017;68:1023–1037.e15. doi: 10.1016/j.molcel.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri C., Santoro R. Genome organization in and around the nucleolus. Cells. 2019;8:579. doi: 10.3390/cells8060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizhanova A., Yan A., Yu J., Zhu L.J., Kaufman P.D. Distinct features of nucleolus-associated domains in mouse embryonic stem cells. Chromosoma. 2020;129:121–139. doi: 10.1007/s00412-020-00734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., Sajini A., Tanna H., Cortes-Garrido R., Gkatza N. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodak M., Yu J., Ciaudo C. Regulation of LINE-1 in mammals. Biomol. Concepts. 2014;5:409–428. doi: 10.1515/bmc-2014-0018. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.P., Tanay A. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth D.G., Takagi M., Sanchez-Pulido L., Petfalski E., Vargiu G., Samejima K., Imamoto N., Ponting C.P., Tollervey D., Earnshaw W.C. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. Elife. 2014;3:e01641. doi: 10.7554/eLife.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A., Macrae T.A., Oses-Prieto J.A., Covarrubias S., Percharde M., Ku G., Diaz A., McManus M.T., Burlingame A.L., Ramalho-Santos M. The transcriptionally permissive chromatin state of embryonic stem cells is acutely tuned to translational output. Cell Stem Cell. 2018;22:369–383.e8. doi: 10.1016/j.stem.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho C., Pereira H.M., Ferreira J., Pina C., Mendonça D., Rosa A.C., Carmo-Fonseca M. Chromosomal G-dark bands determine the spatial organization of centromeric heterochromatin in the nucleus. Mol. Biol. Cell. 2001;12:3563–3572. doi: 10.1091/mbc.12.11.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Widmer R.M., Koller T., Sogo J.M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Corsini N.S., Peer A.M., Moeseneder P., Roiuk M., Burkard T.R., Theussl H.C., Moll I., Knoblich J.A. Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell. 2018;22:543–558.e12. doi: 10.1016/j.stem.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Dalcher D., Tan J.Y., Bersaglieri C., Peña-Hernández R., Vollenweider E., Zeyen S., Schmid M.W., Bianchi V., Butz S., Kuzyakiv R. TIP5 safeguards genome architecture of ground-state pluripotent stem cells. bioRxiv. 2019 doi: 10.1101/2019.12.20.882282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesch J., Bywater M.J., Sanij E., Cameron D.P., Schierding W., Brajanovski N., Son J., Sornkom J., Hein N., Evers M. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun. Biol. 2019;2:39. doi: 10.1038/s42003-019-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillinger S., Straub T., Nemeth A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS One. 2017;12:e0178821. doi: 10.1371/journal.pone.0178821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D., Buetow K.H. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Fedoriw A.M., Calabrese J.M., Mu W., Yee D., Magnuson T. Differentiation-driven nucleolar association of the mouse imprinted Kcnq1 locus. G3 (Bethesda) 2012;2:1521–1528. doi: 10.1534/g3.112.004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw A.M., Starmer J., Yee D., Magnuson T. Nucleolar association and transcriptional inhibition through 5S rDNA in mammals. PLoS Genet. 2012;8:e1002468. doi: 10.1371/journal.pgen.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan L.E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J.R., Bickmore W.A. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka H., Langerova A. Nucleoli in embryos: a central structural platform for embryonic chromatin remodeling? Chromosome Res. 2019;27:129–140. doi: 10.1007/s10577-018-9590-3. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golob J.L., Paige S.L., Muskheli V., Pabon L., Murry C.E. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev. Dyn. 2008;237:1389–1398. doi: 10.1002/dvdy.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Gasser S.M. On TADs and LADs: spatial control over gene expression. Trends Genet. 2016;32:485–495. doi: 10.1016/j.tig.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., Hottiger M.O., Fussenegger M., Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C., Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–814. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C., Scheifele F., Rosenthal F., Hottiger M.O., Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M., Sachs M., Koh F.M., Onodera C., Bulut-Karslioglu A., Lin C.J., Wong P., Nitta R., Song J.S., Ramalho-Santos M. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015;142:118–127. doi: 10.1242/dev.114843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N., Ciesla M., Ngoc P.C.T., Lang S., Arora S., Dimitriou M., Pimkova K., Sommarin M.N.E., Munita R., Lubas M. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–1216.e26. doi: 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdane N., Stefanovsky V.Y., Tremblay M.G., Nemeth A., Paquet E., Lessard F., Sanij E., Hannan R., Moss T. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014;10:e1004505. doi: 10.1371/journal.pgen.1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein N., Hannan K.M., George A.J., Sanij E., Hannan R.D. The nucleolus: an emerging target for cancer therapy. Trends Mol. Med. 2013;19:643–654. doi: 10.1016/j.molmed.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Herdman C., Mars J.C., Stefanovsky V.Y., Tremblay M.G., Sabourin-Felix M., Lindsay H., Robinson M.D., Moss T. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet. 2017;13:e1006899. doi: 10.1371/journal.pgen.1006899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D., Roussel P., Thiry M., Sirri V., Lafontaine D.L. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip. Rev. RNA. 2010;1:415–431. doi: 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., Weissman J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H., Simonsson S. Core transcription factors, Oct4, Sox2 and Nanog, individually form complexes with nucleophosmin (Npm1) to control embryonic stem (ES) cell fate determination. Aging (Albany NY) 2010;2:815–822. doi: 10.18632/aging.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempfer R., Pombo A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 2019;21:207–226. doi: 10.1038/s41576-019-0195-2. [DOI] [PubMed] [Google Scholar]
- Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S.S., Janssen H., Amendola M., Nolen L.D., Bickmore W.A., van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Kresoja-Rakic J., Santoro R. Nucleolus and rRNA gene chromatin in early embryo development. Trends Genet. 2019;35:868–879. doi: 10.1016/j.tig.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran R.I., Spector D.L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone S., Bar D., Slabber C.F., Dalcher D., Santoro R. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep. 2017;18:1248–1262. doi: 10.15252/embr.201744330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wang J. Ribosome heterogeneity in stem cells and development. J. Cell Biol. 2020;219:e202001108. doi: 10.1083/jcb.202001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Santoro R., Koberna K., Grummt I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 2005;24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.Y., Shao W., Chang L., Yin Y., Li T., Zhang H., Hong Y., Percharde M., Guo L., Wu Z. Genomic repeats categorize genes with distinct functions for orchestrated regulation. Cell Rep. 2020;30:3296–3311.e5. doi: 10.1016/j.celrep.2020.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson T.D., Kaufman P.D. The p150N domain of chromatin assembly factor-1 regulates Ki-67 accumulation on the mitotic perichromosomal layer. Mol. Biol. Cell. 2017;28:21–29. doi: 10.1091/mbc.E16-09-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Schmitz K.M., Li J., Grummt I., Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B., Nourse A., Deniz A.A., Kriwacki R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Mars J.C., Tremblay M.G., Sabourin-Felix M. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019;27:31–40. doi: 10.1007/s10577-018-09603-9. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Smetana K., Busch H. Quantitative aspects of isolation of nucleoli of the walker carcinosarcoma and liver of the rat. Cancer Res. 1963;23:510–518. [PubMed] [Google Scholar]
- Murano K., Okuwaki M., Hisaoka M., Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., Oie S., Daitoku H., Okuwaki M., Nagata K. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Muscarella D.E., Vogt V.M., Bloom S.E. The ribosomal RNA gene cluster in aneuploid chickens: evidence for increased gene dosage and regulation of gene expression. J. Cell Biol. 1985;101:1749–1756. doi: 10.1083/jcb.101.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Peterfia B., Solovei I., Cremer T., Dopazo J., Langst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Padeken J., Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr. Opin. Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Thomas G., Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- Percharde M., Bulut-Karslioglu A., Ramalho-Santos M. Hypertranscription in development, stem cells, and regeneration. Dev. Cell. 2017;40:9–21. doi: 10.1016/j.devcel.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lin C.J., Yin Y., Guan J., Peixoto G.A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–405 e319. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W., Solovei I., Brugman W., Graf S., Flicek P., Kerkhoven R.M., van Lohuizen M. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Carpentier M.C., Durut N., Pavlistova V., Jaske K., Schorova S., Parrinello H., Rohmer M., Pikaard C.S., Fojtova M. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 2016;16:1574–1587. doi: 10.1016/j.celrep.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postepska-Igielska A., Krunic D., Schmitt N., Greulich-Bode K.M., Boukamp P., Grummt I. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 2013;14:704–710. doi: 10.1038/embor.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–757.e24. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Sampath P., Pritchard D.K., Pabon L., Reinecke H., Schwartz S.M., Morris D.R., Murry C.E. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T.P., Quin J., Robb E., Wong L.H., Thomas W.G., Stefanovsky V. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R., Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- Santoro R., Li J., Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- Santoro R., Schmitz K.M., Sandoval J., Grummt I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010;11:52–58. doi: 10.1038/embor.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savić N., Bär D., Leone S., Frommel Sandra C., Weber Fabienne A., Vollenweider E., Ferrari E., Ziegler U., Kaech A., Shakhova O. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 2014;15:720–734. doi: 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Schlesinger S., Selig S., Bergman Y., Cedar H. Allelic inactivation of rDNA loci. Genes Dev. 2009;23:2437–2447. doi: 10.1101/gad.544509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer R.A., Magee J.A., Salic A., Morrison S.J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I., Contreras A., Cordero J., Rubio K., Dobersch S., Gunther S., Jeratsch S., Mehta A., Kruger M., Graumann J. MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization. Nat. Genet. 2018;50:990–1001. doi: 10.1038/s41588-018-0139-3. [DOI] [PubMed] [Google Scholar]
- Smith C.L., Matheson T.D., Trombly D.J., Sun X., Campeau E., Han X., Yates J.R., 3rd, Kaufman P.D. A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli. Mol. Biol. Cell. 2014;25:2866–2881. doi: 10.1091/mbc.E14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I., Lucchini R., Koller T., Sogo J.M. Chromatin structure and methylation of rat rRNA genes studied by formaldehyde fixation and psoralen cross-linking. Nucleic Acids Res. 1997;25:1727–1735. doi: 10.1093/nar/25.9.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S., Amiri M., Sonenberg N. Translational control in stem cells. Front. Genet. 2018;9:709. doi: 10.3389/fgene.2018.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchurikov N.A., Fedoseeva D.M., Sosin D.V., Snezhkina A.V., Melnikova N.V., Kudryavtseva A.V., Kravatsky Y.V., Kretova O.V. Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation. J. Mol. Cell Biol. 2015;7:366–382. doi: 10.1093/jmcb/mju038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S., Gierlinski M., Schofield P., Martin D., Barton G.J., Ariyurek Y., den Dunnen J.T., Lamond A.I. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Belmont A.S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A., Ou J., Yu J., Yan A., Pages H., Liu H., Zhu L.J., Kaufman P.D. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019;29:1235–1249. doi: 10.1101/gr.247072.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu X., Nguyen C.M., Liu Y., Gao Y., Lin X., Daley T., Kipniss N.H., La Russa M., Qi L.S. CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell. 2018;175:1405–1417.e14. doi: 10.1016/j.cell.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Susaki K., Takada H., Enomoto K., Miwata K., Ishimine H., Intoh A., Ohtaka M., Nakanishi M., Sugino H., Asashima M. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells. 2014;32:3099–3111. doi: 10.1002/stem.1825. [DOI] [PubMed] [Google Scholar]
- Weeks S.E., Metge B.J., Samant R.S. The nucleolus: a central response hub for the stressors that drive cancer progression. Cell Mol. Life Sci. 2019;76:4511–4524. doi: 10.1007/s00018-019-03231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierich C., Brero A., Stein S., von Hase J., Cremer C., Cremer T., Solovei I. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 2003;11:485–502. doi: 10.1023/a:1025016828544. [DOI] [PubMed] [Google Scholar]
- Woolnough J.L., Atwood B.L., Liu Z., Zhao R., Giles K.E. The regulation of rRNA gene transcription during directed differentiation of human embryonic stem cells. PLoS One. 2016;11:e0157276. doi: 10.1371/journal.pone.0157276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Ling T., Zhou Y., Feng W., Zhu Q., Stunnenberg H.G., Grummt I., Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. U S A. 2012;109:8161–8166. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Deng X., Ma W., Berletch J.B., Rabaia N., Wei G., Moore J.M., Filippova G.N., Xu J., Liu Y. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Lemos B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018;14:e1007258. doi: 10.1371/journal.pgen.1007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu Z., Lu J.Y., Huang B., Zhou H., Xie W., Wang J., Shen X. DEAD-box helicase 18 counteracts PRC2 to safeguard ribosomal DNA in pluripotency regulation. Cell Rep. 2020;30:81–97.e7. doi: 10.1016/j.celrep.2019.12.021. [DOI] [PubMed] [Google Scholar]
- Zhang L.F., Huynh K.D., Lee J.T. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhang S., Hemmerich P., Grosse F. Nucleolar localization of the human telomeric repeat binding factor 2 (TRF2) J. Cell Sci. 2004;117:3935–3945. doi: 10.1242/jcs.01249. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Santoro R., Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V., Chichkov V., Colangelo V., Jamet S., Wang S., Syme A., Koromilas A.E., Crist C. Phosphorylation of eIF2alpha is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell. 2016;18:79–90. doi: 10.1016/j.stem.2015.09.020. [DOI] [PubMed] [Google Scholar]