Summary
The multifunctional histone chaperone, SET, is essential for embryonic development in the mouse. Previously, we identified SET as a factor that is rapidly downregulated during embryonic stem cell (ESC) differentiation, suggesting a possible role in the maintenance of pluripotency. Here, we explore SET's function in early differentiation. Using immunoprecipitation coupled with protein quantitation by LC-MS/MS, we uncover factors and complexes, including P53 and β-catenin, by which SET regulates lineage specification. Knockdown for P53 in SET-knockout (KO) ESCs partially rescues lineage marker misregulation during differentiation. Paradoxically, SET-KO ESCs show increased expression of several Wnt target genes despite reduced levels of active β-catenin. Further analysis of RNA sequencing datasets hints at a co-regulatory relationship between SET and TCF proteins, terminal effectors of Wnt signaling. Overall, we discover a role for both P53 and β-catenin in SET-regulated early differentiation and raise a hypothesis for SET function at the β-catenin-TCF regulatory axis.
Keywords: chromatin, pluripotency, Wnt, embryonic stem cells, differentiation, p53, β-catenin, epigenetics
Highlights
-
•
Loss of SET impairs proliferation of ESCs and disrupts the pluripotent state
-
•
SET directly interacts with P53 and β-catenin
-
•
P53 partially contributes to disturbed expression of lineage markers in SET-KO ESCs
-
•
Loss of SET transactivates β-catenin and enhances Wnt gene expression
Previously, Meshorer and colleagues identified the rapid downregulation of SETα following ESC differentiation. This study reveals that interaction of SET with P53 contributes to defects in lineage marker expression observed in SET-KO ESCs. Paradoxically, although canonical Wnt signaling is activated in the absence of SET, lower levels of active β-catenin are observed, suggesting a SET-mediated redistribution of nuclear β-catenin.
Introduction
To develop into the complete adult organism, lineage specification and restriction events occur to direct differentiation of embryonic stem cells (ESCs) into specific cell types. Three days after fertilization in mouse, the inner cell mass of the pre-implantation blastocyst segregates into the hypoblast and pluripotent epiblast stem cells, the latter possessing the dual ability to self-renew and differentiate into any somatic cell type (Schlesinger and Meshorer, 2019). Understanding early embryonic development involves deciphering the journey taken by an epiblast stem cell toward one of three germ layers: mesoderm, ectoderm, and endoderm (Gadue et al., 2005). Although this process remains cryptic, some progress has been made, owing to ESC culturing techniques and the ability to control their pluripotent state in vitro (Evans and Kaufman, 1981; Murry and Keller, 2008; Ying et al., 2003, 2008). In the last two decades, many of the participants that maintain the balance between ESC self-renewal and differentiation have been revealed; in particular, core pluripotency factors, including NANOG, OCT4, and SOX2 (Boyer et al., 2005) in concert with other transcription factors (Ivanova et al., 2006; Thomson et al., 2011), epigenetic modifiers (Ang et al., 2011; Liang and Zhang, 2013; Loh et al., 2007), chromatin remodelers (Alajem et al., 2015; Gaspar-Maia et al., 2009; Gatchalian et al., 2018), signaling cascades (Lee et al., 2012; Niwa et al., 2009), and overall three-dimensional chromatin structure (Dixon et al., 2015; Ricci et al., 2015).
Earlier attempts in our lab to screen for pluripotency regulators in mouse ESCs (mESCs) identified SET (TAF-I/IPP2A) as a protein that was significantly downregulated during ESC differentiation (Edupuganti et al., 2017; Harikumar et al., 2017). A closer look revealed that only one of two SET isoforms, SETα, decreases during differentiation, whereas its counterpart, SETβ, produced from an alternative promoter, is increased (Edupuganti et al., 2017). SET has been found to participate in different cellular processes, including cell-cycle regulation (Canela et al., 2003; Estanyol et al., 1999), cell migration (Klooster et al., 2007), DNA damage repair (Kalousi et al., 2015; Kim et al., 2014; Mandemaker et al., 2020), linker histone H1 incorporation (Kato et al., 2011), and as an inhibitor of phosphatase 2A (Li et al., 1996). Furthermore, we have previously demonstrated SET's involvement in pluripotency and early neuronal differentiation: SET contributes to the hyperdynamic association of histone H1 with chromatin in ESCs, and SET depletion causes ESCs to favor an endoderm lineage instead of neuroectoderm (Edupuganti et al., 2017). Despite these findings, a mechanistic role of how SET regulates early differentiation events remains unclear. It is likely that SET has numerous functions that act in concert to regulate stem cell identity or maintain a pluripotent chromatin state.
In this study, we investigate the effect of SET-knockout (KO) in mESCs. We find that SET-KO leads to premature differentiation, proliferation defects, and misregulation of developmental genes during differentiation. Next, following immunoprecipitation pull-down of the SET complex, we performed individual protein quantitation by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to identify additional binding partners of SET in ESCs. Among the putative SET-interacting proteins, we validate a previously characterized interaction of SET with P53 (Wang et al., 2016), and further reveal that SET binds to β-catenin.
Despite its well-known roles in the regulation of multiple cellular processes, including DNA repair, apoptosis, cell cycle, and senescence (Ko and Prives, 1996; Riley et al., 2008), the function of p53 in ESCs remains largely elusive. Several studies have proposed a role for p53 in somatic cell reprogramming and pluripotency, as well as in ESC self-renewal (Hong et al., 2009; Kawamura et al., 2009; Marión et al., 2009; Sarig et al., 2010; Utikal et al., 2009). SET has been reported to inhibit p53 transcriptional activity under normal physiological conditions, an inhibition that is relieved upon stress-induced CTD acetylation of p53. p53 mutants mimicking an unacetylated state (p53KR) display strong interaction toward SET, and mutants mimicking the acetylated state (p53KQ) fail to interact with SET (Wang et al., 2016). Moreover, increased expression of p53 targets, p21 and PUMA, was observed in whole-cell spleen extracts of SET-KO mice (Kon et al., 2019). Therefore, it is tempting to speculate that SET might play a similar role in pluripotent cells and inhibit p53 transcriptional activity.
β-Catenin (CTNNB1) was originally known for its role in establishing cell-cell adhesion, and more recently in regulating pluripotency (Huelsken and Birchmeier, 2001; Moon et al., 2002; Wodarz and Nusse, 1998). Importantly, β-catenin is also the primary mediator of the evolutionarily conserved canonical Wnt signaling pathway. Precisely how the Wnt/β-catenin signaling pathway contributes to the maintenance of ESC properties has been the subject of debate within the scientific community. Several studies have found that the pathway is essential for self-renewal (Sato et al., 2004, ten Berge et al., 2011), while others suggest that the activation of the pathway induces mesodermal and/or endodermal differentiation (Bakre et al., 2007; Davidson et al., 2012; Lindsley et al., 2006).
As both p53 and β-catenin are themselves implicated in pluripotency and lineage choice regulation, we chose to investigate their relationship with SET and examine their involvement in early differentiation. To this end, we generated small hairpin RNA (shRNA) knockdown (KD) cell lines for both p53 and β-catenin in a SET-KO background and investigate their involvement in SET-KO-related defects in ESCs. Together, our findings suggest that SET plays a vital role in ESCs via its interaction with p53 and β-catenin and prevents lineage skewing during early differentiation.
Results
Loss of SET Leads to a Premature Differentiation Phenotype
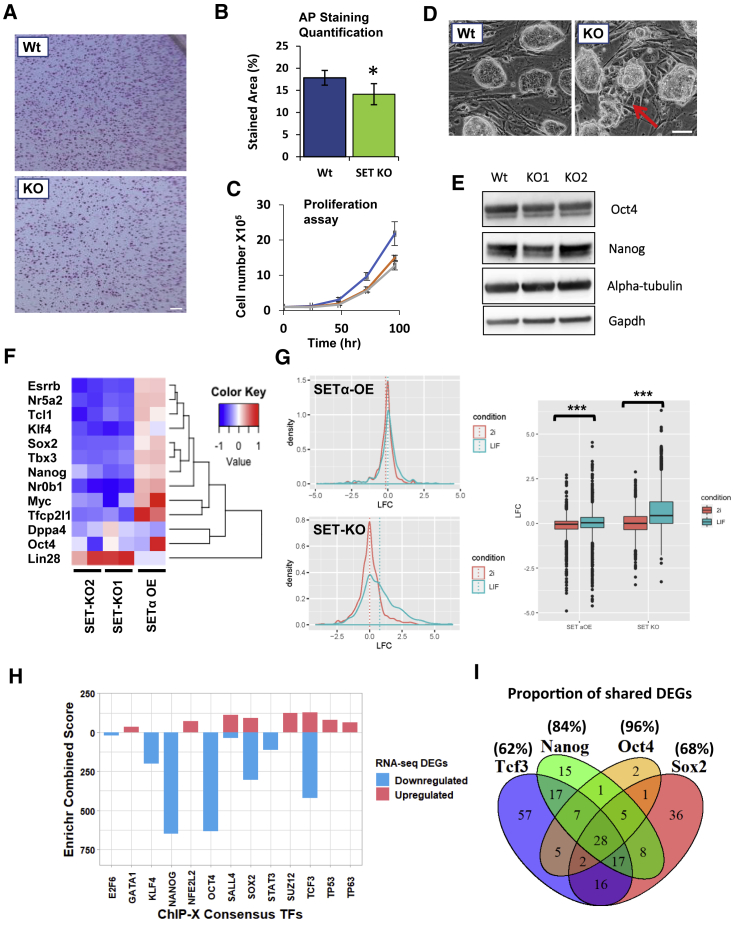
To investigate the role of SET in ESC differentiation, we used CRISPR-Cas9 to generate SET-KO mESCs as described previously (Edupuganti et al., 2017). We ruled out potential off-target effects by using different single-guide RNAs (sgRNAs) targeting two different exonic regions and selected one KO clone from each system. We confirmed the KO both at the RNA and the protein level (Figure S1A). As SET-KD ESCs showed reduced cell proliferation (Edupuganti et al., 2017), we first examined if this was also true for SET-KO cells. Consistent with previous results, SET-KO ESCs showed a reduced staining for alkaline phosphatase (Figures 1A and 1B) and a significant decrease in cell proliferation rates (Figure 1C). We then examined the morphological differences associated with SET-KO ESCs. Unexpectedly, a fraction of SET-KO ESC colonies exhibited partially differentiated morphology (Figure 1D). Western blot for pluripotency markers, NANOG and OCT4, revealed a slight reduction in protein levels for both factors (Figures 1E and S1E).
Figure 1.
SET Is Required to Maintain the Naive Undifferentiated ESC State
(A) Representative images of clone formation assay of WT (top) and SET-KO (bottom). Colonies were detected with alkaline phosphatase staining (pink). Scale bar, 500 μm.
(B) Quantification of staining (percent stained area) was performed in ImageJ. Four (WT) or six (three from SET-KO1 and three from SET-KO2) biological replicates were performed. Values for SET-KO1 and SET-KO2 were adjusted by inflating to account for lower proliferation (60% and 56% of WT, respectively) following 3 days of culture as determined by proliferation assay (C). Error bars represent ±SD; ∗p < 0.05, two-tailed t test.
(C) ESC proliferation assay for WT (blue) and SET-KO (red, green) ESCs. Cells were counted every 24 h, up to 96 h (n = 3 independent biological repeats; error bars represent ±SD).
(D) Phase contrast images of WT (left) and SET-KO (right) undifferentiated ESCs. Arrow (red) points to a partially differentiated colony observed in the SET-KO ESCs. Scale bar, 20 μm.
(E) Western blot of OCT4 and NANOG in WT and SET-KO cell lines. α-Tubulin was used as a loading control.
(F) Gene expression heatmap of normalized counts of pluripotency genes of the indicated cell line relative to WT.
(G) Density histograms (left) and boxplots (right) showing the log-fold change (LFC) in gene expression of LIF/serum versus 2i-specific genes in SET-KO and SETα-OE compared with WT ESCs. Dotted lines in histograms indicate mean LFC. ∗∗∗p < 0.001, t test.
(H) Gene set enrichment analysis of DEGs following SET-KO. Gene lists for transcription factor target genes were obtained from the ChIP-X enrichment analysis database in all cases except for E2F6 (ENCODE target library). Data were generated with Enrichr (https://amp.pharm.mssm.edu/Enrichr/). See Table S1 for list of genes and adjusted p values.
(I) Venn diagram showing the number of genes in common from SET-KO DEGs present in the corresponding transcription factor target gene list. Percentage indicates the proportion of genes shared by at least one other factor. ∗p < 0.05, ∗∗p < 0.001, or ∗∗∗p < 0.0001, t test.
To investigate this phenomenon, we performed RNA sequencing (RNA-seq) on SET-KO and wild-type (WT) ESCs. SET-KO ESCs displayed 754 differentially expressed genes (DEGs) compared with WT (Figure S2A). To ensure that the precocious phenotype was not simply due to misregulated SET expression, we further generated a doxycycline (Dox)-inducible SETα cell line in KH2 ESCs (see Supplemental Experimental Procedures) and performed RNA-seq following overexpression of SETα (1 μg/mL Dox, 24 h) (Figure S1B). SET-over-expression (OE) ESCs had 605 fewer DEGs than SET-KO when compared with WT ESCs (Figure S2A). Of the 13 genes we chose as markers of pluripotency, almost all were downregulated in SET-KO cells when compared with WT, with the exception of Lin28, and supporting the role of SET in this response, the opposite trend was seen in SET-OE ESCs for the same gene set (Figure 1F).
Among our chosen pluripotency markers, LIN28, an RNA-binding protein, was upregulated following SET-KO (Figure 1F). LIN28 is expressed at low amounts in naive ESCs and has been shown to facilitate the transition from the “naive” to the “primed” pluripotent state (Zhang et al., 2016). Furthermore, expression of FGF5, a marker of primed pluripotency, was slightly upregulated in SET-KO ESCs, although this was non-significant (p = 0.08) (Figure S1D). With this is mind, to determine the extent to which SET depletion affects pluripotency, we generated a list of 2i (“ground-state” naive)-, leukemia inhibitory factor (LIF) (“confused” naive)-, and epiblast stem cell (EpiSC) (primed)-specific genes using data from transcriptomic analyses comparing culture conditions of mESCs (Ghimire et al., 2018) (Table S1). Using these lists of genes, we then generated gene expression histograms of log2-fold changes (LFC) in SET-KO and SET-OE mESCs compared with WT controls. Based on these expression criteria, SET-KO ESCs show a shift toward a primed or a more differentiated state, supporting the SET-KO premature differentiation phenotypes (Figures 1A and 1D). Along the same lines, considering the LFC of genes in SET-KO relative to WT ESCs, the mean LFC of LIF-specific genes is significantly higher than of 2i-specific genes (p < 0.001) (Figure 1G). This trend is also evident when comparing EpiSC- and LIF-specific genes; SET-KO ESCs show a higher LFC of genes specific to the more differentiated epiblast state (p < 0.001) (Figure S2B). On average, 2i-specific genes were not significantly differentially expressed in SET-KO ESCs but were slightly decreased in SET-OE ESCs (p < 0.001) (Figure 1G). We note that, since the cells were cultured in LIF/serum, we cannot exclude that some SET-related changes for 2i-specific gene expression may have been suppressed as a result of the relatively heterogeneous culture conditions (Schlesinger and Meshorer, 2019).
Interestingly, a significant number of downregulated DEGs following SET-KO are target genes of transcription factors involved in pluripotency regulation (OCT4, SOX2, NANOG, KLF4, and TCF3) (Figure 1H; Table S2). In fact, of the four most significant putative regulatory transcription factors of SET-KO DEGs, the majority of each set of genes overlaps with at least one other pluripotency factor (Figure 1I). Furthermore, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs in SET-KO ESCs reveals a significant enrichment of factors relating to “signaling pathways regulating pluripotency of stem cells” (Figures S5A and S5B). Taken together, these observations point to a role for SET in maintaining pluripotent regulatory gene networks, particularly of the naive pluripotent state.
SET-KO Disrupts Lineage Specification following Differentiation
Our RNA-seq analysis indicated that almost two-thirds of DEGs were upregulated following SET-KO (Figure S2A). KEGG pathway analysis of only upregulated DEGs revealed an enrichment of several morphogenetic and structural related pathways, including “focal adhesion,” “ECM-receptor interaction,” and “regulation of actin cytoskeleton” (Figure S5B, top). Furthermore, included in the top ten enriched gene ontology (GO) terms for biological processes from analysis of SET-KO DEGs were “embryonic axis specification” and “heterochromatin organization,” which are fundamental processes that occur early in embryonic development—particularly during the naive-to-primed pluripotency transition (Boroviak and Nichols, 2014; Probst and Almouzni, 2008). Indeed, almost a quarter (23/100) of the top 100 GO terms featured the words “development” or “differentiation” (Table S3). These results suggested that activation of lineage specifying cellular processes may be occurring in response to loss of SET.
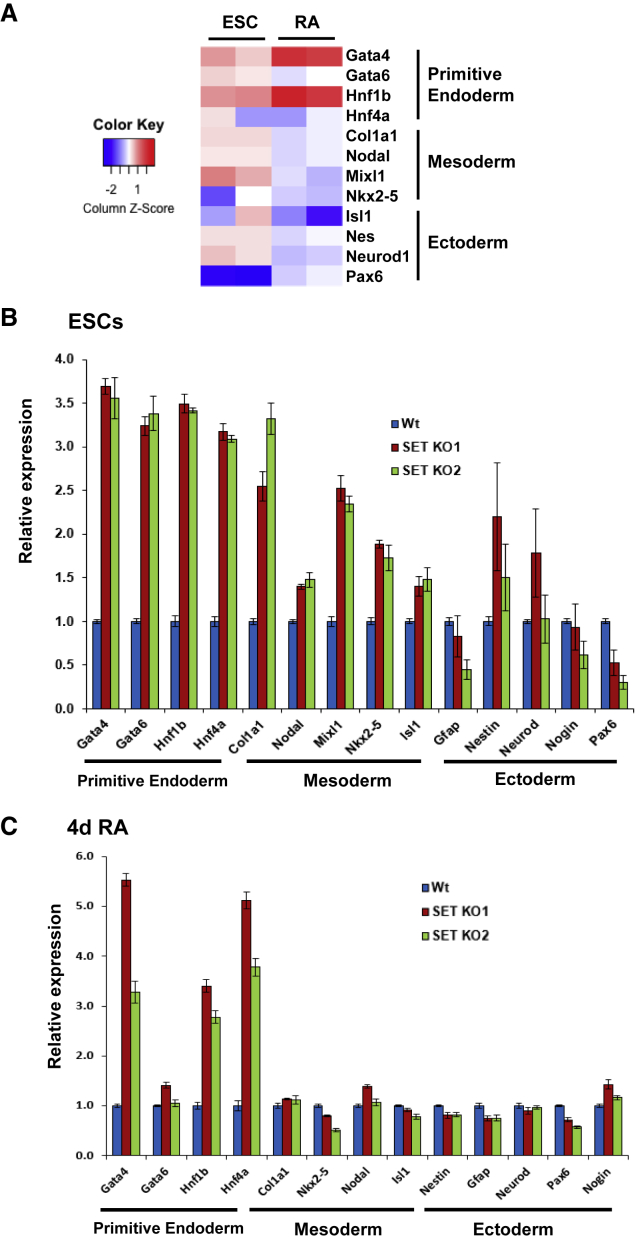
The observation that loss of SET leads to exit from pluripotency and an upregulation of developmental processes prompted us to determine the impact of SET depletion on lineage-specific genes. To test this, we performed RNA-seq and qPCR, and quantified changes in gene expression of markers from all three germ lineages in undifferentiated ESCs (Figure 2A). Consistent with the precocious phenotypic abnormalities, several endoderm/primitive endoderm lineage markers were significantly upregulated, along with a slight upregulation of mesodermal markers (Figure 2B). In contrast, ectodermal markers displayed marginal changes (Figure 2B). These data demonstrate that loss of SET leads to the upregulation of specific differentiation genes. We then examined the effect of SET-KO following retinoic acid (RA) differentiation (4d). Several markers for mesodermal and ectodermal lineages, which were upregulated in undifferentiated KO cells, including Col1a1, Nkx2-5, and Nodal (Figure 2B), reverted to normal levels in the differentiated cells (Figure 2C). Moreover, several primitive endoderm markers remained significantly upregulated in the SET-KO cells following differentiation (Figure 2C). These results demonstrate that SET-KO leads to differentiation defects, highlighting the importance of SET during early ESC differentiation, as well as in maintaining the pluripotent state. Earlier studies have shown that ESCs maintain developmental genes in a poised but repressed state until differentiation cues are received (Bernstein et al., 2006; Harikumar and Meshorer, 2015). The observation that SET-KO cells show upregulation of lineage-specific genes consistently throughout RA differentiation suggests that not only does SET suppress differentiation genes in ESCs, but also that SET is required to prevent skewed differentiation.
Figure 2.
SET-KO Disrupts Lineage Specification Following Differentiation
(A) Heatmap showing expression of linage-specific markers relative to WT following RNA-seq analysis of SET-KO ESCs before and after 4 days of RA differentiation (values scaled by row).
(B) qPCR analysis showing the relative expression of lineage markers for endoderm, mesoderm, and ectoderm in WT and SET-KO ESCs. Expression levels were normalized to GAPDH (n = 3 independent biological experiments; error bars represent ±SEM).
(C) Same as (B) after 4 days of RA differentiation.
P53 Is Partially Responsible for Misregulation of Developmental Genes in SET-KO Cells
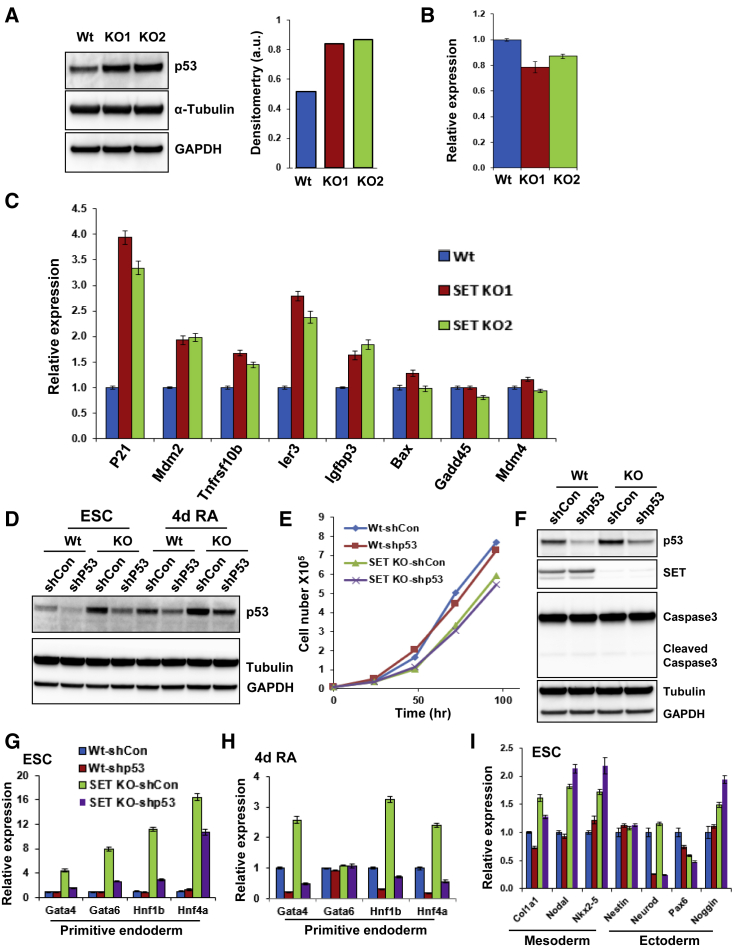
After establishing that SET-KO ESCs have a disrupted pluripotent state, we wished to identify the putative protein complexes SET might belong to or interact with. We began by purifying endogenous SET protein from ESCs and identified its interaction partners using LC-MS/MS (Table S4). We compared our data with existing datasets from MS performed in a human epithelial cell line, and identified several common interacting partners, including p53, β-catenin, α-catenin, δ-catenin, and H1, among others (Krishnan et al., 2017). p53 and its family members were shown to play essential roles during differentiation, development, and reprogramming (Jain and Barton, 2018). We therefore initially focused on the interaction between SET and p53 and tested its potential role in pluripotency and differentiation. First, we confirmed the LC-MS/MS data using immunoprecipitation followed by western blot analysis for endogenous SET protein (Figure S3A). We next examined the effect of SET-KO on p53 levels using western blot and qPCR. Surprisingly, p53 protein level, but not mRNA level, was significantly upregulated in SET-KO ESCs, suggesting that SET-KO leads to the stabilization of p53 protein (Figures 3A and 3B). This is in contrast to a previous study that did not observe stabilization of p53 protein in SET-KO mice: immunohistochemistry analysis of SET-KO embryos demonstrated that, although levels of p21, a gene activated by p53, increase, protein levels of p53 remain unchanged (Kon et al., 2019). qPCR analysis of p53 downstream targets showed increased levels of p21, Mdm2, Ler3, and Igfbp3; however, other target genes, including Bax, Gadd45, and Mdm4, were unaffected (Figure 3C). This suggests that elevated p53 is indeed transcriptionally active and regulates selective genes in SET-KO ESCs. SET-OE ESCs do not have decreased expression of p53; however, p53 target genes, p21 and Mdm4, appeared slightly downregulated, albeit not to a statistically significant degree (Figure S3B).
Figure 3.
SET Functionally Interacts with p53
(A) Western blots for p53 in WT and SET-KO cells. GAPDH and α-Tubulin was used as loading control (left). Densitometry quantification for western blot images of p53 normalized by GAPDH and α-Tubulin (right).
(B) qPCR for p53 in WT and SET-KO clones. Expression level was normalized to GAPDH (n = 3 independent biological experiments; error bars represent ±SEM).
(C) Same as (B) for the relative expression of p53 downstream targets in WT and SET-KO ESCs.
(D) Western blots for p53 in undifferentiated and RA-differentiated ESCs in SET-KO and SET-KO/p53-KD clones. GAPDH and α-Tubulin were used as loading controls.
(E) ESC proliferation assay for WT-shCon, WT-sh-p53, SET-KO-shCon, and SET-KO-sh-p53 ESCs. Cells were counted every 24 h, up to 96 h (n = 3 independent biological repeats).
(F) Western blots of the apoptotic marker cleaved caspase-3 in WT-shCon, WT-sh-p53, SET-KO-shCon, and SET-KO-sh-p53 ESCs.
(G and H) qPCR analysis showing the expression of endodermal lineage markers in ESCs (G) and 4-day RA differentiation (H) in WT-shCon, WT-sh-p53, SET-KO-shCon, and SET-KO-sh-p53 ESCs (expression level was normalized to GAPDH; error bars represent ±SEM; n = 3 independent biological repeats).
(I) Same as (G and H) for mesodermal and ectodermal markers in ESCs.
The increase in p53 stability we observed in SET-KO ESCs led us to investigate whether increased levels of p53 might be responsible for the misregulation of lineage choice in the absence of SET. To this end, we used shRNAs to generate stable KDs for p53 in WT and SET-KO ESCs. qPCR analysis showed that 70%–80% KD efficiency was achieved in both cell lines compared with respective shRNA control cell lines (Figure S3D). Our western blot analysis also confirmed KD of p53 in both WT and SET-KO cells (Figure 3D). As the initial levels of p53 in the SET-KO ESCs are much higher than WT, p53 levels in SET-KO ESCs are almost comparable with those of WT-sh controls following p53 KD (Figure 3D). This confirms our previous observations that p53 protein levels are stabilized in the absence of SET. DNA damage and several other stress conditions have been shown to arrest cell cycle in a p53-dependent manner (Brady et al., 2011; Sullivan et al., 2018). We performed proliferation assays to determine whether p53-induced cell-cycle arrest was contributing to proliferation defects associated with SET-KO ESCs. No significant changes were observed upon p53 KD in WT and SET-KO cells (Figure 3E), suggesting that SET-KO-mediated decrease in proliferation is independent of p53. Likewise, expression of pluripotency factors was not significantly affected by p53 KD in SET-KO cells (Figure S3C). Next, we examined whether upregulated p53 is associated with increased apoptosis. Cleaved caspase-3, a marker of apoptosis (Carthy et al., 1998), showed no induction in WT or SET-KO cells (Figure 3F). This suggests that the decrease in the proliferative capacity of SET-KO ESCs is also independent of apoptosis.
We next tested the expression level of differentiation markers in p53-KD and SET-KO cells. As KO of p53 partially rescues embryonic lethality in SET-KO mice (Kon et al., 2019), we predicted that KD of p53 would correspondingly revert some of the misregulation of lineage-related genes in the absence of SET. Indeed, using qPCR we found that primitive endodermal genes, such as Hnf1a, Hnf1b, and Gata4, which were significantly upregulated in the SET-KO cells by 4- to 16-fold, were largely rescued upon p53 KD in undifferentiated ESCs (Figure 3G). This p53-KD-mediated rescue was also observed in RA-differentiated SET-KO cells—albeit to a lesser degree (Figure 3H). p53 KD further reduced the expression of Hnf1a, Hnf1b, and Gata4 only in RA-differentiated WT cells (Figures 3G and 3H). Interestingly, despite the important role of p53 in regulating mesodermal differentiation genes (Hadjal et al., 2013; Lee et al., 2012), KD of p53 had little effect on mesodermal markers in our system (Figure 3I). Furthermore, ectodermal marker Neuro-d was consistently downregulated in p53-KD ESCs, in both WT and SET-KO ESCs (Figure 3I). These data suggest that p53 upregulation is at least in part responsible for activation of primitive endodermal genes in SET-KO cells.
The Canonical Wnt Signaling Pathway Is Activated in SET-KO ESCs
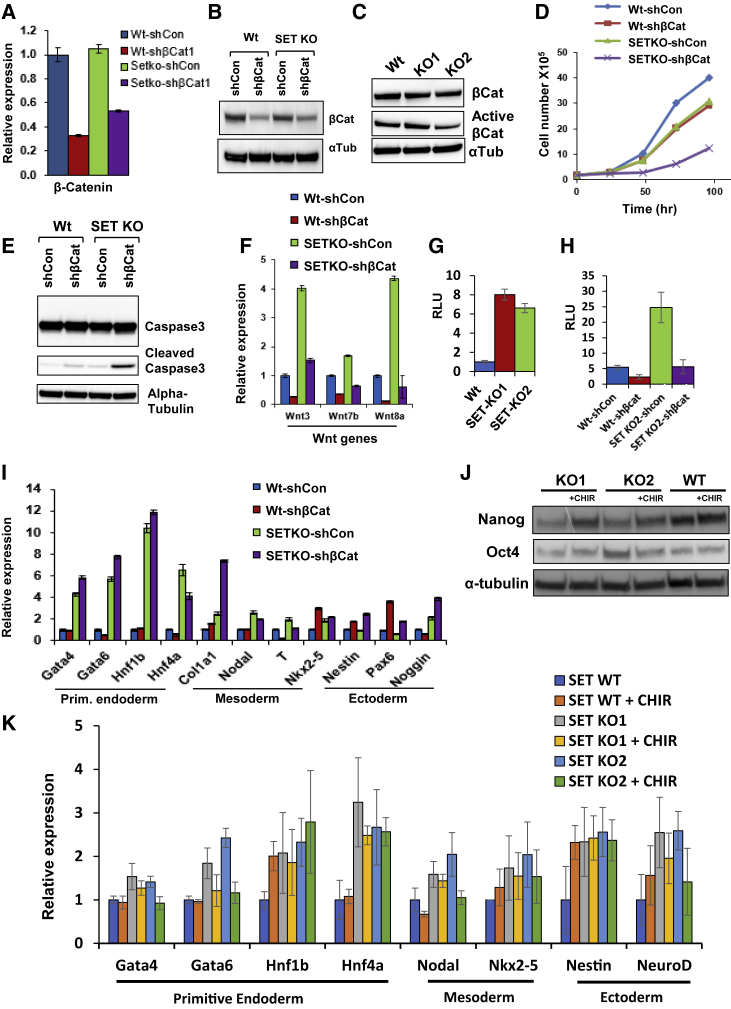
We next focused on the interaction between SET and potential signaling components involved in pluripotency regulation. Having identified an interaction between β-catenin and SET in our MS data, we asked whether β-catenin contributes to the misregulation of differentiation genes in SET-KO cells. To this end, we generated shRNA-stable KD cell lines for β-catenin in WT and SET-KO ESCs. Western blots confirmed ∼50%–70% β-catenin KD efficiency, and both western blots and qPCR analyses demonstrated similar levels of β-catenin WT and SET-KO ESCs (Figures 4A–4C). Interestingly, however, active β-catenin appeared to show a mild decrease in SET-KO ESCs (p = 0.036), which we quantified by densitometry in several other SET-KO clones (Figures 4C and S4A).
Figure 4.
β-catenin KD Induces Apoptosis and Wnt Signaling in SET-KO Cells
(A) qPCR analysis showing β-catenin expression following SET-KO and/or KD in ESCs.
(B) Western blots for β-catenin in WT-shCon, WT-sh-βcat, SET-KO-shCon, and SET-KO-sh-βcat cells. α-Tubulin was used as a loading control.
(C) Western blot of β-catenin and active β-catenin in WT and SET-KO cell lines. α-Tubulin was used as a loading control.
(D) ESC proliferation assay for WT-shCon, WT-sh-βcat, SET-KO-shCon, and SET-KO-sh-βcat ESCs. Cells were counted every 24 h, up to 96 h (n = 3 independent biological repeats).
(E) Western blots of the apoptotic marker cleaved caspase-3 in WT-shCon, WT-sh-βcat, SET-KO-shCon, and SET-KO-sh-βcat ESCs. Tubulin was used as a loading control.
(F) Wnt genes in WT-shCon, WT-sh-βcat, SET-KO-shCon, and SET-KO-sh-βcat ESCs.
(G and H) TOPFlash luciferase assay measuring Wnt signaling/β-catenin activity in WT versus SET-KO ESCs (H) and in WT-shCon, WT-sh-βcat, SET-KO-shCon, and SET-KO-sh-βcat ESCs (I). Relative light units (RLU) is equivalent to the TOPFlash/FOPFlash luminescence ratio.
(I) qPCR expression of lineage markers in WT-shCon, WT-sh-βcat, SET-KO-shCon, and SET-KO-sh-βcat ESCs
(J) Western blot of NANOG and OCT4 in WT and SET-KO cell lines in the absence and presence of 3 μM CHIR. α-Tubulin was used as a loading control.
(K) Expression of lineage markers in SET-KO ESCs compared with SET-WT in the presence and absence of 3 μM CHIR. All qRT-PCR expression levels were normalized to GAPDH (n = 3 independent biological experiments; error bars represent ±SEM).
β-catenin has a well-established role as the key mediating factor in the canonical Wnt signaling pathway (Moon et al., 2002). Intracellular Wnt signaling has been shown to play important roles in proliferation and differentiation of stem cells (Reya and Clevers, 2005). No stark differences in pluripotency factor expression was observed following β-catenin KD in SET-KO ESCs. Although β-catenin KD reduced levels of NANOG in SET-KO ESCs, its levels were comparable with KD in WT cells. A slight reduction of OCT4 was observed following KD β-catenin in SET-KO ESCs (Figure S4B). We then examined the effect of β-catenin KD on cell proliferation. β-catenin KD significantly decreased cell proliferation in both WT and SET-KO ESCs (Figure 4D). The additive effect of SET-KO and β-catenin KD resulted in severely slow proliferating cells. The extent of the reduced proliferation in the SET-KO/β-catenin KD cells prompted us to examine the potential induction of apoptosis. Western blots showed a slight induction of cleaved caspase-3 in WT cells upon β-catenin KD, consistent with previously published data (Raggioli et al., 2014) (Figure 4E, lanes 1 and 2). However, in SET-KO ESCs, the induction of cleaved caspase-3 upon β-catenin KD was significantly higher, suggesting an additive effect on apoptosis by the depletion of both SET and β-catenin, and that β-catenin KD increases apoptosis in both WT and SET-KO ESCs (Figure 4E, lanes 3 and 4).
We next investigated the expression of Wnt genes. To our surprise, we observed that Wnt genes, including Wnt3, Wnt7b, and Wnt8a, were markedly upregulated in SET-KO ESCs, and were all rescued upon β-catenin KD (Figure 4F). Since targets of Wnt signaling include some members of the Wnt signaling pathway itself, this may be due to the self-regulatory mechanisms of Wnt gene expression (MacDonald et al., 2009). Concordantly, β-catenin KD in SET-WT cells also reduced Wnt gene expression. To ensure that this induction of Wnt gene expression corresponded with increased β-catenin-mediated transcription, we used the TOPFlash luciferase assay, which measures the binding of β-catenin/TCF to response elements by activating a reporter gene (Moon et al., 2002). We observed that β-catenin activity is significantly higher in both SET-KO clones compared with WT ESCs—similar to the observed increase in Wnt gene expression (Figure 4G). KD of β-catenin in SET-KO cells returned TOPFlash reporter levels to their original WT state, indicating that β-catenin is indeed a functionally relevant component of SET-KO-induced Wnt signaling activation (Figure 4H).
Next, we investigated the effect of β-catenin KD on lineage markers with qPCR. We found that upregulation of differentiation markers, including Gata4, Gata6, and Noggin, was increased in SET-KO/β-catenin KD ESCs, despite β-catenin KD alone not leading to upregulation of these genes (Figure 4I). The opposite effect was seen only for Nodal in SET-KO ESCs, where β-catenin KD was able to rescue its aberrant upregulation, although to a lesser degree. Moreover, in genes such as Hnf4 and T, β-catenin KD alone caused downregulation, which was also the case in SET-KO ESCs. Overall, the extent to which β-catenin KD worsens or rescues SET-KO induced lineage marker dysregulation does not appear to be germ layer specific (Figure 4I). To see whether activation of canonical Wnt signaling could rescue the lineage specification defects in SET-KO cells, we treated WT and KO cells with 3 μM of CHIR99021 (CHIR), a small molecule that stabilizes β-catenin via inhibition of glycogen synthase kinase 3 (GSK3). As expected, addition of CHIR led to strong increases in expression of Sp5 and Axin2, well-established Wnt target genes (Figure S4C) (Ramakrishnan and Cadigan, 2017). CHIR treatment, however, had little effect on active β-catenin levels in SET-KO cells (Figure S4D). CHIR has been shown to increase expression of pluripotency factors in ESCs by facilitating translocation of β-catenin into the nucleus, we therefore examined protein levels of NANOG and OCT4 following addition of CHIR (Wu et al., 2013). NANOG levels increased following Wnt activation by CHIR in SET-KO cells but no similar increase in OCT4 levels was observed, consistent with previous reports in WT ESCs (Ai et al., 2016) (Figure 4J). This indicates that β-catenin retains nuclear activity SET-KO ESCs despite decreased levels of its active form.
To test whether CHIR could rescue lineage specification or proliferation defects in SET-KO cells, we assayed SET-KO ESCs in the presence and absence of CHIR. Apart from a slight reduction in Gata4 and Gata6 expression, CHIR treatment did not appreciably affect lineage marker expression (Figure 4K). Likewise, CHIR treatment was unable to rescue proliferation defects associated with SET-KO cells (Figure S4E). These results imply that, despite a clear role for β-catenin in SET-KO in TCF/LEF1 activation, canonical β-catenin activity or protein stability by themselves are not wholly responsible for abnormal SET-KO phenotypes. Interestingly, well-characterized targets of the canonical Wnt signaling pathway, Axin2 and Sp5, were not upregulated in SET-KO cells, although Sp5 was significantly upregulated in SET-OE cells (Figure S4C). Nevertheless, SET-KO cells do remain responsive to CHIR-induced Wnt target activation (data not shown).
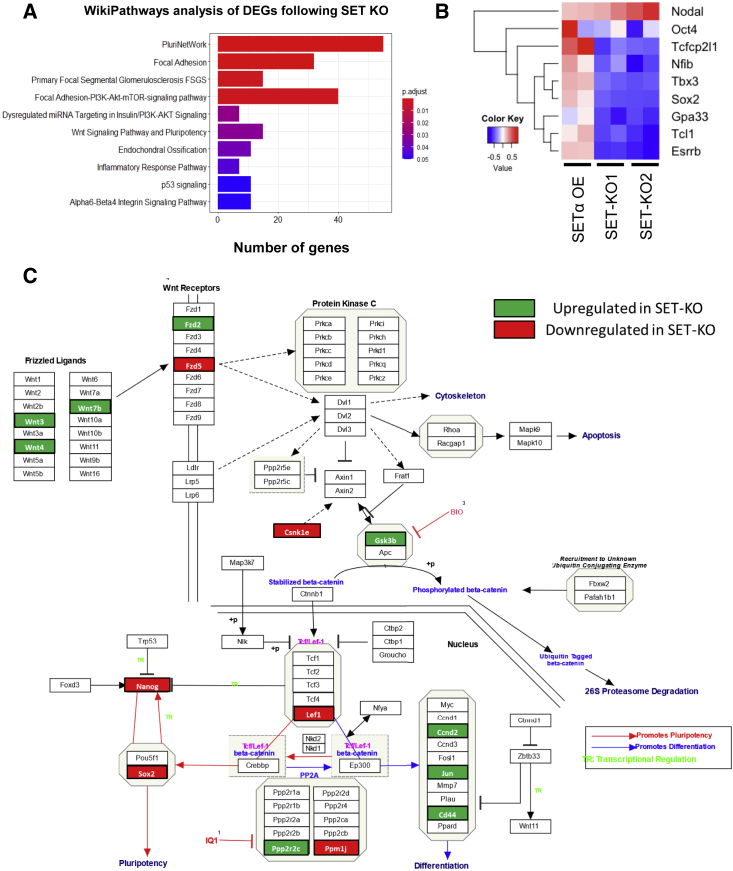
Revisiting our RNA-seq data we found that, after performing gene enrichment analysis on SET-KO DEGs using the WikiPathways database (Kutmon et al., 2016), we found a significant number of genes relating to “Wnt signaling pathway and pluripotency” (Figure 5A). Members of the TCF/LEF1 family of transcription factors are the primary downstream effectors of the canonical Wnt signaling pathway (Cadigan and Waterman, 2012). In particular, TCF3 has been implicated in the exit from pluripotency due to its role in the repression of many pluripotency-related genes (Yi et al., 2011). With this in mind, we analyzed our RNA-seq data in SET-KO and SET-OE ESCs and focused on the expression of genes shown to be specifically regulated by a TCF3-β-catenin interaction, based on TCF3 mutants unable to bind β-catenin (Yi et al., 2011). We found a slight correlation between levels of SET and expression of these β-catenin-regulating TCF3 target genes (Figure 5B). However, this was not the case for all TCF3 target genes: 3 genes, Nodal, Eomes, and Tnfrsf19, were upregulated in SET-KO and not in SET-OE ESCs (Figure S6).
Figure 5.
SET Differentially Regulates Subsets of TCF/LEF1-β-catenin Target Genes
(A) Functional enrichment analysis of DEGs in SET-KO ESCs.
(B) Expression relative to WT of SETα-OE and SET-KO ESCs for subset of genes confirmed to require an interaction between TCF3 and β-catenin.
(C) “Wnt signaling pathway and pluripotency” map from WikiPathways database (wikipathways.org). Highlighted genes in green or red correspond to upregulated or downregulated DEGs, respectively, in SET-KO ESCs.
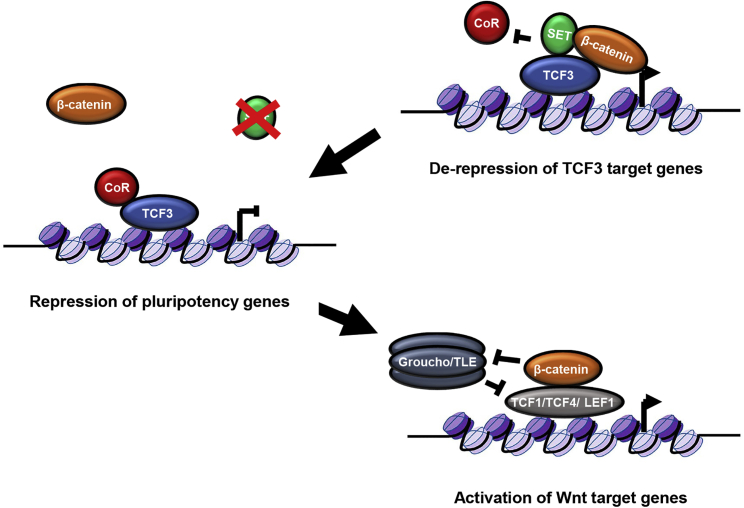
Finally, utilizing the WikiPathways enrichment analysis, we mapped the implicated DEGs on the “Wnt signaling pathway and pluripotency” pathway network (Figure 5C). Interestingly, we find that three genes (Ccnd2, Jun, and Cd44) downstream of differentiation-promoting TCF/LEF1-β-catenin activity are significantly upregulated in SET-KO ESCs. This may reflect the greater level of Wnt signaling in SET-KO ESCs or their more differentiated state. Alternatively, the interaction of SET and β-catenin may also be involved in mediating downstream TCF/LEF1 effector function, particularly as TCF3-β-catenin targets and pluripotency-related genes are downregulated in SET-KO ESCs (Figures 1F and 5B). We hypothesize that SET may have a facilitatory effect on downstream β-catenin effector function, thereby arbitrating the suppression of Wnt signaling and differentiation programs to sustain the pluripotent state of ESCs (Figure 6).
Figure 6.
Model of the SET-β-catenin-TCF3 Regulatory Axis
TCF3 constitutively represses target genes in association with a co-repressive complex (CoR). In the presence of SET, β-catenin may now be able to associate with TCF3 and displace the CoR, thereby derepressing TCF3 action on target genes. In the absence of SET, this derepression does not occur and instead shifts the stoichiometric ratio in favor of β-catenin's interactions with the other members of the TCF/LEF1 family. This interaction involves the activation of target genes through the displacement of the Groucho/TLE repressive complex by β-catenin.
Discussion
SET was initially characterized as a nuclear proto-oncogene following its discovery in patients with acute undifferentiated leukemia (Lindern et al., 1992). In addition, important functions of SET as a molecular chaperone and as an inhibitor of histone acetyltransferase activity have been previously established (Kato et al., 2011; Seo et al., 2001). In previous work, we identified a transcriptional switch early during ESC differentiation, where the predominant SET isoform expressed in ESCs, SETα, is replaced by the isoform predominantly expressed in differentiated cells, SETβ. This conversion is regulated by alternative promoters, each of which is bound by a distinct set of transcription factors (Edupuganti et al., 2017). SET depletion is embryonically lethal (Edupuganti et al., 2017; Kon et al., 2019), indicating that SET plays a crucial role in early development. Furthermore, although SET is highly conserved in nature and significantly expressed in the early embryo (Bayarkhangai et al., 2018), the precise role of SET in regulating early differentiation and pluripotency has not yet been adequately addressed.
In this study, our experiments have revealed a role for SET in maintaining the pluripotent state. In addition to spontaneously differentiating in culture, SET-KO ESCs have a lower overall expression of pluripotency markers and a higher expression of genes specific to a less naive or a more primed pluripotent state. This corresponds to the disturbed expression of markers of endoderm, ectoderm, and mesoderm germ lineages. Following induced differentiation with RA treatment, the abnormal upregulation of primitive endoderm markers was retained. To explore this further, we sought to uncover new interactors of SET by coimmunoprecipitation LC-MS/MS experiments. P53 was among the putative SET binding partners identified in our screen, validating reports that SET binds the unacetylated C terminus of p53 (Kim et al., 2012; Wang et al., 2016). In addition, we found a novel interaction of SET with β-catenin, a multipotential protein with a clear but inconclusive role in the regulation of pluripotency (Yi et al., 2011). Here, we have investigated the contributions of these two proteins to the disturbance of lineage specification in SET-KO ESCs and propose a model for how SET may regulate pluripotency.
In accordance with previous results, we observed that several p53 downstream targets were upregulated in SET-KO ESCs, along with upregulation of differentiation programs (Wang et al., 2016). Interestingly, we found that p53 protein levels are increased in SET-KO cells, with no significant effect on p53 mRNA levels. Post-translational modifications or a pluripotency-specific p53 interaction network may account for this discrepancy and warrants further investigation. In addition, p53 KD in SET-KO ESCs and RA-differentiated cells show rescue of primitive endoderm and endodermal genes, without prominent effects on mesodermal genes and ESC proliferation. These results are consistent with previous studies showing that, in ESCs, p53-regulated target genes are enriched in differentiation-associated genes rather than apoptosis- and cell-cycle-associated genes (Lee et al., 2012; Li et al., 2012; Liang and Zhang, 2013).
These data suggest that a major role of SET in ESC pluripotency is to inhibit p53 transactivation and subsequently prevent the upregulation of primitive/endoderm differentiation genes. As developmental defects in SET-KO mice are only partially rescued by KO of p53 (Kon et al., 2019), it is likely that additional p53-independent interactions of SET with other factors are necessary to maintain normal ESC proliferation. Interestingly, although a previous study established a connection between the p53 family and Wnt inputs during mesoendodermal differentiation, we observed that neither β-catenin nor p53 are involved in the regulation of mesoderm genes in SET-KO ESCs (Wang et al., 2016). Nonetheless, it would be of interest to further explore the role of SET during mesodermal differentiation and the function of β-catenin in this process.
Post-translational modifications (PTMs) affect the function and activity of both p53 (Meek and Anderson, 2009) and β-catenin (Gao et al., 2014; Valenta et al., 2012). For example, Aurora kinase A (AURKA) phosphorylates p53 (Lee et al., 2012) and GSK3β phosphorylates β-catenin (Q.-L. Ying et al., 2008) while also regulating the pluripotent state of mESCs. Adding another layer of complexity, SET itself was shown to undergo phosphorylation (Bayarkhangai et al., 2018; Yin et al., 2019) and was shown to interact with acetylated p53 (Wang et al., 2016). Unlike AURKA or GSK3β, SET has not been reported to possess kinase activity or methyltransferase activity and is not directly involved in catalyzing PTMs. However, data from the current study and others show that SET is heavily associated with protein phosphatases (Table S4) (Krishnan et al., 2017) and inhibits PP2A activity (Li et al., 1996). As SET forms complexes with p53 and β-catenin, and since SET-KO transactivates both of them, we speculate that the loss of SET activates protein phosphatases. For instance, relief of phosphatase inhibition could disturb the balance between PTMs and thereby affect the function of p53 and GSK3β. This is further reflected in transcriptomic and cellular perturbations observed in SET-KO ESCs.
The multipotential nature of both SET and β-catenin, and the detection of their interaction in our MS data, led us to speculate about the relationship between the two proteins in the early embryo. To study this relationship, we depleted β-catenin in SET-KO cells. We found that, unlike the situation in WT ESCs, where knocking down β-catenin has a moderate effect on proliferation and cell survival, β-catenin KD induces apoptosis and a severe survival phenotype in SET-KO ESCs, indicating that β-catenin is indispensable for the survival of ESCs in the absence of SET. Curiously though, KD of β-catenin did not have a discernible impact on the expression of lineage-specific markers in SET-KO ESCs. Similarly, treatment with CHIR, a Wnt signaling activator, was not able to rescue lineage-gene misregulation in SET-KO cells. Instead, we found that SET depletion resulted in the upregulation of several Wnt genes and that β-catenin KD in SET-KO ESCs rescued expression of several Wnt genes, with a mild effect on other developmental genes. In the canonical Wnt signaling pathway, following the binding of a Wnt ligand to its receptor, β-catenin is protected from phosphorylation and subsequent degradation by ubiquitination (Clevers, 2006). Downstream signaling causes β-catenin to translocate into the nucleus where it ultimately associates with TCF/LEF family transcription factors to regulate target genes (Schuijers et al., 2014). Given that Wnt signaling stabilizes β-catenin protein levels in the canonical pathway, we were initially surprised to find that β-catenin protein levels remain largely unaltered in SET-KO cells (Figure 4C). Intriguingly, we found that, instead, a mild decrease in active β-catenin occurred following SET-KO. Furthermore, we did not find an increase in expression of canonical Wnt signaling target genes, such as Axin2 or Sp5. We later observed that β-catenin transactivation was significantly higher in SET-KO ESCs, suggesting that the increase in Wnt gene expression in the absence of SET is consistent with increased nuclear transactivation, despite ostensibly decreased active β-catenin levels and expression of canonical Wnt genes.
mESCs treated with GSK3β inhibitor or WNT3a promote the pluripotent state and prevent differentiation of ESCs (Q.-L. Ying et al., 2008). In contrast, other studies have shown that sustained Wnt activation promotes mesodermal differentiation (Bakre et al., 2007; Gadue et al., 2006; Lindsley et al., 2006). It has been proposed that the influence of Wnt signaling on cell fate decision depends on context, developmental time, and interaction with other signaling pathways (Hoppler and Kavanagh, 2007; MacDonald et al., 2009; Sokol and Melton, 1992; ten Berge et al., 2008; van Amerongen and Nusse, 2009). Although we find that the SET-β-catenin interaction is implicated in mESC Wnt signaling, we also observe that, paradoxically, activation of canonical Wnt signaling is accompanied by premature differentiation and proliferation defects in SET-KO ESCs. These results suggest that there are overlapping and non-overlapping genes that may be differentially regulated by β-catenin depending on the presence of SET. We speculate that SET-dependent β-catenin not only activates Wnt genes but also regulates other targets. Further genome-wide studies will identify the effects of SET-KO on β-catenin redistribution its influence on gene activity. Indeed, SET may play a role in targeting β-catenin specifically to TCF3, instead of other TCF/LEF1 proteins, at pluripotency-related genes as the consensus binding sequence of TCF/LEF1 transcription factors are highly conserved and TCF/LEF1 target genes have a large degree of overlap (Brantjes et al., 2002).
Several of the TCF3 target genes we retrieved from the ChIP-X enrichment analysis database, which were upregulated in SET-OE ESCs, were reciprocally downregulated in SET-KO ESCs (Table S5). Among these genes was Fzd5. The FZD5 protein is a receptor for WNT5a, a ligand for the non-canonical Wnt signaling pathway. WNT5a has been shown to reduce TOPFlash β-catenin reporter activation without affecting the stability of β-catenin protein itself (Flores-Hernández et al., 2020; Mikels and Nusse, 2006; J. Ying et al., 2008). Interestingly, we also found a broadly inverse pattern, with downregulation of β-catenin-dependent TCF3 targets observed in SET-KO cells and a consistent reciprocal increase in expression of the same genes in SET-OE cells (Figure 5B). These results hint that, in SET-KO ESCs, redistribution of β-catenin on chromatin toward a subset of Wnt targets leads to activation of the β-catenin TOPFlash reporter, rather than an increase in β-catenin levels per se.
As β-catenin itself is capable of relieving TCF3-induced repression, our data allude to a synergy between SET and β-catenin in the positive regulation of TCF3 target genes. For example, SET, together with β-catenin, may function to prevent TCF3 from recruiting repressive complexes to promoters, thereby allowing expression of key pluripotency-related genes (Figure 6). However, if a SET-TCF3 interaction exists, it may be indirect as TCF3 was not present in our MS data (Table S4), nor were we able to confirm a direct interaction of SET with TCF3 by coimmunoprecipitation experiments (data not shown). Loss of SET leads to the upregulation of other downstream Wnt signaling target genes, such as Nodal, Tnfrsf19, and Nr6a1 (Figure S6), which are less coupled to TCF3 activity. Notably, β-catenin-TCF1 activity is implicated in promoting differentiation of mESCs (Chatterjee et al., 2015). Indeed, this may contribute to the premature differentiation phenotype we detect in SET-KO ESCs. Based on our analysis, we speculate that TCF3-β-catenin interaction could be a major player in SET-KO ESCs; however, we cannot rule out the involvement of other TCF factors in regulating the β-catenin activity. To validate these hypotheses, further efforts should be made to purify a TCF1/TCF3/β-catenin-SET complex on chromatin. In addition, SET mutants unable to bind β-catenin could be generated to observe how Wnt signaling, lineage specification, and maintenance of self-renewal, are affected following the disturbance of the SET-β-catenin interaction.
To conclude, this study reveals an interaction of SET with p53 and β-catenin in regulating early ESC differentiation. Our data suggest that a major role of SET in ESCs is to exert an inhibitory effect on p53 transactivation. We also hypothesize that, by preventing the formation of a subset of β-catenin-TCF/LEF complexes, SET may function to fine-tune the balance between initiating differentiation programs and maintaining stem cell identity. Our findings warrant deeper investigation of these interactions in early development and disease.
Experimental Procedures
Cells
R1 and KH2 mESCs were cultured using standard conditions. RA was used for differentiation. CRISPR KO cells were generated as described previously (Edupuganti et al., 2017; Ran et al., 2013) using sgRNAs targeting SET exon3. Alkaline phosphatase assay was performed using a kit (Sigma, cat. no. 86R-1KT) following the manufacturer's instructions.
Antibodies
The following antibodies were used: SET (A302-261A, Bethyl), OCT4 (sc-805, Santa Cruz), NANOG (A300-397A, Bethyl), GAPDH (ab8245, Abcam), p53 (2,524, Cell Signaling), β-catenin (610153, BD Transduction), active β-catenin (8814, Cell Signaling), α-tubulin (ab4074, Abcam), and HA (ab9110, Abcam).
LC-MS/MS
Cell pellets were lysed (10% glycerol, 150 mM NaCl, 20 mM HEPES [pH 7.9], 0.9% Triton X-100, 0.1% NP40, 0.2 mM EDTA, 0.5 mM PMSF, 10 μL protease inhibitors cocktail) for 30 min at 4°C and centrifuged at maximum speed. SET antibodies were added to the supernatant and incubated overnight at 4°C, followed by 1 h incubation with protein-A magnetic beads. The beads were washed, boiled, and the supernatants were used for LC-MS/MS and western blotting.
TOPFlash Reporter Assay
Cells were transfected with plasmids containing seven TCF/LEF (TOPFlash, Addgene, cat. no. 12456) or seven mutant TCF/LEF (FOPFlash, Addgene, cat. no. 12457) using TransIT (Mirus). Twenty-four hours after transfection, cells were lysed, and luciferase activity was measured using a Nano-Glo Dual-Luciferase Reporter Assay System (Promega) in a Synergy H1 Hybrid Multi-Mode Reader (BioTek). Cells were co-transfected with 1 ng of plasmid containing NanoLuc luciferase as an internal control.
RNA-Seq and Bioinformatics
RNA extraction was performed using RNeasy Mini Kit (QIAGEN). RNA (500 ng) was used to generate libraries with a QuantSeq 3′ mRNA-Seq Kit (Lexogen). Reads were aligned with STAR (v.2.5.4b) using default settings against the mm10 mouse genome obtained from Ensembl and reads were counted with featureCounts.
Full experimental can be found in the supplementary information.
Data and CodeAvailability
Sequencing data performed for this study were deposited in the NCBI Gene Expression Omnibus, accession number GSE154607.
Author Contributions
A.H. and P.S.L.L. performed all experiments and analyzed the data. J.E.P. and S.K.S. performed and analyzed all MS-related experiments. M.N.-R. performed qRT-PCR experiments. A.H., P.S.L.L., and E.M. conceived the study and designed the analysis. P.S.L.L., A.H., and E.M. wrote the manuscript. E.M. supervised the study.
Acknowledgments
This work was supported by the Israel Science Foundation (ISF 1140/2017 to E.M.). E.M. is the incumbent of the Arthur Gutterman Professor Chair for Stem Cell Research. P.S.L.L. is supported by the European Union's Horizon-2020 Marie Skłodowska Curie EpiSyStem ITN Network (765966).
Published: December 8, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.11.004
Supporting Citations
The following references appear in the supplemental information: Gallart-Palau et al., 2019; Veeman et al., 2003.
Supplemental Information
Exponentially modified protein abundance index (emPAI) is used for label-free protein quantitation
List of DEGs in SET-KO and SET-OE mutants with adjusted p < 0.05. Highlighted in red are genes that have a reciprocally differentially expressed in SET-KO and SET-OE ESCs
References
- Ai Z., Shao J., Wu Y., Yu M., Du J., Shi X., Shi X., Zhang Y., Guo Z. CHIR99021 enhances Klf4 expression through β-catenin signaling and miR-7a regulation in J1 mouse embryonic stem cells. PLoS One. 2016;11:e0150936. doi: 10.1371/journal.pone.0150936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alajem A., Biran A., Harikumar A., Sailaja B.S., Aaronson Y., Livyatan I., Nissim-Rafinia M., Sommer A.G., Mostoslavsky G., Gerbasi V.R. Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Re. 2015;10:2019–2031. doi: 10.1016/j.celrep.2015.02.064. [DOI] [PubMed] [Google Scholar]
- van Amerongen R., Nusse R. Towards an integrated view of Wnt signaling in development. Dev. Camb. Engl. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- Ang Y.-S., Tsai S.-Y., Lee D.-F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakre M.M., Hoi A., Mong J.C.Y., Koh Y.Y., Wong K.Y., Stanton L.W. Generation of multipotential mesendodermal progenitors from mouse embryonic stem cells via sustained Wnt pathway activation. J. Biol. Chem. 2007;282:31703–31712. doi: 10.1074/jbc.M704287200. [DOI] [PubMed] [Google Scholar]
- Bayarkhangai B., Noureldin S., Yu L., Zhao N., Gu Y., Xu H., Guo C. A comprehensive and perspective view of oncoprotein SET in cancer. Cancer Med. 2018;7:3084–3094. doi: 10.1002/cam4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Brugmann S.A., Helms J.A., Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Dev. Camb. Engl. 2008;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boroviak T., Nichols J. The birth of embryonic pluripotency. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130541. doi: 10.1098/rstb.2013.0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C.A., Jiang D., Mello S.S., Johnson T.M., Jarvis L.A., Kozak M.M., Broz D.K., Basak S., Park E.J., McLaughlin M.E. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantjes H., Barker N., van Es J., Clevers H. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 2002;383:255–261. doi: 10.1515/BC.2002.027. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Waterman M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012;4:a007906. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela N., Rodriguez-Vilarrupla A., Estanyol J.M., Díaz C., Pujol M.J., Agell N., Bachs O. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 2003;278:1158–1164. doi: 10.1074/jbc.M207497200. [DOI] [PubMed] [Google Scholar]
- Carthy C.M., Granville D.J., Watson K.A., Anderson D.R., Wilson J.E., Yang D., Hunt D.W.C., McManus B.M. Caspase activation and specific cleavage of substrates after coxsackievirus B3-induced cytopathic effect in HeLa cells. J. Virol. 1998;72:7669–7675. doi: 10.1128/jvi.72.9.7669-7675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S.S., Saj A., Gocha T., Murphy M., Gonsalves F.C., Zhang X., Hayward P., Akgöl Oksuz B., Shen S.S., Madar A. Inhibition of β-catenin-TCF1 interaction delays differentiation of mouse embryonic stem cells. J. Cell Biol. 2015;211:39–51. doi: 10.1083/jcb.201503017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Davidson K.C., Adams A.M., Goodson J.M., McDonald C.E., Potter J.C., Berndt J.D., Biechele T.L., Taylor R.J., Moon R.T. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. 2012;109:4485–4490. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti R.R., Harikumar A., Aaronson Y., Biran A., Sailaja B.S., Nissim-Rafinia M., Azad G.K., Cohen M.A., Park J.E., Shivalila C.S. Alternative SET/TAFI promoters regulate embryonic stem cell differentiation. Stem Cell Reports. 2017;9:1291–1303. doi: 10.1016/j.stemcr.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estanyol J.M., Jaumot M., Casanovas O., Rodriguez-Vilarrupla A., Agell N., Bachs O. The protein SET regulates the inhibitory effect of p21Cip1 on cyclin E-cyclin-dependent kinase 2 activity. J. Biol. Chem. 1999;274:33161–33165. doi: 10.1074/jbc.274.46.33161. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Flores-Hernández E., Velázquez D.M., Castañeda-Patlán M.C., Fuentes-García G., Fonseca-Camarillo G., Yamamoto-Furusho J.K., Romero-Avila M.T., García-Sáinz J.A., Robles-Flores M. Canonical and non-canonical Wnt signaling are simultaneously activated by Wnts in colon cancer cells. Cell. Signal. 2020;72:109636. doi: 10.1016/j.cellsig.2020.109636. [DOI] [PubMed] [Google Scholar]
- Gadue P., Huber T.L., Nostro M.C., Kattman S., Keller G.M. Germ layer induction from embryonic stem cells. Exp. Hematol. 2005;33:955–964. doi: 10.1016/j.exphem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Gadue P., Huber T.L., Paddison P.J., Keller G.M. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallart-Palau X., Serra A., Hase Y., Tan C.F., Chen C.P., Kalaria R.N., Sze S.K. Brain-derived and circulating vesicle profiles indicate neurovascular unit dysfunction in early Alzheimer’s disease. Brain Pathol. Brain Pathol. Zurich Switz. 2019;29:593–605. doi: 10.1111/bpa.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Xiao G., Hu J. Regulation of Wnt/β-catenin signaling by posttranslational modifications. Cell Biosci. 2014;4:13. doi: 10.1186/2045-3701-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E., Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J., Malik S., Ho J., Lee D.-S., Kelso T.W.R., Shokhirev M.N., Dixon J.R., Hargreaves D.C. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 2018;9:5139. doi: 10.1038/s41467-018-07528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimire S., Van der Jeught M., Neupane J., Roost M.S., Anckaert J., Popovic M., Nieuwerburgh F.V., Mestdagh P., Vandesompele J., Deforce D. Comparative analysis of naive, primed and ground state pluripotency in mouse embryonic stem cells originating from the same genetic background. Sci. Rep. 2018;8:5884. doi: 10.1038/s41598-018-24051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjal Y., Hadadeh O., Yazidi C.e., Barruet E., Binétruy B. A p38mapk-p53 cascade regulates mesodermal differentiation and neurogenesis of embryonic stem cells. Cell Death Dis. 2013;4:e737. doi: 10.1038/cddis.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A., Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015;16:1609–1619. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A., Edupuganti R.R., Sorek M., Azad G.K., Markoulaki S., Sehnalová P., Legartová S., Bártová E., Farkash-Amar S., Jaenisch R. An endogenously tagged fluorescent fusion protein library in mouse embryonic stem cells. Stem Cell Reports. 2017;9:1304–1314. doi: 10.1016/j.stemcr.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppler S., Kavanagh C.L. Wnt signalling: variety at the core. J. Cell Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- Huelsken J., Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr. Opin. Genet. Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jain A.K., Barton M.C. p53: emerging roles in stem cells, development and beyond. Development. 2018;145:dev158360. doi: 10.1242/dev.158360. [DOI] [PubMed] [Google Scholar]
- Kalousi A., Hoffbeck A.-S., Selemenakis P.N., Pinder J., Savage K.I., Khanna K.K., Brino L., Dellaire G., Gorgoulis V.G., Soutoglou E. The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Rep. 2015;11:149–163. doi: 10.1016/j.celrep.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Kato K., Okuwaki M., Nagata K. Role of Template Activating Factor-I as a chaperone in linker histone dynamics. J. Cell Sci. 2011;124:3254–3265. doi: 10.1242/jcs.083139. [DOI] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Belmonte J.C.I. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-Y., Lee K.-S., Seol J.-E., Yu K., Chakravarti D., Seo S.-B. Inhibition of p53 acetylation by INHAT subunit SET/TAF-Iβ represses p53 activity. Nucleic Acids Res. 2012;40:75–87. doi: 10.1093/nar/gkr614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-B., Kim D.-W., Park J.W., Jeon Y.-J., Kim D., Rhee S., Chae J.-I., Seo S.-B. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-Iβ regulates Ku70-mediated DNA damage response. Cell. Mol. Life Sci. 2014;71:2731–2745. doi: 10.1007/s00018-013-1525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster J.P., Leeuwen I.V., Scheres N., Anthony E.C., Hordijk P.L. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007;26:336–345. doi: 10.1038/sj.emboj.7601518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L.J., Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kon N., Wang D., Gu W. Loss of SET reveals both the p53-dependent and the p53-independent functions in vivo. Cell Death Dis. 2019;10:237. doi: 10.1038/s41419-019-1484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S., Smits A.H., Vermeulen M., Reinberg D. Phospho-H1 decorates the inter-chromatid axis and is evicted along with shugoshin by SET during mitosis. Mol. Cell. 2017;67:579–593.e6. doi: 10.1016/j.molcel.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A., Mélius J., Waagmeester A., Sinha S.R., Miller R. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-F., Su J., Ang Y.-S., Carvajal-Vergara X., Mulero-Navarro S., Pereira C.F., Gingold J., Wang H.-L., Zhao R., Sevilla A. Regulation of embryonic and induced pluripotency by Aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Makkinje A., Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- Li M., He Y., Dubois W., Wu X., Shi J., Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang Y. Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective. Cell Res. 2013;23:49–69. doi: 10.1038/cr.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindern M.V., Breems D., Baal S.V., Adriaansen H., Grosveld G. Characterization of the translocation breakpoint sequences of two DEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and a SET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes. Chromosomes Cancer. 1992;5:227–234. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Kyba M., Murphy T.L., Murphy K.M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- Loh Y.-H., Zhang W., Chen X., George J., Ng H.-H. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., Tamai K., He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandemaker I.K., Zhou D., Bruens S.T., Dekkers D.H., Verschure P.J., Edupuganti R.R., Meshorer E., Demmers J.A.A., Marteijn J.A. Histone H1 eviction by the histone chaperone SET reduces cell survival following DNA damage. J. Cell Sci. 2020;133:jcs235473. doi: 10.1242/jcs.235473. [DOI] [PubMed] [Google Scholar]
- Marión R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W., Anderson C.W. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels A.J., Nusse R. Purified Wnt5a protein activates or inhibits β-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon R.T., Bowerman B., Boutros M., Perrimon N. The promise and perils of Wnt signaling through β-catenin. Science. 2002;296:1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Probst A.V., Almouzni G. Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation. 2008;76:15–23. doi: 10.1111/j.1432-0436.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- Raggioli A., Junghans D., Rudloff S., Kemler R. Beta-Catenin is vital for the integrity of mouse embryonic stem cells. PLoS One. 2014;9:e86691. doi: 10.1371/journal.pone.0086691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan A.-B., Cadigan K.M. Wnt target genes and where to find them. F1000Research. 2017;6:746. doi: 10.12688/f1000research.11034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Ricci M.A., Manzo C., García-Parajo M.F., Lakadamyali M., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- Riley T., Sontag E., Chen P., Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Sarig R., Rivlin N., Brosh R., Bornstein C., Kamer I., Ezra O., Molchadsky A., Goldfinger N., Brenner O., Rotter V. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med. 2010;207:2127–2140. doi: 10.1084/jem.20100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Schlesinger S., Meshorer E. Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev. Cell. 2019;48:135–150. doi: 10.1016/j.devcel.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Schuijers J., Mokry M., Hatzis P., Cuppen E., Clevers H. Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. EMBO J. 2014;33:146–156. doi: 10.1002/embj.201385358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., McNamara P., Heo S., Turner A., Lane W.S., Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Sokol S.Y., Melton D.A. Interaction of Wnt and activin in dorsal mesoderm induction in Xenopus. Dev. Biol. 1992;154:348–355. doi: 10.1016/0012-1606(92)90073-p. [DOI] [PubMed] [Google Scholar]
- Sullivan K.D., Galbraith M.D., Andrysik Z., Espinosa J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018;25:133–143. doi: 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M., Liu S.J., Zou L.-N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M.T., Slusarski D.C., Kaykas A., Louie S.H., Moon R.T. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Wang D., Kon N., Lasso G., Jiang L., Leng W., Zhu W.-G., Qin J., Honig B., Gu W. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538:118–122. doi: 10.1038/nature19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Nusse R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- Wu Y., Ai Z., Yao K., Cao L., Du J., Shi X., Guo Z., Zhang Y. CHIR99021 promotes self-renewal of mouse embryonic stem cells by modulation of protein-encoding gene and long intergenic non-coding RNA expression. Exp. Cell Res. 2013;319:2684–2699. doi: 10.1016/j.yexcr.2013.08.027. [DOI] [PubMed] [Google Scholar]
- Yi F., Pereira L., Hoffman J.A., Shy B.R., Yuen C.M., Liu D.R., Merrill B.J. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 2011;13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Zeng Y., Xiao Y., Chen Y., Shen H., Dong J. Cyclin-dependent kinase 1-mediated phosphorylation of SET at serine 7 is essential for its oncogenic activity. Cell Death Dis. 2019;10:385. doi: 10.1038/s41419-019-1621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying J., Li H., Yu J., Ng K.M., Poon F.F., Wong S.C.C., Chan A.T.C., Sung J.J.Y., Tao Q. WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/β-catenin signaling, and is frequently methylated in colorectal cancer. Clin. Cancer Res. 2008;14:55–61. doi: 10.1158/1078-0432.CCR-07-1644. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ratanasirintrawoot S., Chandrasekaran S., Wu Z., Ficarro S.B., Yu C., Ross C.A., Cacchiarelli D., Xia Q., Seligson M. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016;19:66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exponentially modified protein abundance index (emPAI) is used for label-free protein quantitation
List of DEGs in SET-KO and SET-OE mutants with adjusted p < 0.05. Highlighted in red are genes that have a reciprocally differentially expressed in SET-KO and SET-OE ESCs