Abstract
RNA in situ hybridization is a powerful technique used to identify the spatial localization of a specific RNA in a tissue section or whole tissue. In this protocol, we describe a reliable method for two-color in situ hybridization that can be used to accurately assess the expression of multiple genes with contrasting or overlapping expression patterns in whole mouse embryos.
Keywords: In situ hybridization, Development, Embryo, RNA probe, Color detection, Antibody
1. Introduction
In situ hybridization is a method of detecting specific mRNA sequences in fixed cell populations and whole tissue. The basic technique involves the use of an enzymatically labeled probe that anneals to complementary sequences in the target tissue, and is visualized with a chromogenic substrate. This technique can be routinely used to visualize the expression patterns of multiple genes in whole mammalian embryos.
The following whole-mount in situ hybridization (WISH) protocol is modified from the protocols of Wilkinson et al. [1], Conlon et al. [2], and Parr et al. [3]. The procedure described here provides a detailed step-by-step method for two-color analysis using Digoxigenin (DIG)-labeled and Fluorescein (FLU)-labeled probes. Simultaneous detection of two or more probes is achieved with two chromogenic substrates, producing beautiful contrasting patterns of gene expression. This protocol has been optimized for the analysis of gene expression in the gastrulating embryo [4–6], but has been readily applied to fetal stages, as well as isolated organs and cultured tissue explants (Fig. 1).
Fig. 1.
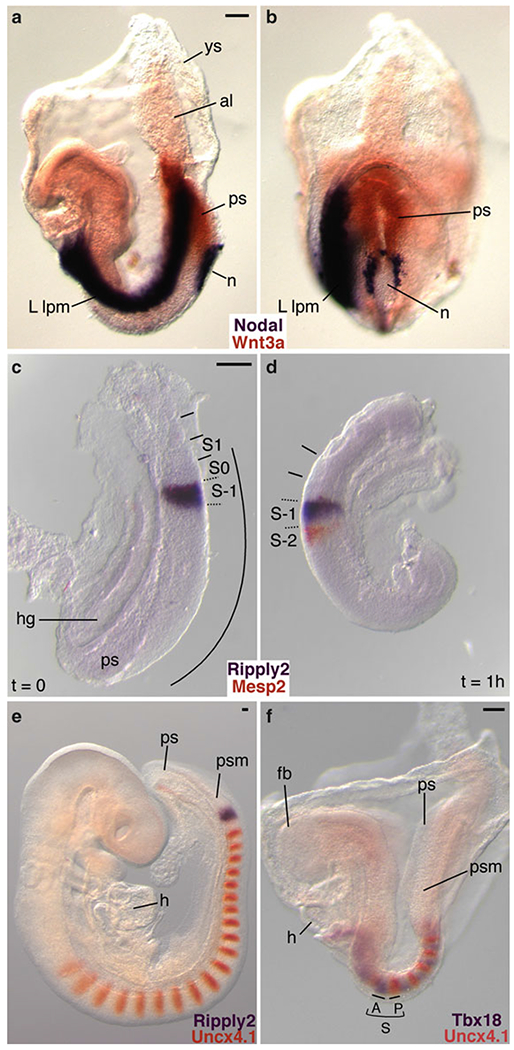
Examples of two-color WISH performed on mouse embryos and cultured embryo explants. In all cases, DIG-labeled RNA probes were detected with BM purple, and FLU-labeled RNA probes were visualized with INT/BCIP. (a, b) Two-color WISH analysis of a 4 somite stage (E8.2) embryo demonstrating the complementary expression of genes encoding the secreted signaling molecules Nodal (purple) and Wnt3a (orange). Nodal is asymmetrically expressed in the node periphery and in the (left) lateral plate mesoderm [9,10], while Wnt3a mRNA is restricted to the midline primitive streak [11] (Reproduced from ref. 5 with permission from Development). (c, d) Embryo-half culture experiments illustrate how dynamic, oscillating gene expression patterns can be visualized with two-color WISH. The bHLH transcription factor, Mesp2 (orange), and the transcriptional corepressor Ripply2 (purple), are important for segment boundary formation [4, 5,12,13]. In the uncultured half-embryo explant (t = 0), both genes are coexpressed in S-1 in the anterior presomitic mesoderm (c). After culturing the complementary half-embryo explant for 1 h, Ripply2 expression remains in S-1 while Mesp2 expression is activated in S-2 (d). (e) Analysis of E9.5 embryos shows that the paired type homeodomain transcription factor Uncx4,1 (orange) is expressed in the caudal half of segmented somites [14–16], and in the posterior half of SO immediately adjacent to the broad somite-wide stripe of Ripply2 expression (purple) in S-1 [4]. (f) Two-color WISH clearly illustrates the complementary expression patterns of segment polarity markers. Expression of the anterior half-somite marker Tbx18 (purple) complements Uncx4,1 expression in the caudal half [5, 17]. All embryo views are lateral with the exception of (b), which is ventroposterior. ys yolk sac, al allantois, ps primitive streak, n node, L lpm left lateral plate mesoderm, hg hindgut, S somite, S-1 presumptive somite, SO forming somite, S1 first newly formed somite, curved line or psm presomitic mesoderm, h heart, fb forebrain, A anterior half somite, Pposterior half somite. Scale bars: 100 μm
2. Materials
Fixatives, hybridization solution, and reagents may be prepared in advance and stored in aliquots, while remaining wash and incubation solutions are prepared fresh on the day of use. All procedures up to the hybridization step must be carried out under RNase-free conditions to preserve the integrity of the RNA. Prepare solutions with RNase-free water purchased from the manufacturer. Autoclave all glassware, plastic ware, and pipette tips, and clean all equipment and work area with RNaseZAP® decontamination solution or similar solution (see Note 1). It is recommended that sterile plastic transfer pipettes are used for changing solutions, and that they are replaced frequently.
2.1. Preparation of Embryos
Phosphate-buffered saline (PBS), pH 7.4.
Micro dissecting tweezers pattern #5 and #55, light superfine points, tip 0.05 × 0.01 mm, material: Dumostar alloy (Roboz RS4978 and RS4984; see Note 2).
VWR® 2 mL screw-cap microcentrifuge tubes (VWR 89004-302; see Note 3).
4 % paraformaldehyde (PFA) in PBS. Dissolve in PBS and stir at 65 °C on a hot plate for 1 h. Filter and aliquot into 15 mL and 50 mL tubes and store at −20 °C until use.
PBT: PBS with 0.1 % SigmaUltra Tween® 20 (Sigma P7949).
Methanol–PBT series (25, 50, 75 % methanol in PBT). Methanol is considered hazardous and should be handled and disposed of using established laboratory procedures. Store in a sealed flammable storage cabinet.
RNaseZap® RNase decontamination solution (Sigma R2020).
2.2. Embryo Powder Preparation and Storage
Mortar and pestle.
Tissue homogenizer.
Acetone. Follow established laboratory procedures when handling and disposing of this chemical. Store in a sealed flammable storage cabinet.
Whatman filter paper.
2.3. Generation of Template
Purified plasmid DNA.
Restriction enzymes.
Phenol–Chloroform–Isoamyl Alcohol 25:24:1 saturated with 10 mM Tris–HCl, pH 8.0, 1 mM EDTA. Phenol can cause severe burns. Handle in a fume hood and wear gloves.
Phase Lock Gel® Heavy, 2 mL tubes (PLG tubes) (Eppendorf 0032005.152).
RNase-free absolute ethanol.
RNase-free 3 M Sodium Acetate (NaOAc), pH 5.2.
RNase-free 70 % ethanol.
10 mM Tris–HCl, pH 7.6.
2.4. Synthesis of Digoxigenin or Fluorescein Labeled RNA Probes
Sterile Dnase, RNase, Protease-free water.
10× transcription buffer (Roche 1277073).
0.1 M dithiothreitol (DTT). Store in aliquots at −20 °C.
10× DIG RNA labeling mix 10 mM ATP, 10 mM CTP, 10 mM GTP, 6.5 mM TTP, 3.5 mM DIG-11-UTP, pH 7.5 (Roche 1277073).
10× Fluorescein RNA labeling mix 10 mM ATP, 10 mM CTP, 10 mM GTP, 6.5 mM UTP, 3.5 mM FLU-12-UTP, pH 7.5 (Roche 1685619).
Linearized DNA template (1 μg/μL).
Protector RNase inhibitor (40 U/μL) (Roche 03335399001).
Sp6, T3, or T7 RNA polymerase (20 U/μL) (Roche 10810274001, 11031163001, 10881767001).
RNase-free DNasel recombinant (10 U/μL) (Roche 04716728001).
TE: 50 mM Tris-HCl, 1 mM EDTA, pH 8.0.
7.5 M LiCl precipitation solution (Ambion AM9480).
Prehybridization/hybridization solution: 50 % formamide (Ambion AM9342), 5× standard saline sodium citrate (SSC), pH 4.5 (Ambion AM9763), 1 % sodium dodecyl sulfate (SDS) (Ambion AM9823), 100 μg/mL tRNA (Sigma R8505), 100 μg/mL heparin (Sigma H3149; see Note 4). Formamide is a hazardous material and should be handled using established laboratory procedures.
2.5. Prehybridization and Hybridization of Embryos
Methanol–PBT series (25, 50, 75 % methanol in PBT).
100 % Methanol.
PBT: PBS with 0.1 % Tween-20.
30 % hydrogen peroxide solution (Sigma H1009).
4 % PFA in PBS.
25 % glutaraldehyde solution (Sigma G5882). Glutaraldehyde is packaged in 10 mL glass ampoules. Upon opening, aliquot into tubes and store at −20 °C.
10 mg/mL proteinase K (Roche 03115836001) in RNase-free water. Store in 25 μL aliquots at −80 °C.
Glycine, OmniPur* (VWR EM4810).
Prehybridization/hybridization solution.
DIG-labeled RNA probe and FLU-labeled RNA probes. Store at −80 °C.
Hybaid Shake “n” Stack Hybridization Oven (Thermo Scientific 6241).
2.6. Posthybridization Washes and Antibody Incubation
Posthybridization wash solution I: 50 % formamide, 4× SSC, 1 % SDS.
Posthybridization wash solution II: 0.5 M NaCl, 10 mM Tris–HCl, pH 7.5, 0.1 % Tween-20.
Posthybridization wash solution III: 50 % formamide, 2× SSC.
RNase A (Sigma R6513).
MABTL: 0.1 M Maleic acid pH 7.5, 0.15 M NaCl, 0.1 % Tween-20, and 2 mM levamisole, adjust pH with NaOH. Filter before use (see Note 5).
Blocking reagent solution: 2 % w/v blocking reagent (Roche 11096176001) in MABTL.
Sheep serum (Sigma S2263; see Note 6).
2.7. Preadsorption of Antibody
Sheep anti-Digoxigenin (Roche 1093274), and anti-Fluorescein (Roche 1426338) Fab fragments conjugated to alkaline phosphatase (AP). Store at 4 °C.
2.8. Antibody Washes and Color Detection
NTMT: 100 mM NaCl, 100 mM Tris–HCl pH 9.5, 50 mM MgCl2, 0.1 % Tween-20.
NTMTL: NTMT with 2 mM levamisole.
BM Purple: AP substrate precipitating (NBT/BCIP ready-to-use solution) (Roche 11442074001). Store at 4 °C protected from light.
INT/BCIP Stock Solution (Roche 11681460001). Store at 4 °C protected from light.
2.9. Embryo Storage and Photography
Glycerol SigmaUltra (Sigma G6279).
Leica MZFLIII high-performance stereomicroscope.
Zeiss AxioCam high-resolution digital camera.
NCL150 Fiber Optic Illuminator.
3. Methods
Subheadings 3.1–3.4 should be performed in advance. Embryos should be gently rocked on a single speed orbital mixer or in a hybridization oven equipped with a variable speed rocking platform during the prehybridization, hybridization, and washing steps. At the beginning of each day of the protocol, prepare and warm solutions in advance. All steps should be performed at room temperature unless otherwise stated.
3.1. Preparation of Embryos
Dissect embryos in ice-cold PBS. Reflect extraembryonic membranes and remove the amnion. Puncture the fourth ventricle in the hindbrain to prevent trapping of reagents and consequent background in the neural tube (see Note 7).
Place the embryos in a 2 mL screw-cap tube, and fix in 4 % PFA in PBS overnight at 4 °C.
Wash three times with PBT for 5 min each.
Dehydrate the embryos in a Methanol series (25, 50, 75 % Methanol–PBT) for 5 min each. Wash twice with 100 % Methanol for 10 min each, and store indefinitely in 100 % Methanol at −20 °C.
3.2. Embryo Powder Preparation and Storage
Dissect wild-type embryos (embryonic day (E) 12.5–14.5 mouse embryos) in cold PBS.
Homogenize embryos in a 50 mL conical tube on ice in a minimum volume of PBS.
Add 4 volumes of ice-cold acetone, gently mix, and then incubate on ice for 30 min.
Centrifuge at 10,000 ×gfor 10 min.
Wash the pellet with ice-cold acetone and centrifuge again.
Spread pellet out on a piece of Whatman filter paper. Then, cover with a second piece of filter paper and let dry overnight at room temperature.
Grind into a fine powder with a newly autoclaved mortar and pestle.
Store dry in an eppendorf tube at 4 °C.
3.3. Generation of Template
Quantify circular plasmid DNA with a standard spectrophotometer.
Prepare template DNA by linearizing 6 μg of DNA with the appropriate restriction enzymes (see Note 8 and Fig. 2). Digest at 37 °C for 3 h in a 100 μL volume. Remove a 5 μL aliquot and run out on a 1 % agarose gel to ensure that the digestion reaction is complete and the plasmid is completely linearized.
Dilute the remaining 95 μL digest with 105 μL RNase-free water, then phenol–chloroform extract the DNA using PPG tubes following manufacturer’s recommendations (see Note 9).
Add 2.5 volumes of 100 % ethanol and 1/10 volume of 3 M NaOAc to precipitate the DNA. Mix well and incubate at −80 °C for a minimum of 1 h.
Spin samples at 4 °C for 15 min, then wash pellet with 70 % ethanol and air-dry.
Resuspend pellet in 5 μL 10 mM Tris–HCl pH 7.6. Use 1 μL (1 μg) as the template for RNA probe synthesis and store remaining template at −20 °C.
Fig. 2.
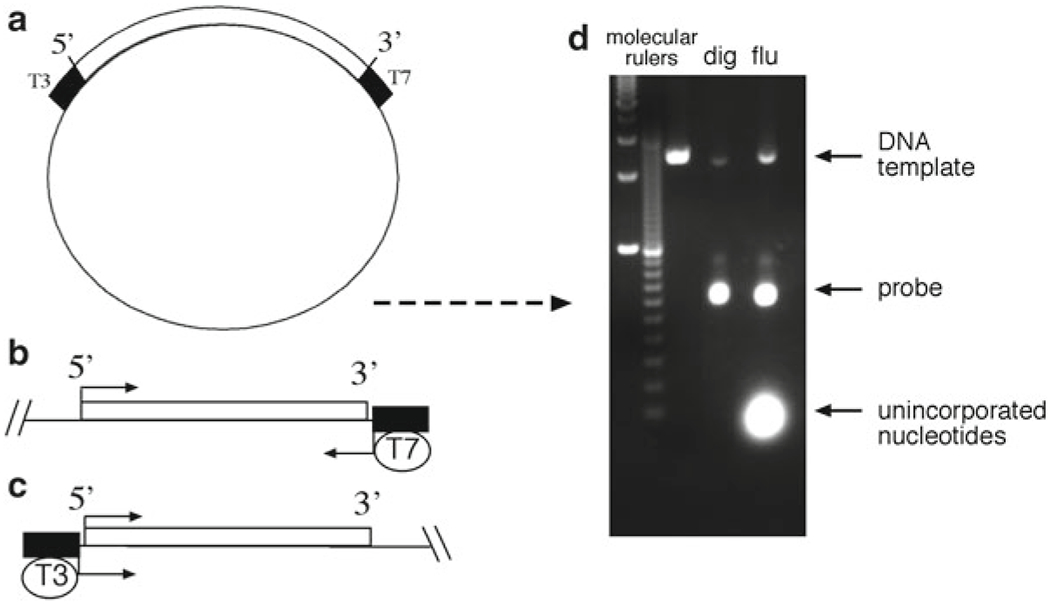
Schematic illustrating the generation of the linear cDNA template and RNA probes. (a) The plasmid must contain unique restriction enzyme sites and RNA polymerase initiation sites for T7, T3, or SP6, flanking the cDNA of interest. (b) In this example, the 5′ end of the insert is digested with an appropriate restriction enzyme and then transcribed with T7 RNA polymerase to generate an antisense probe. (c) Similarly, a sense probe is generated by digesting the 3′ end with an appropriate enzyme, and transcribing with T3 polymerase. (d) Gel electrophoresis of freshly synthesized RNA probes. RNA should migrate to a position that roughly correlates with the size of the cloned insert, while fainter, slowly migrating DNA template bands should also be apparent
3.4. Synthesis of Digoxigenin or Fluorescein Labeled RNA Probes
- Mix these reagents in the following order at room temperature:
Sterile Dnase, RNase, Protease-free water 11.5 μL 10× transcription buffer 2.0 μL 0.1 M DTT 2.0 μL 10× DIG or FLU RNA labeling mix 2.0 μL Linearized template (1 μg) 1.0 μL RNase inhibitor (40 U/μL) 0.5 μL Sp6, T7, or T3 RNA polymerase (20 U/μL) 1.0 μL Incubate at 37 °C for 2 h.
To estimate the transcript amount, a 1 μL aliquot is removed and electrophoresed on a 1 % agarose gel containing 0.5 μg/mL ethidium bromide (see Note 10). A faint template band and an RNA band ten times more intense than the plasmid band should be observed, indicating that approximately 10 μg of probe has been synthesized. Two or more RNA bands are commonly observed.
To the transcription reaction, add 2 μL Dnase I (10 U/μL), and incubate at 37 °C for 15 min.
Add 100 μL TE, 5.3 μL of 7.5 M LiCl, and 300 μL 100 % ethanol, mix, and precipitate RNA at −80 °C for 1 h.
Spin in a microcentrifuge at 4 °C for 30 min.
Wash the pellet twice with 70 % ethanol and air-dry.
Dissolve pellet in 200 μL of warm hybridization solution at approximately 25–50 ng/μL, and store at −80 °C. Probes can withstand multiple freeze-thaw cycles and are stable for years.
3.5. Prehybridization and Hybridization of Embryos
Rehydrate the embryos by washing through descending Methanol–PBT series (75, 50, 25 % Methanol–PBT) followed by washes in PBT for 5 min each.
Bleach embryos with 6 % hydrogen peroxide solution in PBT for 1 h.
Wash three times with PBT for 5 min each.
Treat embryos with 10 μg/mL proteinase K in PBT. Timing is important and depends upon tissue size and the batch of proteinase K used. We recommend the following digestion times as a guideline: digest embryos younger than E7.5 for 1 min or less, E7.5 for 3 min, E8.5 for 7.5 min, E9.5 for 15–20 min if internal tissues are to be probed, or 5 min if gene expression is assessed in superficial ectoderm, and E10.5 for at least 25–30 min (see Note 11).
Inactivate proteinase K with freshly prepared 2 mg/mL glycine in PBT for 5 min.
Wash twice with PBT for 5 min each.
Refix the embryos with 4 % PFA/0.2 % glutaraldehyde in PBS for 20 min.
Wash twice with PBT for 5 min each. Remove as much of the last wash as possible, before adding the prehybridization solution.
Add prehybridization solution to each tube and hybridize at 70 °C for 1 h. Incubate with the tubes vertical in a microcentrifuge tube rack.
Preheat the RNA probes to 70 °C for 10 min prior to the hybridization step. Resuspend with gentle agitation.
Replace the prehybridization solution with fresh hybridization solution, and add 5 μL of 25–50 nĝL of DIG-labeled or FLU-labeled RNA probe for each 1 mL of hybridization solution for a total of 125–250 ng of RNA probe per 1 mL of hybridization solution. For two-color in situ hybridization, add 125–250 ng of each labeled RNA probe per 1 mL hybridization solution. Make sure that the probe(s) is/are in solution by gently flicking the tube.
Place tubes vertically in a tray and incubate at 70 °C overnight.
3.6. Posthybridization Washes and Antibody Incubation
Prepare posthybridization wash solutions I–III. Filter solution 1 through a 0.45 μm filter or equivalent, then pre-warm solutions 1 and 3 at 65 °C.
Wash embryos twice with solution I for 30 min each at 70 °C (see Note 12).
Wash embryos with a 1:1 mix of with solution I: solution II for 10 min at 70 °C.
Wash three times with solution II for 5 min each.
Incubate with 100 μg/mL RNase A in solution II for 1 h at 37 °C (see Note 13).
Prepare the MABTL and blocking reagent solution during the 1 h RNase step. Heat the MABTL in a microwave until the solution becomes cloudy, about 30–45 s. Avoid boiling the solution. Dissolve the blocking reagent by vortexing until completely in solution. Cool on ice before using.
Wash one time with solution II, followed by one wash with solution III for 5 min each.
Wash twice with solution III for 30 min each at 65 °C. Let embryos settle by standing tubes vertically in the Hybaid oven before changing wash solutions. Near the end of the second wash, begin the antibody preadsorption with the embryo powder (see Subheading 3.7).
Wash embryos three times with MABTL for 5 min each.
Block the embryos with 10 % sheep serum in blocking reagent solution (2 % w/v blocking reagent in MABTL) for 1–3 h. Longer blocking times are acceptable.
Replace blocking solution with the preadsorbed antibody (see Subheading 3.7, step 5). Rock tubes on an orbital mixer overnight at 4 °C.
3.7. Preadsorption of Antibody
Place 3 mg of embryo powder into a 1.5 mL tube containing 1 mL block reagent stock (see Note 14). Briefly vortex, and heat inactivate for 30 min at 70 °C.
Cool on ice, and add 10 μL sheep serum and appropriate amount of anti-DIG-AP or anti-FLU-AP antibody for a final dilution of 1:4,000 dilution of anti-DIG-AP Fab fragment, or a 1:2,000 dilution of anti-FLU-AP Fab fragment.
Rock gently at 4 °C for the duration of the blocking procedure. This should be no less than 2 h.
Centrifuge for 10 min at 4 °C.
Recover the supernatant containing the antibody and dilute to desired final concentration with chilled 1 % sheep serum in blocking solution. Keep the antibody mixture at 4 °C until ready to use.
3.8. Antibody Washes
Remove the antibody and wash embryos three times with MABTL for 5 min each.
Wash embryos eight to ten times for 1 h each with MABTL, and continue overnight for a total of 24 h. Decreased background may be obtained by washing for a second or third day especially if large tissues or embryos are being analyzed.
3.9. Color Detection for Single and Two-Color WISH
Wash three times with NTMTL for 10 min each.
Remove the NTMTL and incubate with 1–2 mL of chromogenic substrate of your choice. If using BM Purple, add substrate to the embryos directly. Alternatively, if INT/BCIP is desired, dilute substrate in NTMT (7.5 μL INT/BCIP per 1 mL of NTMT). Cover in foil, then stand the tubes in a rack and develop protected from light (see Note 15).
Monitor the reaction frequently until the desired signal intensity is produced, or background is observed in sense strand controls. The signal usually becomes visible within 1–2 h, but the color reaction may need to proceed overnight at room temperature or 4 °C for low-abundance transcripts.
Stop the color reaction with two rinses in PBT, followed by three washes in PBT for 5 min each. Proceed directly to Subheading 3.11 if only a single color is desired. For two-color analysis, photograph embryos in PBT to document the first color singly, or proceed directly to Subheading 3.10.
3.10. Antibody Incubation and Washes for Two-Color WISH
Briefly post-fix embryos in 4 % PFA in PBS for 20 min.
Wash three times with PBT for 5 min each.
Block embryos with 10 % sheep serum in blocking reagent solution (2 % w/v blocking reagent in MABTL) for 1–3 h.
Begin the preadsorption of the second antibody (see Subheading 3.7).
After the blocking step, remove the blocking solution and replace with the preadsorbed antibody. Rock tubes on an orbital mixer overnight at 4 °C.
Remove the antibody and wash embryos three times with MABTL for 5 min each followed by a minimum of eight to ten washes over a 24 h period. Incubate with the desired AP color reagent, usually INT/BCIP, until the desired signal intensity is produced.
3.11. Embryo Storage
Wash three times with PBT for 5 min each.
Fix embryos in 4 % PFA/0.2 % glutaraldehyde in PBS for 1 h.
Wash three times with PBT for 5 min each.
Wash embryos through a Glycerol/PBT series (50 and 80 % glycerol PBT), and store in 80 % glycerol/PBT. Photograph embryos as soon as possible if using INT/BCIP as a chromogenic substrate as the orange signal fades rapidly. Note also that this precipitate is soluble in alcohols.
3.12. Photography
Photograph embryos in 80 % glycerol using a stereo microscope equipped with a high quality digital camera. We have attained excellent results with the Zeiss Axiocam and Axiovision Imaging software.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
4 Notes
Caution should be taken to minimize the introduction of contaminating RNases which can result in RNA degradation and loss of signal. Always wear gloves when working with RNA or when handling RNA probes as human skin contains copious amounts of RNases.
A “biologie” tip style is recommended for dissecting mouse embryos as it is finer then standard tips. The soft alloy material used in the construction of these tools gives the tweezer flexibility making it an excellent choice for dissecting mouse embryos. Be warned however, that the tip is easily damaged by inadvertent contact with hard surfaces. For guidelines on how to stage embryos refer to ref. 7.
Clear, screw-cap, 2 mL tubes with conical bottoms work best for embryos younger than E10.5. Glass scintillation vials work well for larger samples.
Hybridization solution can be made in advance and stored at −20 °C for up to 1 year. This solution becomes cloudy and viscous when stored at −20 °C. Pre-heat at 65 °C prior to the prehybridization step to clear.
Levamisole is an inhibitor of endogenous AP activity and is used to reduce background staining.
Heat-inactivate sheep serum for 30 min at 70 °C. Aliquot into small volumes (1–3 mL) and store at −20 °C for up to 1 year. Keep in mind that the serum should always match the species from which the antibody is made.
Background frequently arises when the probe or other reagents become trapped in the ventricles of the forebrain and hindbrain of E9.5 or older embryos. Membranes should be punctured before fixation to ensure thorough and even fixation of all tissues. See ref. 8 for practical tips on embryo dissection, and removal of extraembryonic tissue from E6.5 to E13.5 embryos.
Templates generated with a restriction enzyme that leaves a 5′ overhang or blunt end give the best results. Avoid using enzymes that generate 3′ overhangs as they may give rise to unusual transcripts. When making complementary antisense strand probes, choose a unique restriction site at the 5′ end of the insert. The antisense transcript is synthesized using a promoter located at the 3′ end of the insert (Fig. 2b). Conversely, for the sense strand probe, choose a unique restriction site at the 3′ end, and transcribe with the promoter found at the 5′ end (Fig. 2c). A sense strand probe serves as a negative control that should not hybridize with the target mRNA and can be used to monitor background activity.
Phase Lock Gel columns efficiently separate the water soluble (aqueous) phase from the organic phase preventing contamination while increasing recovery. The procedure is faster than traditional phenol-chloroform extraction methods, and offers added protection from exposure to hazardous compounds (see Subheading 2.3, item 4).
When analyzing the amount of RNA, a clean dedicated gel apparatus is recommended to avoid RNA degradation during electrophoresis. Ethidium bromide should always be disposed of safely, in a manner according to regulations set forth by your Office of Environmental Health and Safety.
Permeabilization of the tissues is necessary to increase the accessibility of the target RNA. Proteinase treatment times vary depending on the stage and type of tissue, and should be empirically determined. It should be emphasized that digestion times are important as over-digestion may result in loss of tissue integrity and decreased signal, whilst under-digestion may result in high background.
Embryos become translucent in formamide solution, so allow embryos to settle to the bottom of the tube before changing the solution.
The RNase A digestion step reduces background, but decreases signal as well. This step can sometimes be omitted altogether, but this is probe-dependent. Two-color in situ hybridizations generally benefit from RNase A treatments as the reduced background makes it easier to visualize overlapping gene expression patterns.
1 mL of embryo powder containing blocking solution is sufficient to preadsorb up to 10 μL of anti-DIG antibody or 20 μL anti-FLU antibody.
Development of chromogenic substrate can be performed in any order, however, BM purple is recommended first since the signal develops more quickly. The overall color of the substrate solution should be monitored regularly and replaced if a color change is observed.
References
- 1.Wilkinson DG (1992) Whole mount in situ hybridization of vertebrate embryos, in situ hybridization: a practical approach. IRL, Oxford [Google Scholar]
- 2.Conlon RA, Hermann BG (1993) Detection of messenger RNA by in situ hybridization to postimplantation embryo whole mounts. Methods Enzymol 225:373–783 [DOI] [PubMed] [Google Scholar]
- 3.Parr BA, Shea MJ, Vassileva G, McMahon AP (1993) Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development 119: 247–261 [DOI] [PubMed] [Google Scholar]
- 4.Bins KK, Dunty WC Jr, Yamaguchi TP (2007) Mouse Ripply2 is downstream of Wnt3a and is dynamically expressed during somitogenesis. Dev Dyn 236:3167–3172 [DOI] [PubMed] [Google Scholar]
- 5.Dunty WC Jr, Biris KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP (2008) Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135:85–94 [DOI] [PubMed] [Google Scholar]
- 6.Nakaya MA, Biris K, Tsukiyama T, Jaime S, Rawls JA, Yamaguchi TP (2005) Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development 132:5425–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufman MH (1999) The atlas of mouse development. Academic, London [Google Scholar]
- 8.Hogan B, Beddington R, Costantini F, Lacy E (1994) Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 9.Collignon J, Varlet I, Robertson EJ (1996) Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381:155–158 [DOI] [PubMed] [Google Scholar]
- 10.Lowe LA, Supp DM, Sampath K, Yokoyama T, Wright CV, Potter SS, Overbeek P, Kuehn MR (1996) Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381:158–161 [DOI] [PubMed] [Google Scholar]
- 11.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP (1994) Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev 8:174–189 [DOI] [PubMed] [Google Scholar]
- 12.Morimoto M, Sasaki N, Oginuma M, Kiso M, Igarashi K, Aizaki K, Kanno J, Saga Y (2007) The negative regulation of Mesp2 by mouse Ripply2 is required to establish the rostro-caudal patterning within a somite. Development 134: 1561–1569 [DOI] [PubMed] [Google Scholar]
- 13.Saga Y, Hata N, Koseki H, Taketo MM (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 11: 1827–1839 [DOI] [PubMed] [Google Scholar]
- 14.Mansouri A, Yokota Y, Wehr R, Copeland NG, Jenkins NA, Gruss P (1997) Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev Dyn 210:53–65 [DOI] [PubMed] [Google Scholar]
- 15.Neidhardt LM, Kispert A, Herrmann BG (1997) A mouse gene of the paired-related homeobox class expressed in the caudal somite compartment and in the developing vertebral column, kidney and nervous system. Dev Genes Evol 207:330–339 [DOI] [PubMed] [Google Scholar]
- 16.Leitges M, Neidhardt L, Haenig B, Herrmann BG, Kispert A (2000) The paired homeobox gene Uncx4.1 specifies pedicles, transverse processes and proximal ribs of the vertebral column. Development 127:2259–2267 [DOI] [PubMed] [Google Scholar]
- 17.Kraus F, Haenig B, Kispert A (2001) Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev 100:83–86 [DOI] [PubMed] [Google Scholar]


