Abstract
Background
Intestinal microbial dysbiosis is evident in chronic HIV-infected individuals and may underlie inflammation that persists even during antiretroviral therapy (ART). It remains unclear, however, how early after HIV infection gut dysbiosis emerges and how it is affected by early ART.
Methods
Fecal microbiota were studied by 16s rDNA sequencing in 52 Thai men who have sex with men (MSM), at diagnosis of acute HIV infection (AHI), Fiebig Stages 1–5 (F1-5), and after 6 months of ART initiation, and in 7 Thai MSM HIV-uninfected controls. Dysbiotic bacterial taxa were associated with relevant inflammatory markers.
Results
Fecal microbiota profiling of AHI pre-ART vs HIV-uninfected controls showed a mild dysbiosis. Transition from F1-3 of acute infection was characterized by enrichment in pro-inflammatory bacteria. Lower proportions of Bacteroidetes and higher frequencies of Proteobacteria and Fusobacteria members were observed post-ART compared with pre-ART. Fusobacteria members were positively correlated with levels of soluble CD14 in AHI post-ART.
Conclusions
Evidence of gut dysbiosis was observed during early acute HIV infection and was partially restored upon early ART initiation. The association of dysbiotic bacterial taxa with inflammatory markers suggests that a potential relationship between altered gut microbiota and systemic inflammation may also be established during AHI.
Keywords: ART, HIV, inflammation, immune activation, microbiome
Gut dysbiosis was detected during early acute HIV infection in Thai MSM and was partially restored after 6 months of antiretroviral therapy. Bacterial changes were subtle but significant correlations were observed between pathogenic taxa and systemic inflammatory biomarkers.
During the acute phase of HIV infection, high levels of viral replication occur in the gut-associated lymphoid tissue (GALT) [1]. This leads to a substantial depletion of gut mucosal CD4+ T cells with a preferential loss of T helper 17 (Th17) and T helper 22 (Th22) cells, whose main effector cytokines are interleukin (IL)-17, IL-21, and IL-22. These cytokines are normally enriched in mucosal tissue and play a central role in protecting the integrity of the epithelial barrier and maintaining immune homeostasis at mucosal sites [2–4]. As a consequence of this altered intestinal microenvironment, there is increased permeability with microbial translocation [5, 6], contributing to chronic immune activation and inflammation [7, 8]. The etiology, however, of the persistent immune activation in chronic treated HIV infection is still incompletely understood and is probably multifactorial, encompassing residual HIV replication, co-infections, mucosal alterations, and incomplete immune restoration [9, 10].
Current studies in HIV infection, similar to other chronic inflammatory states [11–13], suggest that resident gut microbiota and their metabolic products are drivers of inflammation that can persist even during antiretroviral therapy (ART) [14, 15]. Initial studies by Brenchley et al. described elevated levels of lipopolysaccharide (LPS) in HIV-positive people that were associated with peripheral blood T-cell activation, providing an indication of a link between intestinal microbial translocation and systemic immune activation [5]. During the last few years, several studies have compared the gut microbiota of HIV‐infected individuals with that of HIV‐uninfected controls in order to evaluate differences in microbial abundance [16–23]. Many studies have included cross-sectional comparisons between HIV-infected individuals and controls and have suggested that HIV infection is associated with alterations in gut communities, in particular an enrichment in Enterobacteriaceae members and Erysipelotricaceae with concurrent depletion of Bacteroides and Clostridia [24]. These dysbiotic bacterial communities are often linked to key markers of inflammation and cellular activation [25], including the kynurenine pathway of tryptophan metabolism and plasma concentrations of the inflammatory cytokine IL-6 in treated HIV-infected individuals [20]. It remains unclear, though, how early after HIV infection gut dysbiosis is established and how early ART treatment may affect it.
Initiation of ART during early acute HIV infection (AHI) has been previously shown to be associated with preservation of immune function, enhanced recovery of CD4+ T-cell numbers and function in blood as well as in the gastrointestinal (GI) tract, and significant reduction in HIV reservoir size [26]. Despite reversing several key features of early GI tract pathology, however, mucosal injury and inflammation may not resolve completely by early ART [27, 28].
In this study, we profiled the intestinal microbiota during AHI, Fiebig Stages 1–5 (F1-5), at the time of diagnosis, and 6 months after ART initiation using HIV-uninfected individuals as controls. Furthermore, we assessed the relationship of dysbiosis with systemic inflammatory and gut epithelial integrity markers.
METHODS
Study Participants and Study Design
RV254/SEARCH010 is a clinical trial (NCT00796146) that was conducted in Bangkok, Thailand, enrolling persons with early acute HIV infection (F1-5) who were identified from the Thai Red Cross Anonymous Clinic through nucleic acid testing and sequential immunoassays [29]. The RV254/SEARCH 010 study was approved by the institutional review boards (IRBs) of Chulalongkorn University in Thailand and the Walter Reed Army Institute of Research in the United States. Participation in this study involved extensive evaluation including rectosigmoid gut biopsies, anal sample collection, and storage of peripheral blood mononuclear cells (PBMCs) and serum/plasma. Initiation of ART was voluntary and done as part of the enrollment in an accompanying protocol (clinicaltrials.gov NCT00796263), which was approved by the Chulalongkorn University IRB. For all the studies mentioned above, subjects gave written informed consent. We profiled the intestinal microbiota during AHI, F1-5, at the time of diagnosis, and 6 months after ART initiation.
Microbiome Study Methods
Rectal samples from 52 men who have sex with men (MSM) at either baseline (n = 37) or 6 months after ART (2NRTI + efavirenz) initiation (n = 31; termed post-ART) were collected, including 16 paired samples at both visits. Samples (n = 7) from HIV-uninfected age-, sex-, and risk group–matched MSM Thai volunteers were obtained from a different protocol (RV304, clinicaltrials.gov NCT01397669) at the same site.
DNA Extraction and PCR Amplification
Bacterial profiles of study participants were generated by broad-range amplification and sequence analysis of bacterial 16S rRNA genes. Microbiome specimen analysis is described in the Supplementary Data and [30].
Laboratory Methods for Blood Measurements
Plasma, serum, and PBMC specimens were processed within 30 minutes of collection. Intestinal fatty acid binding protein (I-FABP) and sCD14 (R&D Biosystems, Minneapolis, MN) were measured by enzyme-linked immunosorbent assay; C-reactive protein (CRP) was measured by electrochemiluminescence (Meso Scale Discovery, Rockville, MD); high-sensitivity IL-6 was measured using the Luminex (Austin, TX) platform according to the manufacturer’s instructions (Millipore, Darmstadt, Germany). All assays used cryopreserved acid citrate dextrose (ACD) plasma matching rectal sampling.
Immunohistochemistry
Rectosigmoid gut biopsies were sampled and processed as described earlier [28]. In brief, subjects underwent a routine sigmoidoscopy procedure under moderate conscious sedation. Approximately 30 endoscopic biopsies were randomly collected from the sigmoid colon using Radial Jaw 3 biopsy forceps (Boston Scientific, Natick, MA), with 20–25 processed for flow cytometry analysis within 30 minutes of collection. Biopsies from AHI pre-ART (n = 23) and post-ART (n = 14) were used for immunohistochemistry (IHC) analysis. Eight patients had sampling of gut biopsies at both visits.
IHC staining and quantitative image analysis (QIA) for myeloperoxidase (MPO) were performed as previously described [27] and are detailed in the Supplementary Data.
Metabolomic Analysis
The plasma metabolic profile of a subset of 17 AHI individuals’ paired samples pre- and post-ART (F1-5) was assessed. Data were acquired at Metabolon, as previously described [31], and are detailed in the Supplementary Data and [30].
Statistical Analyses
In this study, we performed both cross-sectional and paired analysis in order to maximize utilization of all available data. Alpha and beta diversity were calculated using the phyloseq (version 1.19.1) package in R. Boxplots and other visualizations were created using the ggplot2 package in R (https://www.r-project.org). A PERMANOVA test was performed on beta diversity distance matrices using the Adonis function in the vegan package with 999 permutations. The relative abundance of bacterial taxa was calculated based on the total operational taxonomic units (OTUs) of each sample. Subsequenctly, families from all samples were ranked based on their average relative abundance. The top 30 most abundant bacterial families were selected to test for differences between AHI pre-ART (n = 37), post-ART (n = 31), and HIV-uninfected controls (n = 7) using the Mann-Whitney nonparametric test. Two-way P values were calculated, and statistical significance was defined as P < .05. Linear discriminant analysis effect size (LEfSe), based on the nonparametric Kruskal Wallis rank test, was used to detect features with significant differential bacterial abundance beween AHI pre- and post-ART. The paired Wilcoxon signed rank test was also applied to identify OTUs with differential abundance between AHI who had both pre- and post-ART samples (n = 16).
The top 20 most abundant families were imported in Spice [33] to create the pie charts for comparisons of bacterial taxa in HIV-uninfected controls (n = 7), HIV+ pre-ART (n = 37), and HIV+ post-ART (n = 31; permutation test, 20 000 iterations). Statistical significance was defined as P < .05.
Associations between markers of tissue or systemic inflammation (sCD14, IL-6, CRP) and intestinal gut epithelial integrity (I-FABP) and bacterial taxa were assessed using Spearman’s rho nonparametric test in AHI pre- (n = 37) and post-ART (n = 31).
Statistical significance of polymorphonuclear neutrophil (PMN) infiltration in AHI pre-ART (n = 37) compared with post-ART (n = 31) was assessed using Prism, version 8.0, and 2-sided P values <.05 were considered significant.
For statistical analyses and data display of plasma metabolites, any missing values were assumed to be below the limits of detection; these values were imputed with the compound minimum (minimum value imputation); data for each metabolite were also median-scaled for display. Statistical tests were performed in ArrayStudio (Omicsoft) to compare data between experimental groups; P < .05 was considered significant. An estimate of the false discovery rate (q-value) was also calculated, taking into account the multiple comparisons that normally occur in metabolomic-based studies, with q < 0.05 used as an indication of high confidence in a result. Principal coordinates analysis (PCoA) was performed using the ggbiplot, vegan, and pairwise adonis packages in R (https://www.r-project.org).
RESULTS
Mild Dysbiosis Observed in Early Acute HIV Infection
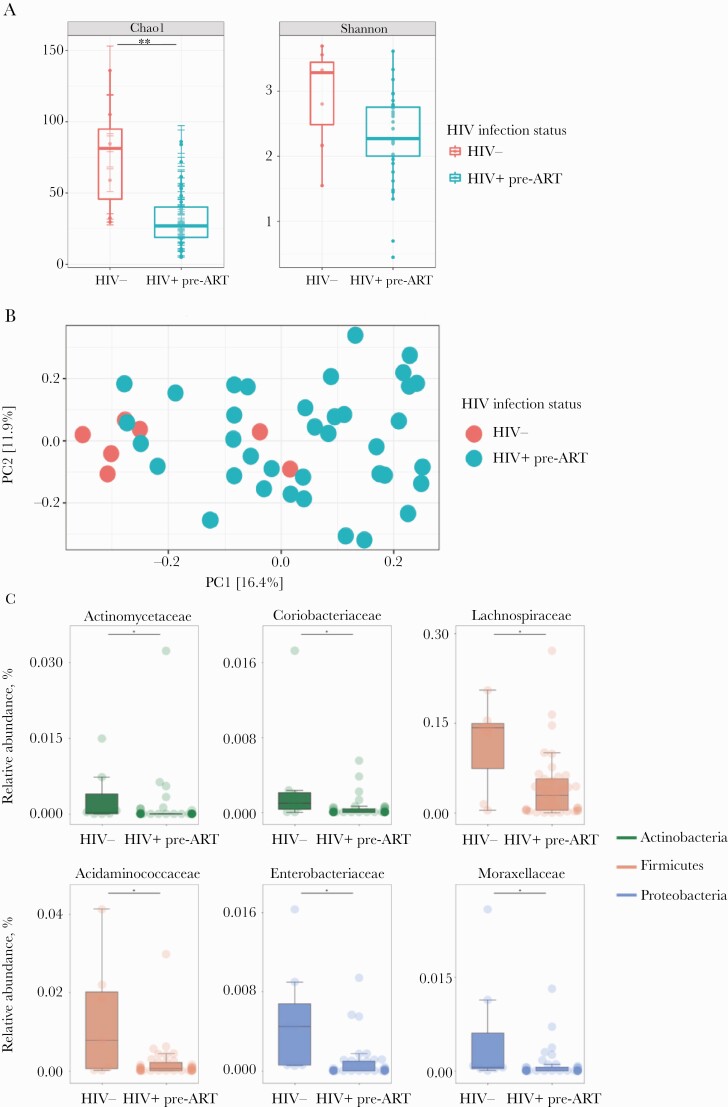
Study participants were MSM with a median age of 28 years. The median CD4 T-lymphocyte count was 386 cells/µL at baseline and 600 cells/µL after 6 months of ART. Median plasma HIV RNA was 334 889 copies/mL at baseline and <50 copies/mL after 6 months of ART (Table 1). To examine whether alteration in the bacterial composition of the fecal microbiota (dysbiosis) was established during the very early acute phase of HIV infection, we first compared AHI participants pre-ART (n = 37) with HIV-uninfected controls (n = 7). HIV infection was associated with a decrease in α-diversity (P = .003, Chao1; P = .06, Shannon Index) (Figure 1A). PCoA revealed clustering of AHI pre-ART and HIV-uninfected controls, which was verified by permutational multivariate analysis of variance (P = .001, PERMANOVA) (Figure 1B). We then evaluated the relative abundance of bacterial taxa belonging to the main phyla. Among the 20 top abundant families, the relative abundance of Actinomycetaceae, Coriobacteriaceae, Acidaminococcacea, and Lachnospiraceae were significantly lower in AHI pre-ART compared with HIV-uninfected controls (P < .005), in agreement with previous data that showed a decrease in Lachnospiraceae in chronically HIV-infected individuals [16, 18, 20–23]. Members of the Proteobacteria phylum, such as Enterobacteriaceae, and Moraxellaceae—previously reported enriched in chronically HIV-infected individuals [16–19, 21–23]—were more abundant in HIV-uninfected controls compared with AHI pre-ART (Figure 1C). Several recent studies [16, 18, 19, 21, 22] have shown a linkage between Prevotella abundance and HIV infection–related inflammation, whereas others have reported a Prevotella-rich microbiota associated with sexual practice, rather than with HIV infection status per se [34]. In our cohort, we corroborated this observation and found no differences in the relative abundance of Prevotellaceae in MSM HIV-uninfected controls and AHI MSM individuals pre-ART (Supplementary Figure 1). Overall, these data suggest that a mild dysbiosis is present during early acute HIV infection.
Table 1.
Study Participants Characteristics
| HIV-Positive Individuals | |||
|---|---|---|---|
| Pre-ART | Post-ART | HIV-Seronegative Controls | |
| No. of subjects | 52a | 7 | |
| 37 | 31 | ||
| Fiebig Stage at diagnosis | - | ||
| 1 | 5 | 3 | |
| 2 | 10 | 10 | |
| 3 | 17 | 16 | |
| 4 | 2 | - | |
| 5 | 3 | 2 | |
| Median age (IQR), y | 28 | 29 | |
| (23.0–34.0) | (22.0–32.0) | (23.5–34.9) | |
| Sex | Male | Male | |
| Risk | MSM | MSM | |
| Median CD4 count (IQR), cells/µL | 386 (267.0–542.5) | 600 (556.0–864.0) | - |
| Median viral load (IQR), HIV-1 RNA copies/mL | 334 889 (161 770–1 042 075) | <50 | - |
| cART | 2NRTI + efavirenz | - | |
Abbreviations: ART, antiretroviral therapy; cART, combined antiretroviral therapy; IQR, interquartile range; MSM, men who have sex with men.
aSixteen had paired pre- and post-ART samples.
Figure 1.
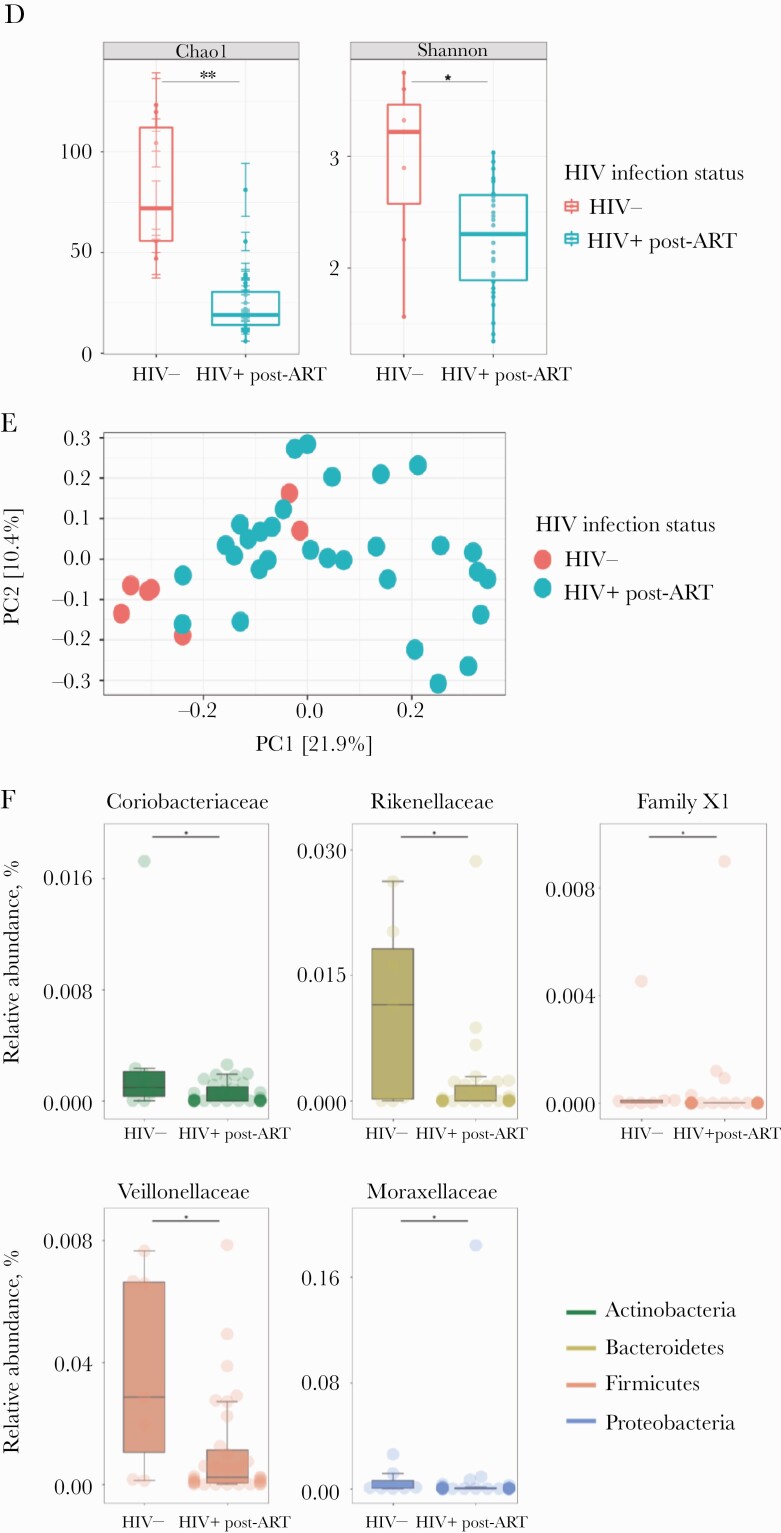
Stool bacterial community shifts in HIV-infected vs HIV-uninfected controls. A, Alpha diversity in HIV+ pre–antiretroviral therapy (ART) vs HIV-uninfected controls by Chai1 Index (P = .003) and Shannon Index (P = .06). B, Principal coordinates analysis (PCoA) shows clustering of acute HIV infection (AHI) pre-ART and HIV-uninfected controls (P = .001, PERMANOVA). C, Top 30 most abundant families in HIV- and HIV+ pre-ART measured and compared using the Mann-Whitney test (P < .05). The color of each family corresponds with the color of its respective phylum. D, Alpha diversity in HIV+ post-ART vs HIV-uninfected controls by Chai1 Index (P = .002) and Shannon Index (P = .05). E, PCoA shows clustering of AHI post-ART and HIV-uninfected controls (P = .001, PERMANOVA). F, Top 30 most abundant families in HIV- and HIV+ post-ART measured and compared using the Mann-Whitney test (P < .05). The color of each family corresponds with the color of its respective phylum.
Effects of Early Antiretroviral Therapy on the Gut Microbiota
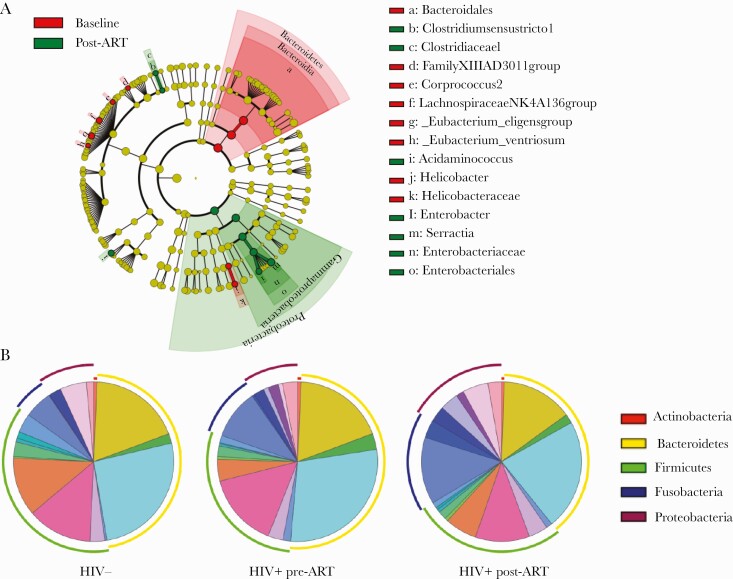
The effects of ART for 6 months on the fecal microbiota of acute HIV participants was then evaluated. Alpha diversity was significantly lower in AHI post-ART (n = 31) compared with HIV-uninfected controls (n = 7; P = .002, Chao1; P = .05, Shannon Index) (Figure 1D). PCoA revealed clustering of AHI post-ART and HIV-uninfected controls, which was verified by permutational multivariate analysis of variance (P = .001, PERMANOVA) (Figure 1E). The nonparametric Mann-Whitney test was then used to identify gut bacterial taxa that differed in abundance between AHI post-ART and HIV-uninfected controls. Among the bacterial taxa depleted in AHI post-ART were Rikenellaceae members, whose frequency has been previously observed to be reduced in chronically HIV-infected individuals [17–21], Coriobacteriaceae, family XI, and Veillonellaceae. Moraxelaceae members were still enriched in HIV-uninfected controls compared with AHI post-ART (Figure 1F). The linear discriminative analysis effect size biomarker discovery tool was subsequently used in unpaired samples to investigate the potential divergence of gut bacterial communities between the pre-ART (n = 37) and post-ART (n = 31) groups. The LEfSe algorithm identified enrichment in Enterobacteriaceae members in AHI post-ART compared with pre-ART, whereas Bacteroidales members were more abundant in AHI pre-ART compared with post-ART (Figure 2A). Although we observed minor changes in the relative abundance of specific bacterial taxa between HIV-uninfected controls, AHI pre-ART, and AHI post-ART, overall gut microbial composition did not appear dramatically different among the 3 groups (permutation test: HIV- vs HIV+ pre-ART, P = .6; HIV- vs HIV+ post-ART, P = .4; HIV+ pre-ART vs HIV+ post-ART, P = .3) (Figure 2B).
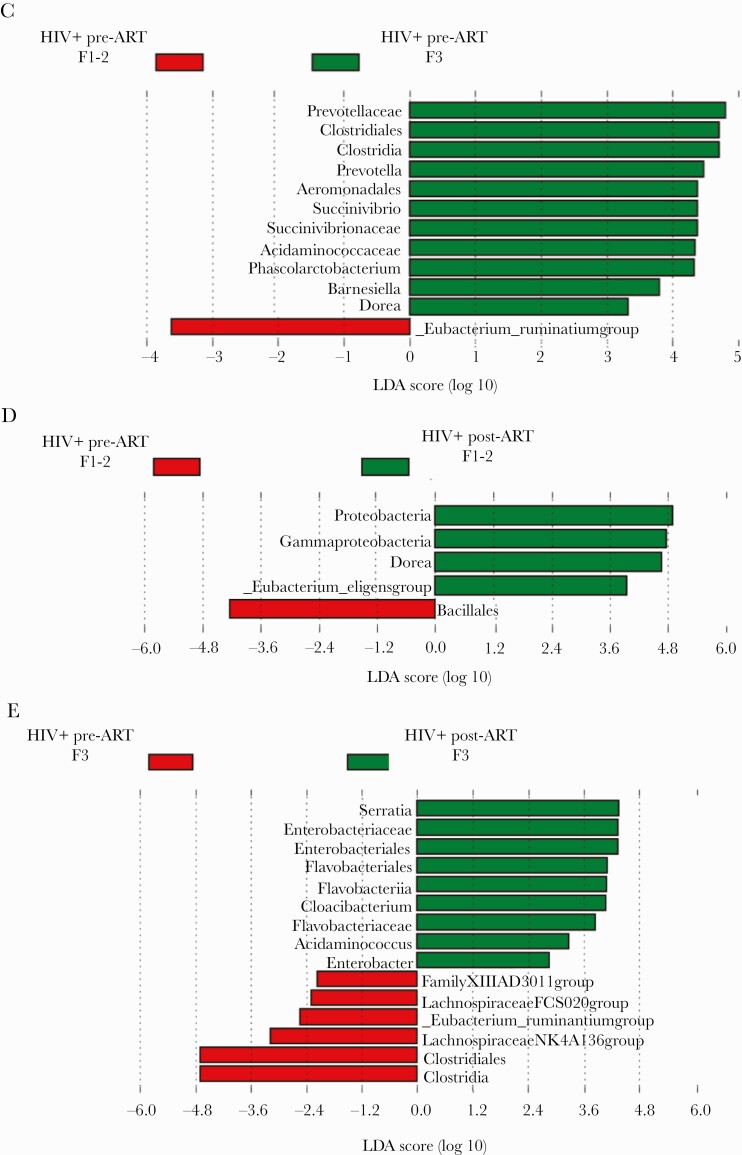
Figure 2.
Gut bacterial changes in HIV+ pre–antiretroviral therapy (ART) and HIV+ post-ART. A, Cladogram of linear discriminant analysis (LDA) effect size highlights discriminatory taxa, with an LDA score >2, between HIV+ pre-ART and HIV+ post-ART. B, Percentage of the top 20 most abundant families in HIV-, HIV+ pre-ART, and HIV+ post-ART (permutation test: HIV- vs HIV+ pre-ART, P = .6; HIV- vs HIV+ post-ART, P = .4; HIV+ pre-ART vs HIV+ post-ART, P = .3). C, LDA scores of differentially abundant taxa among HIV+ pre-ART at F1-2 and HIV+ pre-ART at F3. D, LDA scores of differentially abundant taxa among HIV+ pre-ART at F1-2 and HIV+ post-ART at F1-2. E, LDA scores of differentially abundant taxa among HIV+ pre-ART at F3 and HIV+ post-ART at F3.
To explore if and how the microbiome changes with progression of HIV infection, we compared AHI at the time of diagnosis at Fiebig Stages 1 and 2 (n = 15) vs F3 (n = 17). Linear discriminant analysis scores of differentially abundant taxa in AHI individuals at F1-2 vs F3 pre-ART showed a higher representation of pro-inflammatory bacterial taxa, such as Succinivibrionaceae and Prevotellaceae, in F3 (Figure 2C). Comparison of F1-2 pre-ART (n = 15) vs F1-2 post-ART (n = 13) (Figure 2D), and F3 pre-ART (n = 17) vs F3 post-ART (n = 16) (Figure 2E), showed an increase in Proteobacteria members post-ART, indicating a continuous change in the microbiome under ART. Further analysis was then performed on paired samples, AHI pre-ART (n = 16) vs AHI post-ART (n = 16), to evaluate longitudinally potential microbiota-associated changes within these subjects. As assessed by Wilcoxon signed rank test, there was a significant decrease in Bacteroidetes (P = .007) and an increase in Fusobacteria frequencies (P = .01) in AHI post-ART compared with pre-ART. Proteobacteria and Firmicutes frequencies did not appear to change in AHI post-ART compared with pre-ART (P = .5 and P = .7, respectively) (Supplementary Figure 2). Overall these data reveal a slight shift in the frequencies of bacterial communities within the 3 groups, which is partially restored after 6 months of ART.
Associations of GI Tissue Inflammation and Inflammatory Markers With Bacterial Taxa
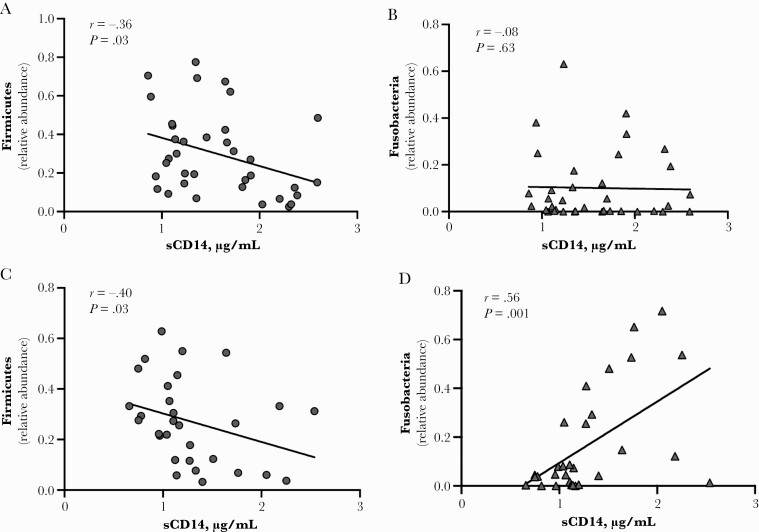
Given that the microbiota of AHI individuals were enriched or depleted in specific bacterial taxa, we assessed potential correlations between the relative abundance of these taxa and markers of tissue or systemic inflammation (sCD14, IL-6, CRP) and intestinal gut epithelial integrity (I-FABP) (Supplementary Table 1). The median sCD14 level in AHI individuals at baseline (n = 37) was 1.40 µg/mL and decreased to 1.14 µg/mL after 6 months of ART (n = 31). Levels of sCD14 inversely correlated with the relative abundance of Firmicutes both pre-ART (r = –.36; P = .03) (Figure 3A) and after 6 months of ART (r = –.40; P = .03) (Figure 3C). No association was observed between Fusobacteria and sCD14 at baseline (r = –.08; P = .63) (Figure 3B); however, after 6 months of treatment, their relative abundance increased and positively correlated with sCD14 levels (r = .55; P = .001) (Figure 3D). These data suggest that a potential relationship between altered gut bacterial communities and systemic inflammation is established during the acute phase of HIV infection and may persist during ART.
Figure 3.
Bacterial taxa frequencies and inflammatory markers. A, Relative abundance of Firmicutes in HIV+ pre–antiretroviral therapy (ART) negatively correlated with sCD14 (r = –.36; P = .03). B, Relative abundance of Fusobacteria in HIV+ pre-ART do not correlate with sCD14 (r = –.08; P = .63). C, Relative abundance of Firmicutes in HIV+ post-ART negatively correlated with sCD14 (r = –.40; P = .03). D, Relative abundance of Fusobacteria in HIV+ post-ART positively correlated with sCD14 (r = .56; P = .001).
It has been previously demonstrated that a local tissue response consisting of infiltration of PMNs in the lamina propria (LP) of the GI tract is a surrogate marker for epithelial damage and local microbial translocation [4]. Using immunohistochemistry and quantitative image analysis, we assessed the extent of PMN infiltration by measuring myeloperoxidase+ (MPO+) within the LP in AHI pre-ART and AHI post-ART. Cross-sectional analysis revealed no significant changes in PMN infiltration in AHI pre-ART (n = 23) compared with post-ART (n = 14; P = .4, Mann-Whitney test). Paired analysis of pre- vs post-ART (n = 8) also showed no significant changes in PMN infiltration (P = .5, Wilcoxon rank test), as reported in Supplementary Figure 3A and B.
Metabolomics Analysis
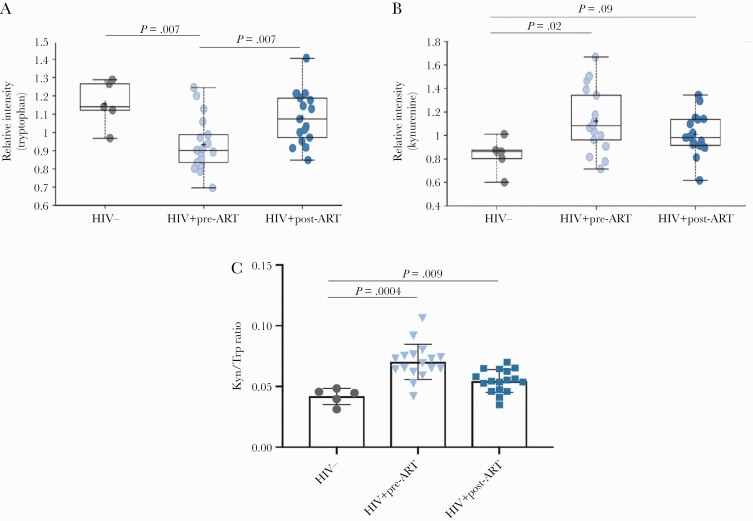
The plasma metabolic profile was assessed in a subset of 17 AHI participants who had paired samples at the time of diagnosis and after 6 months of ART and in 5 HIV-uninfected controls. PCoA revealed clustering of HIV-uninfected controls and HIV+ pre- and post-ART that was verified by pairwise Adonis Euclidean matrices (HIV- vs HIV+ pre-ART, P = .01; HIV- vs HIV+ post-ART, P = .04; HIV+ pre-ART vs HIV+ post-ART, P = .06) (Supplementary Figure 4). A paired t test and Welch’s 2-sample t test were used to identify biochemicals that differed significantly between experimental groups. AHI at the time of diagnosis exhibited several alterations in aminoacid metabolism when compared with the HIV-uninfected controls. Among the aminoacids found to be repressed in AHI pre-ART compared with HIV-uninfected controls, tryptophan was detected (P = .007; q = .004) (Figure 4A). After 6 months of ART, tryptophan levels increased compared with the baseline (P = .007; q = .003), and no significant difference was observed between AHI post-ART and HIV-uninfected controls (P = .28; q = .14). Furthermore, AHI pre-ART displayed significant higher levels of kynurenine compared with HIV-uninfected controls (P = .02; q = .07), whereas no significant difference was detected between HIV+ post-ART and HIV-uninfected controls (P = .09; q = .06) (Figure 4B). Kynurenine is a metabolite that can be produced from tryptophan via the enzyme indoleamine 2,3 dioxygenase (IDO) under proinflammatory stimuli [35]. Interestingly, the IDO1 activity measured by the ratio of plasma concentration of the downstream product, kynurenine, to the parent compound, tryptophan (Kyn:Trp), was significantly higher in AHI pre-ART compared to HIV-uninfected controls (P = .0004) and in AHI post-ART compared to HIV-uninfected controls (P = .009). The Kyn/Trp ratio did not differ significantly between AHI pre-ART and AHI post-ART (P = .1) (Figure 4C). AHI post-ART showed changes in many metabolites with links to microbial activity including several secondary bile acids and metabolites that can be generated via microbial encoded deconjugation/dehydroxylation reactions in the intestine by gut bacteria (Supplementary Table 2).
Figure 4.
Perturbation in amino acid abundance and utilization in HIV-infected vs HIV-uninfected controls. A, Relative intensity of tryptophan in HIV- vs HIV+ pre–antiretroviral therapy (ART; P = .007; q = .04) and HIV+ pre- vs post-ART (P = .007; q = .03). B, Relative intensity of kynurenine in HIV- vs HIV+ pre-ART (P = .023; q = .07) and in HIV- vs HIV+ post-ART (P = .09; q = .06). C, Kyn/Trp ratio significantly differs between HIV- and HIV+ pre-ART (P = .0004) and between HIV- and HIV+ post-ART (P = .009).
DISCUSSION
HIV infection is associated with an altered composition of the fecal microbiota, yet questions remain about how quickly bacterial dysbiosis is established and how antiretroviral treatment can further shape gut microbial communities. Several recent studies have investigated the composition of the gut microbiota in HIV-infected individuals and its potential role in the pathogenesis of the infection. The majority of these studies, however, were cross-sectional and focused on chronically HIV-infected individuals, and controls were not always well matched for confounding variables [16–21]. Interestingly, longitudinal studies using simian immunodeficiency virus (SIV) nonhuman primate (NHP) models have shown that dysbiosis is not consistently observed [36]. Furthermore, experimentally induced intestinal dysbiosis in SIV-infected macaques did not accelerate disease progression [37].
In this study, we reported differences in the composition of the gut microbiota from a longitudinal study of Thai MSM with AHI at the time of HIV diagnosis (F1-5) and after 6 months of ART with ethnicity-, age-, and sex-matched HIV-uninfected individuals as controls. Our data show a dysbiotic gut environment establishing at the earliest acute phase of HIV infection. Many studies have shown an increase in the proportion of Proteobacteria phylum, in particular Enterobacteriaceae members, in treated and untreated chronically HIV-infected individuals compared with HIV-uninfected controls [16–19, 21–23]. Interestingly, we observed a slightly higher abundance of Proteobacteria members in HIV-uninfected controls compared with AHI pre-ART, which could be attributed to the fact that we studied MSM with AHI with age- and sex-matched HIV-uninfected controls and that Thai individuals may have distinct genetics or dietary habits; however, the small sample size precludes definitive conclusions.
After 6 months of ART, we observed an increase in the relative abundance of Proteobacteria phylum, particularly in Enterobacteriaceae, compared with the time of AHI diagnosis, which may suggest a partial restoration of the microbiota. Furthermore, we did not find any significant correlations between Proteobacteria members and markers of systemic inflammation. These data suggest that a modest increase in the relative abundance of Proteobacteria phylum members is not necessarily associated with disease progression.
Consistent with previous observations [20, 21, 34], the relative abundance of Fusobacteria phylum members, which include many potential inflammagenic pathogens, was higher in AHI individuals at the time of diagnosis compared with HIV-uninfected controls. After 6 months of ART, these bacterial taxa positively correlated with levels of sCD14, a well-established marker of disease progression in AHI, which we have recently reported to remain elevated despite early treatment [6, 38, 39]. This may suggest a potential relationship between altered gut bacterial communities and systemic inflammation. Gut tissue inflammation measured by MPO did not change significantly after 6 months of ART, consistent with persistence of local tissue inflammation.
In addition, we investigated the composition of the gut microbiota through stages F1-3 of acute infection, which we previously found to be linked to a progressive loss of CD4, in particular Th17, that occurred predominantly in stage F3 [27]. Our findings suggested an enrichment in pro-inflammatory bacteria during the transition from F1 to F3, which seems to correspond with the progression of disease; however, our small sample size precludes definitive conclusions.
Evaluation of plasma metabolites revealed significant changes in the secondary bile acids linked with microbial activity among AHI pre- and post-ART and HIV-uninfected controls. Bile acids are crucial metabolites in the gastrointestinal tract and contribute to maintaining intestinal immune homeostasis through cross-talk with the gut bacterial residents. The conversion of bile acids by the gut microbiome is identified as a factor impacting both host metabolism and immune responses [40].
The IDO1, the first enzyme of the kynurenine pathway, was significantly higher in AHI pre-ART compared with HIV-uninfected controls and AHI post-ART. Previous studies in treated HIV-infected individuals have shown that dysbiosis characterized by an enrichment in Proteobacteria and a depletion in Bacteroidia correlated with activity of the kynurenine pathway of tryptophan metabolism [18]. We did not observe significant correlations between specific gut bacterial taxa and these metabolites; however, further analysis would be critical to better elucidate the linkage between gut dysbiosis with secondary bile acid abundance and with activity of the kynurenine pathway of tryptophan metabolism during HIV infection.
It is important to note that our study had several limitations, including the small sample size and the lack of HIV controls receiving antiretrovirals, people on pre-exposure prophylaxis, to better decipher the role of medications in microbiome alterations. Metabolomics analysis was conducted only in the plasma and not in the gut. Despite these limitations, our study provided a unique view of microbiome composition during the earliest phases of acute infection in a highly homogeneous and well-defined group of participants, with longitudinal sampling.
In summary, in this study of the microbiome in early acute HIV infection, we found evidence of dysbiosis that was only partially restored after 6 months of ART. Overall, dysbiosis was characterized by subtler, rather than profound, individual changes in the bacterial structure of the gut microbiota, but significant correlations between pathogenic taxa and markers of HIV disease progression were evident. These findings warrant further investigation on the potential role of microbiota alterations in HIV-associated inflammation as well as the potential effects of specific drugs used to treat HIV.
Data Availability
Raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under the BioProject ID number PRJNA510435.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The study team is grateful to the individuals who volunteered to participate in this study, the staff at the Thai Red Cross AIDS Research Centre, and the Armed Forces Research Institute of Medical Sciences.
Financial support. The study was supported by the intramural research program of National Institute of Allergy and Infectious Diseases/National Insitutes of Health and the National Institute of Allergy and Infectious Diseases Microbiome Program. This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The RV254 study was supported by cooperative agreements (W81XWH-07-2-0067, W81XWH-11-2-0174) between The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the US Department of the Army and by an intramural grant from the Thai Red Cross AIDS Research Center. The Thai Government Pharmaceutical Organizations, Gilead, Merck, and ViiV Healthcare provided support for antiretroviral medications.
Disclaimer. The views expressed are those of the authors and should not be construed to represent the positions of the US Army, the Department of Defense, or the Department of Health and Human Services. The investigators have adhered to the policies for protection of human subjects as prescribed in AR-70. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Potential conflicts of interest. J.A. has received honoraria for participating in advisory meetings for ViiV Healthcare, Merck, AbbVie, Gilead, and Roche. The authors declare that they have no competing interests. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. I.S., J.B., J.A., J.B. designed the study. O.S., J.D., B.I., performed experiments. O.S., A.M.O., M.Q., H.M., S.P., B.I., C.D. performed data analysis. N.P., N.C., A.S. performed samples and data collection. O.S., I.S. wrote a draft of the manuscript that was reviewed and enriched by contributions from all authors.
The RV254/SEARCH 010 Study Group team members. From SEARCH/TRC-ARC/HIV-NAT: Nipat Teeratakulpisarn, Supanit Pattanachaiwit, Mark de Souza, James Fletcher, Eugene Kroon, Ponpen Tantivitayakul, Duanghathai Suttichom, Somprartthana Rattanamanee, Kultida Poltavee, Jintana Intasan, Tassanee Luekasemsuk, Hathairat Savadsuk, Somporn Tipsuk, Suwanna Puttamsawin, Khunthalee Benjapornpong, Nisakorn Ratnaratorn, Patcharin Eamyoung, Sasiwimol Ubolyam; from AFRIMS: Robert O’Connell, Siriwat Akapirat, Bessara Nuntapinit, Nantana Tantibul, Nampueng Churikanont, Saowanit Getchalarat, Sandhya Vasan, Rapee Trichavaroj, Chayada Sajiaweerawan, Yuwadee Phuang-Ngern, Surat Jongrakthaitae, Suchada Sukhumvittaya, Putida Saetun, Weerawan Chuenarom; MHRP: Nelson Michael, Ellen Turk, Corinne McCullough, Oratai Butterworth, Mark Milazzo. We also thank the National Institute of Allergy and Infectious Diseases Microbiome Program and sequencing facility.
Contributor Information
Ornella Sortino, Clinical Research Directorate, Frederick National Laboratory for Cancer Research, sponsored by the National Cancer Institute.
Nittaya Phanuphak, SEARCH/Thai Red Cross AIDS Research Centre, Bangkok, Thailand.
Alexandra Schuetz, The Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, Maryland; United States Military HIV Research Program, Walter Reed Army Institute of Research, Silver Spring, Maryland; Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Alexandra M Ortiz, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Nitiya Chomchey, SEARCH/Thai Red Cross AIDS Research Centre, Bangkok, Thailand.
Yasmine Belkaid, Metaorganism Immunity Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland; Microbiome Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Jacquice Davis, Microbiome Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Harry A Mystakelis, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Mariam Quiñones, Bioinformatics and Computational Biosciences Branch, Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Claire Deleage, AIDS and Cancer Virus Program, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research Inc., Frederick, Maryland, USA.
Brian Ingram, Metabolon, Inc., Research Triangle Park, North Carolina.
Rungsun Rerknimitr, SEARCH/Thai Red Cross AIDS Research Centre, Bangkok, Thailand.
Suteeraporn Pinyakorn, The Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, Maryland; United States Military HIV Research Program, Walter Reed Army Institute of Research, Silver Spring, Maryland.
Adam Rupert, Clinical Research Directorate, Frederick National Laboratory for Cancer Research, sponsored by the National Cancer Institute.
Merlin L Robb, The Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, Maryland; United States Military HIV Research Program, Walter Reed Army Institute of Research, Silver Spring, Maryland.
Jintanat Ananworanich, The Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, Maryland; United States Military HIV Research Program, Walter Reed Army Institute of Research, Silver Spring, Maryland; Department of Global Health, University of Amsterdam, Amsterdam, the Netherlands.
Jason Brenchley, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Irini Sereti, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
RV254/SEARCH010 Study Group:
Nipat Teeratakulpisarn, Supanit Pattanachaiwit, Mark de Souza, James Fletcher, Eugene Kroon, Ponpen Tantivitayakul, Duanghathai Suttichom, Somprartthana Rattanamanee, Kultida Poltavee, Jintana Intasan, Tassanee Luekasemsuk, Hathairat Savadsuk, Somporn Tipsuk, Suwanna Puttamsawin, Khunthalee Benjapornpong, Nisakorn Ratnaratorn, Patcharin Eamyoung, Sasiwimol Ubolyam, Robert O’Connell, Siriwat Akapirat, Bessara Nuntapinit, Nantana Tantibul, Nampueng Churikanont, Saowanit Getchalarat, Sandhya Vasan, Rapee Trichavaroj, Chayada Sajiaweerawan, Yuwadee Phuang-Ngern, Surat Jongrakthaitae, Suchada Sukhumvittaya, Putida Saetun, Weerawan Chuenarom, Nelson Michael, Ellen Turk, Corinne McCullough, Oratai Butterworth, and Mark Milazzo
References
- 1. Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol 2008; 1:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenchley JM, Paiardini M, Knox KS, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 2008; 112:2826–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cecchinato V, Trindade CJ, Laurence A, et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol 2008; 1:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Estes JD, Harris LD, Klatt NR, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog 2010; 6:e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006; 12:1365–71. [DOI] [PubMed] [Google Scholar]
- 6. Sandler NG, Wand H, Roque A, et al. ; INSIGHT SMART Study Group . Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 2011; 203:780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pandrea I, Gaufin T, Brenchley JM, et al. Cutting edge: experimentally induced immune activation in natural hosts of simian immunodeficiency virus induces significant increases in viral replication and CD4+ T cell depletion. J Immunol 2008; 181:6687–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paiardini M, Frank I, Pandrea I, et al. Mucosal immune dysfunction in AIDS pathogenesis. AIDS Rev 2008; 10:36–46. [PubMed] [Google Scholar]
- 9. Krishnan S, Wilson EM, Sheikh V, et al. Evidence for innate immune system activation in HIV type 1-infected elite controllers. J Infect Dis 2014; 209:931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilson EM, Sereti I. Immune restoration after antiretroviral therapy: the pitfalls of hasty or incomplete repairs. Immunol Rev 2013; 254:343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Viaud S, Daillère R, Boneca IG, et al. Harnessing the intestinal microbiome for optimal therapeutic immunomodulation. Cancer Res 2014; 74:4217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsai F, Coyle WJ. The microbiome and obesity: is obesity linked to our gut flora? Curr Gastroenterol Rep 2009; 11:307–13. [DOI] [PubMed] [Google Scholar]
- 13. Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014; 146:1513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saxena D, Li Y, Yang L, et al. Human microbiome and HIV/AIDS. Curr HIV/AIDS Rep 2012; 9:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salas JT, Chang TL. Microbiome in human immunodeficiency virus infection. Clin Lab Med 2014; 34:733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dillon SM, Lee EJ, Kotter CV, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dinh DM, Volpe GE, Duffalo C, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015; 211:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013; 5:193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McHardy IH, Li X, Tong M, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8:760–72. [DOI] [PubMed] [Google Scholar]
- 23. Monaco CL, Gootenberg DB, Zhao G, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016; 19:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ortiz AM, Brenchley JM. Microbial translocation: translating simian immunodeficiency virus to HIV. Curr Opin HIV AIDS 2018; 13:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity 2010; 32:737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schuetz A, Deleage C, Sereti I, et al. ; RV254/SEARCH 010 and RV304/SEARCH 013 Study Groups . Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation. PLoS Pathog 2014; 10:e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deleage C, Schuetz A, Alvord WG, et al. Impact of early cART in the gut during acute HIV infection. JCI Insight 2016; 1:e87065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sereti I, Estes JD, Thompson WL, et al. Decreases in colonic and systemic inflammation in chronic HIV infection after IL-7 administration. PLoS Pathog 2014; 10:e1003890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Souza MS, Phanuphak N, Pinyakorn S, et al. ; RV254SEARCH 010 Study Group . Impact of nucleic acid testing relative to antigen/antibody combination immunoassay on the detection of acute HIV infection. AIDS 2015; 29:793–800. [DOI] [PubMed] [Google Scholar]
- 30. Weber N, Liou D, Dommer J, et al. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 2018; 34:1411–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long T, Hicks M, Yu HC, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet 2017; 49:568–78. [DOI] [PubMed] [Google Scholar]
- 32. Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2010; 2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 2011; 79:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noguera-Julian M, Rocafort M, Guillén Y, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016; 5:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y, Guillemin GJ. Kynurenine pathway metabolites in humans: disease and healthy states. Int J Tryptophan Res 2009; 2:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mudd JC, Brenchley JM. Gut mucosal barrier dysfunction, microbial dysbiosis, and their role in HIV-1 disease progression. J Infect Dis 2016; 214(Suppl 2):S58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ortiz AM, Flynn JK, DiNapoli SR, et al. Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat Med 2018; 24:1313–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hunt PW, Sinclair E, Rodriguez B, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 2014; 210:1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sereti I, Krebs SJ, Phanuphak N, et al. ; RV254/SEARCH 010, RV304/SEARCH 013 and SEARCH 011 protocol teams . Persistent, albeit reduced, chronic inflammation in persons starting antiretroviral therapy in acute HIV infection. Clin Infect Dis 2017; 64:124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 2016; 24:41–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under the BioProject ID number PRJNA510435.