Summary
Methyl-CpG binding protein 2 (MeCP2) has historically been linked to heterochromatin organization, and in mouse cells it accumulates at pericentric heterochromatin (PCH), closely following major satellite (MajSat) DNA distribution. However, little is known about the specific function of MeCP2 in these regions. We describe the first evidence of a role in neurons for MeCP2 and MajSat forward (MajSat-fw) RNA in reciprocal targeting to PCH through their physical interaction. Moreover, MeCP2 contributes to maintenance of PCH by promoting deposition of H3K9me3 and H4K20me3. We highlight that the MeCP2B isoform is required for correct higher-order PCH organization, and underline involvement of the methyl-binding and transcriptional repression domains. The T158 residue, which is commonly mutated in Rett patients, is directly involved in this process. Our findings support the hypothesis that MeCP2 and the MajSat-fw transcript are mutually dependent for PCH organization, and contribute to clarify MeCP2 function in the regulation of chromatin architecture.
Keywords: chromocenter clustering, major satellite transcripts, MeCP2, pericentric heterochromatin, Rett syndrome
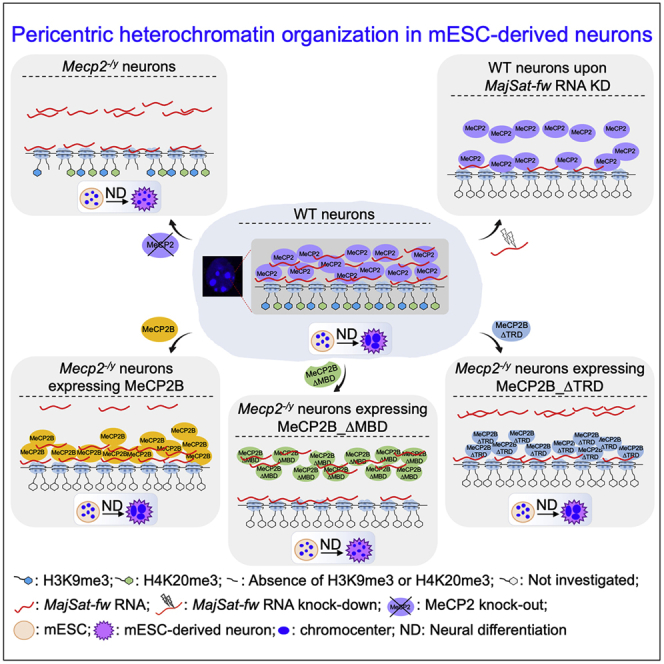
Graphical Abstract
Highlights
-
•
MeCP2 and MajSat-fw transcript targeting to chromocenters is mutually dependent
-
•
MeCP2B and its MBD domain are required for correct higher-order PCH organization
-
•
The TRD domain of the MeCP2 protein binds the MajSat-fw transcript
-
•
The Rett syndrome T158M mutation of MeCP2 impairs higher-order PCH organization
Della Ragione and colleagues describe a novel function of MeCP2 and MajSat forward transcript in terms of their reciprocal targeting to chromocenters in neurons through their physical interaction. Moreover, MeCP2B has a prominent role in higher-order PCH organization, with both its MBD and TRD involved in this process. These findings further clarify MeCP2 function in the regulation of chromatin architecture.
Introduction
Since its identification (Lewis et al., 1992), the number of biological functions ascribed to methyl-CpG binding protein 2 (MeCP2) has grown exponentially. The X-linked MECP2 gene is mutated in 95% of patients with Rett syndrome (RTT; OMIM 312750) (Amir et al., 1999; D'Esposito et al., 1996; Rett, 1966), a severe neurodevelopmental disease that affects mainly female individuals. Expression of MeCP2 increases during neuronal differentiation (Jung et al., 2003). Constitutive and brain-specific ablation of Mecp2 in mouse reproduces the RTT phenotypes and highlights the importance of MeCP2 for brain function (Guy et al., 2001).
The MECP2 gene encodes two splicing isoforms that are highly expressed in brain: MeCP2A and MeCP2B. These both have a methyl-binding domain (MBD) and a transcriptional repression domain (TRD), which, in turn, contains the NCoR/SMRT interaction domain (NID); they differ in terms of their N-terminus (Della Ragione et al., 2016). These MeCP2 isoforms show similar nuclear localization (Kumar et al., 2008); however, MeCP2B is the predominant form expressed in brain (Dragich et al., 2007) and appears to be relevant for RTT pathogenesis (Gianakopoulos et al., 2012). Moreover, MeCP2A and MeCP2B bind unique partners and show different genomic binding site preferences (Martinez de Paz et al., 2019).
The main function of MeCP2 has been linked to DNA methylation-mediated transcriptional repression (Klose and Bird, 2003), although it also activates gene expression in specific brain regions (Ben-Shachar et al., 2009; Chahrour et al., 2008). Multiple roles of MeCP2 might be due to interactions with different molecular partners (for review, see Della Ragione et al., 2016), such as histone deacetylases and switch-independent 3A (Sin3A) for transcriptional repression, or cAMP response element-binding protein (CREB 1) for gene activation.
MeCP2 has a role in genome-wide transcriptional silencing. In mouse neurons, it is important for repression of spurious transcription of repetitive elements, and its genomic binding tracks the methyl-CpG distribution (Skene et al., 2010). In mouse cells, MeCP2 accumulates at pericentric heterochromatin (PCH) (Lewis et al., 1992), which is constitutive heterochromatin organized in higher-order chromatin structures, known as chromocenters. These originate from aggregation of PCH of different chromosomes, undergo dynamic reorganization during differentiation, and are visible using DAPI staining (Almouzni and Probst, 2011). PCH is involved in repression of transposons, chromosomal segregation, genome integrity, and in-cis and in-trans silencing of euchromatic genes. In mouse, PCH is composed of hypermethylated 234-bp major satellite (MajSat) AT-rich repeats, which form complexes with hypo-acetylated and trimethylated histones H3 and H4 (e.g., H3K9me3, H4K20me3). These histones, in turn, promote anchoring of specific factors, such as members of the heterochromatin protein 1 (HP1) family and the cohesin complex, respectively (Fioriniello et al., 2020). The interactions between PCH regions appear to promote genomic compartmentalization in the nucleus (Falk et al., 2019), and recent findings support the importance of PCH in the formation of repressive compartments where silenced genes are positioned (Wijchers et al., 2015). However, whether MeCP2 directly regulates this process remains unclear. MeCP2 mediates chromocenter clustering during both myogenic and neural differentiation, with the contribution of α-thalassemia/mental retardation syndrome X-linked protein (ATRX) (Bertulat et al., 2012; Brero et al., 2005; Marano et al., 2019).
Interestingly, a role for RNAs in higher-order PCH organization has been highlighted (Maison et al., 2002, 2011; Marano et al., 2019; Park et al., 2018). Moreover, noncoding RNAs (ncRNAs) of heterogeneous lengths from both strands of MajSat DNA have been detected (Lehnertz et al., 2003), despite the heterochromatic nature of this genomic region. However, their biological significance remains unclear. MajSat RNA levels increase during neuronal differentiation in the developing mouse brain (Kishi et al., 2012). Also, they participate in the recruitment to PCH of scaffold attachment factor B (SAFB) (Huo et al., 2020) and suppressor of variegation 3-9 homolog (Suv39h), the enzyme responsible for trimethylation of H3K9 (Velazquez Camacho et al., 2017), and they have roles in chromocenter condensation during myogenic differentiation (Park et al., 2018). Moreover, at the two-cell stage during early mouse development, MajSat transcripts are crucial for developmental progression and formation of chromocenters. The functions of the two strands appear to be independent (Casanova et al., 2013; Probst et al., 2010), and the forward transcript (MajSat-fw) is directly involved in de novo targeting of small ubiquitin-like modifier (SUMO)-modified HP1α to PCH (Maison et al., 2011). Whether MajSat RNAs or other RNA components cooperate with MeCP2 for PCH organization is still elusive.
Here, we investigated the role of MajSat RNAs in MeCP2-mediated higher-order PCH organization in neurons. Furthermore, we dissected out the specific role of the MeCP2A and MeCP2B isoforms in this process, as well as the contribution of the MBD and TRD domains. Finally, we examined whether two common RTT mutations, T158M and R306C, affect PCH organization.
Results
MeCP2 Binding to Chromocenters Is Dependent on RNA Components
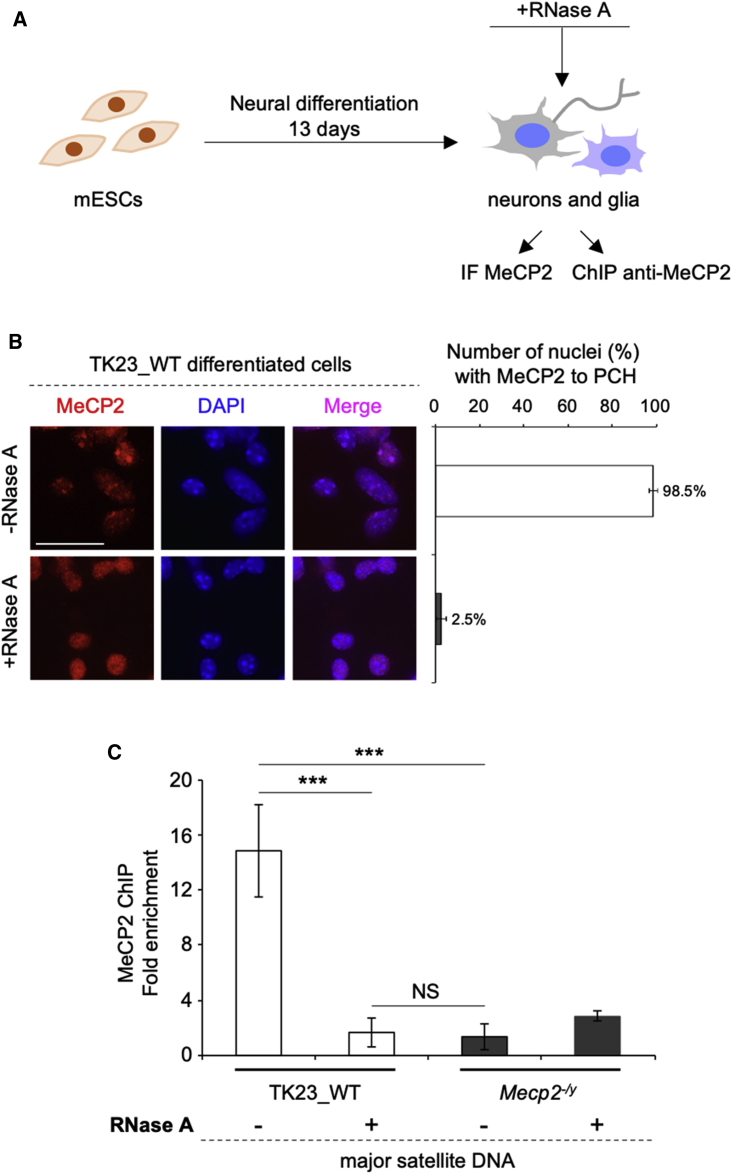
To determine whether an RNA component promotes accumulation of MeCP2 at PCH in neurons, we used TK23 wild-type (TK23_WT) and MeCP2 deficient (Mecp2−/y tEG; here as Mecp2−/y) murine embryonic stem cells (mESCs), which were previously used to demonstrate the role for MeCP2 in chromocenter clustering (Bertulat et al., 2012). Neural differentiation of these cells gives rise to a cell population that is enriched in neurons and astroglia (Bertulat et al., 2012; Fico et al., 2008) (Figure 1A). Immunofluorescence performed after RNase A treatment on terminally differentiated TK23_WT neurons showed that MeCP2 is dispersed in the nucleus, and thus loses its usual PCH accumulation, which was seen for only 2.5% of the nuclei. Conversely, in mock-treated TK23_WT neurons, the majority of the nuclei (98.5%) showed strong spotting of MeCP2 at chromocenters, as expected (Figure 1B). Of note, the dense DAPI staining that corresponds to chromocenters was still visible.
Figure 1.
MeCP2 Accumulation at PCH Depends on an RNA Component
(A) Scheme of the experimental procedure.
(B) Left: Representative immunofluorescence of MeCP2 in terminally differentiated TK23_WT neurons without and with RNase A treatment. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of MeCP2 enrichment at chromocenters without and with RNase A treatment. Data are means ± SD, with ≥100 cells analyzed per condition, from two independent experiments.
(C) ChIP–qPCR of MeCP2 at MajSat DNA in mESC-derived Mecp2−/y (negative control) and TK23_WT neurons without and with RNase A treatment. Data are means ± SD of four independent qPCR replicates from two ChIP experiments. ∗∗∗p < 0.001; NS, not significant (one-tailed Student’s t test).
MeCP2 strongly binds MajSat DNA in mESC-derived TK23_WT neurons, as shown by chromatin immunoprecipitation (ChIP); however, RNase A treatment displaced MeCP2 from these genomic regions (Figure 1C), thus confirming the immunofluorescence data. These findings suggest that an RNA component contributes to the recruitment of MeCP2 to PCH in mESC-derived neurons.
MeCP2 Contributes to Major Satellite Forward Transcript Targeting to Chromocenters
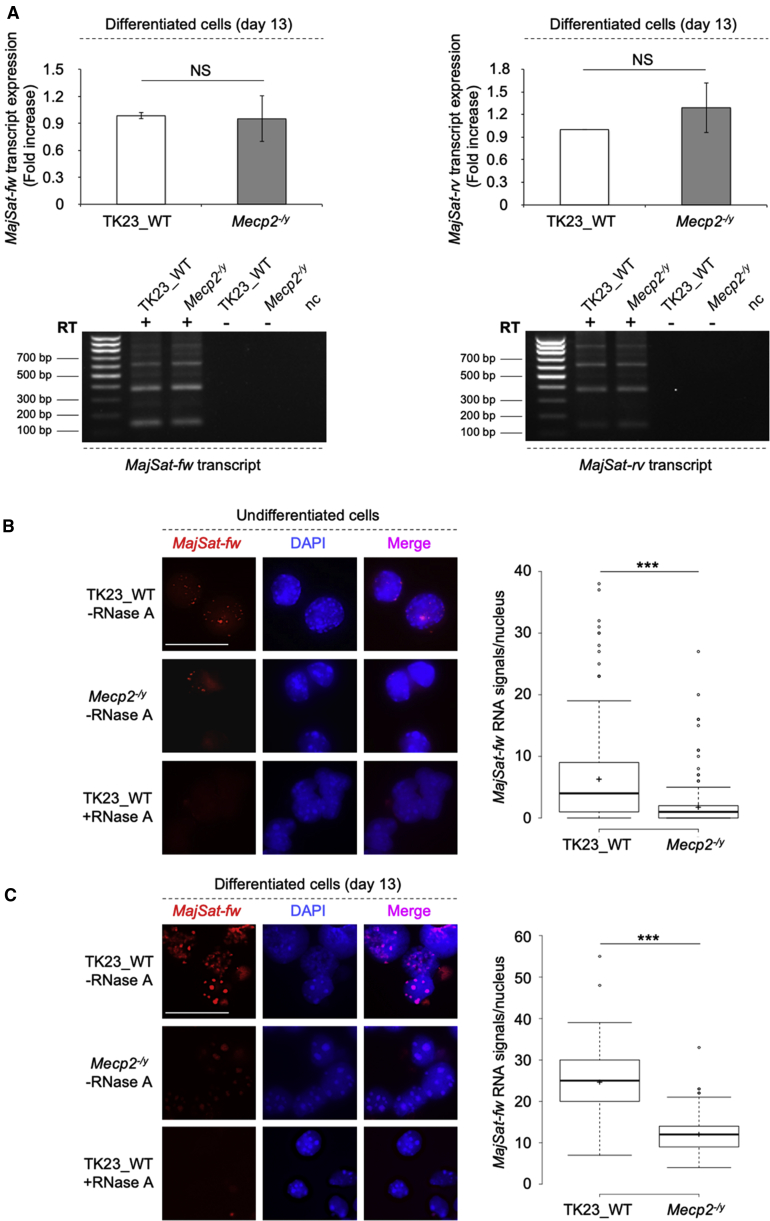
MajSat ncRNAs represent ideal candidates for molecular partners of MeCP2 (Casanova et al., 2013; Maison et al., 2011; Park et al., 2018). We first analyzed the expression of both forward and reverse MajSat (MajSat-fw, MajSat-rv, respectively) RNAs by qPCR after strand-specific reverse transcription (RT-qPCR) in terminally differentiated TK23_WT and Mecp2−/y neurons. MajSat-fw and MajSat-rv RNAs were both expressed in TK23_WT and Mecp2−/y neurons, without any significant differences between these two cell lines (Figure 2A, top). Moreover, the RT-qPCR products of both MajSat-fw and MajSat-rv RNAs had different lengths (Figure 2A, bottom), which suggested the expression of multiple repeated units, as previously reported in other cellular contexts (Maison et al., 2011).
Figure 2.
MeCP2 Has a Role in MajSat Forward Transcript Targeting to PCH
(A) Top: Strand-specific RT-qPCR analysis of MajSat-fw and MajSat-rv transcripts in mESC-derived TK23_WT and Mecp2−/y neurons. Data are normalized to Gapdh and are means ± SD of transcript levels relative to TK23_WT, from three independent experiments. NS, not significant (two-tailed Student's t test). Bottom: Electrophoretic analyses of endpoint strand-specific RT-qPCR for the MajSat-fw and MajSat-rv RNAs. RT, reverse transcriptase; nc, negative control. (B, C) Left: Representative images of i3D-RNA FISH using the MajSat-fw LNA probe in TK23_WT and Mecp2−/y undifferentiated cells (B) and mESC-derived neurons (C) without and with RNase A treatment. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of MajSat-fw RNA signals in the three-dimensional space of each nucleus. Data are shown as box and whisker plots with 200 nuclei analyzed per condition, from two replicate slides. ∗∗∗p < 0.001 (two-sample Kolmogorov-Smirnov test). See also Figure S1.
We next analyzed the nuclear localization of MajSat RNAs in both the TK23_WT and Mecp2−/y undifferentiated cells and in the neurons, using interphase three-dimensional (i3D)-RNA fluorescence in situ hybridization (FISH) with strand-specific locked nucleic acid (LNA) probes. In the undifferentiated cells, the MajSat-fw transcripts were visible as small nuclear spots in both cell lines, and were not detected after RNase A treatment, which ruled out cross-reactions with DNA. However, the number of MajSat-fw RNA signals/nucleus was significantly greater in TK23_WT nuclei compared with Mecp2−/y nuclei (Figure 2B), even though the expression of MajSat-fw RNA was similar in the two cell lines (Figure S1A).
Following the neural differentiation, the sizes and numbers of MajSat-fw RNA signals were increased in both TK23_WT and Mecp2−/y neurons (Figure 2C), compared with undifferentiated cells (Figure 2B), and in both cell lines they were associated with chromocenters. Furthermore, the number of signals was significantly greater for TK23_WT neurons compared with Mecp2−/y neurons, and when the images were captured under constant image-acquisition parameters, the RNA signals for Mecp2−/y nuclei appeared less bright compared to TK23_WT nuclei (Figure 2C).
Interphase three-dimensional-RNA FISH performed at an early differentiation stage (day 8) showed spatial distribution of the MajSat-fw transcripts similar to that for the terminally differentiated neurons, with significantly greater numbers of signals in the TK23_WT cells (Figure S1B). Moreover, comparison of the number of RNA signals/nucleus over the three time points analyzed showed a gradual increase in both TK23_WT and Mecp2−/y cells (Figure S1C), which suggested progressive accumulation of the transcript at the chromocenters during neural differentiation.
Altogether, these findings let us hypothesize a remarkable contribution of MeCP2 in MajSat-fw transcript targeting to PCH during neural differentiation.
In contrast with the MajSat-fw transcript, the MajSat-rv RNA did not show any accumulation as fluorescent spots in the TK23_WT or the Mecp2−/y undifferentiated and differentiated cells (Figure S1D), although its expression was detected in both of these cell lines (Figure 2A).
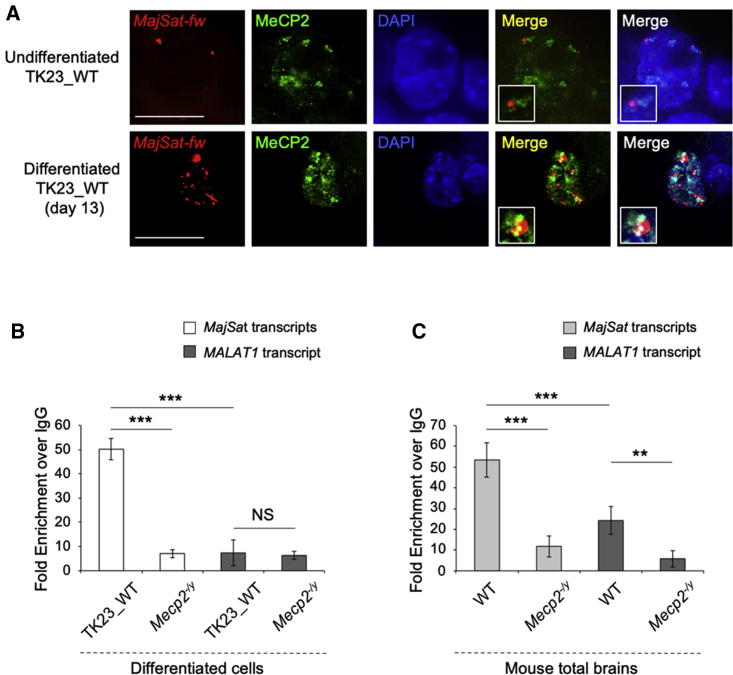
MeCP2 Associates with the MajSat-fw Transcript to Chromocenters and Physically Interacts with It
The reciprocal nuclear localization of MeCP2 and MajSat-fw RNA was assayed by immuno-RNA FISH in TK23_WT cells. In undifferentiated cells, the MajSat-fw transcript was localized to the periphery of the chromocenters, where MeCP2 strongly accumulated. In terminally differentiated TK23_WT neurons, the number of MajSat-fw RNA signals was greater and partially colocalized with the MeCP2 signals to chromocenters (Figure 3A). Furthermore, native RNA immunoprecipitation (RIP) carried out on mESC-derived TK23_WT neurons revealed physical interactions between MeCP2 and the MajSat transcripts. Conversely, binding of metastasis-associated lung-adenocarcinoma transcript 1 (MALAT1) (which shows weak binding with MeCP2 in cerebellum; Maxwell et al., 2013), was comparable between the TK23_WT and negative control neurons (Figure 3B), which confirmed the specificity of this binding. Similar data were obtained in total brain of WT mice, even though weak binding of MeCP2 with the MALAT1 transcript was detected (Figure 3C).
Figure 3.
MeCP2 Spatially and Physically Interacts with the MajSat Transcripts
(A) Representative images of undifferentiated TK23_WT cells and mESC-derived TK23_WT neurons examined by immuno-RNA FISH using MajSat-fw LNA probe and anti-MeCP2 antibody. Chromocenters were stained with DAPI. Merge: MeCP2/MajSat-fw RNA signals and MeCP2/MajSat-fw RNA/DAPI signals. Insets show magnifications of a chromocenter. Scale bar, 15 μm.
(B and C) RNA immunoprecipitation of MajSat transcripts in mESC-derived Mecp2−/y (negative control) and TK23_WT neurons (B), and in total brain from Mecp2−/y (negative control) and WT mice (C), using an anti-MeCP2 antibody.
Data are means ± SD of four independent qPCR replicates from two RIP experiments. ∗∗∗p < 0.001; ∗∗p < 0.01; NS, not significant (one-tailed Student’s t test). See also Figure S2.
We further confirmed a direct interaction between MeCP2 and MajSat-fw transcript in terminally differentiated neurons by fluorescence resonance energy transfer (FRET). Here, we carried out RNA FISH to specifically label the MajSat-fw transcript with an LNA probe in mESC-derived Mecp2[WT]EGFP (referred to as WT-GFP) neurons expressing endogenous MeCP2 fused with EGFP at its C-terminus (Lyst et al., 2013). This assay demonstrated that the donor EGFP conjugated to MeCP2 and the acceptor TYE563, which labels MajSat-fw LNA, are in close proximity (Figure S2).
Altogether, these findings highlight the highly specific binding of MeCP2 to the MajSat transcripts in neurons, both in vitro and in vivo, in agreement with the immuno-RNA FISH data.
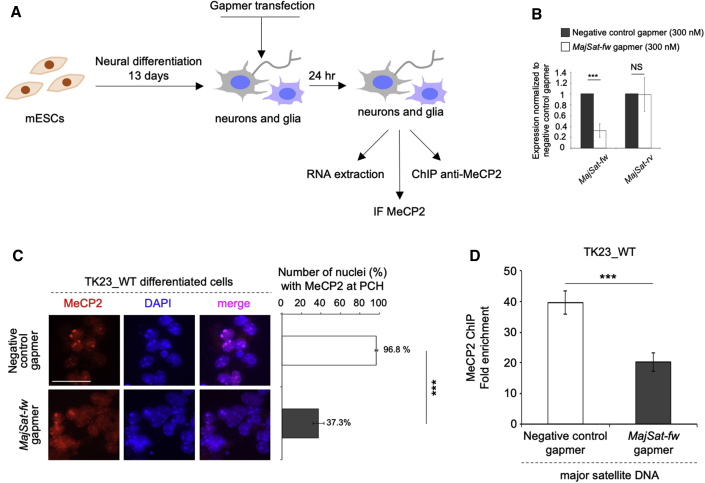
MajSat-fw Transcript Contributes to MeCP2 Targeting to PCH
To understand whether the MajSat-fw transcript is required for targeting of MeCP2 to PCH (see Figure 1), we silenced this ncRNA in mESC-derived TK23_WT neurons by transfection of a specific antisense LNA gapmer (Figure 4A). First, we verified that mESC-derived neurons can be transfected with gapmers (Figure S3A), and then we optimized the protocol to efficiently knock-down MajSat-fw RNA in mESC-derived TK23_WT neurons (Figures 4B and S3B). Here, 300 nM MajSat-fw gapmer was sufficient to deplete ∼80% of the MajSat-fw transcript, without any effects on MajSat-rv transcript levels (Figure 4B). Immunofluorescence carried out in terminally differentiated TK23_WT neurons transfected with the MajSat-fw gapmer showed that MeCP2 was located to chromocenters in <40% of the nuclei, while, in the rest of the nuclei, the majority of MeCP2 signals were widespread in the nucleoplasm. Conversely, in TK23_WT neurons transfected with the negative control gapmer, MeCP2 was strongly accumulated at PCH (Figure 4C). These data were confirmed by ChIP in terminally differentiated TK23_WT neurons, which showed ∼50% lower MeCP2 binding to MajSat DNA after MajSat-fw RNA knock-down (Figure 4D). Of note, major satellite DNA repeats were similarly hypermethylated in TK23_WT neurons transfected with either the MajSat-fw or the negative control gapmer (Figure S3C), which suggests that the reduced accumulation of MeCP2 at PCH is not a result of loss of DNA methylation.
Figure 4.
MeCP2 Targeting to Chromocenters Is Partly Dependent on the MajSat-fw Transcript
(A) Scheme of the experimental procedure.
(B) Strand-specific RT-qPCR analysis of MajSat-fw and MajSat-rv transcripts in mESC-derived TK23_WT neurons transfected with 300 nM negative control or MajSat-fw gapmers. Data are normalized to Gapdh and are means ± SD of transcript levels relative to cells transfected with negative control gapmer, from three biological replicates, with each amplified twice. ∗∗∗p < 0.001; NS, not significant (one-tailed Student’s t test).
(C) Left: Representative immunofluorescence of MeCP2 in mESC-derived TK23_WT neurons transfected with negative control or MajSat-fw gapmers. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of MeCP2 enrichment at PCH following transfection of negative control or MajSat-fw gapmers. Data are means ± SD, with ≥100 cells analyzed per condition, from four independent experiments. ∗∗∗p < 0.001 (one-tailed Student’s t test).
(D) ChIP–qPCR of MeCP2 at MajSat DNA in mESC-derived TK23_WT neurons transfected with negative control or MajSat-fw gapmers. Data are means ± SD of four independent qPCR replicates from two ChIP experiments. ∗∗∗p < 0.001; NS, not significant (one-tailed Student’s t test). See also Figure S3.
Overall, these data highlighted a specific contribution of the MajSat-fw transcript in the targeting of MeCP2 to chromocenters in mESC-derived neurons.
MeCP2 Contributes to the Organization of PCH Architecture by Preserving the Correct Deposition of H3K9me3 and H4K20me3
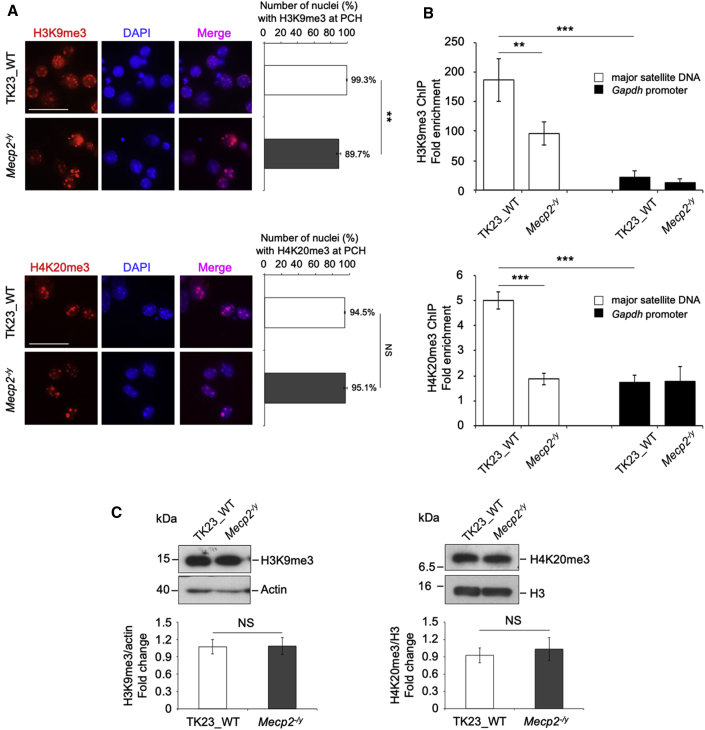
To understand the role of MeCP2 in the global organization of PCH, we analyzed the nuclear distribution of H3K9me3 and H4K20me3 in terminally differentiated TK23_WT and Mecp2−/y neurons. These repressive histone modifications appeared to predominantly accumulate at chromocenters in both of these cell lines. However, quantitative analysis performed by counting the fractions of the nuclei that showed H3K9me3 spotted to chromocenters highlighted a slightly, but significantly, lower proportion (∼10%) of spotted nuclei in Mecp2−/y compared with TK23_WT neurons. The same analysis performed for H4K20me3 did not show any significant differences (Figure 5A); however, ChIP assays, which are a more sensitive and quantitative method, highlighted remarkably lower (∼50%) accumulation of both histone marks at MajSat DNA in the absence of MeCP2 (Figure 5B), despite their unchanged protein levels (Figure 5C).
Figure 5.
H3-Lys9 and H4-Lys20 Trimethylation Is Lower in Terminally Differentiated Mecp2-null Neurons
(A) Left: Representative immunofluorescence of H3K9me3 and H4K20me3 marks in terminally differentiated TK23_WT and Mecp2−/y neurons. Chromocenters were stained with DAPI. Scale bars, 25 μm. Right: Quantification of H3K9me3 and H4K20me3 enrichment at chromocenters in TK23_WT and Mecp2−/y nuclei. Data are means ± SD, with ≥100 nuclei analyzed for each cell line, from three independent experiments. ∗∗p < 0.01; NS, not significant (one-tailed Student’s t test).
(B) ChIP–qPCR of H3K9me3 and H4K20me3 marks at MajSat DNA and Gapdh promoter (negative control region) in mESC-derived TK23_WT and Mecp2−/y neurons. Data are means ± SD of four independent qPCR replicates from two ChIP experiments. ∗∗∗p < 0.001; ∗∗p < 0.01 (one-tailed Student’s t test).
(C) Top: Western blots of H3K9me3 and H4K20me3 marks in mESC-derived TK23_WT and Mecp2−/y neurons. Actin and histone H3 were used for normalization. Bottom: Quantification of H3K9me3 and H4K20me3 protein expression. Data are means ± SD for H3K9me3/actin and H4K20me3/H3 ratios, with Mecp2−/y neurons normalized to TK23_WT neurons, from three independent experiments. NS, not significant (one-tailed Student’s t test).
These data demonstrated that MeCP2 has a role in correct deposition of the H3K9me3 and H4K20me3 marks to PCH in terminally differentiated neurons, thus contributing to the organization of PCH architecture.
MeCP2B Rescues Chromocenter Clustering during Neural Differentiation and Contributes to MajSat-fw Transcript Targeting to PCH
We previously highlighted a key role for MeCP2 in chromocenter clustering during neural differentiation (Bertulat et al., 2012). Furthermore, we have here revealed a reciprocal contribution of MeCP2 and MajSat-fw RNA for their targeting to PCH. Next, we asked which MeCP2 isoform is responsible for this higher-order PCH organization, and what are the contributions of their two main domains, MBD and TRD.
Starting from the Mecp2−/y mESCs, we generated stable clones that selectively and constitutively expressed MeCP2A (Mecp2−/y_MeCP2A) or MeCP2B (Mecp2−/y_MeCP2B), and also MeCP2B lacking MBD (Mecp2−/y_MeCP2B_ΔMBD) or TRD (Mecp2−/y_MeCP2B_ΔTRD) (Figures S4 and S5). MBD or TRD were deleted from MeCP2B because in the brain it is 10-fold more abundant than MeCP2A (Kriaucionis and Bird, 2004).
As the subsequent experiments were performed in differentiated cells, we selected the mESC clones that showed MeCP2 protein levels comparable with those of the TK23_WT neurons (Figures S4A, S4B, S5A, and S5B) and, then, we confirmed their differentiation toward a neural fate (Figures S4C and S5C). Moreover, side-by-side comparison of MeCP2 protein levels in all of these selected clones confirmed their similar expression (Figure S6A).
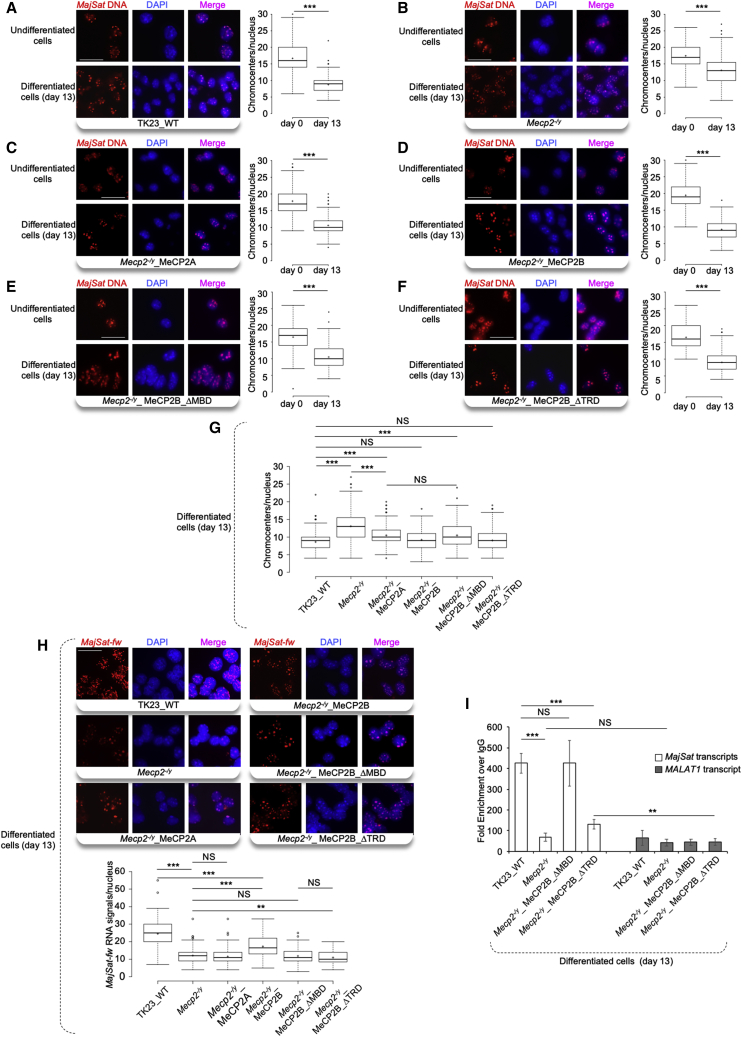
To investigate which isoform and which domain of MeCP2 is involved in chromocenter clustering, we performed i3D-DNA FISH in the selected clones, in comparison with TK23_WT and Mecp2−/y cells, at days 0 (undifferentiated cells) and 13 of neural differentiation. Chromocenters were detected using the MajSat-fw LNA probe, which can reveal even small differences. We found that the MajSat DNA distribution overlapped with the intense DAPI staining in the nuclei of all of the cell lines. Chromocenter clustering was evaluated in each cell line by comparing the number of chromocenters at days 0 and 13. We highlighted significantly fewer chromocenters/nucleus in each cell line at day 13 in comparison with day 0, due to aggregation of chromocenters (clustering), although to different extents (Figures 6A–6F).
Figure 6.
MeCP2B and Its MBD Are the Major Players for Chromocenter Clustering and Both MBD and TRD Contribute to MajSat-fw Transcript Targeting to PCH
(A–F) Left: Representative images of i3D-DNA FISH in TK23_WT, Mecp2−/y, Mecp2−/y_MeCP2A, Mecp2−/y_MeCP2B, Mecp2−/y_MeCP2B_ΔMBD, and Mecp2−/y_MeCP2B_ΔTRD undifferentiated cells (day 0) and terminally differentiated neurons (day 13) using the MajSat-fw LNA probe. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of chromocenters in three-dimensional space of each nucleus in undifferentiated mESCs and terminally differentiated neurons.
(G) Direct comparisons of the data reported in (A–F) for terminally differentiated neurons.
(H) Top: Representative images of i3D-RNA FISH in terminally differentiated TK23_WT, Mecp2−/y, Mecp2−/y_MeCP2A, Mecp2−/y_MeCP2B, Mecp2−/y_MeCP2B_ΔMBD, and Mecp2−/y_MeCP2B_ΔTRD neurons, using the MajSat-fw LNA probe. Chromocenters were stained with DAPI. Scale bar, 25 μm. Bottom: Quantification of MajSat-fw RNA signals in three-dimensional space of each nucleus in terminally differentiated neurons. Data for TK23_WT and Mecp2−/y neurons are also reported in Figure 2C. For (A–H), data are shown as box and whisker plots with 200 nuclei analyzed per condition, from two replicate slides. ∗∗p < 0.01; ∗∗∗p < 0.001; NS, not significant (two-sample Kolmogorov-Smirnov test, without (A–F) and with (G, H) Benjamini–Hochberg correction).
(I) RNA immunoprecipitation of MajSat transcripts in mESC-derived Mecp2−/y_MeCP2B_ΔMBD, Mecp2−/y_MeCP2B_ΔTRD, TK23_WT (positive control), and Mecp2−/y (negative control) neurons, using an anti-MeCP2 antibody. Data are means ± SD of three independent qPCR replicates from one representative RIP experiment. Additional biological replicates yielded similar results. ∗∗∗p < 0.001; ∗∗p < 0.01; NS, not significant (one-tailed Student’s t test). See also Figures S4–S6.
To highlight possible defects in chromocenter clustering, we compared the number of chromocenters/nucleus among the different cell lines as the terminally differentiated neurons. In comparison with TK23_WT neurons, Mecp2−/y neurons showed impaired chromocenter clustering, as previously reported (Bertulat et al., 2012). Mecp2−/y_MeCP2A neurons showed a partial rescue of the chromocenter clustering, whereas MeCP2B expression completely recovered PCH condensation (Figure 6G), which suggested a prominent role of MeCP2B in chromocenter clustering. Interestingly, Mecp2−/y_MeCP2B_ΔMBD neurons showed a defect in chromocenter clustering similar to that of Mecp2−/y_MeCP2A neurons, whereas MeCP2B_ΔTRD expression induced complete rescue (Figure 6G). This suggested that MBD was required for correct chromocenter clustering, whereas TRD was dispensable for this process.
Next, we dissected out the role of the different MeCP2 isoforms and of their two main domains in the targeting of the MajSat-fw RNA to PCH, by performing i3D-RNA FISH in the terminally differentiated TK23_WT, Mecp2−/y, Mecp2−/y_MeCP2A, Mecp2−/y_MeCP2B, Mecp2−/y_MeCP2B_ΔMBD, and Mecp2−/y_MeCP2B_ΔTRD neurons. As already observed for TK23_WT and Mecp2−/y neurons (Figure 2C), in all of these clones, the MajSat-fw RNA predominantly colocalized with chromocenters. Mecp2−/y_MeCP2A neurons showed fewer spots of MajSat-fw RNA associated with PCH as compared with TK23_WT neurons and similar to those of Mecp2−/y neurons, whereas Mecp2−/y_MeCP2B neurons showed an intermediate number between TK23_WT and Mecp2−/y neurons. Furthermore, both Mecp2−/y_MeCP2B_ΔMBD and Mecp2−/y_MeCP2B_ΔTRD neurons showed similar numbers of RNA signals/nucleus to those for Mecp2−/y neurons (Figure 6H). Of note, the number of RNA signals/nucleus in Mecp2−/y_MeCP2B_ΔTRD neurons was even lower than in Mecp2−/y neurons, although this was only a small difference. Altogether, these findings indicated the prevalent contribution of MeCP2B in targeting of MajSat-fw transcript to PCH in terminally differentiated neurons, and underlined the involvement of both MBD and TRD in this function. Moreover, native RIP performed in mESC-derived Mecp2−/y_MeCP2B_ΔMBD and Mecp2−/y_MeCP2B_ΔTRD neurons revealed that MeCP2B_ΔMBD bound MajSat RNAs similar to the WT MeCP2, whereas in the absence of TRD, this interaction was strongly affected, although residual binding was detected (Figure 6I). These data highlighted a key role for TRD in the binding of MeCP2 with MajSat RNAs in mESC-derived neurons.
Finally, immunofluorescence showed that MeCP2A, MeCP2B, and MeCP2B_ΔTRD accumulated at chromocenters (Figure S6B), similar to the endogenous MeCP2 protein, although there were small amounts of MeCP2B_ΔTRD in the cell cytoplasm, which might be due to the absence of one nuclear localization signal (Kumar et al., 2008). Conversely, MeCP2B_ΔMBD appeared to be widespread throughout the nucleoplasm, which was broadly confirmed by ChIP, even though residual binding of MeCP2B_ΔMBD to MajSat DNA was detected (Figures S6B and S6C).
The RTT Mutation T158M Impairs Chromocenter Clustering and Targeting of the MajSat-fw Transcript to PCH
We tested the effects on higher-order PCH organization of two common RTT missense mutations (Neul et al., 2008) located within NID and MBD: R306C and T158M, respectively (Lyst et al., 2013; Nan et al., 1993).
For this, we used Mecp2[R306C]EGFP, Mecp2[T158M]EGFP, and Mecp2[WT]EGFP knock-in mESCs (referred to as R306C-GFP, T158M-GFP, and WT-GFP, respectively) in which the MeCP2 proteins were fused in-frame with EGFP at their C-terminus (Lyst et al., 2013). Importantly, both MeCP2A and MeCP2B were affected by the knock-in.
First, we confirmed that the neural differentiation of R306C-GFP and T158M-GFP was similar to WT-GFP mESCs (Lyst et al., 2013) (Figure S7A). Furthermore, the MeCP2WT−GFP and MeCP2R306C−GFP proteins accumulated at PCH in terminally differentiated neurons, whereas MeCP2T158M−GFP partially lost this chromocenter accumulation, as previously reported (Lyst et al., 2013). Moreover, MeCP2R306C−GFP and MeCP2T158M−GFP protein levels were similar to those of MeCP2WT−GFP (Figure S7B).
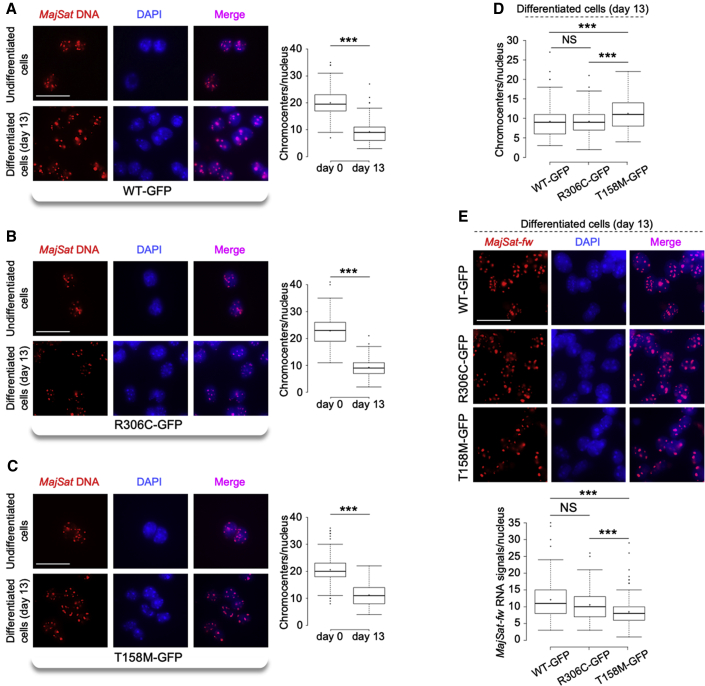
Interphase three-dimensional-DNA FISH performed in undifferentiated WT-GFP, R306C-GFP, and T158M-GFP mESCs (day 0) and in terminally differentiated neurons (day 13) highlighted that the MajSat DNA distribution overlapped with intense DAPI staining in the nuclei of all of the cell lines (Figures 7A–7C). Comparisons of the numbers of chromocenters/nucleus at day 0 and day 13 for each cell line showed that the chromocenters were clustered in all of these cells (Figures 7A–7C). Nonetheless, when we compared the terminally differentiated neurons with each other, T158M-GFP, but not R306C-GFP, neurons showed a significant defect in chromocenter clustering, as compared with WT-GFP neurons (Figure 7D). Furthermore, analysis of the nuclear distribution of MajSat-fw RNA by i3D-RNA FISH in mESC-derived R306C-GF, T158M-GFP, and WT-GFP neurons highlighted accumulation of the MajSat-fw transcript primarily at PCH in all of the cell lines. Moreover, WT-GFP and R306C-GFP neurons showed similar numbers of RNA signals/nucleus, whereas for T158M-GFP neurons, these were significantly lower (Figure 7E).
Figure 7.
The T158M Mutation Affects Both Chromocenter Clustering and Targeting of the MajSat-fw Transcript to Chromocenters
(A–C) Left: Representative images of i3D-DNA FISH in WT-GFP, R306C-GFP, and T158M-GFP undifferentiated cells (day 0) and terminally differentiated neurons (day 13), using the MajSat-fw LNA probe. Chromocenters were stained with DAPI. Scale bar, 25 μm. Right: Quantification of chromocenters in three-dimensional space of each nucleus in undifferentiated mESCs and in terminally differentiated neurons.
(D) Direct comparisons of the data reported in (A–C) for terminally differentiated neurons.
(E) Top: Representative images of i3D-RNA FISH in terminally differentiated WT-GFP, R306C-GFP, and T158M-GFP neurons, using the MajSat-fw LNA probe. Chromocenters were stained with DAPI. Scale bar, 25 μm. Bottom: Quantification of MajSat-fw RNA signals in the three-dimensional space of each nucleus in terminally differentiated neurons. For both i3D-DNA FISH and i3D-RNA FISH, data are shown as box and whisker plots with 200 nuclei analyzed per condition, from two replicate slides. ∗∗∗p < 0.001; NS, not significant (two-sample Kolmogorov-Smirnov test, without (A–C) and with (D, E) Benjamini–Hochberg correction). See also Figure S7.
Altogether, these data underlined the relevance of the T158 amino acid for both PCH condensation during neural differentiation and targeting of MajSat-fw RNA to chromocenters in neurons.
Discussion
Chromocenters appear to form a repressive environment that is enriched in silencing factors that contribute to maintenance of heterochromatic status (Almouzni and Probst, 2011; Wijchers et al., 2015). We previously highlighted a role for MeCP2 in PCH condensation during neural differentiation (Bertulat et al., 2012; Marano et al., 2019), a context of particular interest considering the neurological nature of Rett syndrome. However, the mechanisms underlying higher-order PCH organization in neurons remain unclear.
Several ncRNAs are involved in chromatin silencing, through recruitment of repressive factors to specific genomic regions (Saxena and Carninci, 2011). We show here that MeCP2 targeting to chromocenters in neurons is dependent on an RNA component, as previously reported for other PCH-related proteins (Huo et al., 2020; Maison et al., 2002; Marano et al., 2019). MajSat ncRNAs are optimal candidates as partners of MeCP2 in the organization of PCH architecture (Casanova et al., 2013; Maison et al., 2011; Park et al., 2018). We showed that MajSat-fw and MajSat-rv transcripts are expressed in TK23_WT and Mecp2−/y neurons with similar expression levels in the two cell lines, thus excluding any MeCP2-mediated transcriptional regulation. Of note, only the MajSat-fw RNA is condensed into spots at chromocenters, as previously reported (Maison et al., 2011), which suggests diffusion of MajSat-rv RNA into the nucleoplasm, and that the two strands act independently of each other.
Our data strongly suggest that MeCP2 and the MajSat-fw transcript are mutually dependent for their targeting to chromocenters in neurons. In the absence of MeCP2, there were fewer spots of MajSat-fw RNA associated with PCH at each time of neural differentiation, in comparison with the WT condition, which suggested a contribution of MeCP2 in MajSat-fw RNA targeting to PCH. Considering the unchanged levels of MajSat-fw transcript between TK23_WT and Mecp2−/y cells, we hypothesized intranuclear diffusion of part of this transcript in the MeCP2-deficient cells. Furthermore, the MajSat-fw transcript knock-down in terminally differentiated TK23_WT neurons strongly reduced the accumulation of MeCP2 at chromocenters, despite unchanged MajSat DNA methylation. In addition, MeCP2 binds MajSat transcripts, as reported for HP1α and SAFB (Huo et al., 2020; Maison et al., 2011), and according to the interactions of MeCP2 with RNA (Maxwell et al., 2013). These findings allow hypothesis of a direct role for the MajSat-fw transcript in MeCP2 targeting to PCH, although we cannot rule out the involvement of additional factors.
We highlighted a reduction in H3K9me3 and H4K20me3 deposition to PCH in the absence of MeCP2, without any consequent increase in MajSat expression. We believe that decreased accumulation of the MajSat-fw transcript at PCH caused by MeCP2 depletion leads to defective recruitment of Suv39h and suppressor of variegation 4-20 homolog (Suv4-20h) histone methyltransferases (HMTs), and then to reduced H3K9me3 and H4K20me3 deposition at chromocenters. This hypothesis is in agreement with the contribution of satellite RNAs for targeting Suv39h to PCH (Velazquez Camacho et al., 2017). Alternatively, MeCP2 might directly recruit HMTs to PCH, in line with the known interplay between MeCP2 and HMTs (Fuks et al., 2003; Lehnertz et al., 2003).
Overall, these data underlined an important contribution of satellite transcripts in higher-order PCH organization in neurons. We hypothesize that MeCP2 and MajSat-fw RNA might cooperate for the organization of silent compartments, by acting as structural organizing factors. In support of this, MajSat RNAs have been linked to de novo chromocenter formation (Casanova et al., 2013; Probst et al., 2010).
At present, the redundancy of MeCP2A and MeCP2B isoforms is still controversial (Jugloff et al., 2008; Kerr et al., 2012). We highlighted that MeCP2B is the major player for both chromocenter clustering and MajSat-fw RNA targeting to PCH during neural differentiation. MeCP2B expression in the Mecp2-null context was indeed sufficient to rescue the chromocenter clustering, whereas MeCP2A induced only partial PCH condensation. In addition, MeCP2B expression partly recovered targeting of the MajSat-fw RNA to PCH, whereas MeCP2A-expressing cells showed similar defects in RNA accumulation to those in Mecp2-null neurons. Nevertheless, the incomplete rescue of RNA targeting in the MeCP2B-expressing cells suggested a small, but significant, contribution of MeCP2A.
Recent data have underlined the relevance to RTT pathogenesis of MBD and NID (Tillotson et al., 2017). MBD promotes binding of MeCP2 to PCH (Brero et al., 2005; Kumar et al., 2008), whereas TRD mediates transcriptional silencing (Lyst et al., 2013); however, their roles in higher-order PCH organization remain to be better clarified. We show that MBD is required for chromocenter clustering during neural differentiation, whereas TRD appears to be dispensable, as previously observed in myoblasts (Brero et al., 2005). Interestingly, the similar defect of chromocenter clustering in MeCP2B_ΔMBD or MeCP2A-expressing cells indicated comparable relevance of MBD and the MeCP2B N-terminus in PCH condensation. Furthermore, MajSat-fw RNA targeting to PCH is impaired in the absence of MBD or TRD, which underlines the relevance of both of these domains in this process. Our data support the inclusion of a MajSat RNA binding site in TRD. Thus, we hypothesize that MeCP2B_ΔTRD binds chromocenters and promotes their clustering, while the lack of the MajSat RNA binding site is responsible for reduced targeting of MajSat-fw RNA to PCH. Conversely, the impairment of both chromocenter clustering and targeting of MajSat-fw RNA to PCH in MeCP2B_ΔMBD-expressing neurons might be ascribed to the strongly reduced interaction with the heterochromatic foci, even though the binding of the transcript is preserved.
Some MeCP2 RTT mutations affect its DNA-binding capacity and chromatin-binding kinetics (Agarwal et al., 2011; Kumar et al., 2008). However, whether the alterations in chromatin structure derived from MeCP2 defects contribute to RTT pathogenesis remains unclear. We show that the T158M, but not the R306C, mutation moderately impairs both chromocenter clustering and MajSat-fw RNA targeting to PCH. These defects might be due to partial delocalization of MeCP2T158M−GFP from PCH, considering that the T158M mutation reduces its affinity for DNA (Ho et al., 2008). We hypothesize that the fraction of MeCP2T158M−GFP bound to PCH is responsible for both partial chromocenter clustering and MajSat-fw RNA targeting to PCH.
Overall, these data underline the importance of the T158 amino acid in higher-order PCH organization, as observed for MBD.
In conclusion, it is tempting to speculate that defects in PCH organization caused by MeCP2 alterations are responsible for inappropriate localization of specific genes with respect to silent compartments. This might give rise to their incorrect expression, which will have a pathogenic role in RTT. However, further studies are needed to more clearly correlate alterations of higher-order PCH organization and the RTT-related phenotype.
Experimental Procedures
Antisense LNA Gapmer Transfection
mESC-derived WT neurons were transfected with 10 nM LNA gapmer MajSat-fw-6FAM (Exiqon), TK23_WT neurons were transfected with 25 nM–300 nM LNA gapmer MajSat-fw or the negative control A gapmer (scrambled) (Exiqon), using Lipofectamine RNAiMax (Life Technologies), according to the manufacturer’s protocol. Twelve hours after transfection, the medium was replaced, and after a further 12 h, the cells were collected.
Interphase Three-Dimensional-DNA FISH
Cells were fixed with 4% paraformaldehyde (PFA) for 10 min, permeabilized with 0.2% Triton X-100/PBS for 10 min, and stored in 75% ethanol overnight at 4°C. Following dehydration in 90% and 100% ethanol, and denaturation in 2× saline sodium citrate buffer (SSC), 50% formamide (Sigma-Aldrich) at 80°C for 30 min, hybridization was carried out with 0.1 μM major 1 LNA fluorescent probe (Exiqon) (Probst et al., 2010) in 30% formamide, 1.6 mg/mL salmon sperm DNA (Sigma-Aldrich), 10% dextran sulfate (Fluka), 1 mg/mL BSA, 2× SSC, for 35 min at 37°C. After three washes in 0.1× SSC for 5 min at 60°C, the slides were mounted using Vectashield (Vector Laboratories)/DAPI.
Interphase Three-Dimensional-RNA FISH and Immuno-RNA FISH
nterphase three-dimensional-RNA FISH was performed as previously reported (Maison et al., 2011). Briefly, cells were permeabilized with 0.5% Triton X-100/CSK buffer (10 mM Pipes, pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) supplemented with 10 mM vanadyl ribonucleoside complex (VRC; New England Biolabs) for 5 min on ice, washed with CSK buffer and PBS. For RNase A treatment, there was an additional incubation with 1 mg/mL RNase A (Roche)/PBS or PBS (mock) for 10 min at room temperature. Cells were fixed with 3% PFA for 12 min and stored in 70% ethanol overnight at −20°C. Following dehydration with 80%, 95%, and 100% ethanol, cells were incubated with 0.2 μM major 1 or major 2 LNA fluorescent probes (Exiqon) (Probst et al., 2010) in 50% formamide, 10% dextran sulfate, 2 mg/mL BSA, 10 mM VRC, 2× SSC, for 35 min at 37°C. After three washes in 0.1× SSC for 5 min at 60°C, the slides were mounted using Vectashield/DAPI.
Immuno-RNA FISH was performed as previously reported (Chaumeil et al., 2002), with minor changes. Briefly, cells were fixed with 3% PFA for 10 min, washed with PBS and CSK buffer, permeabilized with 0.5% Triton X-100/CSK buffer supplemented with 2 mM VRC for 5 min on ice, and washed with CSK buffer and PBS. After blocking with 10% normal goat serum, 1% BSA, 0.4 U RNaseOUT (Life Technologies) in PBS for 15 min, the cells were incubated with the anti-MeCP2 antibody (Table S3) in blocking solution for 45 min.
After washes in PBS, the cells were incubated with the secondary antibody (Alexa Fluor 488 donkey anti-rabbit; Thermofisher Scientific). The cells were then post-fixed in 3% PFA for 10 min, washed with 2× SSC, and incubated with 0.4 μM major 1 LNA fluorescent probe in 30% formamide, 1.6 mg/mL salmon sperm DNA, 10% dextran sulfate, 1 mg/mL BSA, 20 mM VRC, 2× SSC, for 35 min at 37°C. After three washes in 0.1× SSC for 5 min at 60°C, the slides were mounted using Vectashield/DAPI.
Sequence of Primers, Probes, and Gapmers
The sequences of the primers, LNA fluorescent probes, and antisense LNA gapmers used in this study are provided in Tables S1 and S2.
Microscopy and Image Analysis
For all imaging experiments, except for FRET, the images were captured with a fluorescence microscope (DM6000B; Leica) run with the LAS AF 2.6 software (Leica), using a ×63 (NA1.4) objective lens (Leica) and a digital camera (DFC 360FX; Leica).
The proportions of nuclei with MeCP2, H3K9me3, and H4K20me3 enrichment at chromocenters were obtained by analyzing different focal planes along the z axis. For i3D-DNA FISH and i3D-RNA FISH, multichannel z stack images were acquired with a step size of 0.18 μm with the LAS AF 2.6 software, and then the chromocenters (for i3D-DNA FISH) and RNA signals (for i3D-RNA FISH) per nucleus were manually counted along the z axis. The RStudio software was used to generate the box plots. For immuno-RNA FISH, multichannel z stack images were acquired with a step size of 0.18 μm, and then subjected to deconvolution using the LAS AF 2.6 software.
Nuclei of undifferentiated cells were selected at random, and the analysis of mESC-derived neurons was performed on cells with a neuronal morphology.
Statistical Analysis
For the i3D-DNA FISH and i3D-RNA FISH experiments, statistical analysis was carried out using two-sample Kolmogorov-Smirnov test without or with Benjamini–Hochberg correction (for multiple comparisons). Data are shown as box and whisker plots, where the whiskers show the 5%–95% percentiles of the confidence intervals, the horizontal lines within each box show the medians, the crosses indicate the means, and outliers are shown as dots.
For the Western blot reported in Figure S6A, one-way ANOVA followed by Tukey post-hoc test was applied. For all of the other experiments, one-tailed or two-tailed paired Student's t tests were used, as reported in the Figure legends. One-tailed paired Student's t test was used when unidirectional changes were expected. Data are presented as means ± SD (for all experiments, except for FRET assay) or as means ± SEM (for FRET). p values <0.05 are considered significant.
Author Contributions
F.D.R. conceived the study; F.D.R., M.D.E, S.F., E.C., and D.M. planned and designed the experiments with the help of M.R.M.; A.B. helped in the earlier experimental phase; F.M. and M.M. managed the animal colonies; F.D.R, S.F., E.C., D.M., C.Z., and A.R. performed the experiments; F.D.R. and S.F. wrote the manuscript with the help of D.M.; M.D.E. and M.R.M. gave conceptual advice and edited the manuscript; all of the authors have reviewed and approved the final manuscript.
Conflict of Interests
The authors declare no competing interests.
Acknowledgments
The authors are grateful to Prof. Bird and Dr. J. Selfridge for providing the Mecp2[WT]EGFP, Mecp2[R306C]EGFP, and Mecp2[T158M]EGFP knock-in mESCs; Dr. C. Schanen for providing the MTmmMeCP2AEGFP and MTmmMeCP2BEGFP plasmids; Dr. S. Filosa for providing assistance for the generation of the mESC stable clones’; Dr. I. De Feis and M. Gagliardi for statistical suggestions; and Dr. C. Berrie for scientific English editing of the manuscript. They also thank the Integrated Microscopy Facility of IGB-CNR and the Advanced Light Microscopy Facility of CEINGE. This work was supported by the Epigenomics Flagship Project (EPIGEN) MIUR-CNR, Italy; the UE Initial Training Network Project ‘DISCHROM’ (238242), European Union; the Jérôme Lejeune Foundation (1545), France; the Project MIUR PON03_PE00060_7, Italy; the RETT Italian Association (AIRETT, National Grant 2019), Italy; and the Project PON/MISE 2014-2020 FESR F/050011/01-02/X32, Italy.
Published: December 8, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.11.006.
Supplemental Information
References
- Agarwal N., Becker A., Jost K.L., Haase S., Thakur B.K., Brero A., Hardt T., Kudo S., Leonhardt H., Cardoso M.C. MeCP2 Rett mutations affect large scale chromatin organization. Hum. Mol. Genet. 2011;20:4187–4195. doi: 10.1093/hmg/ddr346. [DOI] [PubMed] [Google Scholar]
- Almouzni G., Probst A.V. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus. 2011;2:332–338. doi: 10.4161/nucl.2.5.17707. [DOI] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S., Chahrour M., Thaller C., Shaw C.A., Zoghbi H.Y. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum. Mol. Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertulat B., De Bonis M.L., Della Ragione F., Lehmkuhl A., Milden M., Storm C., Jost K.L., Scala S., Hendrich B., D'Esposito M. MeCP2 dependent heterochromatin reorganization during neural differentiation of a novel Mecp2-deficient embryonic stem cell reporter line. PLoS One. 2012;7:e47848. doi: 10.1371/journal.pone.0047848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brero A., Easwaran H.P., Nowak D., Grunewald I., Cremer T., Leonhardt H., Cardoso M.C. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M., Pasternak M., El Marjou F., Le Baccon P., Probst A.V., Almouzni G. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep. 2013;4:1156–1167. doi: 10.1016/j.celrep.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J., Okamoto I., Guggiari M., Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet. Genome Res. 2002;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
- D'Esposito M., Quaderi N.A., Ciccodicola A., Bruni P., Esposito T., D'Urso M., Brown S.D. Isolation, physical mapping, and northern analysis of the X-linked human gene encoding methyl CpG-binding protein, MECP2. Mamm. Genome. 1996;7:533–535. doi: 10.1007/s003359900157. [DOI] [PubMed] [Google Scholar]
- Della Ragione F., Vacca M., Fioriniello S., Pepe G., D'Esposito M. MECP2, a multi-talented modulator of chromatin architecture. Brief Funct. Genomics. 2016;15:420–431. doi: 10.1093/bfgp/elw023. [DOI] [PubMed] [Google Scholar]
- Dragich J.M., Kim Y.H., Arnold A.P., Schanen N.C. Differential distribution of the MeCP2 splice variants in the postnatal mouse brain. J. Comp. Neurol. 2007;501:526–542. doi: 10.1002/cne.21264. [DOI] [PubMed] [Google Scholar]
- Falk M., Feodorova Y., Naumova N., Imakaev M., Lajoie B.R., Leonhardt H., Joffe B., Dekker J., Fudenberg G., Solovei I. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 2019;570:395–399. doi: 10.1038/s41586-019-1275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fico A., Manganelli G., Simeone M., Guido S., Minchiotti G., Filosa S. High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cells Dev. 2008;17:573–584. doi: 10.1089/scd.2007.0130. [DOI] [PubMed] [Google Scholar]
- Fioriniello S., Marano D., Fiorillo F., D'Esposito M., Della Ragione F. Epigenetic factors that control pericentric heterochromatin organization in mammals. Genes (Basel) 2020;11:595. doi: 10.3390/genes11060595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F., Hurd P.J., Wolf D., Nan X., Bird A.P., Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Gianakopoulos P.J., Zhang Y., Pencea N., Orlic-Milacic M., Mittal K., Windpassinger C., White S.J., Kroisel P.M., Chow E.W., Saunders C.J. Mutations in MECP2 exon 1 in classical Rett patients disrupt MECP2_e1 transcription, but not transcription of MECP2_e2. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B:210–216. doi: 10.1002/ajmg.b.32015. [DOI] [PubMed] [Google Scholar]
- Guy J., Hendrich B., Holmes M., Martin J.E., Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Ho K.L., McNae I.W., Schmiedeberg L., Klose R.J., Bird A.P., Walkinshaw M.D. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Huo X., Ji L., Zhang Y., Lv P., Cao X., Wang Q., Yan Z., Dong S., Du D., Zhang F. The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol. Cell. 2020;77:368–383.e7. doi: 10.1016/j.molcel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Jugloff D.G., Vandamme K., Logan R., Visanji N.P., Brotchie J.M., Eubanks J.H. Targeted delivery of an Mecp2 transgene to forebrain neurons improves the behavior of female Mecp2-deficient mice. Hum. Mol. Genet. 2008;17:1386–1396. doi: 10.1093/hmg/ddn026. [DOI] [PubMed] [Google Scholar]
- Jung B.P., Jugloff D.G., Zhang G., Logan R., Brown S., Eubanks J.H. The expression of methyl CpG binding factor MeCP2 correlates with cellular differentiation in the developing rat brain and in cultured cells. J. Neurobiol. 2003;55:86–96. doi: 10.1002/neu.10201. [DOI] [PubMed] [Google Scholar]
- Kerr B., Soto C.J., Saez M., Abrams A., Walz K., Young J.I. Transgenic complementation of MeCP2 deficiency: phenotypic rescue of Mecp2-null mice by isoform-specific transgenes. Eur. J. Hum. Genet. 2012;20:69–76. doi: 10.1038/ejhg.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi Y., Kondo S., Gotoh Y. Transcriptional activation of mouse major satellite regions during neuronal differentiation. Cell Struct. Funct. 2012;37:101–110. doi: 10.1247/csf.12009. [DOI] [PubMed] [Google Scholar]
- Klose R., Bird A. Molecular biology. MeCP2 repression goes nonglobal. Science. 2003;302:793–795. doi: 10.1126/science.1091762. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S., Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 2004;32:1818–1823. doi: 10.1093/nar/gkh349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kamboj S., Malone B.M., Kudo S., Twiss J.L., Czymmek K.J., LaSalle J.M., Schanen N.C. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J. Cell Sci. 2008;121:1128–1137. doi: 10.1242/jcs.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B., Ueda Y., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Lyst M.J., Ekiert R., Ebert D.H., Merusi C., Nowak J., Selfridge J., Guy J., Kastan N.R., Robinson N.D., de Lima Alves F. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Roche D., Montes de Oca R., Probst A.V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.P. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat. Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- Marano D., Fioriniello S., Fiorillo F., Gibbons R.J., D'Esposito M., Della Ragione F. ATRX contributes to MeCP2-mediated pericentric heterochromatin organization during neural differentiation. Int. J. Mol. Sci. 2019;20:5371. doi: 10.3390/ijms20215371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Paz A., Khajavi L., Martin H., Claveria-Gimeno R., Tom Dieck S., Cheema M.S., Sanchez-Mut J.V., Moksa M.M., Carles A., Brodie N.I. MeCP2-E1 isoform is a dynamically expressed, weakly DNA-bound protein with different protein and DNA interactions compared to MeCP2-E2. Epigenetics Chromatin. 2019;12:63. doi: 10.1186/s13072-019-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell S.S., Pelka G.J., Tam P.P., El-Osta A. Chromatin context and ncRNA highlight targets of MeCP2 in brain. RNA Biol. 2013;10:1741–1757. doi: 10.4161/rna.26921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Meehan R.R., Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neul J.L., Fang P., Barrish J., Lane J., Caeg E.B., Smith E.O., Zoghbi H., Percy A., Glaze D.G. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee H., Han N., Kwak S., Lee H.T., Kim J.H., Kang K., Youn B.H., Yang J.H., Jeong H.J. Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization. Nucleic Acids Res. 2018;46:11759–11775. doi: 10.1093/nar/gky923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Okamoto I., Casanova M., El Marjou F., Le Baccon P., Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Rett A. [On a unusual brain atrophy syndrome in hyperammonemia in childhood] Wien Med. Wochenschr. 1966;116:723–726. [PubMed] [Google Scholar]
- Saxena A., Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene P.J., Illingworth R.S., Webb S., Kerr A.R., James K.D., Turner D.J., Andrews R., Bird A.P. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson R., Selfridge J., Koerner M.V., Gadalla K.K.E., Guy J., De Sousa D., Hector R.D., Cobb S.R., Bird A. Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature. 2017;550:398–401. doi: 10.1038/nature24058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez Camacho O., Galan C., Swist-Rosowska K., Ching R., Gamalinda M., Karabiber F., De La Rosa-Velazquez I., Engist B., Koschorz B., Shukeir N. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife. 2017;6:e25293. doi: 10.7554/eLife.25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers P.J., Geeven G., Eyres M., Bergsma A.J., Janssen M., Verstegen M., Zhu Y., Schell Y., Vermeulen C., de Wit E. Characterization and dynamics of pericentromere-associated domains in mice. Genome Res. 2015;25:958–969. doi: 10.1101/gr.186643.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.