Abstract
Aims
Preliminary evidence from animal and human studies shows that gut microbiota composition and levels of microbiota-derived metabolites, including short-chain fatty acids (SCFAs), are associated with blood pressure (BP). We hypothesized that faecal microbiota composition and derived metabolites may be differently associated with BP across ethnic groups.
Methods and results
We included 4672 subjects (mean age 49.8 ± 11.7 years, 52% women) from six different ethnic groups participating in the HEalthy Life In an Urban Setting (HELIUS) study. The gut microbiota was profiled using 16S rRNA gene amplicon sequencing. Associations between microbiota composition and office BP were assessed using machine learning prediction models. In the subgroups with the largest associations, faecal SCFA levels were compared in 200 subjects with lower or higher systolic BP. Faecal microbiota composition explained 4.4% of the total systolic BP variance. Best predictors for systolic BP included Roseburia spp., Clostridium spp., Romboutsia spp., and Ruminococcaceae spp. Explained variance of the microbiota composition was highest in Dutch subjects (4.8%), but very low in South-Asian Surinamese, African Surinamese, Ghanaian, Moroccan and Turkish descent groups (explained variance <0.8%). Faecal SCFA levels, including acetate (P < 0.05) and propionate (P < 0.01), were lower in young Dutch participants with low systolic BP.
Conclusions
Faecal microbiota composition is associated with BP, but with strongly divergent associations between ethnic groups. Intriguingly, while Dutch participants with lower BP had higher abundances of several SCFA-producing microbes, they had lower faecal SCFA levels. Intervention studies with SCFAs could provide more insight in the effects of these metabolites on BP.
Keywords: Blood pressure, Gut microbiota, Short-chain fatty acids, Hypertension, Ethnicity
Graphical Abstract
See page 4268 for the editorial comment on this article (doi: 10.1093/eurheartj/ehaa760)
Introduction
Hypertension is the leading modifiable risk factor for cardiovascular morbidity and mortality, and thereby the most important risk factor for preventable death worldwide. The pathogenesis of essential hypertension remains incompletely understood and is currently attributed to a complex interplay of genetic and cardiovascular risk factors.1 However, recent studies have shown that only 3–4% of the variance in systolic blood pressure (SBP) can be explained by common genetic risk variants.2 Lifestyle factors such as diet and obesity are known to be important for the pathogenesis of hypertension.3 Analysis of population data from the UK Biobank revealed that lifestyle factors can modify blood pressure (BP) by up to 4–5 mmHg depending on genetic risk.4
The gut microbiota is a reflection of both genetic make-up and life-long exposure to dietary risk factors and could play a key role in mediating the development of essential hypertension.5 Key metabolites produced by gut microbiota are short-chain fatty acids (SCFAs), which are end-products of intestinal fermentation of otherwise indigestible dietary fibres.6 Animal studies point towards a direct link between faecal SCFAs and BP, mediated by SCFAs receptors in kidneys and blood vessels.7 In humans, evidence of the relation between faecal SCFA levels and BP is scarce and conflicting. Both higher and lower faecal SCFAs have been associated with higher BP.8–10 Assuming that the gut microbiota and SCFAs are indeed associated with BP, this would provide new perspectives on both the pathogenesis and treatment of hypertension.
Earlier studies have identified important differences in both the prevalence, pathogenesis, and treatment responses of hypertension among ethnic groups.11–13 In addition, we found substantial differences in gut microbiota composition between ethnic groups within the population-based HEalthy Life In an Urban Setting (HELIUS) cohort that were only partly explained by sociodemographic, lifestyle, or dietary influences.14 Therefore, ethnic differences should be taken into account when studying associations between the gut microbiota composition and BP. Hence, in this cross-sectional study, we aim to investigate associations between the gut microbiota, faecal SCFA levels, and BP across different ethnic groups using data from the HELIUS cohort study.
Methods
Study population
We used cross-sectional data obtained during baseline visits between 2011 and 2015 of the ongoing HELIUS prospective cohort study. The aims and design of this study have been described previously.14 In brief, based on the municipality registry of Amsterdam, people aged between 18 and 70 were randomly sampled, stratified by ethnicity (Dutch, South Asian Surinamese, African Surinamese, Ghanaian, Turkish, or Moroccan). For the present analysis, we included participants with available BP measurements, body mass index (BMI), and faecal samples. All participants provided written informed consent and the study was approved by the medical ethical review board of the Amsterdam UMC, location AMC. This study followed the principles of the Declaration of Helsinki.
Data were collected during morning study visits at local research sites. Prior to these visits, all participants were asked to refrain from smoking. Body mass index was calculated from height and weight. Blood pressure was measured after at least 5 min of rest in the supine position, using the average of two consecutive measurements obtained with a validated semi-automatic oscillometric device (Microlife WatchBP Home; Microlife AG, Switzerland). Fasting glucose and creatinine levels were measured in venous blood samples, and estimated glomerular filtration rate (eGFR) was calculated using the CKD-EPI formula. In addition, urinary albumin-to-creatinine ratio was determined from early morning spot urine samples. Albuminuria was defined as a ratio ≥30 mg/mmol.15 Participants were asked to bring all current medication, from which current use of BP-lowering and glucose-lowering medication was determined. Diabetes was defined based on elevated fasting glucose levels (≥7 mmol/L) or the use of glucose-lowering medication. Hypertension was defined according to guidelines as an elevated SBP >140 mmHg or diastolic BP (DBP) >90 mmHg or self-reported use of BP-lowering medication.15
Gut microbiota composition
Participants received faecal collection tubes either prior or during the study visit. They were asked to bring a fresh faecal sample within 6 h after collection, or, if not possible, to store the sample overnight in a freezer. Samples were stored at −20°C at the study visit location for a maximum of 1 day before transportation to the central freezer (−80°C). Samples obtained from participants who either had diarrhoea in the week prior to collection or used antibiotics within 3 weeks prior to collection were excluded. Samples were shipped to the Wallenberg Laboratory (Sahlgrenska Academy at University of Gothenburg, Sweden) for sequencing. DNA was extracted from 150 mg aliquot of faecal samples using a repeated bead-beating protocol.16 Faecal microbiota composition was determined by sequencing the V4 region of the 16S rRNA gene on an Illumina MiSeq (llumina RTA v1.17.28; MCS v2.5, San Diego, CA, USA) using 515 F and 806 R primers designed for dual-indexing17 and the V2 Illumina kit (2 × 250 bp paired-end reads). PCR was performed in duplicate reactions as previously described.14 Pre-processing of the raw sequencing data, as described in Supplementary material online, Supplement S1 resulted in a dataset containing 4672 samples.
Faecal short-chain fatty acid measurements
Faecal SCFA levels were measured using high-performance liquid chromatography (HPLC) with UV detection according to the method of De Baere et al.18 In addition, for all samples, dry weights were determined after freeze-drying a homogenized faecal aliquot for 24 h. All concentrations resulting from HPLC measurements were corrected for the difference in the wet and dry weight per sample.
Statistical analysis
We used machine learning models to assess the association between gut microbiota composition and BP. Analyses were performed for the total study population and for subgroups stratified by age (≤50, >50 years), sex, and ethnicity. A separate set of models was performed using adjusted SBP and DBP values. Adjustments were made by determining the residuals after fitting a linear regression model for each of the subgroups with SBP/DBP as the dependent variable and age, sex, and BMI as independent variables. For age, we used sex-specific restricted cubic splines, of which the order was chosen based on the Akaike Information Criterion. Machine learning models were build aiming to predict SBP, corrected SBP, DBP, and corrected DBP from the gut microbiota composition (i.e. from the relative abundance of microbial 16S rRNA amplicon sequence variants; ASVs). Gradient boosted tree models were used in a nested cross-validation structure to prevent overfitting and ensure robustness of results (Supplementary material online, Supplement S2). Models were built using an iterative flow. In each iteration, the dataset was randomly split into a test set containing 20% of the participants and a training set containing the remaining 80%. Thereafter, five-fold cross-validation was performed strictly within the train set in order to fit and optimize the model hyperparameters. The resulting model was finally evaluated on the test set. Two random variables were added to the predictor data during each iteration to serve as a benchmark. Explained variance was determined as the proportion of variance of the outcome (SBP or DBP) explained by the model-predicted values (predicted SBP or DBP), using:
Explained variance and the ranked list of predictor importance were recorded for each iteration and were averaged across 100 iterations. If the explained variance was negative, we concluded that the model did not have any predictive power.
Spearman rank correlation coefficients were calculated between the top 10 best SBP-predicting ASVs found by the machine learning models and both SBP and DBP. Furthermore, participants were categorized into tertiles of the relative abundance of each of the ASVs. Effect sizes for the effect of each ASV on SBP were estimated for every tertile using linear regression in a crude model correcting only for age and sex, and in a full model with additional correction for BMI, smoking, use of antihypertensive medication, and history of diabetes.
For the analyses of faecal SCFAs, we used a subgroup of 200 participants selected from Dutch participants aged ≤50, as the explained variance was highest in the Dutch and the young subgroups. Based on age-specific percentiles (<30, 30–40, 40–50 years), 50 men and 50 women with the highest SBP were selected. Using sex, age, and BMI, these 100 participants were matched to 100 other participants from the lowest 35th percentile of SBP. Faecal SCFA concentrations and abundance of the top predicting ASV’s were compared between the high and low BP group using Mann–Whitney U tests. In addition, the relation with microbiota composition was examined using a correlation matrix of SCFA concentration, BP, and the top 10 predicting ASVs.
Machine learning was implemented in Python (v. 3.7.4) using the XGBoost (v. 0.90), numpy (v. 1.16.4), pandas (v. 0.25.1), and scikit-learn (v. 0.21.2) packages. Statistical analyses were performed using R version 3.6.2, using the Regression Modelling Strategies (rms) version 5.1-4 and Nonparametric Pre-processing for Parametric Causal Interference (MatchIt) version 3.0.2 packages. Figures were created using R with the corrplot version 0.84 package, and Graphpad version 8.3.0.
Results
Population characteristics
Characteristics of the included 4672 participants are shown in Table 1. Younger participants (≤50 years) had a lower prevalence of hypertension (24.2%) compared with older participants (57.1%), and a lower use of antihypertensive medication (8.8% vs. 33.4%). Dutch and Moroccan groups consisted of relatively more men than the other groups. South Asian Surinamese, African Surinamese, and Ghanaian groups had higher BP and higher proportions of participants with hypertension than other ethnic groups. Body mass index was lowest in Dutch (25.5 ± 4.4 kg/m2) and in the South Asian Surinamese groups (26.6 ± 4.5 kg/m2). Diabetes prevalence was highest in South Asian Surinamese participants (23.9%) and lowest in Dutch participants (4.8%).
Table 1.
Population characteristics
| Overall | Younger (≤50) | Older (>50) | Female | Male | ||
|---|---|---|---|---|---|---|
| N | 4672 | 2217 | 2455 | 2429 | 2243 | |
| Female | 2429 (52.0) | 1220 (55.0) | 1209 (49.2) | — | — | |
| Age (years) | 49.8 ± 11.7 | 39.9 ± 8.3 | 58.8 ± 5.2 | 49.2 ± 11.7 | 50.5 ± 11.6 | |
| SBP (mmHg) | 129.9 ± 18.2 | 123.7 ± 16.0 | 135.4 ± 18.1 | 127.2 ± 18.5 | 132.7 ± 17.3 | |
| DBP (mmHg) | 81.1 ± 10.6 | 79.1 ± 10.5 | 83.0 ± 10.4 | 78.6 ± 10.4 | 83.9 ± 10.2 | |
| BMI (kg/m2) | 27.2 ± 4.9 | 26.7 ± 5.0 | 27.7 ± 4.8 | 27.8 ± 5.4 | 26.6 ± 4.2 | |
| eGFR (mL/min/1.73 m2) | 97.1 ± 17.0 | 105.6 ± 14.8 | 89.3 ± 15.0 | 98.7 ± 17.2 | 95.3 ± 16.5 | |
| Hypertension | 1937 (41.5) | 536 (24.2) | 1401 (57.1) | 924 (38.0) | 1013 (45.2) | |
| Antihypertensive drugs | 1016 (21.7) | 196 (8.8) | 820 (33.4) | 559 (23.0) | 457 (20.4) | |
| Lipid-lowering drugs | 580 (12.4) | 86 (3.9) | 494 (20.1) | 240 (9.9) | 340 (15.2) | |
| Albuminuria | 196 (4.2) | 67 (3.0) | 129 (5.3) | 91 (3.8) | 105 (4.7) | |
| Diabetes | 507 (10.9) | 96 (4.3) | 411 (16.8) | 218 (9.0) | 289 (12.9) | |
| Antidiabetic drugs | 367 (7.9) | 61 (2.8) | 306 (12.5) | 178 (7.3) | 189 (8.4) | |
| Smoking | 941 (20.1) | 456 (20.6) | 485 (19.8) | 349 (14.4) | 592 (26.4) | |
| Dutch | SAS | Afr Sur | Ghanaian | Turkish | Moroccan | |
| N | 1328 | 575 | 1128 | 462 | 436 | 605 |
| Female | 633 (47.7) | 300 (52.2) | 672 (59.6) | 255 (55.2) | 224 (51.4) | 281 (46.4) |
| Age (years) | 51.43 ± 12.7 | 51.6 ± 11.2 | 51.9 ± 10.5 | 48.2 ± 9.0 | 44.2 ± 11.0 | 45.5 ± 11.4 |
| SBP (mmHg) | 127.6 ± 17.2 | 132.4 ± 19.6 | 132.9 ± 17.9 | 137.2 ± 18.2 | 124.1 ± 16.2 | 125.3 ± 17.6 |
| DBP (mmHg) | 79.5 ± 10.2 | 81.6 ± 10.4 | 83.4 ± 10.4 | 85.8 ± 11.0 | 79.5 ± 10.3 | 77.6 ± 9.8 |
| BMI (kg/m2) | 25.5 ± 4.4 | 26.6 ± 4.5 | 28.2 ± 5.4 | 28.2 ± 4.5 | 28.9 ± 4.8 | 27.9 ± ± 4.7 |
| eGFR (mL/min/1.73 m2) | 91.2 ± 14.9 | 91.8 ± 16.8 | 99.1 ± 18.3 | 100.4 ± 17.7 | 104.6 ± 13.4 | 104.5 ± 14.1 |
| Hypertension | 455 (34.3) | 280 (48.7) | 598 (53.0) | 272 (58.9) | 127 (29.1) | 146 (24.1) |
| Antihypertensive drugs | 210 (15.8) | 174 (30.3) | 353 (31.3) | 136 (29.4) | 64 (14.7) | 49 (8.1) |
| Lipid-lowering drugs | 135 (10.2) | 160 (27.8) | 130 (11.5) | 40 (8.7) | 53 (12.2) | 44 (7.3) |
| Albuminuria | 26 (2.0) | 45 (7.8) | 42 (3.7) | 27 (5.9) | 19 (4.4) | 28 (4.6) |
| Diabetes | 63 (4.8) | 137 (23.9) | 143 (12.7) | 47 (10.2) | 39 (9.0) | 64 (10.6) |
| Antidiabetic drugs | 25 (1.9) | 116 (20.2) | 104 (9.2) | 35 (7.6) | 30 (6.9) | 45 (7.4) |
| Smoking | 263 (19.8) | 139 (24.2) | 300 (26.6) | 17 (3.7) | 117 (26.8) | 74 (12.2) |
Data are presented as mean ± SD or n (%).
Afr Sur, African Surinamese; BMI, body mass index; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate (CKD-EPI); SAS, South Asian Surinamese; SBP, systolic blood pressure.
Microbiota composition and blood pressure
The BP variance that was explained by gut microbiota composition is shown in Table 2, stratified for the different subgroups. In the total population, the explained variance of BP levels by microbiota composition was 4.4% for SBP and 4.3% for DBP. Explained variance was higher in younger subjects (5.3% for both SBP and DBP) than in older subjects (2.5% for SBP; 1.4% for DBP), and higher in women (3.9% for SBP; 2.2% for DBP) than in men (1.8% for SBP; 0.3% for DBP). There was a clear difference in explained variance between Dutch (4.8% for SBP; 0.4% for DBP) and other ethnic groups (<0.8% for SBP; <0.5% for DBP). The correlations between alpha diversity of gut microbiota and BP (Supplementary material online, Supplement S3) showed the same pattern as the explained variance with stronger correlations in young, female, and Dutch subgroups.
Table 2.
Explained variance of blood pressure by microbiota composition for different subgroups
| Microbiota composition (explained variance in %) |
||||
|---|---|---|---|---|
| Group | SBP | Res SBP | DBP | Res DBP |
| All subjects | 4.44 | 2.22 | 4.30 | 2.05 |
| Younger (≤50) | 5.31 | 3.11 | 5.34 | 2.81 |
| Older (>50) | 2.51 | 1.75 | 1.40 | 0.87 |
| Men | 1.82 | 1.41 | 0.32 | 1.41 |
| Women | 3.89 | 1.90 | 2.23 | 1.30 |
| Dutch | 4.76 | 0.60 | 0.40 | n.a. |
| SA Surinamese | n.a. | 0.64 | n.a. | 0.09 |
| Afr Surinamese | n.a. | 0.74 | n.a. | 0.08 |
| Ghanaian | n.a. | n.a. | n.a. | n.a. |
| Moroccan | 0.77 | 0.42 | n.a. | 0.64 |
| Turkish | n.a. | n.a. | 0.48 | 0.62 |
Explained variance in % of the gut microbiome composition for blood pressure. Colours indicate levels of explained variance.
Afr Surinamese, African Surinamese; DBP, diastolic blood pressure; n.a., explained variance in these models was negative, meaning that these models had no predictive power; res, residuals adjusted for age, sex, BMI; SA Surinamese, South Asian Surinamese; SBP, systolic blood pressure.
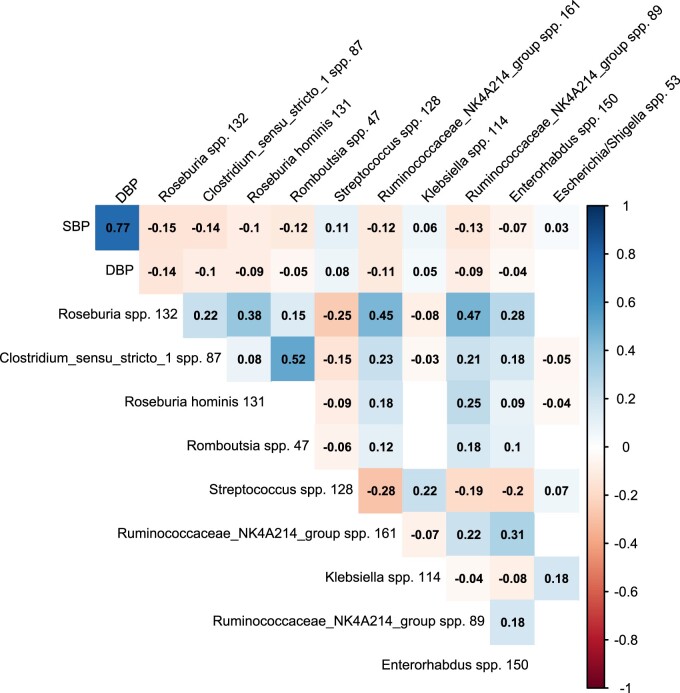
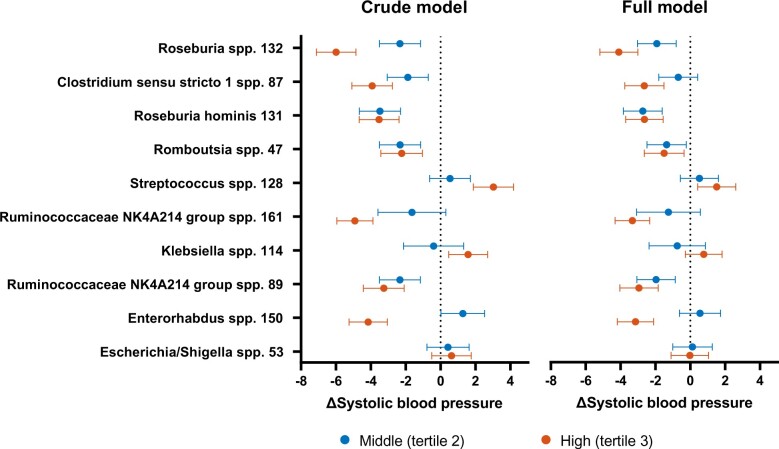
In the total study population, the best-predicting ASVs were Roseburia spp., Clostridium sensu stricto spp., Roseburia hominis, Romboutsia spp., Streptococcus spp., and Ruminococcaceae NK4A214 spp. (Supplementary material online, Supplement S4). The abundance of the best-predicting ASVs was negatively associated with both SBP and DBP, except for Streptococcus spp. and Klebsiella spp., as shown in Figure 1. In addition, the correlation plot showed significant collinearity between the best-predicting ASVs. In the regression analyses, the effect of these ASVs ranged between a 6.0 mmHg lower and 3.0 mmHg higher SBP (Figure 2, Supplementary material online, Supplement S5 ), with increasing effect sizes for higher abundance in most of the ASVs. Roseburia spp. was both the best predictor from the machine learning model and had the largest absolute effect on BP: the middle and highest tertile were associated with a lower SBP of, respectively, 2.3 mmHg (95%CI 1.2–3.5) and 6.0 mmHg (95%CI 4.9–7.1). The effect of the ASV abundance on BP was attenuated when adjusting for confounders, including use of medication, but remained significant for most predictors, ranging between a 4.1 mmHg lower and 1.5 mmHg higher SBP. In this model, we found for the second tertile of Roseburia spp., a 1.9 mmHg (95%CI 0.8–3.0) lower SBP, while participants in the upper tertile had a 4.1 mmHg (95%CI 3.0–5.2) lower SBP.
Figure 1.
Correlation plot for top 10 predictors of systolic blood pressure from gut microbiota composition. Only significant (P < 0.05) Spearman correlation coefficients between the relative abundance of each of the microbes, systolic (SBP) and diastolic (DBP) blood pressure are shown. Colours indicate direction and strength of each correlation.
Figure 2.
Linear regression coefficients with 95%-confidence intervals per tertile of amplicon sequence variant counts for top 10 predictors of systolic blood pressure (SBP) derived from gut microbiota composition, with the lowest tertile as reference. Left side: crude model (correcting for age and sex); right side: additional correction for body mass index, smoking, use of antihypertensive medication, and history of diabetes.
Faecal short-chain fatty acid levels and blood pressure
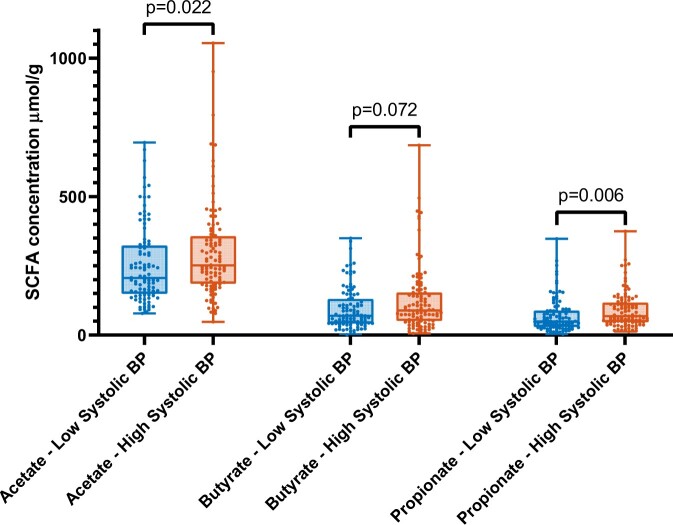
Matching of Dutch subjects on age, sex, and BMI resulted in 100 subjects with low SBP and 100 subjects with high SBP (Supplementary material online, Supplement S6). Consistent with the data from the full cohort, subjects with low BP had higher abundance of Roseburia spp. (P = 0.0047), Roseburia hominis (P = 0.047), and Ruminococcaceae spp. (P = 0.045). Differences of faecal SCFA levels are shown in Figure 3. Low SBP subjects had significantly lower faecal levels of acetate (P = 0.022) and propionate (P = 0.006), and there was a trend of lower butyrate levels (P = 0.072). In addition, faecal SCFA levels were negatively correlated with the top 10 ASVs, and positively with SBP and DBP (Supplementary material online, Supplement S7).
Figure 3.
Comparison of short-chain fatty acid (SCFA) levels between high vs. low blood pressure (BP) subgroups (boxes: median with interquartile range; bars: minimum and maximum). Differences tested with Mann–Whitney U tests.
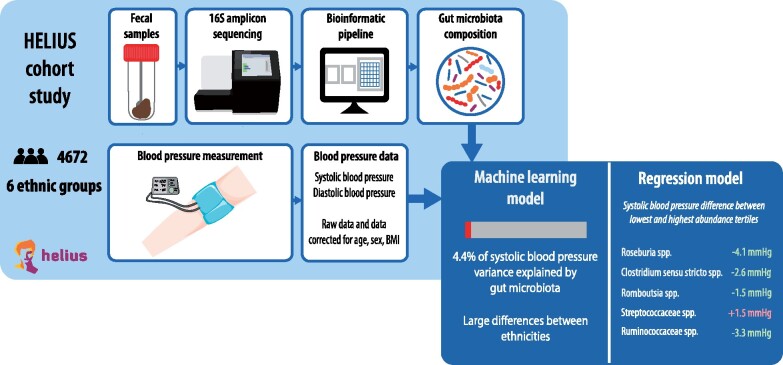
Take Home Figure.
Analysis of the association between gut microbiome composition and blood pressure using a machine learning model. Left panel shows data processing pipeline; right panel the variance in systolic blood pressure explained by microbiota composition, and the microbial predictors for blood pressure.
Discussion
Our main finding is that gut microbiota composition is associated with BP and that the explained BP variance was widely divergent between ethnic groups. Associations between the gut microbiota and BP remained essentially unchanged after correcting for possible confounders, including BMI. Remarkably, while SCFA-producing microbes were associated with lower BP, increased faecal SCFA levels were associated with higher BP. In line with this finding, SCFA-producing microbes were negatively correlated with faecal SCFA levels. The current study extends previous findings in cohort studies by evaluating the association between gut microbiota composition and BP in a large multi-ethnic cohort using machine learning prediction models.19 , 20
We found that, in the complete cohort, machine learning models based on gut microbiota composition explained 4.4% and 4.3% of the variance in SBP and DBP. Regression models showed that in the top tertiles of microbiota predictors SBP was 4–6 mmHg lower compared with the lowest tertile. This is a similar effect size compared with the effect of genetic or lifestyle risk factors on SBP in the UK Biobank.4 Based on recent meta-analyses of randomized controlled trials, this corresponds to an overall cardiovascular risk reduction of 8–12%.21 After correction for BMI, explained variance of SBP and DBP was attenuated to 2.2% and 2.1%. In line with these findings, in the regression analyses the effects were attenuated towards a difference of 2–4 mmHg after correction for BMI and other covariates, suggesting that the effect is only partly driven by BMI.
In the analysis of the top predictors, the finding that SCFA-producing microbes are associated with lower BP is in line with two studies of microbiota composition and hypertension that found lower abundances of either Ruminococcaceae spp. or Roseburia spp. in subjects with higher BP.10 , 20 Moreover, comparable to our results, higher abundances of Klebsiella spp. and Streptococcaceae spp. have been previously associated with higher BP.10 , 22
Previous analyses of HELIUS and other cohorts have shown significant ethnic differences in gut microbiota composition.14 , 23 We add that there are substantial differences in the association of gut microbiota and BP between ethnicities, sexes, and ages, as we observed the highest explained variance in the young, female, and Dutch subgroups. In addition to differences in microbiota composition, this could relate to age, sex, and ethnic-specific effects in the underlying aetiology of hypertension. At younger age, lifestyle and genetic factors are important determinants, while at older age SBP increases and DBP decreases as a consequence of arterial stiffness.24 In addition, multiple studies have shown that older individuals and individuals of African descent are more salt-sensitive, suggesting that they have a more volume-dependent hypertension phenotype.25 Earlier findings from animal models pointed towards a relation between the gut microbiota and salt-sensitive BP driven by abundance of Lactobacillus spp.26 In contrast, we observed a lower explained variance in older, Ghanaian, and African Surinamese subjects, and Lactobacillus spp. was not among the top predictors in these models nor in the model with all subjects. We therefore could not confirm the association between gut microbiota composition and salt-sensitivity in susceptible populations.
Faecal SCFA levels were higher in subjects with higher BP, which is in line with previous results from other cohort studies that examined the relation between faecal SCFAs and BP.8 , 9 These positive associations between BP and faecal SCFA levels in our cohort seem to conflict with the negative associations found between BP and SCFA-producing microbes. However, faecal SCFA levels are not a direct measure of intestinal SCFA production but rather a net result of SCFA production after subtracting SCFA absorption.27 We found consistent correlations in the subgroup with faecal SCFA levels in which SCFA-producing bacteria were both negatively correlated with BP and with faecal SCFA levels. Therefore, we hypothesize that higher microbial SCFA production up-regulates intestinal SCFA absorption resulting in relatively lower levels of SCFAs excretion in faeces.28
The observed differences in SCFA levels between subjects with high and low BP and the multiple SCFA-producing microbes provide further evidence for the hypothesis that SCFAs have a role in BP regulation. Animal studies have shown that SCFAs can have disparate effects on BP depending on the receptors involved. Free fatty acid receptors (FFAR) are G-protein coupled receptors that can be found in a variety of tissues, including the kidney and renal artery, and causes arterial vasodilation in response to propionate, acetate, and butyrate.7 In contrast, a BP elevating effect is mediated by the SCFA receptor Olfr78 in mice through renin release from granules in the renal juxtaglomerular apparatus.29 The human analogue of this olfactory receptor is OR51E2, which responds to propionate and acetate, but not butyrate.7 It has been suggested that Olfr78 and OR51E2 serve as a negative feedback loop for the BP-lowering effects of the FFARs, specifically FFAR3.29 Future intervention studies with oral SCFAs could further unravel the cross-talk between the different SCFA and BP regulation in humans.
To our knowledge, this is the first study to assess the relation between gut microbiota composition and BP across different ethnic groups. We used a large population-based sample with standardized BP measurements for our analysis. Faecal samples were obtained using a standardized protocol from participants without diarrhoea and prior antibiotics use and analysed using 16S rRNA sequencing, which is a widely used and reproducible method to determine microbiota composition.30 For the main analysis, we used machine learning prediction models with nested cross-validation, which enabled us to simultaneously include the complete processed sequencing results in the models while minimizing the risk of overfitting. We corrected for BMI using residuals after fitting a regression model, which could lead to an additional random error in the corrected values. However, both correction for covariates in the regression analyses and the correlations between alpha diversity and BP yielded similar results. While the machine learning results could be hampered by the use of BP-lowering drugs or glucose-lowering medication, effects remained significant after correction in the regression model. In the analyses of SCFA levels, we matched the low and high BP groups for age, sex, and BMI. However, significant differences in BMI remained after matching, which could have affected our results. Lastly, the cross-sectional design of this study complicates causal interpretation of the observed associations. In that regard, we expect that prospective data from the HELIUS cohort study will enable us to assess the longitudinal relation between gut microbiota composition and the development of hypertension. If a longitudinal relation can be confirmed such that changes in microbiota are found to precede and be proportional to changes in BP, potential therapeutic strategies that could be considered include supplementation of (a combination of) specific bacterial strains, modulating gut metabolites such as SCFAs, faecal microbiota transplantation, or antibiotic treatment.
In conclusion, we found a consistent association between gut microbiota composition and BP, with large differences in explained variance between age and ethnic subgroups. Future studies should take ethnic differences into account when studying the gut microbiota in relation to BP. The observed associations between SCFA-producing microbes and BP provide further evidence for the hypothesis that SCFAs play a role in BP regulation. Intervention studies with SCFAs could provide more insight in the underlying mechanism of these metabolites on BP.
Supplementary Material
Acknowledgements
The authors wish to acknowledge V. Tremaroli, R. Jakubowicz, M. Krämer for their help with the DNA extraction, PCR amplification, and sequencing. We would like to thank the AMC Biobank for their support with sample storage and the participants, research nurses and HELIUS staff for their help in data collection. The take home figure contains illustrations that are adapted from Servier Medical Art (https://smart.servier.com/) under a Creative Commons Attribution 3.0 Unported License.
Funding
The Academic Medical Center (AMC) of Amsterdam and the Public Health Service of Amsterdam (GGD) provided core financial support for HELIUS. The HELIUS study is also funded by research grants of the Dutch Heart Foundation [Hartstichting; 2010T084], the Netherlands Organization for Health Research and Development [ZonMw; 200500003], the European Integration Fund [EIF; 2013EIF013], and the European Union [Seventh Framework Programme, FP-7; 278901]. B.V. is appointed on an Amsterdam Cardiovascular Sciences [ACSPhD2019P003] and an Alzheimer Nederland grant [WE.03-2017-12]. M.N. is supported by a ZONMW-VIDI grant 2013 [016.146.327], a Dutch Heart Foundation CVON IN CONTROL-2 grant. The study reported here was additionally supported by Fondation Leducq [17CVD01], JPI [A healthy diet for a healthy life; 2017-01996_3], and Novo Nordisk Foundation [NNF15OC0016798, NNF17OC0028232] to M.N. and F.B.
Conflict of interest: M.N. is on the scientific advisory board of Caelus Health, the Netherlands. F.B. is on the scientific advisory board of MetaboGen AB, Sweden and received grants from BioGaia AB, Sweden. None of these conflicts of interest bear direct relation to the outcomes of this study. All other authors declare that they have no competing interests.
Data availability
The 16S rRNA gene amplicon raw sequence data and associated metadata have been deposited at the European Genome-phenome Archive under study number EGAD00001004106. The HELIUS data are owned by the Amsterdam UMC, location AMC in Amsterdam, the Netherlands. Any researcher can request the data by submitting a proposal as outlined at http://www.heliusstudy.nl/.
Contributor Information
Barbara J H Verhaar, Department of Internal Medicine, section Geriatrics, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam Cardiovascular Sciences, De Boelelaan 1117-1118, 1081 HV, Amsterdam, the Netherlands.
Didier Collard, Department of Internal Medicine, section Vascular Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam Cardiovascular Sciences, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands.
Andrei Prodan, Department of Internal Medicine, section Vascular Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam Cardiovascular Sciences, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands.
Johannes H M Levels, Department of Internal Medicine, section Vascular Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam Cardiovascular Sciences, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands.
Aeilko H Zwinderman, Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Amsterdam UMC, University of Amsterdam, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands.
Fredrik Bäckhed, Wallenberg Laboratory, Department of Molecular and Clinical Medicine, Sahlgrenska Academy, University of Gothenburg, Bruna Stråket 16, 413 45 Gothenburg, Sweden.
Liffert Vogt, Department of Internal Medicine, section Nephrology, Amsterdam UMC, University of Amsterdam, Amsterdam Cardiovascular Sciences, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands.
Mike J L Peters, Department of Internal Medicine, section Geriatrics, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam Cardiovascular Sciences, De Boelelaan 1117-1118, 1081 HV, Amsterdam, the Netherlands.
Majon Muller, Department of Internal Medicine, section Geriatrics, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam Cardiovascular Sciences, De Boelelaan 1117-1118, 1081 HV, Amsterdam, the Netherlands.
Max Nieuwdorp, Department of Internal Medicine, section Vascular Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam Cardiovascular Sciences, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands; Wallenberg Laboratory, Department of Molecular and Clinical Medicine, Sahlgrenska Academy, University of Gothenburg, Bruna Stråket 16, 413 45 Gothenburg, Sweden.
Bert-Jan H van den Born, Department of Internal Medicine, section Vascular Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam Cardiovascular Sciences, Meibergdreef 9, 1105 AZ, Amsterdam, the Netherlands; Department of Public Health, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
References
- 1. Poulter NR, Prabhakaran D, Caulfield M. Hypertension. Lancet 2015;386:801–812. [DOI] [PubMed] [Google Scholar]
- 2. Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, Torstenson ES, Kovesdy CP, Sun YV, Wilson OD, Robinson-Cohen C, Roumie CL, Chung CP, Birdwell KA, Damrauer SM, DuVall SL, Klarin D, Cho K, Wang Y, Evangelou E, Cabrera CP, Wain LV, Shrestha R, Mautz BS, Akwo EA, Sargurupremraj M, Debette S, Boehnke M, Scott LJ, Luan J, Zhao J-H, Willems SM, Thériault S, Shah N, Oldmeadow C, Almgren P, Li-Gao R, Verweij N, Boutin TS, Mangino M, Ntalla I, Feofanova E, Surendran P, Cook JP, Karthikeyan S, Lahrouchi N, Liu C, Sepúlveda N, Richardson TG, Kraja A, Amouyel P, Farrall M, Poulter NR, Laakso M, Zeggini E, Sever P, Scott RA, Langenberg C, Wareham NJ, Conen D, Palmer CNA, Attia J, Chasman DI, Ridker PM, Melander O, Mook-Kanamori DO, Harst P. V D, Cucca F, Schlessinger D, Hayward C, Spector TD, Jarvelin M-R, Hennig BJ, Timpson NJ, Wei W-Q, Smith JC, Xu Y, Matheny ME, Siew EE, Lindgren C, Herzig K-H, Dedoussis G, Denny JC, Psaty BM, Howson JMM, Munroe PB, Newton-Cheh C, Caulfield MJ, Elliott P, Gaziano JM, Concato J, Wilson PWF, Tsao PS, Velez Edwards DR, Susztak K, O’Donnell CJ, Hung AM, Edwards TL, Understanding Society Scientific Group. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet 2019;51:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forman JP, Stampfer MJ, Curhan GC. Diet and lifestyle risk factors associated with incident hypertension in women. JAMA 2009;302:401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pazoki R, Dehghan A, Evangelou E, Warren H, Gao H, Caulfield M, Elliott P, Tzoulaki I. Genetic predisposition to high blood pressure and lifestyle factors: associations with midlife blood pressure levels and cardiovascular events. Circulation 2018;137:653–661. [DOI] [PubMed] [Google Scholar]
- 5. Marques FZ, Mackay CR, Kaye DM. Beyond gut feelings: how the gut microbiota regulates blood pressure. Nat Rev Cardiol 2018;15:20–32. [DOI] [PubMed] [Google Scholar]
- 6. Cummings JH, Pomare EW, Branch HWJ, Naylor CPE, MacFarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987;28:1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pluznick JL. Microbial short-chain fatty acids and blood pressure regulation. Curr Hypertens Rep 2017;19:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huart J, Leenders J, Taminiau B, Descy J, Saint-Remy A, Daube G, Krzesinski JM, Melin P, De Tullio P, Jouret F. Gut microbiota and fecal levels of short-chain fatty acids differ upon 24-hour blood pressure levels in men. Hypertension 2019;74:1005–1013. [DOI] [PubMed] [Google Scholar]
- 9. de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 2018;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C, Han X, Huang Y, Zhao L, Li P, Fang Z, Zhou J, Guan X, Ding Y, Wang S, Khan M, Xin Y, Li S, Ma Y. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol 2017;7:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Laer SD, Snijder MB, Agyemang C, Peters RJ, van den Born B-JH. Born BJH van den. Ethnic differences in hypertension prevalence and contributing determinants—the HELIUS study. Eur J Prev Cardiol 2018;25:1914–1922. [DOI] [PubMed] [Google Scholar]
- 12. Tu W, Eckert GJ, Hannon TS, Liu H, Pratt LM, Wagner MA, DiMeglio LA, Jung J, Pratt JH. Racial differences in sensitivity of blood pressure to aldosterone. Hypertension 2014;63:1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta AK, Poulter NR, Dobson J, Eldridge S, Cappuccio FP, Caulfield M, Collier D, Cruickshank JK, Sever PS, Feder G, on behalf of ASCOT Investigators. Ethnic differences in blood pressure response to first and second-line antihypertensive therapies in patients randomized in the ASCOT trial. Am J Hypertens 2010;23:1023–1030. [DOI] [PubMed] [Google Scholar]
- 14. Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, Tremaroli V, Bakker GJ, Attaye I, Pinto-Sietsma SJ, Raalte DH, van Snijder MB, Nicolaou M, Peters R, Zwinderman AH, Bäckhed F, Nieuwdorp M. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med 2018;24:1526–1531. [DOI] [PubMed] [Google Scholar]
- 15. The Task Force for the management of arterial hypertension of the European Society of Cardiology (ESC) and the European Society of Hypertension (ESH). 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur Heart J 2018;39:3021–3104. [DOI] [PubMed] [Google Scholar]
- 16. Mobini R, Tremaroli V, Ståhlman M, Karlsson F, Levin M, Ljungberg M, Sohlin M, Bertéus Forslund H, Perkins R, Bäckhed F, Jansson PA. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: a randomized controlled trial. Diabetes Obes Metab 2017;19:579–589. [DOI] [PubMed] [Google Scholar]
- 17. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 2013;79:5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Baere S, Eeckhaut V, Steppe M, De Maesschalck C, De Backer P, Van Immerseel F, Croubels S. Development of a HPLC-UV method for the quantitative determination of four short-chain fatty acids and lactic acid produced by intestinal bacteria during in vitro fermentation. J Pharm Biomed Anal 2013;80:107–115. [DOI] [PubMed] [Google Scholar]
- 19. Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, Bowyer RCE, Martin T, Williams FMK, Menni C, Bell JT, Spector TD, Steves CJ. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun 2018;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR, Shikany JM, Lloyd-Jones DM, Launer LJ, Fodor AA, Meyer KA. Gut microbiota composition and blood pressure: the CARDIA study. Hypertension 2019;73:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ettehad D, Emdin CA, Kiran A, Anderson SG, Callender T, Emberson J, Chalmers J, Rodgers A, Rahimi K. Blood pressure lowering for prevention of cardiovascular disease and death: a systematic review and meta-analysis. Lancet 2016;387:957–967. [DOI] [PubMed] [Google Scholar]
- 22. Kim S, Goel R, Kumar A, Qi Y, Lobaton G, Hosaka K, Mohammed M, Handberg EM, Richards EM, Pepine CJ, Raizada MK. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci 2018;132:701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, Lucas SK, Beura LK, Thompson EA, Till LM, Batres R, Paw B, Pergament SL, Saenyakul P, Xiong M, Kim AD, Kim G, Masopust D, Martens EC, Angkurawaranon C, McGready R, Kashyap PC, Culhane-Pera KA, Knights D. US immigration westernizes the human gut microbiome. Cell 2018;175:962–972.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gurven M, Blackwell AD, Rodríguez DE, Stieglitz J, Kaplan H. Does blood pressure inevitably rise with age? Hypertension 2012;60:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown MJ. Hypertension and ethnic group. Br Med J 2006;332:835–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mahler A, Balogh A, Marko L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Fratzer C, Krannich A, Gollasch M, Grohme DA, Corte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, Müller DN. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017;551:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 2015;11:577–591. [DOI] [PubMed] [Google Scholar]
- 28. Yang T, Magee KL, Colon-Perez LM, Larkin R, Liao YS, Balazic E, Cowart JR, Arocha R, Redler T, Febo M, Vickroy T, Martyniuk CJ, Reznikov LR, Zubcevic J. Impaired butyrate absorption in the proximal colon, low serum butyrate and diminished central effects of butyrate on blood pressure in spontaneously hypertensive rats. Acta Physiol 2019;226:e13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiotade rived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A 2013;110:4410–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bender JM, Li F, Adisetiyo H, Lee D, Zabih S, Hung L, Wilkinson TA, Pannaraj PS, She RC, Bard JD, Tobin NH, Aldrovandi GM. Quantification of variation and the impact of biomass in targeted 16S rRNA gene sequencing studies. Microbiome 2018;6:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene amplicon raw sequence data and associated metadata have been deposited at the European Genome-phenome Archive under study number EGAD00001004106. The HELIUS data are owned by the Amsterdam UMC, location AMC in Amsterdam, the Netherlands. Any researcher can request the data by submitting a proposal as outlined at http://www.heliusstudy.nl/.