Abstract
Chondroitin sulfate (CS) is the placental receptor for the VAR2CSA malaria protein, expressed at the surface of infected erythrocytes during Plasmodium falciparum infection. Infected cells adhere to syncytiotrophoblasts or get trapped within the intervillous space by binding to a determinant in a 4-O-sulfated CS chains. However, the exact structure of these glycan sequences remains unclear. VAR2CSA-reactive CS is also expressed by tumor cells, making it an attractive target for cancer diagnosis and therapeutics. The identities of the proteoglycans carrying these modifications in placental and cancer tissues remain poorly characterized. This information is clinically relevant since presentation of the glycan chains may be mediated by novel core proteins or by a limited subset of established proteoglycans. To address this question, VAR2CSA-binding proteoglycans were affinity-purified from the human placenta, tumor tissues and cancer cells and analyzed through a specialized glycoproteomics workflow. We show that VAR2CSA-reactive CS chains associate with a heterogenous group of proteoglycans, including novel core proteins. Additionally, this work demonstrates how affinity purification in combination with glycoproteomics analysis can facilitate the characterization of CSPGs with distinct CS epitopes. A similar workflow can be applied to investigate the interaction of CSPGs with other CS binding lectins as well.
Keywords: cancer, chondroitin sulfate, glycopeptides, mass spectrometry, proteoglycans
Introduction
Chondroitin sulfate (CS) proteoglycans are important biomolecules involved in the organization of most extracellular matrices (Lindahl et al. 2015). They are composed of a core protein modified with one or more polyanionic CS chains (Anderson et al. 1965). CS is a high molecular weight linear polysaccharide made up of long stretches of alternating N-acetylgalactosamine (GalNAc) and glucuronic acid (GlcA) residues. The biosynthesis of CS initiates with the assembly of a linkage tetrasaccharide (β4GlcAβ3Galβ3Galβ4Xylβ1-O-) covalently bound to specific serine residues on a small subset of proteins, called proteoglycans (PGs) (Lindahl and Roden 1966; Roden and Smith 1966). Extension of this linkage region entails the attachment of a GalNAc residue followed by an alternating addition of GlcAβ3 and GalNAcβ4 units. While the backbone structure is simple, a high degree of structural variation arises through the action of carbohydrate-specific sulfotransferases that add sulfates to the component disaccharide units (Habuchi 2000; Kusche-Gullberg and Kjellen 2003). The GalNAc residues can be modified by 4-O sulfation (C4S, CSA), 6-O sulfation (C6S, CSC) or both (C4, 6diS, CSE) and/or 2-O sulfation of the GlcA residues (CSD). The biological activities of CS depend on the type and degree of sulfation as well as the length of the chains (Kawashima et al. 2002; Zhou et al. 2014; Koike et al. 2015).
In addition to their structural role in extracellular matrix (ECM) organization, CSPGs mediate specialized functions, such as cellular signaling, cell adhesion, cell migration/invasion, chemokinesis and morphogenesis (Maurel et al. 1994; Hwang et al. 2003; Chanana et al. 2009; Dickendesher et al. 2012; Smith et al. 2015). Multiple pathogens exploit proteoglycans as adhesion receptors, and they modulate cancer cell behavior by shaping the tumor microenvironment (Esko et al. 1988; Uyama et al. 2006; Avirutnan et al. 2007; Cooney et al. 2011). Malaria-infected erythrocytes (IEs) evade host immune surveillance through adhesion to CS in the placenta (Fried and Duffy 1996). Placental sequestration of IEs is mediated by the VAR2CSA protein, a member of the PfEMP1 protein family produced by the malaria pathogen Plasmodium falciparum (Buffet et al. 1999; Reeder et al. 1999; Magistrado et al. 2008). Upon erythrocyte infection, VAR2CSA is expressed at the plasma membranes of IEs, facilitating cytoadherence to distinct subsets of CS chains on CSPGs at the plasma membrane of the placental syncytiotrophoblasts (Ayres Pereira et al. 2016). Placental adhesion is an immune escape mechanism that results in massive sequestration of IEs in the placenta, causing severe complications such as maternal anemia, spontaneous abortion, low birth weight and stillbirth (Brabin et al. 2004).
The specific binding of VAR2CSA-expressing IEs to CS in placental tissue and not to CS in other organs suggests that placental CS is structurally distinct. Interestingly, the same type of CS modification has been found in a large number of tumors, allowing for a broad targeting of human cancer cells with a recombinant VAR2CSA (rVAR2) construct (Salanti et al. 2015). The specific structure of this CS subtype, also called oncofetal CS (ofCS), remains to be fully characterized. Current evidence suggests the presence of discrete structural requirements for binding including, but not limited to, extensive 4-O sulfation (Achur et al. 2000; Alkhalil et al. 2000; Valiyaveettil et al. 2001; Achur et al. 2008; Sugiura et al. 2016a). While some studies have suggested what PGs may carry ofCS (Ayres Pereira et al. 2016; Clausen et al. 2016), the full repertoire of core proteins and their specific protein attachment sites remains unknown. This information is important as the CS chains may be associated with completely novel core proteins or may be restricted to a specific subset of proteoglycans. Elucidating the chemical makeup of CSPGs that can bind VAR2CSA is thus critical to modulate malarial infection and tumor biology.
Structural analysis of proteoglycans is challenging. The CS chains are heterogeneous in terms of size, charge density and degree of sulfation. Multiple CS chains can also be attached to the same core protein, at different sites, and with different degrees of site occupancy. Typically, the CS chains are separated from the core protein either chemically or through extensive proteolytic digestion. The purified CS chains are then enzymatically degraded into disaccharides by bacterial chondroitinases and analyzed by chromatography, NMR or mass spectrometry (Sugahara et al. 1988; Oguma et al. 2007; Lawrence et al. 2008; Sisu et al. 2011; Gill et al. 2013). Application of these methods provides a picture of the general disaccharide composition and location of sulfate groups, but critical information such as the identity of the proteoglycan core proteins and the glycan attachment sites are lost. Expression of core proteins is instead commonly evaluated from mRNA expression data and/or western blots. As a complementary method, we recently reported a novel glycoproteomics protocol to analyze CSPG linkage region glycopeptides in complex samples (Gomez Toledo et al. 2015; Noborn et al. 2015; Nilsson et al. 2017). Application of this method to human plasma, urine and cerebral spinal fluid led to the identification of novel CSPGs and a detailed characterization of the interalpha trypsin inhibitor complex.
In the present study, we combined our previously published glycoproteomics method, with an affinity purification of CSPGs on rVAR2. This adds a level of biological relevance above the general purification of CSPGs by ion exchange chromatography, by allowing the identification of CSPGs that carry distinct structural epitopes. We purified CSPGs from human placental tissue, bladder urothelial carcinoma tumor samples and two cancer cell lines of diverse cellular origins by affinity chromatography on a column containing rVAR2. The fractionated material was analyzed through a glycoproteomics workflow, similar to the previously described (Gomez Toledo, et al. 2015; Noborn et al. 2015; Nilsson, et al. 2017), to define the identity of the enriched CSPGs and the location of their CS attachment sites. Collectively, the data indicates that ofCS can be carried by multiple core proteins, including completely novel CSPGs.
Results
CS proteoglycan extraction and glycoproteomics workflow
To establish the identity of the VAR2CSA-reactive proteoglycans expressed in the human placenta and cancer, rVAR2 was immobilized on columns and used for affinity purification (Materials and Methods). Multiple cell lines and tissues were selected for the analysis, including the human placenta, a bladder urothelial carcinoma tumor sample and two cancer cell lines: C32 (melanoma) and BeWo (choriocarcinoma). Cell and tissue samples were homogenized, and proteins were initially extracted either with 2% sodium dodecylsulfate (SDS) or with 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), to facilitate isolation of membrane-anchored CSPGs. The detergent-extracted material was diluted and subsequently fractionated on a column containing rVAR2 by removing low-affinity material with a low salt buffer and elution of high-affinity material using a high salt buffer (Materials and Methods). The high-affinity material was then analyzed by liquid chromatography-mass spectrometry. The general workflow is depicted in Figure 1. Due to the denaturating properties of SDS, the detergent was removed, and trypsin digestion was done on these samples prior to rVAR2 enrichment, whereas samples solubilized with CHAPS were fractionated first and then processed for mass spectrometry.
Fig. 1.
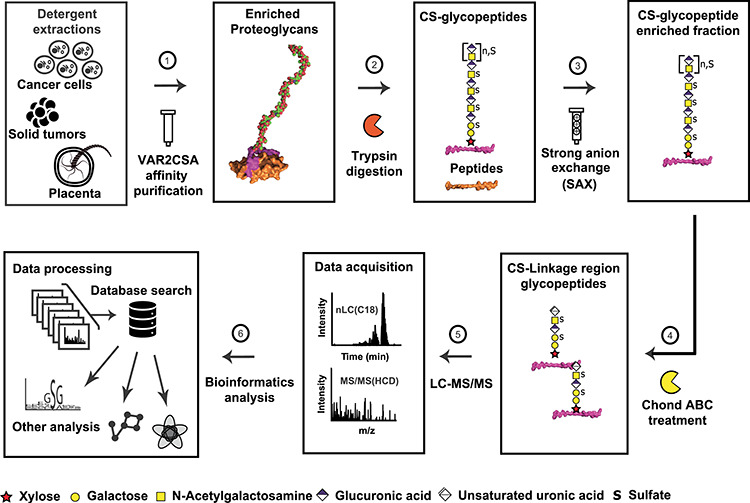
VAR2CSA affinity chromatography coupled to a glycoproteomics pipeline facilitates structural characterization of CS glycopeptides. CSPGs from human placenta, bladder tumor samples and two cancer cell lines were subjected to detergent extractions and fractionated on immobilized recombinant Var2CSA. The pull-down fractions were proteolytically digested with trypsin, and the generated CS glycopeptides were isolated through strong anion exchange (SAX) chromatography. The enriched CS-substituted glycopeptides were further subjected to chondroitinase ABC treatment to generate glycopeptides containing the CS linkage region. After reverse-phase chromatography on C18 column, the CS linkage region glycopeptides were analyzed by high-resolution mass spectrometry to fully characterize the core protein identities, the CS attachment sites and the glycan composition of the linkage region oligosaccharides.
A comparison of glycopeptide preparations from equal amounts of the placenta (100 mg) using SDS or CHAPS showed the successful enrichment of CS glycopeptides in both samples, based on the recovery of the diagnostic oxonium ion at m/z 362.11 (4,5-unsaturated uronate-HexNAc) derived from the linkage region oligosaccharide (Figure 2A and B). Multiple core proteins were identified in both the SDS and the CHAPS samples, including well-known CSPGs, such as decorin (DCN) and versican (VCAN). However, the total ion intensity in the SDS samples (Figure 2A) was generally lower, compared to the CHAPS samples (Figure 2B). This pattern was more pronounced for glycopeptides eluting between 30 and 45 min, presumably derived from different core proteins. Both extraction methods led to similar glycopeptide identifications, with the notable exception of membrane-anchored proteoglycans CD44-antigen (CD44), chondroitin sulfate proteoglycan 4 (CSPG4), glypican 3 (GPC3) and syndecan-1 (SDC1), which were found in the CHAPS extracts but were absent from the SDS extracts (Figure 2B).
Fig. 3.
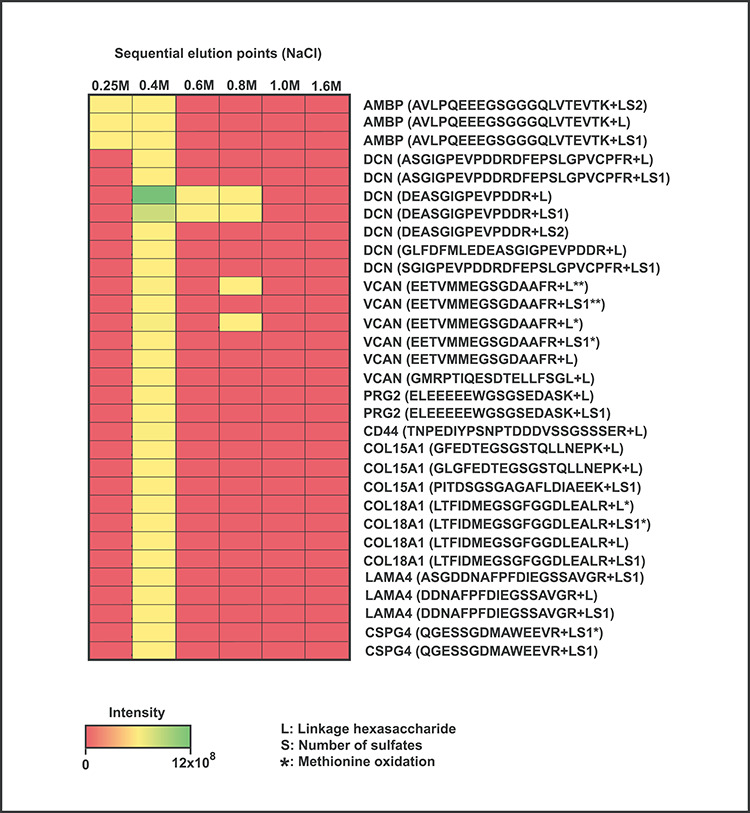
Sequential fractionation of placental CSPGs coupled to glycoproteomics detection. Intensity distribution of CS linkage region glycopeptides derived from VAR2CSA-reactive CSPGs, eluted at different NaCl concentrations from the rVAR2 column. Most glycopeptides were detected at the 0.4 M elution step, except for DCN and VCAN, which were also identified in the high salt fractions suggesting association with CS chains displaying higher-affinity binding.
Fig. 2.
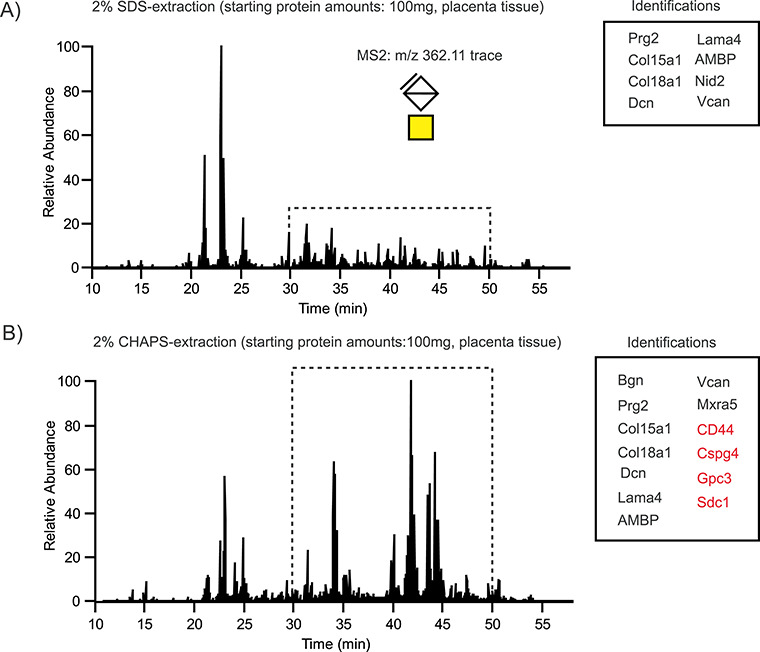
Detergent extraction of placental CSPGs. Intensity traces of the CS-specific oxonium ion m/z 362.11 in human placental samples after extraction either with sodium dodecyl sulfate (SDS) (A) or with 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and fractionation on a column with immobilized rVAR2 (B). The major intensity differences were observed for CS glycopeptides eluting between 30 and 50 min (highlighted with a dashed box) corresponding to glycosites derived from the membrane-anchored proteoglycans CD44, CSPG4, GPC3 and SDC1 (marked in red).
Core proteins and distributions
Based on these findings, subsequent experiments employed the CHAPS extraction method. Each biological source was analyzed in triplicate. In total, the glycoproteomics workflow identified 219 unique glycopeptides, covering 36 unique CS attachment sites derived from 25 nonredundant core proteins. All glycopeptides were identified using a false discovery rate (FDR) cutoff of 1% (Table I and Supplemental File 1). Annotated representative MS/MS spectra for all the identified proteoglycans and their suggested glycan structures are shown in Supplemental Figure 1. The final protein list contains multiple secreted, membrane-anchored and soluble CSPGs, including four novel CS-modified core proteins: B-type brain natriuretic peptides (NPPB), sushi-repeat-containing protein X chromosome (SRPX), laminin subunit gamma 2 (LAMC) and nidogen-2 (NID2).
Table I.
Summary of all human chondroitin sulfate proteoglycans identified in this study: Core proteins (24), unique CS glycopeptides (217) and unique sites (36)
| Protein | # of unique sites | Tissue | Subcellular localization |
|---|---|---|---|
| Aggrecan core protein | 1 | Bladder tumor | ECM |
| Amyloid-like protein 2 | 1 | C32 | Membrane |
| Biglycan | 1 | Placenta | Secreted |
| Bone marrow proteoglycan | 1 | Placenta | Secreted |
| B-type brain natriuretic peptidesa | 1 | BeWo | Secreted |
| CD44 antigen | 1 | C32, placenta | Membrane |
| Chondroitin sulfate proteoglycan 4 | 1 | C32, placenta, bladder tumor | Membrane |
| Collagen alpha-1(XII) chain | 1 | Bladder tumor | ECM |
| Collagen alpha-1(XV) chain | 3 | Placenta, bladder tumor | ECM |
| Collagen alpha-1(XVIII) chain | 2 | Placenta, bladder tumor | ECM |
| Collagen and calcium-binding EGF domain-containing protein 1 | 1 | BeWo | Secreted |
| Decorin | 1 | C32, placenta, bladder tumor | Secreted |
| Endothelial cell-specific molecule 1 | 1 | Bladder tumor | Secreted |
| Glypican-3 | 1 | Placenta | Membrane |
| HLA class II histocompatibility antigen gamma chain | 1 | C32 | Membrane |
| Laminin subunit alpha-4 | 1 | C32, placenta, bladder tumor | ECM |
| Laminin subunit gamma 2a | 1 | BeWo | ECM |
| Matrix remodeling-associated protein 5 | 1 | Placenta, bladder tumor | ECM |
| Nidogen-2a | 2 | Placenta, bladder tumor | ECM |
| Perlecan | 1 | Bladder tumor | ECM |
| Protein AMBP | 1 | C32, placenta, bladder tumor | Secreted |
| Sushi-repeat-containing protein (SRPX)a | 1 | C32 | Secreted |
| Syndecan-1 | 1 | Placenta, bladder tumor, C32, BeWo | Membrane |
| Syndecan-4 | 1 | BeWo, C32 | Membrane |
| Versican core protein | 8 | C32, placenta, bladder tumor | ECM |
aNovel chondroitin sulfate proteoglycans.
Interestingly, a large proportion of the identified glycopeptides were exclusively found as semitryptic species, in close vicinity to sites of endogenous proteolytic processing such as propeptide regions. NPPB is the precursor protein of the B-type natriuretic peptide, a cardiac hormone and a sensitive clinical marker for heart failure (Sergeeva and Christoffels. 2013). Its proteoglycan form was exclusively found in the BeWo choriocarcinoma cells, and the novel CS attachment site was located at Ser34 within the propeptide region. In contrast, the novel proteoglycan SRPX was only detected in C32 cells, but not in BeWo, the placenta or in the bladder tumor samples. The CS attachment site on SRPX was also located on the propeptide region at Ser34, immediately after the cleavage site for the signal peptide. A novel CS attachment site was also identified towards the C-terminal domain of LAMC2, in a region containing two consecutive SG dipeptide repeats that included the potential CS attachment sites S803 and S805. All identified LAMC2 glycopeptides carried a single CS chain, but the MS/MS fragmentation did not distinguish between these two glycosylation sites. NID2 is an important basement membrane component that mediates matrix organization through interactions with perlecan, collagens, laminins and other ECM proteins. The novel proteoglycan form of NID2 was identified in the placenta and bladder tumor samples, but not in any of the tumor cell lines. Two CS glycosylation sites were found at S357 and S452, between the NIDO and the EGF-like 1 domain.
In total, 21 out of the 25 identified core proteins were modified with a single CS chain, whereas 4 proteins carried 2 or more chains. A notable example was VCAN, a large basement membrane proteoglycan that regulates cell motility, growth and differentiation (Wight 2002). VCAN occurs in five isoforms that differ in the amount and content of CS chains distributed along the GAG-α and GAG-β domains of the core protein. Interestingly, eight unique glycosylation sites were found in VCAN from the C32 cells, the placenta and bladder tumor samples, all of them exclusively derived from the GAG-β domain. The V2 and V3 isoforms completely lack the GAG-β domain, while V4 only has a portion of it, directly implicating the V0 and V1 isoforms as the most likely species detected in our samples. Additionally, it needs to be pointed out that VCAN is a well-known CSPG but the exact locations of the CS attachment sites have not been well resolved. Thus, our data also provides a site-specific molecular description of eight novel CS glycosylation sites on the GAG-β domain of VCAN.
A comparative view of the core protein distributions highlights the fact that some proteoglycans were detected in a tissue-specific fashion whereas others were present in multiple samples. The most widely distributed CSPGs were the membrane-anchored CSPG4 (found in C32, the placenta and bladder tumor) and SDC1 (found in C32, BeWo, the placenta and bladder tumor) (Table I). CSPG4 is a well-studied melanoma-associated antigen that becomes heavily upregulated in many kinds of cancer (Wang et al. 2010), whereas SDC1 located at the plasma membrane of the placental syncytiotrophoblasts is the main endogenous receptor for VAR2CSA-expressing erythrocytes (Ayres Pereira et al. 2016).
Multiple proteoglycans were identified during the rVAR2 affinity purification, indicating that VAR2CSA binds to the placenta through protein–carbohydrate interactions, in line with previous reports (Fried and Duffy 1996; Alkhalil et al. 2000; Clausen et al. 2016; Sugiura et al. 2016b). However, since affinity purification was performed using a single elution point, it is possible that VAR2CSA displays different affinities towards discrete CSPGs. For example, some of the core proteins may be modified with CS chains highly enriched in the VAR2CSA-binding motif, which will translate into higher binding affinities. To test this possibility, placental CSPGs were bound to the rVAR2 column and sequentially eluted by stepwise increase in the NaCl concentration (0.25, 0.4, 0.6, 0.8, 1.0 and 1.6 M). The eluted material was desalted and subjected to the glycoproteomics workflow to identify core proteins associated with CS chains displaying high-affinity binding to rVAR2.
As shown in Figure 3, glycopeptides derived from AMBP (bikunin) were detected in the wash fraction at 0.25 M, consistent with the fact that AMBP is an exceptionally low sulfated CSPG and therefore a poorer VAR2CSA ligand (Ly et al. 2011). Most of the glycopeptides derived from VAR2CSA-reactive proteoglycans eluted at 0.4 M NaCl. Glycopeptides derived from DCN and VCAN also were recovered in the high salt fractions (0.6 and 0.8 M NaCl) indicating that these core proteins have a higher affinity for VAR2CSA. Importantly, we did not observe any correlation between linkage region modifications and the retention of the CSPGs on the rVAR2 column, suggesting that the assembly of the VAR2CSA epitope is not linked to the novel modifications of the linker region but rather to epitopes on the extended GAG chains of the enriched proteoglycans. Taken together, our data supports the concept that ofCS chains are displayed by several CSPGs in both placental and cancer cells. Each cell coexpresses different CSPGs, and some, but not all, core proteins are shared between tumor and cancer cell lines of different origin. Additionally, this data confirms that SDC1 is one of the main membrane-anchored proteoglycans in the human placenta and confirms its status as a carrier of VAR2CSA reactive of CS chains, in line with previous reports (Ayres Pereira et al. 2016). However, other placental CSPGs were also identified, including CSPG4, CD44 and GPC3. Furthermore, our data shows that the glycoproteomics approach can be combined with more refined fractionation to identify core proteins carrying CS chains that are enriched for the VAR2CSA-binding CS determinants.
Fig. 6.
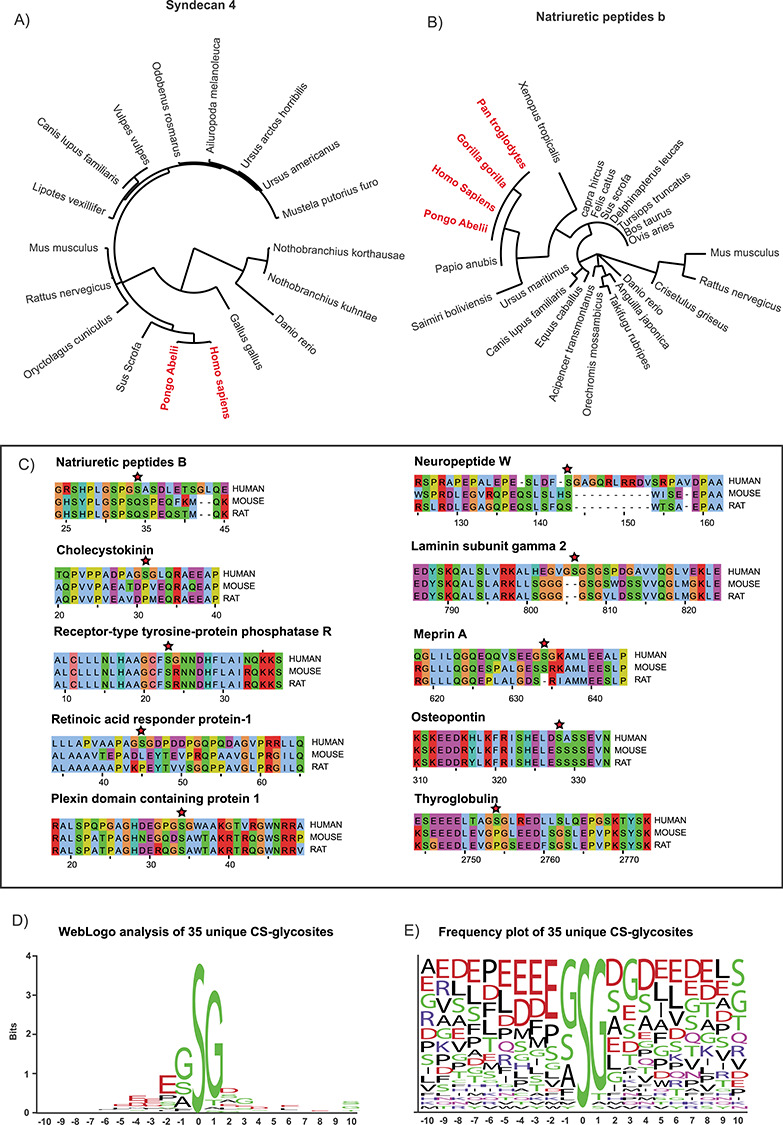
Sequence analysis of the CS glycopeptides. Phylogenetic analysis of the amino acid sequence of SDC4 (A) and NPPB (B) across multiple species indicates that the novel human CS glycosites are conserved (in red) in primates but not in other mammals, fish or reptile protein sequences. Similarly, some previously identified human CS glycosites also display poor sequence conservation when compared with the homologous mouse and rat sequences (C). Sequence motif WebLogo analysis of 35 unique glycosites identified in this study highlights the importance of small amino acids directly adjacent to the glycosylated serine. The presence of acidic residues in the vicinity of the CS attachment site was also commonly found (D-E).
Glycoform heterogeneity
A powerful feature of the glycoproteomics workflow is that it provides insights into the structure of the CS linkage regions. Chondroitinase treatment depolymerizes the chains into disaccharides and a hexasaccharide “stub” attached to the peptide. This “CS stub” includes the linkage tetrasaccharide, plus an additional Δ4,5-unsaturated hexuronic acid-GalNAc disaccharide on the nonreducing end (Figure 1). Several unusual modifications of the linkage region include sialylation, phosphorylation and sulfation (Persson et al. 2018; Tone et al. 2008a). To examine the structure of the linkage regions in the VAR2-binding CSPGs, the raw MS/MS files were subjected to spectral clustering, as previously reported (Nasir et al. 2016, Wang et al. 2016). In this method, all precursor ions were filtered based on the presence of the CS glycopeptide-specific m/z 362.11 oxonium ion. The extracted glycopeptide scans were then clustered based on spectral similarities because similar fragmentation patterns would indicate similar structural features. We have previously shown that the mass difference between the clustered parent ions reflects glycoform variations (Nasir et al. 2016). For example, two identical linkage regions differing only in the presence of a single sulfate substitution (79.96 Da) will fragment similarly and efficiently cluster. The mass difference between the clustered parent ions will then equal the mass of the sulfate group.
A total of 4270 CS glycopeptide spectra from the placental samples, 7243 spectra from the bladder tumor samples and 4380 spectra derived from the cancer cell files were grouped through the online clustering workflow at the Global Natural Products Social Molecular Networking (GNPS): https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp. Histograms depicting the mass differences between the clustered precursors are shown in Figure 4A–C. As expected, some of the major variations among the clustered, and therefore structurally related precursor ions, could be accounted for by the presence of chemical modifications, such as ammonium adducts (+17.03 Da) and carbamidomethyl derivatives (+57.02 Da), introduced artifactually during the proteomics sample preparation and analysis. However, differences reflecting 1 (+79.96 Da) or 2 (+159.92 Da) additional sulfate groups were clearly present, as well as mass shifts corresponding to a HexA-GalNAc disaccharide with or without a sulfate group. The presence of these extra mass values suggests structures containing an additional disaccharide (i.e., an octasaccharide linkage fragment), possibly resulting from incomplete digestion by chondroitinase. Some mass shifts corresponding to Neu5Ac were also detected. As previously shown, these are associated with abundant mucin-type glycosylation on bikunin CS glycopeptides (Gomez Toledo et al. 2015). Accordingly, bikunin was readily identified in the C32, placenta and bladder tumor samples (Table I and Supplemental File 1). Interestingly, we also observed some mass shifts in the cancer cell lines reflecting the presence of fucosylated CS glycoforms. These extra modifications were present specifically in the BeWo cells and traced to glycopeptides derived from the membrane-bound proteoglycan SDC4.
Fig. 7.
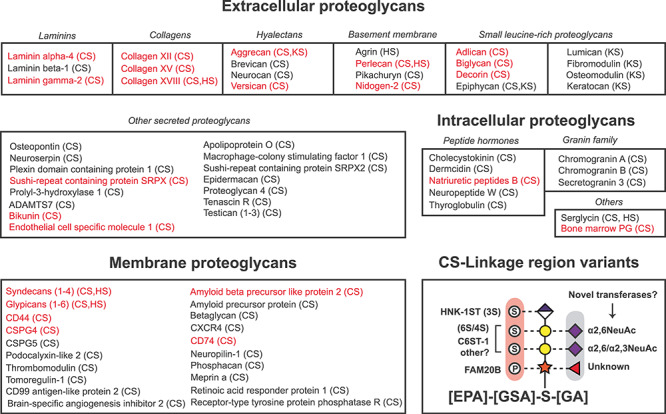
Summary of all 77 mammalian proteoglycan core proteins known to date and all reported CS linkage region variations. Mammalian proteoglycans can be classified based on their cellular localization as extracellular, intracellular and membrane-anchored proteoglycans. The core proteins identified in this study are colored in red and stretch over these three categories. A summary of all CS linkage region variations reported in the literature includes phosphorylation, sulfation, sialylation and fucosylation (lower panel, bottom right). A proposed consensus motif for CS attachment is also presented, based on sequence analysis of the 36 unique glycosites identified in this study. References for the listed proteoglycans are listed in Supplemental File 2.
Fig. 4.
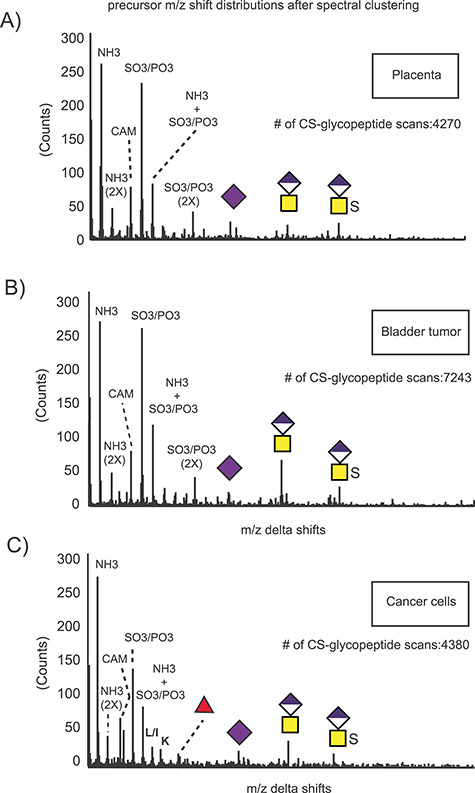
Glycoform analysis of linkage region substitutions through spectral clustering and molecular networking. All MS/MS scans were filtered based on the presence of the CS-specific oxonium ion m/z 362.11, and the selected glycopeptide scans were clustered based on spectral similarities. The m/z precursor shift distributions for the clustered glycopeptides derived from the human placenta (A), bladder tumor (B) and cancer cell lines (C) reveal the presence of structural variations in the samples based on number of sulfate groups, length of the depolymerized linkage regions, sialic acid and fucose as well as artefactual chemical modifications such as ammonium adducts and carbamidomethylation.
The fragmentation pattern of the SDC4 glycopeptides upon higher-energy collisional dissociation (HCD) is shown in Figure 5A–D. The single CS chain is attached to Ser95 and flanked by a glycine residue in a semiconsensus motif for GAG attachment (SG dipeptide flanked by acidic residues) (Kokenyesi and Bernfield 1994; Zhang and Esko 1994). Both hexasaccharide (Figure 5A) and octasaccharide (Figure 5C) cleavage products were detected, consistent with the results from the glycoform networking analysis. As expected, oxonium ions corresponding to the nonreducing disaccharide Δ4,5-HexA-GalNAc (m/z 362.11) were among the most abundant ions in the MS/MS spectra. The octasaccharide yielded internal HexA-GalNAc (m/z 380.12) fragment ions (Figure 5C and D). As mentioned above, several glycopeptides were also found carrying fucose (Fuc) (Figure 5B and D). Glycopeptide fragments at m/z 1289.63 (Pep + Xyl), m/z 1435.68 (Pep + Xyl [Fuc]) and m/z 1597.74 (Pep + Xyl [Fuc] + Gal) indicated that the Fuc moiety was attached to the Xyl residue. The position of this rare substitution agrees with our previous report of a similar modification on human urinary CSPGs (Gomez Toledo et al. 2015).
Fig. 5.
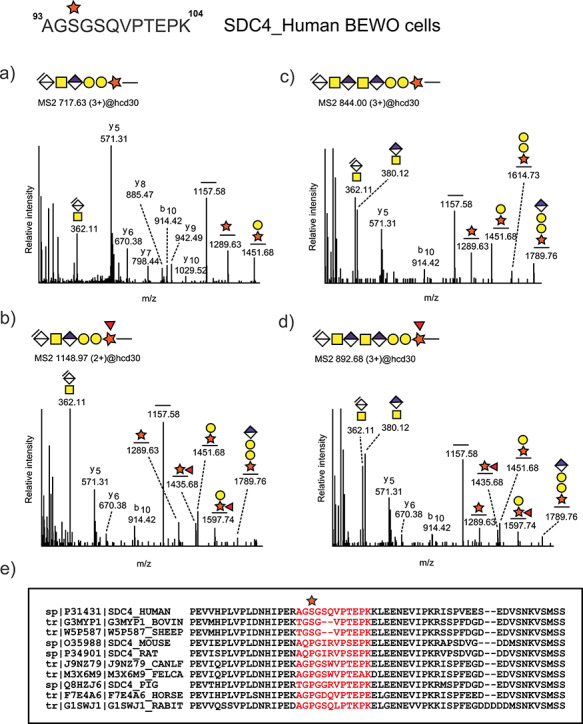
Identification of CS linkage region glycopeptides derived from human syndecan-4 in Var2CSA enriched fractions from BeWo cells. Representative higher-energy collisional dissociation (HCD) fragmentation spectra of CS glycopeptides carrying hexasaccharide (A-B) and octasaccharide (C-D) linkage region fragments. The octasaccharide structures were characterized by the presence of internal disaccharide fragments HexA-GalNAc at m/z 380.12 in addition to the terminal unsaturated fragment Δ4,5-unsaturated HexA-GalNAc at m/z 362.11. Hexasaccharides (B) and octasaccharide (D) glycoforms displaying fucose substitution attached to xylose were also detected. Sequence alignment of the amino acid sequences covering the novel SDC4 glycosylation site shows that this glycosite is not well-conserved across multiple mammalian species (E).
Sequence and homology
To assess if the novel CS glycosylation site on SDC4 was conserved across species, we performed multiple sequence alignment of the SDC4 protein sequence across several mammalian organisms (Figure 5E). The murine SDC4 core protein contains heparan sulfate (HS) and CS chains depending on the cellular context, and the predicted attachment sites at Ser44, Ser65 and Ser67 have been experimentally confirmed (Shworak et al. 1994). These attachment sites are well conserved in the homologous human sequence at Ser39, Ser61 and Ser63. Interestingly, the rVAR2-enriched SDC4 glycopeptides from the BeWo cells were modified at a different site, Ser95. In contrast to the human sequence, the murine and rat sequences contain a Pro at that position, rendering this site unavailable for CS substitution. The same is true for canine, feline, porcine, equine and leporine sequences (Figure 5E).
The phylogenetic analysis of SDC4 was expanded to cover both mammalian and nonmammalian species. Interestingly, all homologous proteins to human SDC4, across multiple phylogenies, were substituted with other amino acids at that particular site and were therefore unavailable for glycosylation (Figure 6A). The primate SDC4 sequence was a notable exception. The CS glycosylation site in NPPB mirrors the sequence pattern, by being available in multiple primates but changed in most other mammalian, fish and reptilian NPPB sequences (Figure 6B). We extracted multiple experimentally determined CS glycosylation sites from previous datasets (Noborn et al. 2015; Nasir et al. 2016). At least eight additional human CS attachment sites were identified as being poorly conserved across species, including mice and rats, two of the most heavily used animals for modeling human disease (Figure 6C). The evolutionary and functional implications of these observations warrant further investigation.
We evaluated the degree of sequence conservation around the glycosylation sites to identify a potential common motif for CS attachment. In total, 35 unique glycosites were subjected to WebLogo motif analysis (Figure 6D and E). In line with previous reports (Kokenyesi and Bernfield 1994; Zhang and Esko 1994), all CS chains were attached to serine residues directly adjacent to small amino acids, typically glycine but occasionally alanine and serine residues. Acidic residues in close proximity to the glycosylation site, alone or in “patches,” were also rather common. These findings provide further support to the idea that preferred polypeptide sequences facilitate site-specific CS glycosylation (Kokenyesi and Bernfield 1994; Zhang and Esko 1994).
Finally, we mined the literature to create a table of all mammalian proteoglycans for which there is biochemical support regarding their proteoglycan status (Figure 7). With the addition of the 4 novel proteoglycans identified in this study, 58 CSPGs have been identified. Some of these are hybrid proteoglycans containing either CS and/or heparan sulfate (HS) or keratan sulfate (KS) chains. In total, 77 proteoglycans distributed across multiple cellular and extracellular compartments are known. One surprising finding was that an increasing number of CS-carrying peptidic hormones have been identified during the last few years, expanding the number of known intracellular proteoglycans from just serglycin (Conte et al. 2006; Noborn et al. 2015). The proteoglycan family stretches over multiple functional and structural protein categories, with diverse physicochemical properties and molecular functions. A summary of all linkage region variations known to date and a potential consensus sequence for GAG attachment is shown in the bottom right panel of Figure 7.
Discussion
In this study we applied a glycoproteomics strategy to analyze affinity-purified ofCS-carrying proteoglycans derived from the human placenta, a bladder urothelial carcinoma tumor sample and two human cancer cell lines from different origins. Detailed analysis of the core proteins, their glycosylation sites and the glycan structures of the CS linkage regions confirmed that a variety of core proteins can be substituted with ofCS chains. A total of 25 ofCS-carrying core proteins were identified in this study. While some CSPGs showed a tissue-restricted expression, others displayed a broad expression pattern. For example, large proteoglycans such as aggrecan and perlecan were only detected in the bladder tumor sample, whereas others, such as the transmembrane proteoglycan SDC1, were found in the placenta, bladder tumor and cancer cells. We have previously shown that CS-modified SDC1 is the primary receptor for VAR2CSA-expressing infected erythrocytes in the placenta (Ayres Pereira et al. 2016). In that study, we isolated the membrane of the syncytiotrophoblasts and found SDC1 as the primary CSPG, whereas a number of other CSPGs were expressed in the whole placental tissue. These findings suggested that while multiple CSPGs are present in placental tissue, only SDC1 was physically available for IE cytoadherence in the intervillous spaces and the lining syncytium. Here, we confirm that multiple ofCS-carrying CSPGs are present in placental tissue. Thus, the assembly of ofCS does not appear to depend on the nature of the core protein. Further, combining a more refined VAR2CSA fractionation with the proteomics workflow, we identified specific core proteins, such as DCN and VCAN, enriched by their VAR2CSA-reactive glycans. In previous studies, we have shown that the CS chains of placental DCN are indeed strong VAR2CSA binders based on proximity ligation (PLA) assays (Ayres Pereira et al. 2016). However, the tissue localization of DCN was found to be limited to the placental villi stroma, thus not favoring DCN as the placental receptor for infected erythrocytes. The significance of these modifications for the endogenous functions of placental DCN as well as their potential impact during placental malaria infections is an important issue that warrants further investigation.
A survey of the literature revealed that the number of biochemically characterized human proteoglycans (CS, HS and KS) amounts to 77 core proteins (Figure 7). This list includes four novel core proteins reported in this study: NPPB, LAMC2, SRPX and NID2. Interestingly, mucin-type O-linked glycosylation on the NPPB cardiac hormone has previously been reported (Shimizu et al. 2002; Schellenberger et al. 2006). The theoretical molecular weight of NPPB is around 12 kDa, but glycosylated high molecular weight variants of 25 kDa and 36 kDa are also common. In these previous reports, posttranslational modification of Ser34 had been suggested from “blank cycle” Edman sequencing, but the glycosylated peptide was not identified by MS. As the methodological approaches used in those studies were not capable of detecting CS modifications, it is possible that the CS attachment was simply missed. However, another possibility is that GAG modification of NPPB only takes place in certain tissues and under specific conditions. Importantly, glycosylation enhances the stability of NPPB and influences the proteolytic processing of its bioactive peptides (Semenov et al. 2009; Jiang et al. 2010). Our findings suggest that the extent and biological importance of the glycosylation status of NPPB, including the novel CS attachment site, should be revisited. In fact, we have previously shown that other prohormones carry CS modifications as well (Noborn et al. 2015). Our finding of CS attachment to NPPB adds further support to the notion that glycosaminoglycans may play a role in the processing, storage and/or biological functions of human prohormones. The general role of glycosylated NPPB in cancer and particularly in choriocarcinoma remains to be determined.
Interestingly we found that the tetrasaccharide linker associated with SDC4 in the BeWo cells was fucosylated. Several studies have shown that the biosynthesis of the CS linkage oligosaccharide is more complicated than previously assumed. To date, xylose phosphorylation, galactose sulfation and galactose sialylation have been demonstrated (Tone et al. 2008b; Gomez Toledo et al. 2015). Furthermore, we have shown that fucose attachment to the linking xylose can be detected in multiple human urinary proteoglycans (Nasir et al. 2016). In this study, the fucose moiety of glycosylated SDC4 was found attached to xylose, at the same position as previously observed (Gomez Toledo et al. 2015; Nasirs et al. 2016). The transferases involved in these modifications as well as their biological functions remain to be determined. Additional studies are also needed to determine if the repertoire of linkage region modifications differs for ofCS containing proteoglycans vs. proteoglycans that contain CS HS or KS.
Using bioinformatics, we demonstrated that the novel identified CS attachment site in SDC4 is not conserved across species. Surprisingly, this was also true for NPPB as well as for eight additional human CSPG core proteins. Phylogenetic analysis indicated that although most primate SDC4 and NPPB sequences show a high level of conservation around the CS glycosylation site, most other mammalian and nonmammalian species lack a serine residue at the homologous position, preventing site-specific xylosylation and GAG attachment. From an evolutionary standpoint, the specific CS substitution of human and primate sequences may reflect novel functions. More importantly, this observation emphasizes the importance of selecting appropriate experimental animal models that accurately recapitulate the cellular and biochemical context of the human system.
In conclusion, we show that high-resolution glycoproteomics can be coupled to affinity chromatography workflows to identify core protein subpopulations carrying particular glycosaminoglycan modifications. Although this current report focuses on the interaction of VAR2CSA with ofCS-carrying proteoglycans, it is possible that other lectin receptors with defined specificities can be used in a similar fashion. Integrating targeted glycomics and glycoproteomics methods as the one described in this study will increase our knowledge of the diversity of proteoglycans and provide the basis for further studies of the relevant biosynthetic enzymes and their physiological function in physiology and disease processes such as infection and cancer.
Methods
Cell cultures
The C32 melanoma and the BeWo choriocarcinoma cells were obtained from ATCC and grown in the suggested media supplemented with 10% FBS, 1% glutamine and 1% penicillin and streptomycin cocktail.
Proteoglycan extraction from cells and tissue
Placental and bladder urothelial carcinoma tissues (100 mg wet weight) were freeze-dried overnight, resuspended in saline solution (10 mM Phosphate, 2.7 mM KCL, 137 mM NaCl pH 7.4) and homogenized. The samples were centrifuged for 10 min at 14,000G to remove cellular debris, and the clear supernatant was collected. Membrane proteins were extracted by incubating the remaining tissue pellet in EBC lysis buffer (120 mM NaCl, 50 mM Tris-HCl, 2.5 mM MgCl2, 1 mM EDTA and a protease inhibitor cocktail) with either 2% SDS (2 h RT) or 2% CHAPS (2 h 4C), after which the samples were briefly spun down and soluble and detergent fractions were collected and combined. For cell culture extractions, cells were harvested from 9 x cell culture plates (145 × 20 MM) per cell line and incubated in EBC lysis buffer with 2% CHAPS for 2 h at 4°C.
Patient samples
Placental tissue was obtained from a pregnant woman delivering at Rigshospitalet University Hospital, Copenhagen, Denmark. The patient provided informed consent for the tissue to be used for research purposes. A patient with muscle invasive bladder cancer, who underwent cystectomy at Vancouver General Hospital, Vancouver, BC, Canada, was selected for extraction of bladder cancer tissue. The patient provided informed consent for the tissue to be used for research purposes (UBC Clinical Research Ethics Board # H09-01628). The collection of human tissue in this study abided by the Helsinki Principles. Optimal cutting temperature (OCT) compound (Sakura Finetek, California, USA) was used for tissue embedding. OCT-embedded frozen tissue blocks were retrieved from the archives of the Molecular Pathology and Cell Imaging Laboratory, Vancouver Prostate Centre (Vancouver, Canada). Six-μm-thick frozen sections were cut using Leica CM1850 Cryostat at −20°C, for H&E staining to confirm the abundance of urothelial carcinoma and to exclude necrotic tissue histologically before proteoglycan extraction.
Production of rVAR2
The minimal binding domain of VAR2CSA FCR3 (DBL1-ID2a) holding a terminal V5 tag, penta-His tag and split protein tag was recombinantly expressed in Shuffle T7 Escherichia coli as previously described (Salanti et al. 2015). In brief, the protein was purified on a HisTrap column (GE Healthcare, Sigma) followed by a size exclusion chromatography step, and the presence of monomeric rVAR2 was verified by SDS-PAGE.
Preparation of rVAR2 affinity column
The immobilization of rVAR2 onto the HiTrap Sepharose matrix was obtained in a two-step manner utilizing the Spytag-Spycatcher fusion system (Zakeri et al. 2012). Here, the addition of a 13 amino acid peptide tag (spyTag) onto the N-terminal of rVAR2 allowed for covalent attachment to the SpyCatcher protein immobilized on the column. The SpyCatcher was produced in B21 Escherichia coli and purified on a HisTrap column (GE Healthcare, Sigma) after which purity and coupling efficiency was verified on SDS-PAGE. For the preparation of the rVAR2 affinity column, 1 mL HiTrap NHS HP column (GE Healthcare, Sigma) was activated with 1 mM HCl and incubated with 50 mM SpyCatcher for 30 min. Deactivation of column matrix and removal of unbound material were done according to manufacturer’s protocol. Subsequently, 8 μM rVAR2 was added on to the column and allowed to incubate for 1 h at RT to facilitate its fusion to the immobilized SpyCatcher. Finally, the column was washed and equilibrated in PBS.
rVAR2 affinity chromatography
Due to the denaturing properties of SDS, extracted proteins were digested prior to rVAR2 enrichment. Briefly, samples were reduced with DTT (5 mM), alkylated with iodoacetamide (15 mM) and digested with trypsin 1:40 w/w (Promega) overnight. SDS was removed from the digested samples using Pierce detergent removal spin columns (Thermo Scientific). The nondenaturing properties of CHAPS allowed the rVAR2 enrichment step to be performed on protein extracts. For these samples, tissue and cell lysates were adjusted to a final concentration of 1% CHAPS and loaded onto the rVAR2 affinity column. The column was extensively washed with 0.25 M NaCl and eluted in 1.6 M NaCl. Fractionated placental samples were sequentially eluted from the rVAR2 column by increasing the salt concentrations to 0.25, 0.4, 0.6, 0.8, 1.0 and 1.6 M NaCl, respectively. Samples were desalted and stored in −20°C until further analysis.
Enrichment and LC-MS/MS analysis of CS-modified glycopeptides
Glycopeptide enrichment and mass spectrometric analysis were conducted as previously described (Noborn et al. 2015). Briefly, the samples where reduced with dithiothreitol (DTT, 5 mM), alkylated with iodoacetamide (15 mM) and trypsinized overnight. Chondroitin sulfate-containing glycopeptides were enriched by SAX chromatography (Vivapure, Q mini H) and eluted in three steps by increasing NaCl concentration (0.4, 0.8 and 1.6 M NaCl). Samples were combined, desalted and subjected to chondroitinase ABC digestion (1 mU chondroitinase ABC (Sigma) in 10 μL 55 mM sodium acetate, pH 8.0) overnight. Digested glycopeptides were desalted using C18 spin columns (Thermo Scientific, Inc, Waltham, MA) according to the manufacturer’s protocol, dried and stored at −20°C until LC-MS/MS analysis.
LC-MS/MS analysis
Mass spectrometry analysis was acquired on an Orbitrap Fusion (Thermo Fisher Scientific) equipped with an EASY n-LC 1000 (Thermo Fisher Scientific). Samples were diluted in 3% acetonitrile and 0.2% formic acid in water and separated using an in-house packed column, 3 μm ReproSil-Pur C18-AQ particles (Dr. Maisch). The gradient elution was run at 150 nL/min with 7–37% B-solvent over 60 min, 37–80% B-solvent over 5 min and a hold at 80% B-solvent for 10 min. A-solvent is 0.2% formic acid in water, and B-solvent is 0.2% formic acid in acetonitrile. MS1 spectra at m/z 600–2000 were recorded in positive ion mode at 120 k resolution. For MS/MS the most abundant precursor ions of each MS1 spectrum were selected for MS2 by higher-energy collision dissociation (HCD) with normalized collision energy (NCE) values of 20% and 35% during 3 s. The isolation window was 2 m/z units, and an exclusion time of 15 s was implemented. The MS2 m/z range was 100–2000, spectra were collected in profile mode, and the resolution was set to 30 k. The maximum injection time was 115 ms and automated gain control was set to 50 k.
MS data processing and database searches
LC-MS/MS files were processed and searched with Mascot distiller (v. 2.6.10) (Matrix Science). Peaks with signal to noise > 5 were deisotoped and tabulated in the MH+ format. Mascot searches were conducted against the UniProt human database (downloaded 03-22-2018), number of entries: 20,315. Search parameters were set as follows: enzyme, trypsin with up to two potential missed cleavages; fixed modifications: carbamidomethyl on cysteines; and variable modifications: oxidation of methionine and proline. Variable modification accounting for the CS hexasaccharide (993.2809 u) on Ser, including one sulfate (1073.2377 u) and two sulfates (1153.1945), was also included. MS1 accuracy was set to 5 ppm, and MS2 accuracy was set to 20 ppm. The false discovery rate (FDR) was set to 1% and was calculated by searching the MS/MS data against a reversed decoy database. All identified glycopeptides were subjected to manual spectral verification as detailed below. In most cases, the presence of sulfate groups was not possible to separate from potential linkage phosphorylation due to insufficient chromatographic and mass resolution.
MS/MS manual spectral verification
All glycoproteomics identifications were manually validated by searching for the presence of CS glycopeptides and generating extracted ion chromatograms filtered for the absolute presence of m/z 362.10–362.13 at the MS/MS level using the Xcalibur software (Thermo Scientific). The final assignments, of both Mascot and cluster analysis-derived hits, were based on the following criteria: (1) deviation from the calculated monoisotopic mass of the precursor ions <5 ppm; (2) the presence of ΔGlcAGalNAc-specific oxonium ions at m/z 362.11 and the presence of oxonium ions at m/z 126.05, m/z 138.06 and m/z 186.08 originating from decomposition of the m/z 204.09 HexNAc B-ion; (3) stepwise glycopeptide fragmentation and presence of the deglycosylated peptide ion confirming the sequence and mass of the CS linkage region; and (4) the presence of at least three peptide fragment ions originating from the intact C-terminal (y-ions) and N-terminal (b-ions), correct within ±0.01 amu, consistent with the proposed amino acid sequence. Monosaccharide identities of CS linkage components were assumed from established biosynthetic rules since isomeric monosaccharides are also isobaric and cannot be distinguished by mass through glycoproteomics analysis.
Molecular networking
A molecular network was created using the online workflow at the Global Natural Products Social Molecular Networking (GNPS): https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp. The data was filtered by removing all MS/MS peaks within +/− 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top six peaks in the +/− 50 Da window throughout the spectrum. The data was then clustered with MS cluster with a parent mass tolerance of 0.1 Da and a MS/MS fragment ion tolerance of 0.1 Da to create consensus spectra. Further, consensus spectra that contained less than one spectrum were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than six matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes.
Sequence analysis
Multiple sequence alignment was performed using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) algorithm. The phylogenetic analysis and tree building were accomplished using PHYML as a component of the online Phylogeny.fr platform (Dereeper et al. 2008). Consensus motif analysis was performed using the online sequence logo generator WebLogo (Crooks et al. 2004).
Supplementary Material
Acknowledgements
The work was supported by grants from the Swedish Research Council (2014-08266, 2017-00955), the Swedish state under the ALF agreement between the Swedish government and the county councils (ALFGBG_721971) (grant P01 HL131474 to JDE), the European Research Council (Grant no 766544), Independent Research Fund Denmark (Grant no 8020-00446A), the Canadian Institutes of Health Research (CIHR# 377771) and the Danish Cancer Benzon Foundation Society.
Conflict of interest
T.M.C, M.D and A.S are shareholders of VAR2 Pharmaceuticals that owns the rights to the use of rVAR2 in cancer therapy.
Data statement
The proteomics raw data and metadata have been deposited in the MassIVE archive, MSV000084677, and it is publicly available.
Author contributions
A.G.T, G.L, A. S and T.M., conceptualization; A.G.T, J.P, C.B.S, A.P, J. N, M.A.P and T.M.C, data curation; A.G.T, J.P, A.P and J. N, formal analysis; A.G.T, J.P, C.B.S, A.P, J. N, M.A.P and T.M.C, investigation; A.G.T, J.P, A.P, J. N, G.L and T.M.C, methodology; T.G, S.C and A.S., recombinant proteins; A.G.T, J.P and T.C.M, writing-original draft; A.G.T, J.P, C.B.S, A.P, J. N, M.A.P, J.D.E, G.L, A.S and T.C.M, writing-review and editing; J.D.E, G.L, A.S and T.M.C, supervision; M.D, J.D.E, G.L, A.S and T.M.C, funding acquisition; H.Z.O, P. C. B and M. D, clinical samples.
References
- Achur RN, Kakizaki I, Goel S, Kojima K, Madhunapantula SV, Goyal A, Ohta M, Kumar S, Takagaki K, Gowda DC. 2008. Structural interactions in chondroitin 4-sulfate mediated adherence of plasmodium falciparum infected erythrocytes in human placenta during pregnancy-associated malaria. Biochemistry. 47:12635–12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achur RN, Valiyaveettil M, Alkhalil A, Ockenhouse CF, Gowda DC. 2000. Characterization of proteoglycans of human placenta and identification of unique chondroitin sulfate proteoglycans of the intervillous spaces that mediate the adherence of plasmodium falciparum-infected erythrocytes to the placenta. J Biol Chem. 275:40344–40356. [DOI] [PubMed] [Google Scholar]
- Alkhalil A, Achur RN, Valiyaveettil M, Ockenhouse CF, Gowda DC. 2000. Structural requirements for the adherence of plasmodium falciparum-infected erythrocytes to chondroitin sulfate proteoglycans of human placenta. J Biol Chem. 275:40357–40364. [DOI] [PubMed] [Google Scholar]
- Anderson B, Hoffman P, Meyer K. 1965. The O-serine linkage in peptides of chondroitin 4- or 6-sulfate. J Biol Chem. 240:156–167. [PubMed] [Google Scholar]
- Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, Malasit P, Atkinson JP, Diamond MS. 2007. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 3:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres Pereira M, Mandel Clausen T, Pehrson C, Mao Y, Resende M, Daugaard M, Riis Kristensen A, Spliid C, Mathiesen L, EK L, et al. 2016. Placental sequestration of plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and chondroitin sulfate a on Syndecan-1. PLoS Pathog. 12:e1005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabin BJ, Romagosa C, Abdelgalil S, Menendez C, Verhoeff FH, McGready R, Fletcher KA, Owens S, D'Alessandro U, Nosten F et al. 2004. The sick placenta-the role of malaria. Placenta. 25:359–378. [DOI] [PubMed] [Google Scholar]
- Buffet PA, Gamain B, Scheidig C, Baruch D, Smith JD, Hernandez-Rivas R, Pouvelle B, Oishi S, Fujii N, Fusai T et al. 1999. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate a: A receptor for human placental infection. Proc Natl Acad Sci U S A. 96:12743–12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanana B, Steigemann P, Jackle H, Vorbruggen G. 2009. Reception of slit requires only the chondroitin-sulphate-modified extracellular domain of Syndecan at the target cell surface. Proc Natl Acad Sci U S A. 106:11984–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TM, Pereira MA, Al Nakouzi N, Oo HZ, Agerbaek MO, Lee S, Orum-Madsen MS, Kristensen AR, El-Naggar A, Grandgenett PM et al. 2016. Oncofetal Chondroitin Sulfate Glycosaminoglycans are Key Players in Integrin Signaling and Tumor Cell Motility. MCR: Molecular Cancer Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte M, Arcaro A, D'Angelo D, Gnata A, Mamone G, Ferranti P, Formisano S, Gentile F. 2006. A single chondroitin 6-sulfate oligosaccharide unit at Ser-2730 of human thyroglobulin enhances hormone formation and limits proteolytic accessibility at the carboxyl terminus. Potential insights into thyroid homeostasis and autoimmunity. J Biol Chem. 281:22200–22211. [DOI] [PubMed] [Google Scholar]
- Cooney CA, Jousheghany F, Yao-Borengasser A, Phanavanh B, Gomes T, Kieber-Emmons AM, Siegel ER, Suva LJ, Ferrone S, Kieber-Emmons T et al. 2011. Chondroitin sulfates play a major role in breast cancer metastasis: A role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast Cancer Res: BCR. 13:R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: A sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M et al. 2008. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickendesher TL, Baldwin KT, Mironova YA, Koriyama Y, Raiker SJ, Askew KL, Wood A, Geoffroy CG, Zheng B, Liepmann CD et al. 2012. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat Neurosci. 15:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Rostand KS, Weinke JL. 1988. Tumor formation dependent on proteoglycan biosynthesis. Science. 241:1092–1096. [DOI] [PubMed] [Google Scholar]
- Fried M, Duffy PE. 1996. Adherence of plasmodium falciparum to chondroitin sulfate a in the human placenta. Science. 272:1502–1504. [DOI] [PubMed] [Google Scholar]
- Gill VL, Aich U, Rao S, Pohl C, Zaia J. 2013. Disaccharide analysis of glycosaminoglycans using hydrophilic interaction chromatography and mass spectrometry. Anal Chem. 85:1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Toledo A, Nilsson J, Noborn F, Sihlbom C, Larson G. 2015. Positive mode LC-MS/MS analysis of chondroitin sulfate modified Glycopeptides derived from light and heavy chains of the human inter-alpha-trypsin inhibitor complex. Mol Cell Proteomics: MCP. 14:3118–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi O. 2000. Diversity and functions of glycosaminoglycan sulfotransferases. Biochim Biophys Acta. 1474:115–127. [DOI] [PubMed] [Google Scholar]
- Hwang HY, Olson SK, Esko JD, Horvitz HR. 2003. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 423:439–443. [DOI] [PubMed] [Google Scholar]
- Jiang JJ, Pristera N, Wang W, Zhang XM, Wu QY. 2010. Effect of sialylated O-glycans in pro-brain natriuretic peptide stability. Clin Chem. 56:959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H, Atarashi K, Hirose M, Hirose J, Yamada S, Sugahara K, Miyasaka M. 2002. Oversulfated chondroitin/dermatan sulfates containing GlcAbeta1/IdoAalpha1-3GalNAc(4,6-O-disulfate) interact with L- and P-selectin and chemokines. J Biol Chem. 277:12921–12930. [DOI] [PubMed] [Google Scholar]
- Koike T, Mikami T, Shida M, Habuchi O, Kitagawa H. 2015. Chondroitin sulfate-E mediates estrogen-induced osteoanabolism. Sci Rep. 5:8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokenyesi R, Bernfield M. 1994. Core protein structure and sequence determine the site and presence of heparan sulfate and chondroitin sulfate on syndecan-1. J Biol Chem. 269:12304–12309. [PubMed] [Google Scholar]
- Kusche-Gullberg M, Kjellen L. 2003. Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol. 13:605–611. [DOI] [PubMed] [Google Scholar]
- Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. 2008. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 283:33674–33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Couchman J, Kimata K, Esko JD. 2015. Proteoglycans and sulfated glycosaminoglycans In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, et al. editors. Essentials of glycobiology. NY: Cold Spring Harbor; p. 207–221. [Google Scholar]
- Lindahl U, Roden L. 1966. The chondroitin 4-sulfate-protein linkage. J Biol Chem. 241:2113–2119. [PubMed] [Google Scholar]
- Ly M, Leach FE 3rd, Laremore TN, Toida T, Amster IJ, Linhardt RJ. 2011. The proteoglycan bikunin has a defined sequence. Nat Chem Biol. 7:827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistrado P, Salanti A, Ndam NGT, Mwakalinga SB, Resende M, Dahlback M, Hviid L, Lusingu J, Theander TG, Nielsen MA. 2008. VAR2CSA expression on the surface of placenta-derived plasmodium falciparum - infected erythrocytes. J Infect Dis. 198:1071–1074. [DOI] [PubMed] [Google Scholar]
- Maurel P, Rauch U, Flad M, Margolis RK, Margolis RU. 1994. Phosphacan, a chondroitin sulfate proteoglycan of brain that interacts with neurons and neural cell-adhesion molecules, is an extracellular variant of a receptor-type protein tyrosine phosphatase. Proc Natl Acad Sci U S A. 91:2512–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir W, Toledo AG, Noborn F, Nilsson J, Wang M, Bandeira N, Larson G. 2016. SweetNET: A bioinformatics workflow for Glycopeptide MS/MS spectral analysis. J Proteome Res. 15:2826–2840. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Noborn F, Gomez Toledo A, Nasir W, Sihlbom C, Larson G. 2017. Characterization of glycan structures of chondroitin sulfate-glycopeptides facilitated by sodium ion-pairing and positive mode LC-MS/MS. J Am Soc Mass Spectrom. 28:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noborn F, Gomez Toledo A, Sihlbom C, Lengqvist J, Fries E, Kjellen L, Nilsson J, Larson G. 2015. Identification of chondroitin sulfate linkage region glycopeptides reveals prohormones as a novel class of proteoglycans. Mol Cell Proteomics: MCP. 14:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguma T, Tomatsu S, Montano AM, Okazaki O. 2007. Analytical method for the determination of disaccharides derived from keratan, heparan, and dermatan sulfates in human serum and plasma by high-performance liquid chromatography/turbo ionspray ionization tandem mass spectrometry. Anal Biochem. 368:79–86. [DOI] [PubMed] [Google Scholar]
- Persson A, Gomez Toledo A, Vorontsov E, Nasir W, Willen D, Noborn F, Ellervik U, Mani K, Nilsson J, Larson G. 2018. LC-MS/MS characterization of xyloside-primed glycosaminoglycans with cytotoxic properties reveals structural diversity and novel glycan modifications. J Biol Chem. 293:10202–10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder JC, Cowman AF, Davern KM, Beeson JG, Thompson JK, Rogerson SJ, Brown GV. 1999. The adhesion of plasmodium falciparum-infected erythrocytes to chondroitin sulfate a is mediated by P. falciparum erythrocyte membrane protein 1. P Natl Acad Sci USA. 96:5198–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden L, Smith R. 1966. Structure of the neutral trisaccharide of the chondroitin 4-sulfate-protein linkage region. J Biol Chem. 241:5949–5954. [PubMed] [Google Scholar]
- Salanti A, Clausen TM, Agerbaek MO, Al Nakouzi N, Dahlback M, Oo HZ, Lee S, Gustavsson T, Rich JR, Hedberg BJ et al. 2015. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell. 28:500–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger U, O'Rear J, Guzzetta A, Jue RA, Protter AA, Pollitt NS. 2006. The precursor to B-type natriuretic peptide is an O-linked glycoprotein. Arch Biochem Biophys. 451:160–166. [DOI] [PubMed] [Google Scholar]
- Semenov AG, Postnikov AB, Tamm NN, Seferian KR, Karpova NS, Bloshchitsyna MN, Koshkina EV, Krasnoselsky M, Serebryanaya DV, Katrukha AG. 2009. Processing of pro-brain natriuretic peptide is suppressed by O-glycosylation in the region close to the cleavage site. Clin Chem. 55:489–498. [DOI] [PubMed] [Google Scholar]
- Sergeeva IA, Christoffels VM. 2013. Regulation of expression of atrial and brain natriuretic peptide, biomarkers for heart development and disease. Biochim Biophys Acta. 1832:2403–2413. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Masuta K, Aono K, Asada H, Sasakura K, Tamaki M, Sugita K, Yamada K. 2002. Molecular forms of human brain natriuretic peptide in plasma. Clin Chim Acta. 316:129–135. [DOI] [PubMed] [Google Scholar]
- Shworak NW, Shirakawa M, Mulligan RC, Rosenberg RD. 1994. Characterization of ryudocan glycosaminoglycan acceptor sites. J Biol Chem. 269:21204–21214. [PubMed] [Google Scholar]
- Sisu E, Flangea C, Serb A, Zamfir AD. 2011. Modern developments in mass spectrometry of chondroitin and dermatan sulfate glycosaminoglycans. Amino Acids. 41:235–256. [DOI] [PubMed] [Google Scholar]
- Smith PD, Coulson-Thomas VJ, Foscarin S, Kwok JC, Fawcett JW. 2015. "GAG-ing with the neuron": The role of glycosaminoglycan patterning in the central nervous system. Exp Neurol. 274:100–114. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Yamashina I, De Waard P, Van Halbeek H, Vliegenthart JF. 1988. Structural studies on sulfated glycopeptides from the carbohydrate-protein linkage region of chondroitin 4-sulfate proteoglycans of swarm rat chondrosarcoma. Demonstration of the structure gal(4-O-sulfate)beta 1-3Gal beta 1-4XYL beta 1-O-Ser. J Biol Chem. 263:10168–10174. [PubMed] [Google Scholar]
- Sugiura N, Clausen TM, Shioiri T, Gustavsson T, Watanabe H, Salanti A. 2016a. Molecular dissection of placental malaria protein VAR2CSA interaction with a chemo-enzymatically synthesized chondroitin sulfate library. Glycoconj J. 33:985–994. [DOI] [PubMed] [Google Scholar]
- Sugiura N, Clausen TM, Shioiri T, Gustavsson T, Watanabe H, Salanti A. 2016b. Molecular dissection of placental malaria protein VAR2CSA interaction with a chemo-enzymatically synthesized chondroitin sulfate library. Glycoconj J. [DOI] [PubMed] [Google Scholar]
- Tone Y, Pedersen LC, Yamamoto T, Izumikawa T, Kitagawa H, Nishihara J, Tamura J, Negishi M, Sugahara K. 2008a. 2-o-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. J Biol Chem. 283:16801–16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y, Pedersen LC, Yamamoto T, Izumikawa T, Kitagawa H, Nishihara J, Tamura J, Negishi M, Sugahara K. 2008b. 2-O-phosphorylation of xylose and 6-O-sulfation of galactose in the protein linkage region of Glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. J Biol Chem. 283:16801–16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyama T, Ishida M, Izumikawa T, Trybala E, Tufaro F, Bergstrom T, Sugahara K, Kitagawa H. 2006. Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J Biol Chem. 281:38668–38674. [DOI] [PubMed] [Google Scholar]
- Valiyaveettil M, Achur RN, Alkhalil A, Ockenhouse CF, Gowda DC. 2001. Plasmodium falciparum cytoadherence to human placenta: Evaluation of hyaluronic acid and chondroitin 4-sulfate for binding of infected erythrocytes. Exp Parasitol. 99:57–65. [DOI] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T et al. 2016. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 34:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Yu L, Sakakura K, Visus C, Schwab JH, Ferrone CR, Favoino E, Koya Y, Campoli MR et al. 2010. CSPG4 in cancer: Multiple roles. Curr Mol Med. 10:419–429. [DOI] [PubMed] [Google Scholar]
- Wight TN. 2002. Versican: A versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol. 14:617–623. [DOI] [PubMed] [Google Scholar]
- Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. 2012. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 109:E690–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LJ, Esko JD. 1994. Amino-acid determinants that drive Heparan-sulfate assembly in a proteoglycan. J Biol Chem. 269:19295–19299. [PubMed] [Google Scholar]
- Zhou ZH, Karnaukhova E, Rajabi M, Reeder K, Chen T, Dhawan S, Kozlowski S. 2014. Oversulfated chondroitin sulfate binds to chemokines and inhibits stromal cell-derived factor-1 mediated signaling in activated T cells. Plos One. 9:e94402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


