Fig. 3.
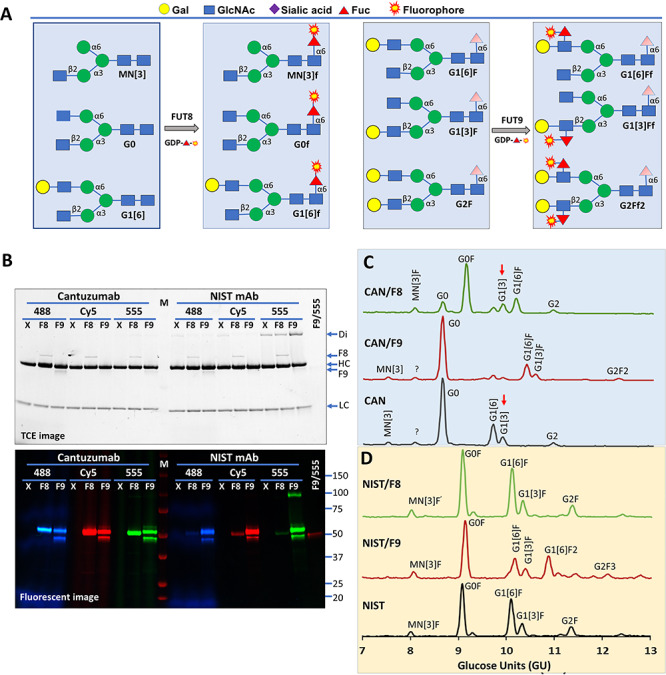
Probing fucosylation on Cantuzumab and NIST antibodies by FUT8 and FUT9. (A) Nomenclature of common antibody glycans and strategies for their detection. M, mannose; N, glucosamine; G, galactose; F, fucose; f, fluorophore-conjugated fucose. Only sugar residues at the nonreducing ends are reflected in the short names. Sugars in shade are flexible. (B) Probing substrate glycans of FUT8 (F8) and FUT9 (F9) on Cantuzumab, an anti-Muc1 therapeutic antibody, and NIST reference mAb 8671 using GDP-Alexa Fluor®555-fucose (555), GDP-Cy5-fucose (Cy5) and GDP-Alexa Fluor®488-Fucose (488) as the donor substrates. Cantuzumab was expressed in FUT8 knockout cells and is devoid of core-6 fucose. M, western molecular marker (Bio-Rad). All reactions were separated on 4–20% gradient SDS–PAGE and imaged with TCE staining (upper panel) and fluorescent imager (lower panel). FUT9 (at 50 kDa) exhibited self-labeling. X, no enzyme control; Di, unreduced dimer of the heavy chain; HC, heavy chain; LC, light chain. (C) GlyQ analysis of the Cantuzumab (CAN) after in vitro modification by FUT8 and FUT9, showing FUT8-modified G0 and G1[6] but not G1[3] (red arrows) and FUT9 modified G1[6] and G1[3]. (D) GlyQ analysis of the NIST mAb after in vitro modification by FUT8 and FUT9, showing that FUT8 had no modification on the glycans and FUT9 modified G1[6]F and G2F.
