Abstract
Acute estrogen deficiency in women can occur due to many conditions including hyperprolactinemia, chemotherapy, GnRH agonist treatment, and removal of hormone replacement therapy. Ovariectomized (OVX) rodent models, often combined with a high-fat diet (HFD), have been used to investigate the effects of decreased estrogen production on metabolism. Since evidence suggests that gut microbes may facilitate the protective effect of estrogen on metabolic dysregulation in an OVX + HFD model, we investigated whether the gut microbiome plays a role in the diet-independent weight gain that occurs after OVX in adult female mice. 16S rRNA gene sequence analysis demonstrated that OVX was not associated with changes in overall gut bacterial biodiversity but was correlated with a shift in beta diversity. Using differential abundance analysis, we observed a difference in the relative abundance of a few bacterial taxa, such as Turicibacter, 3 to 5 weeks after OVX, which was subsequent to the weight gain that occurred 2 weeks postsurgery. A cohousing study was performed to determine whether exposure to a healthy gut microbiome was protective against the development of the metabolic phenotype associated with OVX. Unlike mouse models of obesity, HFD maternal-induced metabolic dysregulation, or polycystic ovary syndrome, cohousing OVX mice with healthy mice did not improve the metabolic phenotype of OVX mice. Altogether, these results indicate that changes in the gut microbiome are unlikely to play a causal role in diet-independent, OVX-induced weight gain (since they occurred after the weight gain) and cohousing with healthy mice did not have a protective effect.
Keywords: ovariectomy, estrogen, gut microbiome, cohousing
The sex steroid hormone estrogen plays a key role in regulating physiological processes involved in reproduction, metabolism, the cardiovascular system, and bone in mammals [1–4]. Of the 3 estrogens (estrone, estradiol, estriol) produced in females, 17-β estradiol (E2) is the primary estrogen circulating in premenopausal women. Estrogens bind to and activate intracellular signaling from 2 related estrogen receptors: estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ) [5]. In animal models, knockout of ERα (αERKO) in female mice caused increased weight and adiposity [6, 7], insulin resistance [7–10], as well as elevated fasting blood glucose (FBG) [8] and fasting blood insulin (FBI) levels [8–10]. In contrast, KO of ERβ in female mice did not cause obesity [8, 10], suggesting that estrogen signaling via ERα primarily regulates metabolism in females.
In addition to the gradual reduction in estrogen levels that occurs during menopause, acute hypoestrogenism or estrogen deficiency in women can occur due to conditions such as hyperprolactinemia, chemotherapy, GnRH agonist treatment, and the removal of hormone replacement therapy [11–14]. Various rodent models of estrogen deficiency have been used to study the role of estrogen in female metabolism. In ovariectomy (OVX) mouse models, OVX resulted in increased weight and adiposity but not insulin resistance [15]. However, OVX mice that were fed a high-fat diet (HFD) developed insulin resistance along with elevated weight and fat mass [15–17]. Interestingly, hormone replacement studies in OVX and OVX + HFD mouse models showed that E2 treatment can attenuate weight gain and protect against insulin resistance [15–17] but only in the presence of ERα [18–20]. These findings indicate that estrogen signaling via ERα is protective against metabolic dysregulation in females.
The gut microbiome, which consists of microbes in the intestinal tract and their metabolites, plays an important role in human health and disease [21, 22], including complex interactions between host metabolism and gut microbes [23–26]. Numerous studies have reported associations between changes in gut microbiota and metabolic disorders such as obesity, type 2 diabetes, and polycystic ovary syndrome in humans and rodent models [27–32]. In addition, studies demonstrated that fecal microbiome transplantation from obese human donors into germ-free mice resulted in an obese phenotype [33, 34], indicating that changes in the gut microbiome may be sufficient to induce metabolic dysregulation. Furthermore, studies showed that cohousing germ-free mice transplanted with stool from lean donors with mice transplanted with stool from obese donors resulted in a protective effect [31, 33, 34], suggesting that the exchange of the gut microbiome between mice due to coprophagy was sufficient to protect the mice from developing obesity.
More recently, studies exploring the relationship between the gut microbiome and estrogen have implicated the significance of the estrobolome, the collection of microbes that can metabolize estrogens and modulate their enterohepatic circulation [35, 36]. While 1 study found that postmenopausal women had lower gut microbial alpha diversity compared with premenopausal women [37], another study reported no differences between these 2 groups of women [38]. Similarly, in rodent models, 1 study found that OVX mice had lower gut microbial alpha diversity than sham-operated mice [39], but other studies indicated that there was no correlation between biodiversity and OVX [40–42]. It is interesting to note, however, that differences in beta diversity were consistently found between sham and OVX mice [16, 39–41].
There is also evidence that OVX + HFD alters the gut microbiome in a unique way compared with OVX alone. It was reported that the gut microbiome of OVX + HFD mice had increased relative abundance (RA) of bacterial taxa from Verrucomicrobia and Proteobacteria, as well as Akkermansia compared with that of OVX mice fed normal chow [39]. Moreover, the presence of Bifidobacterium animalis was only detected in OVX mice compared with OVX + HFD mice [39]. In an OVX + HFD and E2 replacement study, the beta diversity of the gut microbiome of OVX + HFD mice was shown to be distinct from that of HFD mice and OVX + HFD + E2 mice [16]. Interestingly, OVX + HFD + E2 mice had reduced RA of Proteobacteria compared with OVX + HFD mice [16]. Findings from an OVX + HFD + E2 replacement study in leptin mutant (ob/ob) mice also indicated that E2 treatment was associated with an increased abundance of S24-7 bacteria [43]. Altogether, these studies suggest that there may be important host/microbe interactions involved in the protective effect of E2 on OVX + HFD-induced metabolic dysregulation. Since gut microbes appear to play a role in the protective effect of estrogens on the long-term metabolic dysregulation that occurs in OVX females receiving HFD, we investigated whether the gut microbiome also influences the diet-independent weight gain observed shortly after OVX.
1. Materials and Methods
A. Ovariectomy Mouse Model
Eight-week-old C57BL/6N female mice were purchased from Envigo. Mice were housed in a vivarium with a 12-hour light, 12-hour dark cycle (light period: 6:00 am to 6:00 pm). Mice were given ad libitum access to water and food (Teklad S-2335 Mouse Breeder Irradiated Diet; Envigo, Indianapolis, IN). All of the experiments were approved by the University of California San Diego Institutional Animal Care and Use Committee (Protocol S14011). At 8 weeks of age, all mice (n = 8 per group for the 1st cohort, n = 10 per group for the 2nd cohort) underwent either a sham (SHAM) surgery or OVX. Mice were weighed weekly throughout the experiment.
B. Insulin Tolerance Test
Mice were fasted for 5 hours and tail vein blood was collected to measure FBI. Fasting blood glucose was measured with a handheld glucometer (One Touch UltraMini; LifeScan, Inc., Milpitas, CA), and an intraperitoneal insulin tolerance test (ITT) was performed. Fasting blood glucose levels were measured prior to time point 0. At time point 0, an intraperitoneal injection of insulin (0.75 U/kg in sterile saline; Humulin R U-100) was administered. Glucose was measured subsequently at 15, 30, 45, 60, 90, and 120 minutes postadministration of insulin.
C. Tissue Collection
At the end of the experiment, the mice were euthanized with 2.5% isoflurane delivered with a precision vaporizer followed by a physical method. Terminal blood was collected through the inferior vena cava. Parametrial fat pads were dissected and weighed.
D. Hormone Assays
Hormone levels were assessed at week 5. Luteinizing hormone (LH) levels were measured using a radioimmunoassay (range 0.04–75 ng/mL) by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core Facility. Serum insulin was measured with a mouse enzyme-linked immunosorbent assay (ALPO) by the University of California, Davis Mouse Metabolic Phenotyping Center.
E. Statistical Analyses
Data was expressed as the standard error of the mean for each group. Data residuals were checked for normality and data underwent Box Cox transformation if residuals were not normal. If transformation did not result in normality, a nonparametric test was used. The statistical package JMP 14 (SAS) was used to analyze differences between groups by Student t-test or 2-way repeated measures ANOVA followed by post hoc comparisons of individual time points. Statistical significance was defined as P < 0.05.
F. Fecal Sample Collection and DNA Isolation
Fecal samples were collected from 1 cohort of 8-week-old female mice (n = 10 per group) prior to surgery and once per week for 5 weeks. Fecal samples were frozen and stored at -80°C. As described previously [30], DNA was extracted from the fecal samples and amplified via polymerase chain reaction using primers 515F and 806R. Amplicon sequence libraries were prepared at the Scripps Research Institute Next Generation Sequencing Core Facility and the libraries were sequenced on a MiSeq (Illumina, Inc., San Diego, CA).
G. 16S rRNA Gene Sequence Analysis
Raw sequences were imported into QIIME 2 (version 2019.10) with the q2-tools-import script, and sequences were demultiplexed with the q2-demux emp-single script. This procedure resulted in 3.4 million sequences, with an average of 28 000 sequences per sample. DADA2 software was used to obtain a set of observed sequence variants (SVs) [44]. Based on the quality scores, the forward reads were truncated at position 240 with the q2-dada2-denoise script. Taxonomy was assigned with a pretrained naive Bayes classifier (Greengenes 13_8 99% operational taxonomic units) and the q2-feature-classifier plugin [45]. Out of 120 samples, 1 was removed because of insufficient sequence coverage (OVX sample at week 2), resulting in 119 samples. In total, 1126 SVs were identified from 119 fecal samples. The resulting SVs were then aligned in Multiple Alignment using Fast Fourier Transform (MAFFT) [46], and a phylogenetic tree was built in FastTree [47]. Taxonomic distributions of the samples were calculated with the q2-taxa-barplot script. Alpha and beta diversity metrics were computed with the q2-diversity core-metrics script at a rarefied sampling depth of 9638. Two alpha diversity metrics, observed SVs and Faith’s phylogenetic diversity (PD), were used to estimate microbial richness and phylogenetic biodiversity, respectively [48]. UniFrac was used to compare the similarity (beta diversity) between the microbial communities by calculating the shared PD between pairs of microbial communities [49, 50].
H. Statistical Analysis of 16S rRNA Sequences
Statistical calculations were performed in the R statistical package (version 3.6.2) with the phyloseq (version 1.30.0) [51] and vegan package (version 2.5.6). Alpha diversity data were tested for normality via the Shapiro-Wilk test. Principal coordinate analysis (PCoA) plots [52] were constructed in the phyloseq R package. Permutational multivariate analysis of variance (PERMANOVA) used post-treatment weighted and unweighted UniFrac distance measures to assess bacterial community compositional differences (999 permutations, “vegan” package). DESeq2 [53] (version 1.26.0) in Bioconductor (version 3.10.1) was used to identify bacterial genera that were differentially abundant between OVX and SHAM mice. P-values were obtained by the Wald significance test, and a false discovery rate correction was applied using a threshold of P < 0.05.
2. Results
A. Ovariectomy in Mice Results in Metabolic Dysregulation
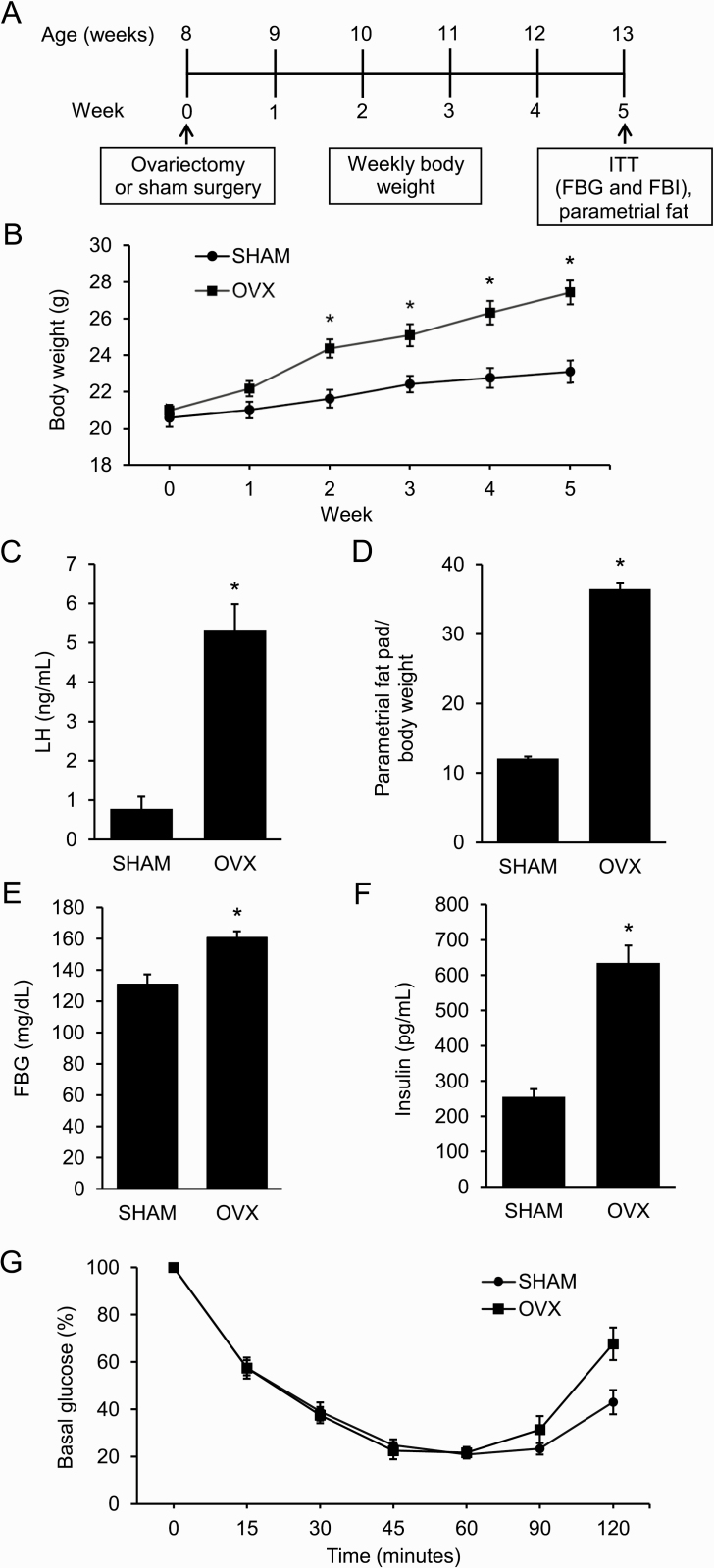
Adult female mice were either OVX or SHAM at 8 weeks of age (Fig. 1A). By 2 weeks postsurgery, OVX mice weighed significantly more than SHAM mice (Fig. 1B). As expected, OVX mice had elevated LH compared with SHAM mice due to a lack of negative feedback (Fig. 1C). In addition, OVX mice had increased parametrial fat relative to body weight at the end of the experiment (Fig. 1D). An ITT, performed in week 5, showed that OVX mice had elevated FBG and FBI compared with SHAM mice (Fig. 1E and 1F). Interestingly, OVX mice were not insulin resistant (Fig. 1G).
Figure 1.
Ovariectomized mice develop distinct metabolic phenotype compared to sham-operated (SHAM) mice. A: Schematic of study design: female mice were either ovariectomized (OVX) or SHAM at 8 weeks of age (n = 8 per group). Experimental procedures included weekly weight assessment, insulin tolerance test (ITT), and parametrial fat collection. B–F: Compared to SHAM mice, OVX mice had increased weight, luteinizing hormone (LH) levels, abdominal adiposity, fasting blood glucose (FBG), and fasting blood insulin (FBI) after 5 weeks. G: OVX mice had similar insulin sensitivity as SHAM mice after 5 weeks. Graph error bars represent standard error of the mean. Student t-test or repeated-measures analysis of variance with post hoc Student t-tests to compare OVX versus SHAM at specific time points were performed; * P < 0.05.
B. Ovariectomy Did Not Alter Species Richness (Alpha Diversity) of the Gut Microbiome
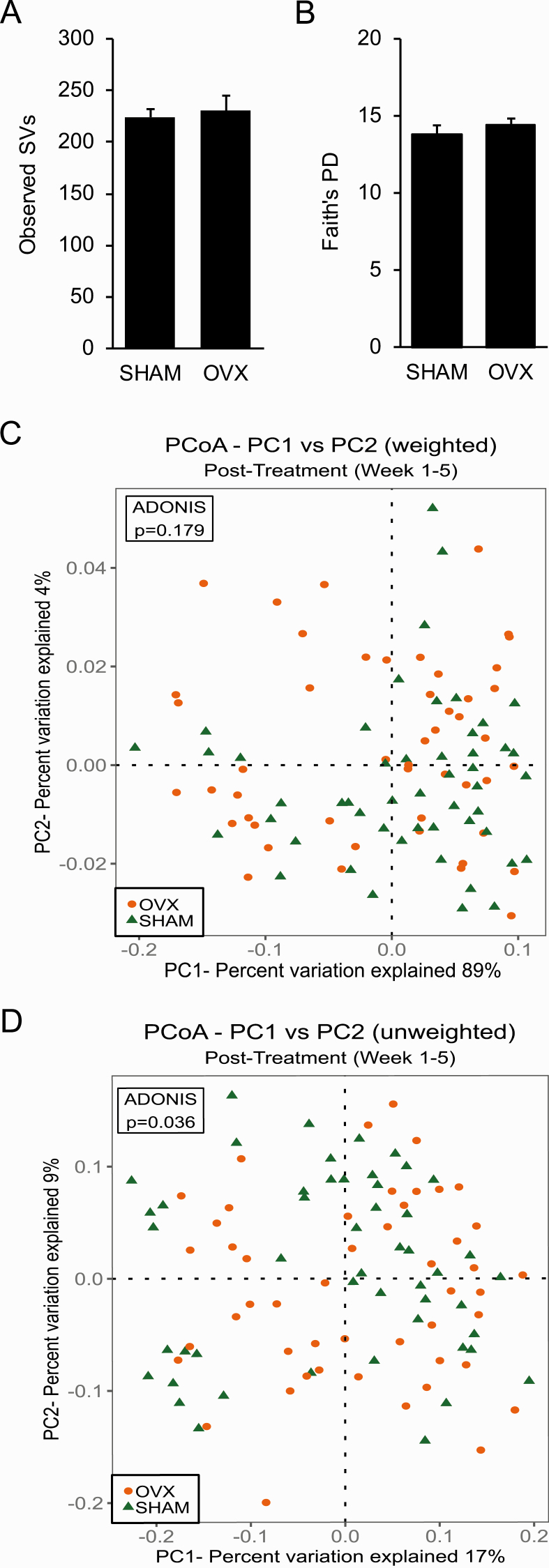
Alpha (within-sample) diversity in fecal samples from SHAM and OVX mice collected 5 weeks after surgery was calculated using observed SVs as an estimate of species richness and Faith’s PD as an estimate of species richness that accounts for phylogenetic relationships (Fig. 2). There was no significant difference in the number of observed SVs or Faith’s PD between SHAM mice and OVX mice (Fig. 2).
Figure 2.
Ovariectomy was not associated with changes in overall biodiversity of gut microbiome or in beta diversity that takes taxa abundance into account. A–B: Alpha diversity in fecal samples from sham-operated (SHAM) and ovariectomized (OVX) mice (n = 10 per group) was calculated using the number of observed sequence variants (SVs) as an estimate of species richness (A) and Faith’s phylogenetic diversity (PD) as an estimate of species richness that takes phylogenetic relationships into account (B). No difference in the number of observed SVs or Faith’s PD was observed between SHAM and OVX mice as determined by a Student t-test. Graph error bars represent standard error of the mean. C–D: Beta diversity was measured using Principle Coordinate Analysis (PCoA) of weighted UniFrac (C) and unweighted UniFrac for fecal samples collected post-treatment (weeks 1–5) (D). Proportion of variance explained by each principle coordinate axis is denoted in the corresponding axis labels. Results of permutational multivariate analysis of variance ADONIS test are shown in box inset.
C. Ovariectomy Shifted the Composition of the Gut Microbiome as Measured by Unweighted, but not Weighted, UniFrac
Beta (intersample) diversity was estimated using weighted (takes SV abundance into account) and unweighted UniFrac analyses to compare the phylogenetic similarity of the gut microbial communities in samples obtained postsurgery in SHAM mice and OVX mice. Visualization of both weighted and unweighted UniFrac distances via PCoA did not reveal distinct clustering of samples by treatment (Fig. 2C and 2D). In addition, a PERMANOVA test did not detect a significant effect of OVX on gut microbial composition for weighted UniFrac (Fig. 2C). However, a PERMANOVA test for unweighted UniFrac indicated that there was an effect of OVX on gut microbial community structure when abundance was not taken into account (P = 0.036) (Fig. 2D).
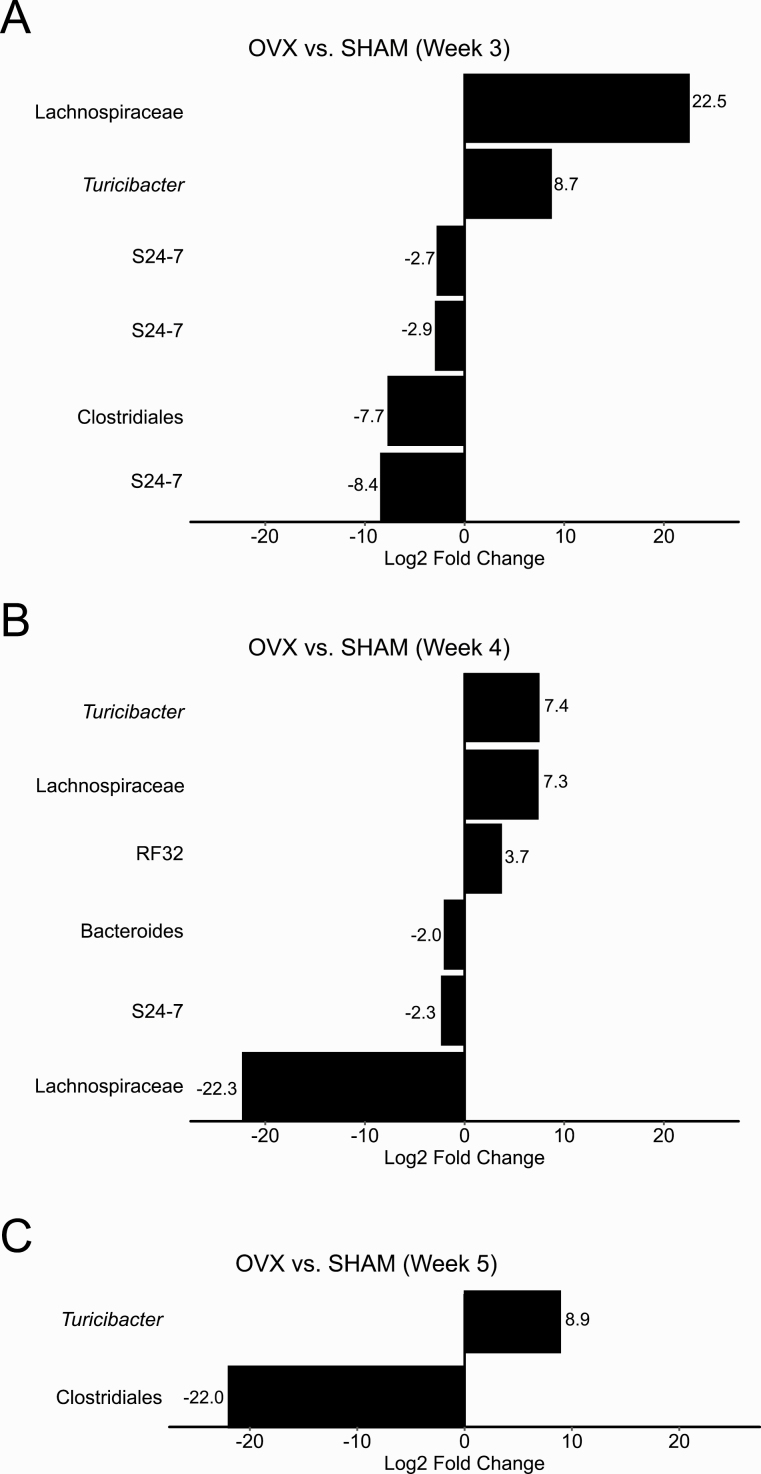
D. Ovariectomy Is Associated with Changes in a Few Bacterial Genera Subsequent to Weight Gain
No significant differences in the RA of any bacterial taxa was found between SHAM mice and OVX mice at 1 to 2 weeks postsurgery despite a significant increase in weight by week 2. Three weeks after surgery, 2 bacterial genera were identified with higher RA (unidentified Lachnospiraceae and Turicibacter) and 4 bacterial genera were identified with lower RA (unidentified Clostridiales and S24-7) in OVX mice compared with SHAM mice (Fig. 3A). By week 4, the unidentified Lachnospiraceae, Turicibacter, and S24-7 as well as members of 3 more additional genera from RF32, Bacteroides, and Lachnospiraceae were differentially abundant (Fig. 3B). Additionally, DESeq2 identified a higher RA of Turicibacter and a lower RA of an unidentified Clostridiales in OVX mice compared with SHAM mice 5 weeks after OVX (Fig. 3C).
Figure 3.
Changes in the relative abundance of S24-7, Turicibacter, unidentified Clostridiales, and Lachnospiraceae occurred after ovariectomy-induced weight gain at 2 weeks. Results from DESeq2 differential abundance analysis were expressed as a log2-fold change for sham-operated (SHAM) versus ovariectomized (OVX) mice for weeks 3 to 5 after surgery (A–C). Bacterial taxa that were significantly different between SHAM mice and OVX mice after multiple comparison correction and had a log2-fold effect of greater than 2.5 were displayed. Positive log2-fold changes represent bacterial genera increased in OVX mice relative to SHAM mice, and negative changes represent bacterial genera increased in SHAM mice relative to OVX mice. No significant differences in the relative abundance of bacterial taxa was found between SHAM mice and OVX mice at weeks 1 to 2 postsurgery.
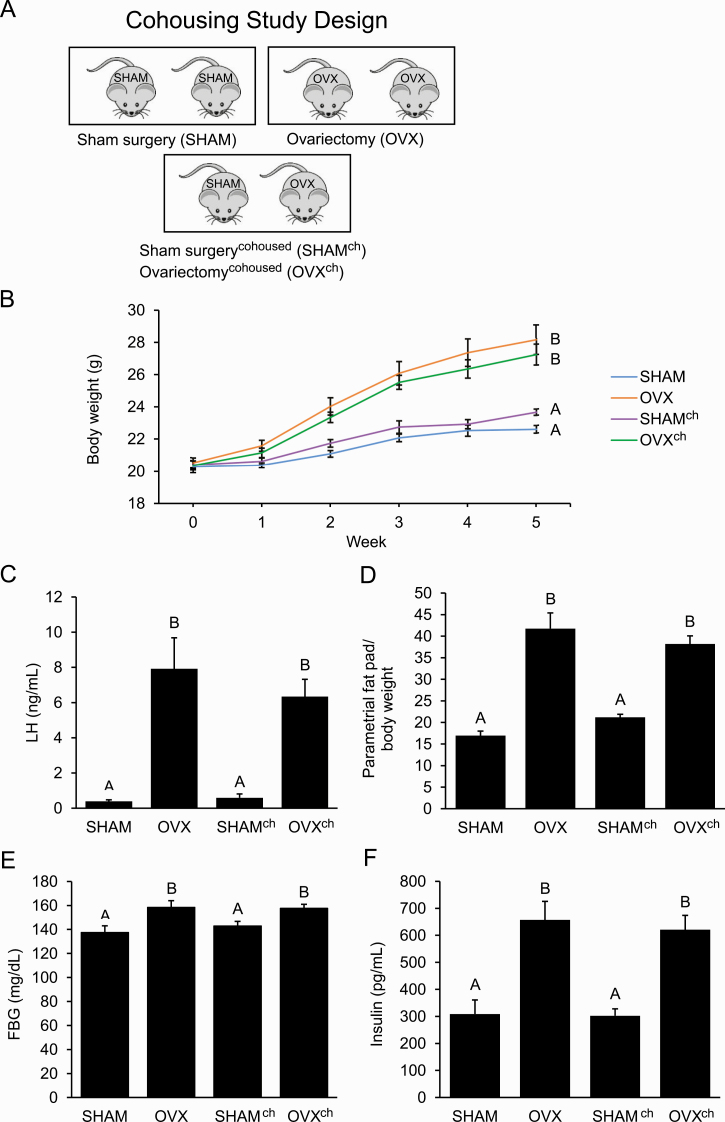
E. Cohousing Ovariectomized Mice with SHAM Mice Did Not Protect against Metabolic Dysregulation
To investigate whether exposure to a healthy gut microbiome can protect against metabolic dysregulation of OVX mice, a cohousing study was performed. The mice were housed 2 mice per cage in 3 different housing arrangements, resulting in 4 groups of mice: SHAM cohoused with SHAM, OVX cohoused with OVX, SHAM cohoused with OVX (SHAMch), or OVX cohoused with SHAM (OVXch) (Fig. 4A). Ovariectomy and OVXch mice had elevated weight compared with SHAM and SHAMch mice after 5 weeks (Fig. 4B). Serum LH levels were significantly elevated in OVX and OVXch mice but not in SHAM or SHAMch mice (Fig. 4C). Both OVX and OVXch mice had greater abdominal adiposity compared with SHAM and SHAMch mice when parametrial fat was measured relative to body weight at the end of the experiment (Fig. 4D). Unlike SHAM and SHAMch mice, OVX and OVXch mice also had increased FBG and FBI (Fig. 4E and 4F).
Figure 4.
Cohousing ovariectomized mice with sham-operated (SHAM) mice did not protect against the development of the ovariectomized (OVX) metabolic phenotype. A: Design of cohousing study with adult female mice housed 2 per cage in 3 different housing arrangements that resulted in 4 groups of mice (n = 10 per group): SHAM, OVX, SHAMch, and OVXch. B–F: Compared with SHAM mice, OVX mice had increased weight, luteinizing hormone (LH) levels, abdominal adiposity, fasting blood glucose (FBG), and fasting blood insulin (FBI) after 5 weeks. Compared with OVX mice, OVXch mice had similar weight, LH levels, abdominal adiposity, FBG, and FBI. Graph error bars represent standard error of the mean. Different letters indicate significant differences in a 1-way analysis of variance (ANOVA) or repeated-measures 2-way ANOVA followed by post hoc comparisons with the Tukey-Kramer honestly significant difference test; P < 0.05.
3. Discussion
Our results demonstrated that OVX had a significant effect on metabolism in adult female mice. In contrast to previous studies that assessed the effect of OVX on metabolic phenotypes 3 months or more postsurgery, we focused on the effect of estrogen deficiency in the short term after surgery. By 2 weeks postsurgery, OVX mice had increased weight gain compared with SHAM mice (Fig. 1B). Five weeks postsurgery, OVX mice had increased weight, abdominal adiposity, FBG, and FBI compared with SHAM mice (Fig. 1D–1F), indicating that OVX has rapid effects on female metabolism. In addition, we found that OVX mice did not have insulin resistance (Fig. 1G), which contrasts with multiple studies that reported insulin intolerance in OVX + HFD mice [15–17] and indicates that short-term E2 deficiency is not sufficient to cause insulin resistance in an adult OVX mouse model.
Our study also demonstrated that OVX had a minimal effect on overall gut microbiome composition in terms of alpha diversity. According to 16S rRNA gene sequencing analysis, there was no difference in alpha diversity of gut bacteria between OVX mice and SHAM mice 5 weeks after surgery (Fig. 2A and 2B). However, another study that investigated changes in the gut microbiome after OVX reported that OVX mice had lower alpha diversity in their gut microbiome 12 weeks after surgery [39]. One possible explanation for this inconsistency is that the duration of our experiment did not allow for sufficient time for the gut microbiome of OVX mice to differentiate from SHAM, although an adult OVX rat model showed that there was no difference in alpha diversity of the gut microbiome between OVX and SHAM rats 13 weeks after surgery [41]. More longitudinal studies will be needed to determine how much time is required for OVX to alter gut microbial composition in rodents.
In contrast with alpha diversity, we demonstrated that there was a difference in beta diversity between SHAM and OVX mice as measured by unweighted UniFrac but not weighted UniFrac analysis (Fig. 2C and 2D). While unweighted UniFrac is based on the presence or absence of observed bacterial species, weighted UniFrac takes abundance into account. Thus, the significant difference in unweighted UniFrac between SHAM and OVX mice may be due to the presence of a few distinct bacterial taxa that differentiate SHAM and OVX gut microbiomes. While other studies reported differences in beta diversity between SHAM and OVX mice using weighted UniFrac analysis [39, 54], the durations of these experiments were much longer than that of our study. Interestingly, 1 study showed a difference in beta diversity between SHAM mice and OVX mice 10 weeks postsurgery but not 6 weeks postsurgery [54], which again suggests that the length of time postsurgery may be important with regards to observing differences in the gut microbiome of OVX rodents compared with SHAM controls.
While changes in gut microbiota were not observed in the first 2 weeks after surgery, OVX resulted in changes after 3 to 5 weeks. In particular, we found increased RA of Turicibacter and unidentified Lachnospiraceae, as well as decreased RA of S24-7 and unidentified Clostridiales in OVX mice compared with SHAM mice 3 to 5 weeks postsurgery (Fig. 3). Similar to our findings, increased RA of unidentified Lachnospiraceae and decreased RA of unidentified S24-7 in the gut microbiome of OVX mice was previously reported [54]. Clostridiales species were found to be enriched in HFD-fed mice and decreased in OVX mice in a OVX + HFD study [39], which is consistent with our findings. While Turicibacter was not previously linked to OVX, lower abundance of Turicibacter was associated with increased body weight in a HFD rodent model [55]. It was also reported that Turicibacter may be involved in short chain fatty acid production [56]. At 4 weeks postsurgery, we found increased RA of RF32 and decreased RA of Bacteroides in the gut microbiome of OVX mice compared with SHAM mice (Fig. 3B). The lower RA of Bacteroides in the gut microbiome of OVX mice is consistent with findings from an OVX + HFD study [39], and a higher abundance of RF32 in the gut microbiome has been linked to HFD in a few mouse models [57, 58]. Altogether, these findings suggest that changes in gut bacteria do not play a causal role in the metabolic dysregulation observed in the short-term after OVX but instead are a symptom.
To further explore whether changes in the gut microbiome post-OVX play a causal role in weight gain, we utilized a cohousing paradigm to test whether exposure to a healthy gut microbiome was protective against development of a metabolic phenotype induced by OVX (Fig. 4A). Overall, we found that cohousing did not attenuate OVX metabolic dysregulation since OVX mice that were cohoused with SHAM mice had similar metabolic symptoms as OVX mice that were not cohoused with SHAM mice, including weight gain, abdominal adiposity, FBG, and FBI (Fig. 4B and 4D–4F). These results are in contrast with other cohousing studies in which protection against metabolic phenotypes was conferred by healthy mice to mouse models of obesity, maternally induced insulin resistance, and polycystic ovary syndrome [33, 59, 60]. Altogether, these studies demonstrate that, while cohousing can be protective against the development of metabolic dysregulation in other contexts, it was unable to alter the development of OVX-induced weight gain.
In conclusion, this study demonstrated that minimal changes in gut microbiota observed after OVX do not appear to play a causal role in OVX-induced metabolic dysregulation since they occurred after weight gain. This finding contrasts with alterations in the gut microbiome associated with metabolic dysregulation in the OVX + HFD model [16, 39]. Additionally, cohousing OVX mice with SHAM-operated mice did not improve the metabolic phenotype of OVX mice, which further supports the idea that changes in the gut microbiome do not cause the metabolic dysregulation associated with OVX. While our findings suggest that the gut microbiome does not play a mechanistic role in the weight gain that occurs rapidly after OVX, additional studies will be needed to understand the relationship between E2 levels and changes in bacteria such as Turicibacter and S24-7, as well as unidentified Lachnospiraceae and Clostridiales that may have long-term effects on the host.
Acknowledgements
We thank members of the Kelley and Thackray laboratories for insightful comments and suggestions. Hormone levels were measured by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core Facility (P50 HD28934) and the University of California, Davis Mouse Metabolic Phenotyping Core (U24 DK092993).
Financial Support: This work was funded by the National Institute of Child Health and Human Development through a cooperative agreement as part of the National Centers for Translational Research in Reproduction and Infertility (Grant P50 HD012303, to V.G.T.). V.G.T. was also funded by Grant R01 HD095412.
Author Contributions: V.G.T. conceived and designed the study; L.S., L.J.C., A.C., and R.S.S. collected samples and performed reproductive and metabolic assessments; L.S. and L.J.C. performed DNA extractions and PCR amplifications; L.S., C.M.O., S.T.K., and V.G.T. analyzed the data; L.S., C.M.O., S.T.K., and V.G.T. wrote and edited the manuscript.
Glossary
Abbreviations
- E2
17-β estradiol
- ERα
estrogen receptor alpha
- ERβ
estrogen receptor beta
- αERKO
ERα knockout
- FBG
fasting blood glucose
- FBI
fasting blood insulin
- HFD
high-fat diet
- ITT
insulin tolerance test
- LH
luteinizing hormone
- OVX
ovariectomy
- OVXch
OVX cohoused with SHAM
- PCoA
principal coordinate analysis
- PD
phylogenetic diversity
- PERMANOVA
permutational multivariate analysis of variance
- RA
relative abundance
- SHAM
sham-operated
- SHAMch
SHAM cohoused with OVX
- SVs
sequence variants
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability
Some or all data generated or analyzed during this study are included in this published article, the European Nucleotide Archive (Study Accession Number PRJEB40) or https://doi.org/10.5281/zenodo.4203456.
References and Notes
- 1. Burns KA, Korach KS. Estrogen receptors and human disease: an update HHS Public Access. Arch Toxicol. 2012;86(10):1491–1504. Doi: 10.1007/s00204-012-0868-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116(3):561–570. Doi: 10.1172/JCI27987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mauvais-Jarvis F, Clegg DJ, Hevener AL. The role of estrogens in control of energy balance and glucose homeostasis. Endocr Rev. 2013;34(3):309–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carr MC. The emergence of the metabolic syndrome with menopause. J Clin Endocrinol Metab. 2003;88(6):2404–2411. [DOI] [PubMed] [Google Scholar]
- 5. Kelley ST, Thackray VG. Phylogenetic analyses reveal ancient duplication of estrogen receptor isoforms. J Mol Evol. 1999;49(5):609–614. Doi: 10.1007/PL00006582 [DOI] [PubMed] [Google Scholar]
- 6. Xu P, Cao X, He Y, et al. Estrogen receptor-α in medial amygdala neurons regulates body weight. J Clin Invest. 2015;125(7):2861–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Handgraaf S, Riant E, Fabre A, et al. Prevention of obesity and insulin resistance by estrogens requires ERα activation function-2 (ERαAF-2), whereas ERαAF-1 is dispensable. Diabetes. 2013;62(12):4098–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bryzgalova G, Gao H, Ahren B, et al. Evidence that oestrogen receptor-alpha plays an important role in the regulation of glucose homeostasis in mice: insulin sensitivity in the liver. Diabetologia. 2006;49(3):588–597. [DOI] [PubMed] [Google Scholar]
- 9. Ribas V, Drew BG, Le JA, et al. Myeloid-specific estrogen receptor α deficiency impairs metabolic homeostasis and accelerates atherosclerotic lesion development. Proc Natl Acad Sci. 2011;108(39):16457–16462. Doi: 10.1073/PNAS.1104533108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, Cooke PS. Increased adipose tissue in male and female estrogen receptor-knockout mice. Proc Natl Acad Sci. 2000;97(23):12729–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cattoni A, Parissone F, Porcari I, et al. Hormonal replacement therapy in adolescents and young women with chemo- or radio-induced premature ovarian insufficiency: Practical recommendations. Blood Rev. 2020. Doi: 10.1016/j.blre.2020.100730 [DOI] [PubMed] [Google Scholar]
- 12. Vuralli D, Ozon ZA, Gonc EN, Alikasifoglu A, Kandemir N. Long-term effects of GnRH agonist treatment on body mass index in girls with idiopathic central precocious puberty. J Pediatr Endocrinol Metab. 2020;33(1):99–105. [DOI] [PubMed] [Google Scholar]
- 13. Chiumello G, Brambilla P, Guarneri MP, Russo G, Manzoni P, Sgaramella P. Precocius puberty and body composition: effects of GnRH analog treatment. In: Journal of Pediatric Endocrinology and Metabolism. Vol 13 London: : Freund Publishing House Ltd; 2000:791–794. [DOI] [PubMed] [Google Scholar]
- 14. Lee SJ, Yang EM, Seo JY, Kim CJ. Effects of gonadotropin-releasing hormone agonist therapy on body mass index and height in girls with central precocious puberty. Chonnam Med J. 2012;48(1):27–31. Doi: 10.4068/cmj.2012.48.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riant E, Waget A, Cogo H, Arnal JF, Burcelin R, Gourdy P. Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology. 2009;150(5):2109–2117. [DOI] [PubMed] [Google Scholar]
- 16. Kaliannan K, Robertson RC, Murphy K, et al. Estrogen-mediated gut microbiome alterations influence sexual dimorphism in metabolic syndrome in mice. Microbiome. 2018;6(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Camporez JPG, Jornayvaz FR, Lee HY, et al. Cellular mechanism by which estradiol protects female ovariectomized mice from high-fat diet-induced hepatic and muscle insulin resistance. Endocrinology. 2013;154(3):1021–1028. Doi: 10.1210/en.2012-1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geary N, Asarian L, Korach KS, Pfaff DW, Ogawa S. Deficits in E2-dependent control of feeding, weight gain, and cholecystokinin satiation in ER-alpha null mice. Endocrinology. 2001;142(11):4751–4757. [DOI] [PubMed] [Google Scholar]
- 19. Thammacharoen S, Geary N, Lutz TA, Ogawa S, Asarian L. Divergent effects of estradiol and the estrogen receptor-α agonist PPT on eating and activation of PVN CRH neurons in ovariectomized rats and mice. Brain Res. 2009;1268:88–96. Doi: 10.1016/j.brainres.2009.02.067 [DOI] [PubMed] [Google Scholar]
- 20. Santollo J, Wiley MD, Eckel LA. Acute activation of ERα decreases food intake, meal size, and body weight in ovariectomized rats. Am J Physiol – Regul Integr Comp Physiol. 2007;293(6):2194–2201. Doi: 10.1152/ajpregu.00385.2007 [DOI] [PubMed] [Google Scholar]
- 21. Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res. 2013;69(1):75–86. [DOI] [PubMed] [Google Scholar]
- 22. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. Doi: 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Den Besten G, Van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. Doi: 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30(3):332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–15723. Doi: 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J, Qin J, Li Y, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. Doi: 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 28. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. [DOI] [PubMed] [Google Scholar]
- 29. Torres PJ, Siakowska M, Banaszewska B, et al. Gut microbial diversity in women with polycystic ovary syndrome correlates with hyperandrogenism. J Clin Endocrinol Metab. 2018;103(4):1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kelley ST, Skarra DV, Rivera AJ, Thackray VG. The gut microbiome is altered in a letrozole-induced mouse model of polycystic ovary syndrome. Plos One. 2016;11(1):e0146509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. Doi: 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. Doi: 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. [DOI] [PubMed] [Google Scholar]
- 35. Chen KL, Madak-Erdogan Z. Estrogen and microbiota crosstalk: should we pay attention? Trends Endocrinol Metab. 2016;27(11):752–755. Doi: 10.1016/j.tem.2016.08.001 [DOI] [PubMed] [Google Scholar]
- 36. Baker JM, Al-Nakkash L, Herbst-Kralovetz MM. Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas. 2017;103:45–53. [DOI] [PubMed] [Google Scholar]
- 37. Zhao H, Chen J, Li X, Sun Q, Qin P, Wang Q. Compositional and functional features of the female premenopausal and postmenopausal gut microbiota. FEBS Lett. 2019;593(18):2655–2664. Doi: 10.1002/1873-3468.13527 [DOI] [PubMed] [Google Scholar]
- 38. Santos-Marcos JA, Rangel-Zuñiga OA, Jimenez-Lucena R, et al. Influence of gender and menopausal status on gut microbiota. Maturitas. 2018;116:43–53. doi: 10.1016/j.maturitas.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 39. Choi S, Hwang YJ, Shin MJ, Yi H. Difference in the gut microbiome between ovariectomy-induced obesity and diet-induced obesity. J Microbiol Biotechnol. 2017;27(12):2228–2236. Doi: 10.4014/jmb.1710.10001 [DOI] [PubMed] [Google Scholar]
- 40. Mendes E, Acetturi BG, Thomas AM, et al. Prophylactic supplementation of Bifidobacterium longum 51A protects mice from ovariectomy-induced exacerbated allergic airway inflammation and airway hyperresponsiveness. Front Microbiol. 2017;8:1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang F, Yu P, Gui X, Wang Y, Xue C, Wang J. Sialoglycoprotein isolated from the eggs of Carassius auratus prevents bone loss: an effect associated with the regulation of gut microbiota in ovariectomized rats. Food Funct. 2016;7(12):4764–4771. Doi: 10.1039/c6fo01103a [DOI] [PubMed] [Google Scholar]
- 42. Tousen Y, Matsumoto Y, Matsumoto C, et al. The combined effects of soya isoflavones and resistant starch on equol production and trabecular bone loss in ovariectomised mice. Br J Nutr. 2016;116(2):247–257. [DOI] [PubMed] [Google Scholar]
- 43. Acharya KD, Gao X, Bless EP, Chen J, Tetel MJ. Estradiol and high fat diet associate with changes in gut microbiota in female ob/ob mice. Sci Rep. 2019;9(1):1–13. Doi: 10.1038/s41598-019-56723-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. Doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bokulich NA, Kaehler BD, Rideout JR, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90 Doi: 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. Doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Price MN, Dehal PS, Arkin AP. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. Doi: 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinforma. 2006;2:121–128. Doi: 10.1177/117693430600200007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. Doi: 10.1038/ismej.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. Doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McMurdie PJ, Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. In: Pacific Symposium on Biocomputing. NIH Public Access; 2012:235–246. /pmc/articles/PMC3357092/?report=abstract. Accessed August 31, 2020. [PMC free article] [PubMed] [Google Scholar]
- 52. Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology. 2003;84(2):511–525. Doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2 [DOI] [Google Scholar]
- 53. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee DH, Kim MJ, Song EJ, et al. Nutrikinetic study of genistein metabolites in ovariectomized mice. Plos One. 2017;12(10):e0186320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo X, Li J, Tang R, et al. High fat diet alters gut microbiota and the expression of paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediators Inflamm. 2017;2017:9474896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhong Y, Nyman M, Fåk F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol Nutr Food Res. 2015;59(10):2066–2076. Doi: 10.1002/mnfr.201500187 [DOI] [PubMed] [Google Scholar]
- 57. Patrone V, Minuti A, Lizier M, et al. Differential effects of coconut versus soy oil on gut microbiota composition and predicted metabolic function in adult mice. BMC Genomics. 2018;19(1):808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hamilton MK, Boudry G, Lemay DG, Raybould HE. Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. Am J Physiol Gastrointest Liver Physiol. 2015;308(10):G840–G851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang M, Zhang Y, Miller D, et al. Microbial reconstitution reverses early female puberty induced by maternal high-fat diet during lactation. Endocrinol (United States). 2020;161(2):bqz041 Doi: 10.1210/endocr/bqz041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Torres PJ, Ho BS, Arroyo P, et al. Exposure to a healthy gut microbiome protects against reproductive and metabolic dysregulation in a PCOS mouse model. Endocrinology. 2019;160(5):1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article, the European Nucleotide Archive (Study Accession Number PRJEB40) or https://doi.org/10.5281/zenodo.4203456.