Abstract
There is urgent therapeutic need for COVID-19, a disease for which there are currently no widely effective approved treatments and the emergency use authorized drugs do not result in significant and widespread patient improvement. The food and drug administration-approved drug ivermectin has long been shown to be both antihelmintic agent and a potent inhibitor of viruses such as Yellow Fever Virus. In this study, we highlight the potential of ivermectin packaged in an orally administrable nanoparticle that could serve as a vehicle to deliver a more potent therapeutic antiviral dose and demonstrate its efficacy to decrease expression of viral spike protein and its receptor angiotensin-converting enzyme 2 (ACE2), both of which are keys to lowering disease transmission rates. We also report that the targeted nanoparticle delivered ivermectin is able to inhibit the nuclear transport activities mediated through proteins such as importin α/β1 heterodimer as a possible mechanism of action. This study sheds light on ivermectin-loaded, orally administrable, biodegradable nanoparticles to be a potential treatment option for the novel coronavirus through a multilevel inhibition. As both ACE2 targeting and the presence of spike protein are features shared among this class of virus, this platform technology has the potential to serve as a therapeutic tool not only for COVID-19 but for other coronavirus strains as well.
Keywords: spike protein, severe acute respiratory syndrome virus, angiotensin-converting enzyme 2, RNA virus
The COVID-19, a disease caused by a novel coronavirus strain called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), poses a unique challenge to domestic and international public health. As of September 21, 2020, there are over 31.1 million cases worldwide, and nearly 6.9 million cases in the United States alone. This viral strain is highly transmittable and infects respiratory tissue, and can cause flu-like symptoms as well as more severe respiratory issues and death by respiratory failure.1,2 Until now, over half a million people have died due to this virus in 2020 alone. The SARS-CoV-2 virus surface spike protein interacts with angiotensin-converting enzyme 2 (ACE2) receptors in the lung and facilitates the entry of the virus into host cells. Most of the tissue damage is a product of the immune response and resulting inflammation.3−11 Therefore, the development of new drugs and treatment modalities is urgently needed to fight the disease.
There are currently at least 15 completed clinical trials conducted around the world to search for various therapeutics to combat COVID-19 infection with no appreciable success (Table S1 in the Supporting Information). Newer studies are focusing more on COVID-19-specific inhibitors from existing natural compounds such as flavonoids.12,13 Many of the experimental therapeutics are meant only to rescue patients in severe respiratory distress or those already undergoing intubation and mechanical ventilation, rather than patients at an early to moderate stage of infection. Among ongoing clinical trials, one aims to use the “wonder drug” ivermectin (IVM)14 in combination with aspirin, dexamethasone, and enoxaparin (Table S2). This trial uses IVM via a single dose of 200 μg/kg and may pose issues of toxicity and accelerated clearance from the bloodstream resulting in low effective dose.15,16 In this article, we report an orally administrable IVM-loaded nanoparticle (NP) and its ability to lower the expression of the ACE2 receptor and the SARS-CoV-2 spike protein.
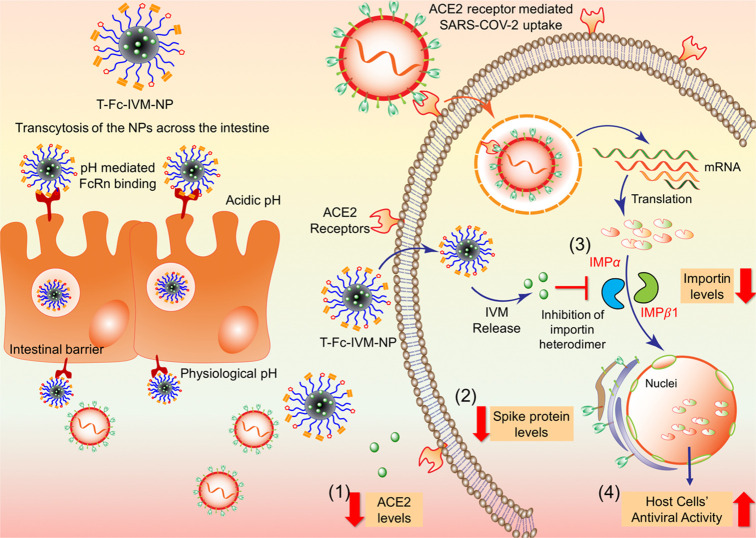
We recently developed a therapeutic IVM-loaded NP to treat Zika virus infection in the blood.17 The developed IVM nanoformulation allows the therapeutic to be gradually released into the bloodstream, which maintains its level in the blood at approximately the minimum effective therapeutic dose while keeping it below the maximum tolerated dose. This NP was constructed using poly(lactide-co-glycolide)-b-poly(ethylene glycol)-maleimide (PLGA-b-PEG-Mal) polymer, and was tagged to an Fc immunoglobulin fragment to take advantage of FcRn-driven crossing of the gut epithelial barrier to reach the bloodstream (Figure 1A). Here, we adopted the synthesis and characterization of these NPs and employed the NPs as a treatment option for COVID-19. A key goal of any new agent or nanoformulation should be not only to reduce levels of proteins that contribute to the virus’ infectious nature, but also to exploit mechanisms to prevent viral entry into cells in the first place. ACE2, which is present in high quantities in respiratory epithelia, allows for viral entry and infection of the lung and alveolar cells. ACE2-expressing cells in the lung are involved in key processes such as blood pressure regulation and interferon production. SARS-CoV-2 binding to this receptor can impede on those processes, making it an important possible target to reduce viral infection. Therefore, we hypothesized that if IVM is found to effectively decrease levels of viral spike protein as well as cellular levels of ACE2, it could theoretically be a better therapeutic when delivered in a controlled fashion using our orally administrable NP. IVM itself is known to be toxic, with an EC50 value between 1 and 10 mM. Furthermore, the maximum plasma concentrations of IVM in human after an injected dose of 150 μg/kg is typically only in the range of 9 to 75 ng/mL, but higher doses of the drug may be needed for it to be effective in controlling viral load and transmission.18 Thus, NP-mediated delivery of IVM will allow the drug to be gradually released into the bloodstream at an effective therapeutic dose while keeping it below the maximum tolerated dose. In particular, IVM-loaded nanoparticles can be engineered to contain a bound Fc immunoglobulin antibody fragment to target FcRn receptors on gut epithelial cells, which should allow for transcytosis of orally delivered nanoparticles into the bloodstream and potential accumulation at respiratory epithelial cells, which are particularly affected by SARS-CoV-2 (Figure 1).
Figure 1.
Graphical representation showing that the targeted-Fc-IVM-NPs in the acidic gut lumen bind to FcRn receptors, allowing NPs to transcytose across the intestinal barrier and release at the physiological pH of blood. IVM delivered via T-Fc-IVM-NPs shows the ability to (1) decrease ACE2 receptor levels, (2) decrease SARS-CoV-2 spike protein levels, and (3) decrease levels of the nuclear transport proteins importin α and β1, which leads to (4) an increase in the antiviral activity of infected cells.
The Centers for Disease Control and Prevention (CDC) recently suggested that the pregnant population are at a higher risk for severe complications from COVID-19 as compared to nonpregnant people, and that adverse pregnancy outcomes can happen among pregnant people with COVID-19, a finding supported by other institutions.19 As a whole, pregnant women are more likely to be admitted to intensive care units and put on mechanical ventilators than are nonpregnant women, placing the fetus at increased risk as well. Thus, it is important to develop therapeutics that can be provided as a method of care to pregnant women without affecting the fetus. Our previous studies have shown that when delivered with the mentioned NP platform, IVM cannot cross the placental barrier.17 Thus, we envision that this platform can serve as a promising potential therapeutic for pregnant individuals affected by COVID-19. The ability of the NP to reduce ACE2 levels in various cell lines and thus reduce viral uptake, paired with its capacity to decrease spike protein expression, will allow this therapeutic to be utilized against other coronavirus strains, strengthening our preparedness for future viral outbreaks.
Results and Discussion
Ability of Ivermectin Nanoformulation to Reduce ACE2 and Spike Protein Expression
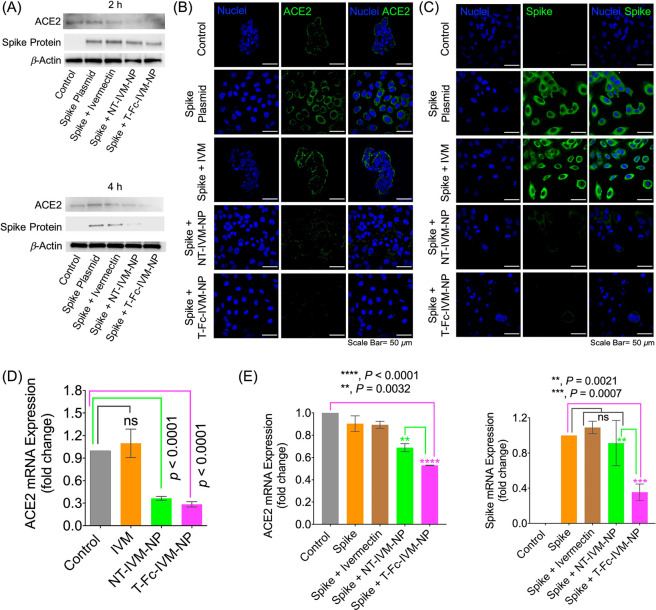
IVM-loaded PLGA-b-PEG-MAL nanoparticles (IVM-NPs) were synthesized by following a nanoprecipitation method. The NPs were characterized using dynamic light scattering (DLS) and were found to have sizes of approximately 70–80 nm and zeta potential of ∼30 mV with 20% feed of IVM (Figure S1). IVM loading was quantified using high performance liquid chromatography (HPLC) (Figure S1). The Fc immunoglobulin fragment targeting moiety was attached using thiol–ene chemistry, creating the targeted T-Fc-IVM-NPs. The conjugation of the Fc fragment was confirmed and quantified through a bicinchoninic acid (BCA) assay. SARS-CoV-2 is a positive sense single-stranded RNA virus, and one of the most crucial components of its structure is the surface spike protein that allows it to enter and infect cells. Thus, a logical method to model the conditions of viral infection in vitro and study the expression of the spike protein is to transfect cells using a plasmid expressing spike protein. This would mimic infectious conditions and allow for measurements of NP-delivered IVM’s ability to inhibit the viral spike protein without requiring the construction of pseudoviruses or other technologies. To test the therapeutic abilities of the T-Fc-IVM-NPs against both ACE2 and the viral spike proteins, HEK293T human embryonic kidney epithelial cells were transfected with a plasmid containing the SARS-CoV-2 viral spike protein. These cells were subsequently treated with IVM, a nontargeted IVM-loaded nanoparticle, NT-IVM-NP, made from an PLGA-b-PEG–OH polymer, or T-Fc-IVM-NPs. The treatments were with 10 μM of free IVM or the nanoformulations with respect to IVM for a period of 4 h followed by incubation for 20 h. The incubation time and dosing were decided based on the uptake kinetics and the IC50 values for the articles in HEK293T cells (Figure S2 for cellular updake data and Figure S3 for cytotoxicity by the MIT assay). Western blot data revealed that the expressions of the spike protein and ACE2 in the HEK293T cells were significantly decreased by IVM nanoformulations but not by free IVM (Figure 2A,B, Figure S4 for quantification). These results were further confirmed by immunofluorescence studies demonstrating the ability of T-Fc-IVM-NP to reduce ACE2 and spike protein levels (Figure 2B,C). We observed a decrease in spike and ACE2 expression in HEK293T cells when cells were treated with free IVM for 24 h (Figure S5A,B). Similarly, we observed a decrease in spike and ACE2 expression at 24 h by immunofluorescence (Figure S5C). At 4 h treatment, we observed a differential effect of IVM and IVM nanoformulations. However, the 24 h treatment did not show any difference in IVM and IVM-NPs. This may indicate that IVM-loaded NPs are able to be taken up into cells more at earlier time points than free IVM to exert their effects. Furthermore, we have shown that bioavailability of IVM nanoformulations is more than free IVM.17 In balb/c mice, free IVM showed only 20% of injected dose in the blood as compared to T-Fc-IVM-NP which was >50% in the blood after 24 h. Our studies also revealed that the HEK293T cells have low basal ACE2 expression. To confirm this further, we treated the cells with increased concentrations of angiotensin II (ANG II) and documented that such treatment increases the ACE2 levels in these cells (Figure S6). We also evaluated the effect of free IVM and its nanoformulation on basal ACE2 gene expression at the transcriptional level (Figure 2D). In addition, both ACE2 as well as spike was studied in HEK293T cells transfected with spike plasmid (Figure 2E). Cells were treated with 10 μM of IVM and its nanoformulation for 4 h and real time PCR was carried out. It was observed that T-Fc-IVM-NP significantly inhibited spike mRNA expression. Free IVM and NT-IVM-NP did not have significant inhibitory effects on spike mRNA expression. Similarly, we observed that ACE2 mRNA expression was decreased by T-Fc-IVM-NP. NT-IVM-NP also decreased ACE2 mRNA expression, but this decrease was less compared to that seen in T-Fc-IVM-NP. Taken together, our data suggest that in a cellular context, T-Fc-IVM-NP is more effective than free IVM and NT-IVM-NP in decreasing spike and ACE2 gene expression at the transcriptional level.
Figure 2.
(A) Western blot showing expression of ACE2 and spike protein in HEK293T cells transfected with plasmid expressing spike protein with treatment of IVM, NT-IVM-NPs, or T-Fc-IVM-NPs. Cells were treated with the articles for 2 or 4 h at a concentration of 10 μM with respect to IVM, after which the media was changed and the cells were further incubated up to a total of 24 h. Immunofluorescence staining showing expression of (B) ACE2 and (C) spike protein in HEK293T cells transfected with a plasmid expressing spike protein with treatment of IVM, NT-IVM-NPs, or T-Fc-IVM-NPs. Cells were treated with the articles for 4 h at a concentration of 10 μM with respect to IVM, followed by an additional 20 h incubation. (D) Fold change in mRNA expression level of ACE2 in HEK293T cells upon treatment with articles at a concentration of 10 μM with respect to IVM. (E) Fold change in mRNA expression levels of ACE2 and spike protein in spike-protein expressing HEK293T cells. The cells transfected with spike plasmid were treated with articles at a concentration of 10 μM with respect to IVM. For panels D and E, the treatment time was 4 h, followed by an additional incubation for 20 h.
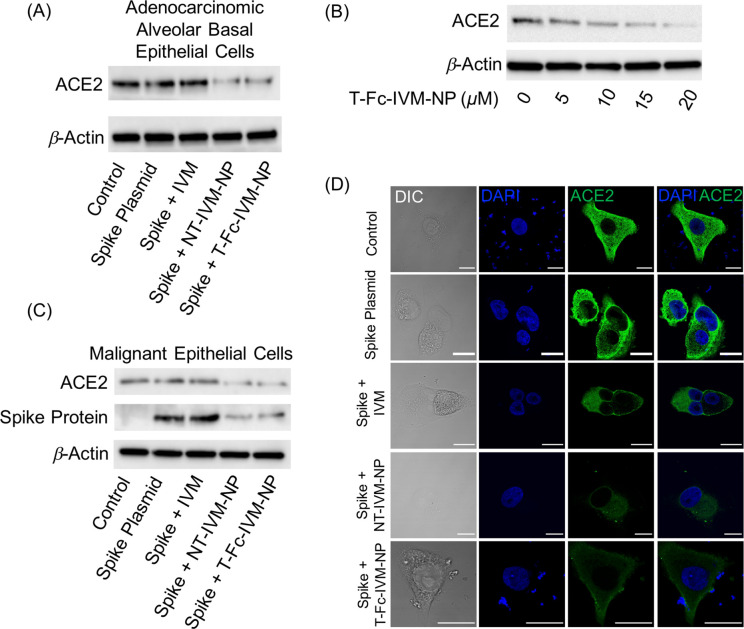
We also analyzed the effects of T-Fc-IVM-NP in two other ACE2-expressing epithelial cell lines, A549 adenocarcinomic alveolar basal epithelial cells and HeLa malignant epithelial cells, to study the potential impact of the therapeutic on ACE2 and spike protein expression in lung cells and other epithelia that may be infected by SARS-CoV-2. In the A549 cells, ACE2 expression was found to decrease after treatment with IVM and the IVM nanoformulations, and the largest decrease in expression was seen after treatment with the IVM-loaded nanoparticles (Figure 3A, Figure S7A for quantification). Accordingly, the A549 cells were treated with increasing doses of T-Fc-IVM-NPs, and the Western blot revealed a dose-dependent effect of the nanoformulation on ACE2 expression (Figure 3B, Figure S7B for quantification). In HeLa cells, treatment with T-Fc-IVM-NP showed a decrease in the expression of both spike protein and ACE2, and the more evident decrease appeared to be in the cells treated with the nanoparticles (Figure 3C, Figure S7C,D for quantification). Furthermore, immunofluorescence staining in A549 cells revealed a decrease in expression of ACE2 following treatment with IVM, NT-IVM-NPs, and T-Fc-IVM-NPs (Figure 3D). The inhibition of both ACE2 and the viral spike protein in a variety of cell lines shows that the IVM-loaded nanoparticle will be effective in treating many different cell types that may be infected.
Figure 3.
(A) Western blot showing expression of ACE2 in A549 adenocarcinomic alveolar basal epithelial cells transfected with plasmid expressing spike protein with treatment of IVM, NT-IVM-NPs, or T-Fc-IVM-NPs. Cells were treated with the articles for 4 h at a concentration of 10 μM with respect to IVM, followed by incubation in normal media for an additional 20 h. (B) Western blot showing a dose-dependent decrease in basal ACE2 expression after treatment with varying concentrations of T-Fc-IVM-NPs with respect to IVM in A549 cells. (C) Western blot showing expression of ACE2 in HeLa malignant epithelial cells transfected with plasmid expressing spike protein with treatment of IVM, NT-IVM-NPs, or T-Fc-IVM-NPs. Cells were treated with the articles for 4 h at a concentration of 10 μM with respect to IVM, followed by incubation in normal media for an additional 20 h. (D) Immunofluorescence staining showing expression of ACE2 in A549 cells transfected with plasmid expressing spike protein with treatment of IVM, NT-IVM-NPs, or T-Fc-IVM-NPs. Cells were treated with the articles for 4 h at a concentration of 10 μM with respect to IVM, followed by incubation in normal media for an additional 20 h.
Pseudovirus Inhibition Study
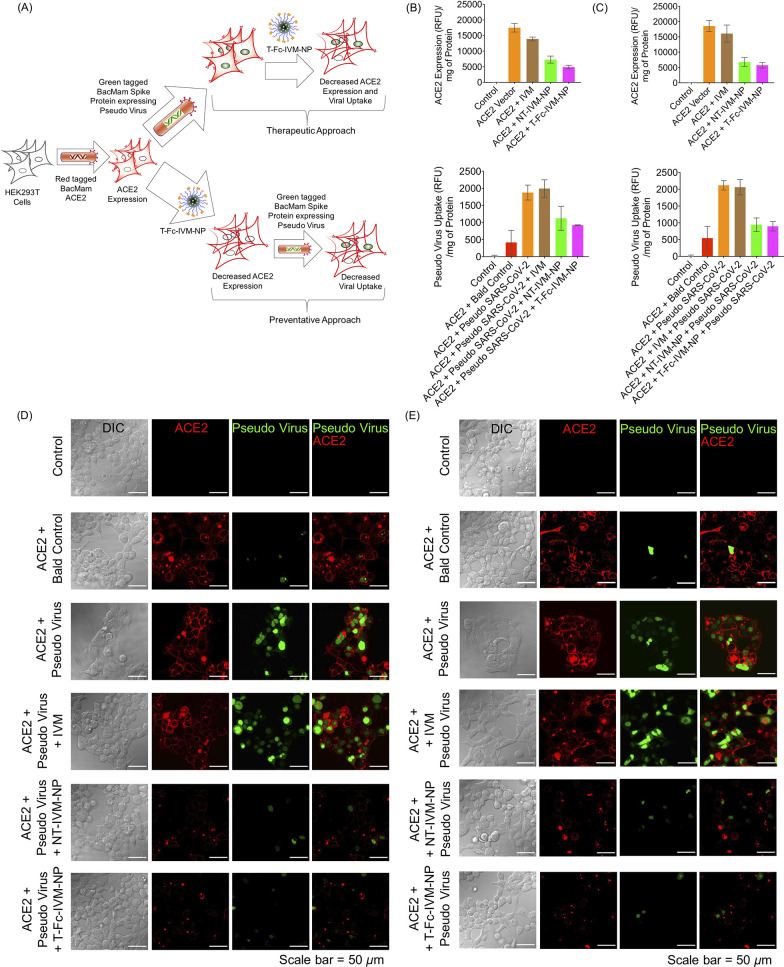
As we observed a decrease in ACE2 protein expression by T-Fc-IVM-NP, we evaluated if this decrease affects the virus uptake in HEK293T cells by carrying out a red-tagged ACE2 reporter and mNeonGreen pseudo-SARS-CoV-2 based assay. This two-step assay allows for the nuclei of transduced cells to fluorescent green if the virus is taken up in an ACE2-driven process. This experiment was carried out using therapeutic and preventative settings (Figure 4A).
Figure 4.
(A) Schematic representation of mNeonGreen pseudovirus reporter protein accumulation in HEK293T cells and the efficacy of T-Fc-IVM-NP showing inhibition of both ACE2 and pseudovirus uptake under (B) therapeutic and (C) preventative treatment methods as measured by the microplate reader. Confocal microscopy images revealing the changes in the expression of red-tagged ACE2 receptor on the cell membrane and mNeonGreen pseudovirus accumulation in the nucleus following the treatment of articles under (D) therapeutic and (E) preventative treatment methods. The article concentration was kept at 10 μM with respect to IVM for 4 h followed by an additional 20 h of incubation.
The therapeutic approach, in which the cells were exposed to the ACE2 reporter and pseudo-SARS-CoV-2 followed by treatment with IVM or its nanoformulation, resulted in significant decrease in the levels of both ACE2 and pseudovirus uptake (Figure 4B). In the preventative approach, in which treatment preceded ACE2 reporter and pseudovirus exposure with the goal of preventing uptake, there were pronounced decreases in both ACE2 expression and pseudovirus uptake after treatment with T-Fc-IVM-NP (Figure 4B). In addition, the effects of the articles were monitored via confocal microscopy, which also revealed decreases in the red and green fluorescence in the cells after treatment with T-Fc-IVM-NP treatment under both the therapeutic (Figure 4C) and preventative (Figure 4D) approaches. The decrease in green fluorescence in cells’ nuclei after the therapeutic approach, in which pseudovirus particles had already entered cells prior to T-Fc-IVM-NP treatment, showed that this nanoformulation may also be effective in decreasing the expression of the viral proteins once they are inside the cells. This assay was also performed in normal human small airway epithelial cells (HSAEC) which are known to get infected by SARS-CoV-2 (Figure S8).
Potential Mechanism of Action of T-Fc-IVM-NP
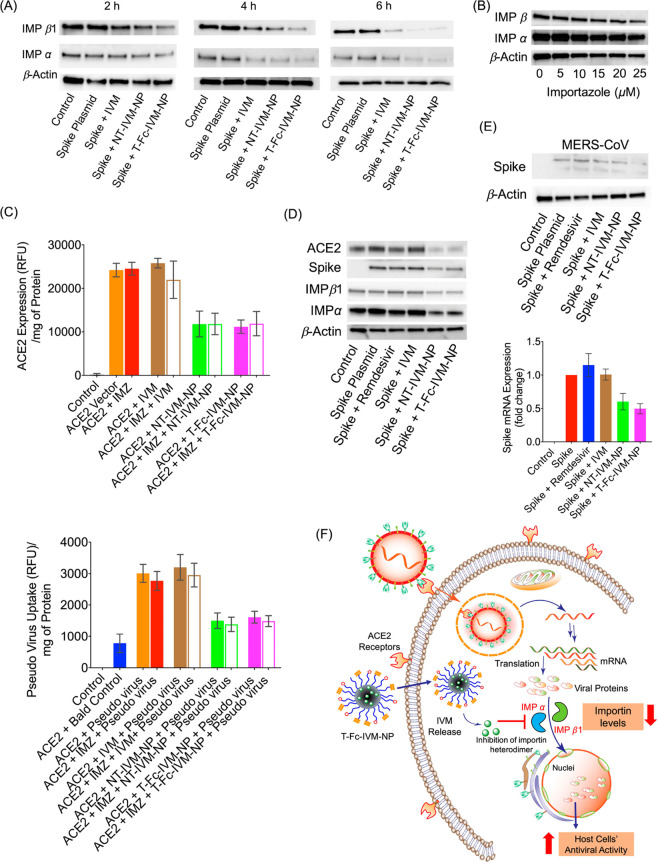
Though the specific mechanism by which the released IVM could inhibit the replication of the SARS-CoV-2 virus and expression of spike protein is yet to be determined, a possibility could be through the inhibition of the nuclear transport activities mediated through proteins such as importin (IMP) α/β1 heterodimer, as IVM was previously shown to inhibit a similar interaction between IMPα/β1 and the human immunodeficiency virus-1 (HIV-1) integrase protein.20−25 The IMP α/β1 heterodimer is a key nuclear transport protein and is believed to play a role in transporting viral proteins to the nucleus of infected cells. IMP α and β1 work through the recognition of nuclear localization signals on proteins, and IMP α and β1 have previously been associated with the nuclear transport of other viral proteins such as HIV-1 integrase and dengue virus nonstructural protein 5 (NS5).24 IMP α and β1 transport of viral proteins to the nucleus allows proteins such as dengue virus’ NS5 protein to diminish cells’ antiviral responses by impacting mRNA splicing and immune signaling.26 Therefore, investigating the IVM-loaded nanoparticle’s potential inhibitory effect on IMP α and β1 is key to fully characterizing the therapeutic’s antiviral properties. To more closely study the potential inhibitory activity of IVM nanoformulations on IMP α/β1 activity, we conducted a time-dependent treatment study in spike protein-expressing HEK293T cells. Cells were treated with T-Fc-IVM-NP for periods of 2, 4, or 6 h, and then incubated in media for an additional 22, 20, or 18 h, respectively. Western blot analyses revealed that T-Fc-IVM-NP showed inhibition of both IMP α and β1 (Figure 5A). These results suggest that the ability of T-Fc-IVM-NPs to inhibit importin expression might lead to a decrease in viral protein transport to the nucleus. The results may also indicate that uptake of NP-delivered IVM into cells may occur more quickly than the uptake of free IVM, as the effects of free IVM on IMP α and β1 expression only begin to be seen at a later time point. In our studies, we observed that T-Fc-IVM-NP-released IVM has a greater efficiency in inhibiting both IMP α and IMP β1 compared to free IVM or NT-IVM-NP (Figure 5A). The time-dependent studies demonstrated that an incubation time of 4 h is most efficient in bringing such inhibition when the cells are treated with T-Fc-IVM-NP (Figure 5).
Figure 5.
(A) Western blot showing the change in expression of IMP α and β1 in HEK293T cells following treatment with IVM and its nanoformulations. Cells were treated with articles for 2, 4, and 6 h at a concentration of 10 μM with respect to IVM, followed by incubation in normal media up to a total of 24 h. (B) Western blot showing dose-dependent changes in IMP α and β1 after treatment with varying concentrations of importazole (IMZ) in HEK293T cells. (C) Efficacy of IMZ in combination with IVM, NT-IVM-NP, and T-Fc-IVM-NP on both ACE2 expression and pseudovirus uptake under preventative treatment method as measured by the microplate reader. (D) Western blot showing the change in expression of ACE2, spike protein, and IMP α and β1 in HEK293T cells following treatment with remdesivir, IVM, or its nanoformulations. Cells were treated with articles for 4 h at a concentration of 10 μM with respect to IVM or remdesivir, followed by incubation in normal media up to a total of 24 h. (E) Western blot (top) and RT-PCR (bottom) data showing the change in expression of MERS-CoV spike protein in HEK293T cells following treatment with remdesivir, IVM, or its nanoformulations. Cells were treated with articles for 4 h at a concentration of 10 μM with respect to IVM or remdesivir, followed by incubation in normal media up to a total of 24 h. (F) Schematic representation of how IVM delivered through T-Fc-IVM-NP inhibits IMP α and β1, thus increasing host cell antiviral activity.
To further probe the mechanism of action of the IVM-loaded targeted NP through the IMPα/β1 system, spike protein-expressing cells were treated with importazole, a commercially available IMPβ1 inhibitor, before pseudovirus infection, with the goal of inhibiting IMPβ1 and observing whether pseudovirus protein transport to the nucleus was diminished. A concentration-dependent study indicated that 25 μM of importazole resulted in a significant decrease in IMPβ1 expression (Figure 5B), but not significant decrease in the nuclear transport of the pseudovirus protein (Figure S9). Subsequent studies conducted by pretreating the cells with 20 μM of importazole followed by T-Fc-IVM-NP treatment indicated that there was still a concurrent decrease in ACE2 and spike protein expression despite importazole treatment (Figure 5C), demonstrating that this NP may have multiple molecular targets and may be inhibiting pseudovirus expression and nuclear transport through other mechanisms, such as by lowering surface ACE2, inhibiting spike protein, and impacting other processes in addition to IMPα/β1 inhibition.
To compare the efficacy of the T-Fc-IVM-NP treatment against an FDA-approved COVID-19 treatment, remdesivir, the expression levels of ACE2, spike protein, and IMP α and β1 were observed after treatment with either remdesivir, IVM, or the IVM nanoformulations in spike expressing HEK293T cells (Figure 5D for Western blot, Figure S10 for RT-PCR). While remdesivir did not show the ability to decrease the levels of these proteins key to SARS-CoV-2 cell entry and virulence, the IVM nanoformulations were able to lower expression of ACE2, spike protein, and IMP β1. This resulting difference in expression may occur because treatments such as remdesivir specifically target the virus’ RNA polymerase, while IVM has multiple molecular targets.
To further test the applicability of T-Fc-IVM-NP treatment against other spike-containing members of the coronavirus family, we observed the ability of T-Fc-IVM-NP to decrease both protein and gene expression of the spike protein of MERS-CoV, a previous 2012 pandemic-causing coronavirus strain (Figure 5E, top for protein expression and bottom for RT-PCR). Remdesivir was unable to lower MERS-CoV spike protein levels. T-Fc-IVM-NPs lowered the expression of MERS-CoV spike at the both protein and gene levels indicating that the IVM nanoformulation’s spike-inhibiting capacity may extend across multiple other coronavirus strains. While the molecular target of remdesivir, the viral RNA polymerase, is known to frequently mutate across coronavirus strains, the structural similarities of spike protein across coronavirus strains means that the IVM nanoformulation might have the potential to serve as treatment for future spike-containing viruses as well. These results altogether indicate that the T-Fc-IVM-NPs are able to be taken up into cells, and not only decrease ACE2 expression and viral spike protein expression, but also inhibit the IMP α/β1 heterodimer (Figure 5F). Because of the IMP α/β1 proteins’ extensive role in the transport of viral proteins to the nucleus for other viruses, the results suggest that the T-Fc-IVM-NP-mediated reduction in transport of SARS-CoV-2 proteins to the nucleus may improve the cells’ antiviral responses.
Mitochondrial Functions and Inflammation of Spike-Infected Cells and Effects of T-Fc-IVM-NP
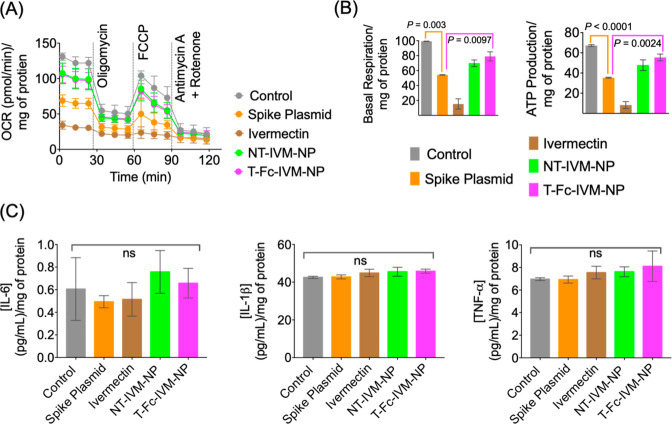
SARS-CoV-2 has been found to impact host mitochondrial functions through ACE2 regulation and open-reading frames that can allow for increased viral replication and evasion of host cell immunity.27 The mitochondrial effects of spike protein expression and the mitochondrial toxicity of treatment using IVM and its nanoformulations was tested using a mitostress assay in HEK293T cells transfected with spike plasmid. Initially, spike protein expression within the HEK293T cells was found to slightly impact mitochondrial bioenergetics through the decrease of basal and maximum respiration as well as ATP production (Figure 6A,B). This effect was further compounded by treatment with free IVM, which more significantly decreased these three metrics and led to further mitochondrial dysfunction. However, the NT-IVM-NP and T-Fc-IVM-NP treatments did not lead to further decreases in basal respiration, maximum respiration, and ATP production over what had already been caused by spike protein, indicating nanoparticle-delivered IVM is less toxic and may even slightly reverse the toxic effects of spike protein on mitochondria (Figure 6). The improvement of these indicators of mitochondrial health supports the use of the therapeutic nanoparticle over free IVM as an antiviral agent to treat SARS-CoV-2 infection. Seeing as the therapeutic nanoparticle increased respiration and ATP production close to the levels of normal untreated cells, future time-dependent treatments using IVM-loaded NPs may show that higher doses of T-Fc-IVM-NPs allow for a full increase of mitochondrial health back up to the levels of control cells (Figure 6A,B). Our preliminary analyses of cytochrome c oxidase and mitochondrial complex V activity in spike expressing HEK293T cells and subsequent treatment with IVM and its T/NT NPs did not demonstrate any significant differences (Figure S11).
Figure 6.
(A) Cellular toxicity of IVM and IVM-loaded NPs as measured by mitochondrial respiration profiles in spike protein-expressing HEK293T cells using the Seahorse analyzer and MitoStress assay: oligomycin, ATP synthase inhibitor; FCCP-carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, an ionophore; rotenone, an inhibitor of mitochondrial complex I; and antimycin A, an inhibitor of mitochondrial complex III. (B) Basal respiration and ATP production from the MitoStress assay. (C) Expression of cytokines IL-6, IL-1β, and TNFα in the media of spike protein-expressing HEK293T cells treated with IVM, NT-IVM-NP, or T-Fc-IVM-NP.
We also analyzed the levels of inflammatory markers such as IL-1β, IL-6, and TNFα before and after spike transfection and subsequent NP treatment in HEK293T cells. We did not observe any change in these inflammatory markers (Figure 6C).
Conclusions
As a whole, these results paint a positive picture for the development of an IVM-loaded nanoparticle therapeutic to treat COVID-19. Future in vivo testing and other toxicity studies need to be conducted, but the ability of an orally deliverable therapeutic to both decrease the infectious capabilities of SARS-CoV-2 through inhibition of its spike protein as well as decrease its entry rates through downregulation of ACE2 expression makes the nanoformulation a promising possible treatment for COVID-19. Considering that the gut is a potential route of entry for SARS-CoV-2, the gut-to-bloodstream entry of our ivermectin-loaded nanoformulation is ideal, as ivermectin is released gradually once in the bloodstream. Furthermore, in the bloodstream the nanoparticles will eventually be able to reach the lung epithelia as part of normal circulation and treat SARS-CoV-2 infection there, a process that will be augmented due to COVID-19-associated inflammation in the lung and resulting leaky endothelia that will allow nanoparticles to accumulate at the infected sites. The nanoparticles are not particularly immunogenic, so their accumulation at infected lung alveolar cells will not result in additional immune cell activation.
The IVM-loaded nanoparticle presents the opportunity for treatment not only of SARS-CoV-2, but of other coronavirus strains as well, due to the main two molecular targets—spike protein and the ACE2 receptor. These targets are common among other strains of coronavirus, which may vary in disease severity but have the potential to respond to IVM nanoformulation treatment. Currently, many treatments such as remdesevir and steroids are used to treat patients in critical condition, many of whom are experiencing severe respiratory distress, but these treatments are not available to or reasonable for the vast majority of less severe cases. Whereas treatments such as remdesivir target the virus’ RNA polymerase, are given intravenously, and may not be applicable to other diseases, the orally delivered IVM nanoformulation can be tested against a variety of coronavirus strains and other respiratory viruses.
Materials and Methods
Description of materials and methods used, cell lines, chemicals, and biochemical are described in the Supporting Information.
Statistics
All the statistical analyses and graphical representations were performed using Prism (GraphPad). As the tests of statistical significance do not provide an estimate of the magnitude of the difference between groups, all levels of significance were described as either significant or not significant within the text of this report. The two-tailed statistical analyses were conducted at significance level P = 0.05.
Acknowledgments
This work was supported by the Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine. We thank Shrita Sarkar for her help for the illustration of Figure 1 and Dr. Nagesh Kolishetti for helpful discussion.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00179.
Description of materials and methods, cell lines, chemicals, biochemicals, tables showing completed and ongoing clinical trial for different therapeutic options of COVID-19, and additional data (PDF)
Author Contributions
S.D. conceptualized the research; S.D., B.S., and A.S.S. designed the research; S.D. supervised experiments, provided directions and resources for all experiments; B.S., M.Z.K., and A.S.S. performed research; B.S. and M.Z.K. contributed reagents; B.S., M.Z.K., A.S.S., and S.D. analyzed the data. All authors discussed the results and commented on the manuscript; B.S., M.Z.K., A.S.S., and S.D. wrote the manuscript.
The authors declare no competing financial interest.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- Li X.; Geng M.; Peng Y.; Meng L.; Lu S. (2020) Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 10 (2), 102–108. 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K.; Fujiogi M.; Koutsogiannaki S. (2020) COVID-19 pathophysiology: A review. Clin. Immunol. 215, 108427. 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M.; Wang K.; Viveiros A.; Nguyen Q.; Zhong J.; Turner A. J.; Raizada M. K.; Grant M. B.; Oudit G. Y. (2020) Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ. Res. 126 (10), 1456–1474. 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; He X.; Zhang L.; Ran Q.; Wang J.; Xiong A.; Wu D.; Chen F.; Sun J.; Chang C. (2020) Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J. Autoimmun. 112, 102463. 10.1016/j.jaut.2020.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonagle D.; Sharif K.; O’Regan A.; Bridgewood C. (2020) The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun. Rev. 19 (6), 102537. 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M.; Martin J. C. (2020) Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 20 (6), 355–362. 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C.; Zhou L.; Hu Z.; Zhang S.; Yang S.; Tao Y.; Xie C.; Ma K.; Shang K.; Wang W.; Tian D. S. (2020) Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 71, 762. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W.; Huang N.; Becavin C.; Berg M.; Queen R.; Litvinukova M.; Talavera-Lopez C.; Maatz H.; Reichart D.; Sampaziotis F.; Worlock K. B.; Yoshida M.; Barnes J. L. (2020) SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687. 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367 (6485), 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Penninger J. M.; Li Y.; Zhong N.; Slutsky A. S. (2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46 (4), 586–590. 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P.; Khan F.; Kumar A.; Srivastava A.; Jha N. K. (2021) Screening of Potent Inhibitors Against 2019 Novel Coronavirus (Covid-19) from Alliumsativum and Allium cepa: An In Silico Approach. Biointerface Res. Appl. Chem. 11 (1), 7981–7993. 10.33263/BRIAC111.79817993. [DOI] [Google Scholar]
- Pandey P.; Khan F.; Rana A. K.; Srivastava Y.; Jha S. K.; Jha N. K. (2021) A Drug Repurposing Approach Towards Elucidating the Potential of Flavonoids as COVID-19 Spike Protein Inhibitors. Biointerface Res. Appl. Chem. 11 (1), 8482–8501. 10.33263/BRIAC111.84828501. [DOI] [Google Scholar]
- Mastrangelo E.; Pezzullo M.; De Burghgraeve T.; Kaptein S.; Pastorino B.; Dallmeier K.; de Lamballerie X.; Neyts J.; Hanson A. M.; Frick D. N.; Bolognesi M.; Milani M. (2012) Ivermectin Is a Potent Inhibitor of Flavivirus Replication Specifically Targeting NS3 Helicase Activity: New Prospects for an Old Drug. J. Antimicrob. Chemother. 67 (8), 1884–1894. 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidary F.; Gharebaghi R. (2020) Ivermectin: a systematic review from antiviral effects to COVID-19 complementary regimen. J. Antibiot. 73 (9), 593–602. 10.1038/s41429-020-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. P.; Du W. J.; Huang L. P.; Wei Y. W.; Wu H. L.; Feng L.; Liu C. M. (2016) The Pseudorabies Virus DNA Polymerase Accessory Subunit UL42 Directs Nuclear Transport of the Holoenzyme. Front. Microbiol. 7, 124. 10.3389/fmicb.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surnar B.; Kamran M. Z.; Shah A. S.; Basu U.; Kolishetti N.; Deo S.; Jayaweera D. T.; Daunert S.; Dhar S. (2019) Orally Administrable Therapeutic Synthetic Nanoparticle for Zika Virus. ACS Nano 13, 11034–11048. 10.1021/acsnano.9b02807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkassaby M. H. (1991) Ivermectin uptake and distribution in the plasma and tissue of Sudanese and Mexican patients infected with Onchocerca volvulus. Trop. Med. Parasitol. 42 (2), 79–81. [PubMed] [Google Scholar]
- Dashraath P.; Wong J. L. J.; Lim M. X. K.; Lim L. M.; Li S.; Biswas A.; Choolani M.; Mattar C.; Su L. L. (2020) Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am. J. Obstet. Gynecol. 222 (6), 521–531. 10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L.; Druce J. D.; Catton M. G.; Jans D. A.; Wagstaff K. M. (2020) The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 179, 104787. 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R. R.; Chauhan V.; Fang Y.; Pekosz A.; Kerrigan M.; Burton M. D. (2005) Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in Vero cells. J. Virol. 79 (17), 11507–12. 10.1128/JVI.79.17.11507-11512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timani K. A.; Liao Q.; Ye L.; Zeng Y.; Liu J.; Zheng Y.; Ye L.; Yang X.; Lingbao K.; Gao J.; Zhu Y. (2005) Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 114 (1–2), 23–34. 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff K. M.; Rawlinson S. M.; Hearps A. C.; Jans D. A. (2011) An AlphaScreen(R)-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screening 16 (2), 192–200. 10.1177/1087057110390360. [DOI] [PubMed] [Google Scholar]
- Wagstaff K. M.; Sivakumaran H.; Heaton S. M.; Harrich D.; Jans D. A. (2012) Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443 (3), 851–6. 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulan W. N.; Heydet D.; Walker E. J.; Gahan M. E.; Ghildyal R. (2015) Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 6, 553. 10.3389/fmicb.2015.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. N. Y.; Atkinson S. C.; Wang C.; Lee A.; Bogoyevitch M. A.; Borg N. A.; Jans D. A. (2020) The broad spectrum antiviral ivermectin targets the host nuclear transport importin alpha/beta1 heterodimer. Antiviral Res. 177, 104760. 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- Singh K. K.; Chaubey G.; Chen J. Y.; Suravajhala P. (2020) Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol Cell Physiol 319 (2), C258–C267. 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.