Abstract
Objectives
Bacillus Calmette–Guérin (BCG) vaccination has been implicated in protection against severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) and as a non‐specific immunisation method against the virus. We therefore decided to investigate T‐cell and B‐cell epitopes within the BCG‐Pasteur strain proteome for similarity to immunogenic peptides of SARS‐CoV‐2.
Methods
We used NetMHC 4.0 and BepiPred 2.0 epitope prediction methods for the analysis of the BCG‐Pasteur proteome to identify similar peptides to established and novel SARS‐CoV‐2 T‐cell and B‐cell epitopes.
Results
We found 112 BCG MHC‐I‐restricted T‐cell epitopes similar to MHC‐I‐restricted T‐cell SARS‐CoV‐2 epitopes and 690 BCG B‐cell epitopes similar to SARS‐CoV‐2 B‐cell epitopes. The SARS‐CoV‐2 T‐cell epitopes represented 16 SARS‐CoV‐2 proteins, and the SARS‐CoV‐2 B‐cell epitopes represented 5 SARS‐CoV‐2 proteins, including the receptor binding domain of the spike glycoprotein.
Conclusion
Altogether, our results provide a mechanistic basis for the potential cross‐reactive adaptive immunity that may exist between the two microorganisms.
Keywords: 2019‐nCoV, Bacillus Calmette–Guérin, BCG, COVID‐19, SARS‐CoV‐2
We analysed the Bacillus Calmette–Guérin (BCG)‐Pasteur strain proteome for immunogenic peptides that are similar to severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) T‐cell and B‐cell epitopes. We found 112 BCG MHC‐I‐restricted T‐cell epitopes similar to an MHC‐I‐restricted T‐cell SARS‐CoV‐2 epitope and 690 BCG B‐cell epitopes similar to a SARS‐CoV‐2 B‐cell epitope. Altogether, our results provide a mechanistic basis for the potential cross‐reactive adaptive immunity that may exist between the two microorganisms.

Introduction
The current pandemic caused by severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) has led to exponentially rising morbidity and mortality worldwide. Apart from aggressive quarantine and hygiene control measures, the most effective way to inhibit SARS‐CoV‐2 spread is a population‐wide vaccination campaign. Since the date of the introduction and overall effectiveness of SARS‐CoV‐2 vaccines are not known, alternative approaches for active immunisation against SARS‐CoV‐2 are under consideration. The role of Bacillus Calmette–Guérin (BCG) vaccination in the prevention of SARS‐CoV‐2 infection and in the epidemiology of COVID‐19 has been frequently implicated. 1 BCG is an attenuated Mycobacterium bovis strain, a species of the Mycobacterium tuberculosis complex, and is used worldwide for vaccination against M. tuberculosis. Interestingly, BCG appears to have multiple effects. Thus, it was observed that BCG immunisation could also induce a so‐called heterologous immune response against viruses such as human papillomavirus, influenza A, live yellow fever vaccine and hepatitis B (HBsAg) as well as organisms such as Candida albicans (C. albicans) and Staphylococcus aureus (S. aureus). 2 , 3 , 4
The adaptive component of heterologous immunity is thought to originate from the epitope similarity between distant microorganisms coupled with the polyspecificity of T‐cell and B‐cell responses. In this study, we decided to explore whether BCG immunisation‐induced heterologous adaptive immunity could theoretically play a role in the protection against SARS‐CoV‐2. We compared the T‐cell and B‐cell epitopes of the BCG strain Pasteur 1173P2 (BCG‐Pasteur) with T‐cell and B‐cell epitopes of SARS‐CoV‐2 and identified numerous highly similar epitopes between the two microorganisms.
Results
General description of peptide similarity between the BCG‐Pasteur and SARS‐CoV‐2 proteomes
To explore the similarity of the BCG‐Pasteur proteome to other proteomes, we compared the 9‐mer and 15‐mer peptides of the BCG‐Pasteur proteome to 40 viral proteomes and the C. albicans and S. aureus proteomes. We chose the 9‐mer and 15‐mer peptide lengths as characteristic lengths for MHC‐I‐restricted epitopes and MHC‐II‐restricted epitopes/ linear B‐cell epitopes. Our data showed that the number of both the 9‐mer and 15‐mer peptides with high identity (≥ 67%) to BCG‐Pasteur peptides was strongly correlated with the proteome sizes of the microorganisms (Figure 1a and b). In addition to SARS‐CoV‐2, the comparison included RSV, HPV, yellow fever virus, influenza A, HSV‐1, C. albicans and S. aureus. These pathogens served as positive controls, as the prevalence/severity of the infections caused have been shown to be reduced by BCG vaccination and/or BCG‐induced cross‐immunity was observed previously. 2 , 4 These pathogens, including SARS‐CoV‐2, correlate very well and follow the trend between genome length and the prevalence of similar peptides across a wide range of proteome sizes. Comparison of the similarity between BCG‐Pasteur proteome and the original SARS‐CoV‐2 or random proteomes showed that the original SARS‐CoV‐2 proteome contained a significantly higher number of similar 9‐mer peptides than all of the 50 random proteomes. For the 15‐mer peptides, the SARS‐CoV‐2 proteome demonstrated higher numbers of similar peptides than most of the random proteomes (Figure 2a). Next, we explored how the peptide length and degree of identity between the peptide pairs influence the number of similar peptides between the BCG‐Pasteur and SARS‐CoV‐2 proteomes (Figure 2b). Very large numbers of 9‐mer and 15‐mer peptides with limited (< 3 aa) identity were found but when using ≥ 67% identity as a threshold level, the numbers of 9‐mer peptides with six or higher identical amino acids were 40 352 and the numbers of 15‐mer peptides with 10 or more identical amino acids were 24 (Figure 2b insets).
Figure 1.
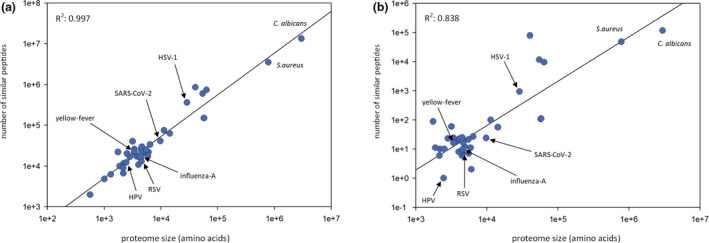
Exploration of peptide similarity between the Bacillus Calmette–Guérin (BCG)‐Pasteur proteome and other proteomes. All possible 9‐mer and 15‐mer BCG‐Pasteur peptides were compared to the same‐length peptides of a variety of 40 viral proteomes and the C. albicans and Staphylococcus aureus proteomes, ranging in size from < 1e+3 to > 1e+6. (a) 9‐mer peptides with ≥ 67% identity. (b) 15‐mer peptides with ≥ 67% identity. Pearson correlation coefficients between the number of similar peptides and the proteome lengths are also shown. HPV, human papillomavirus; HSV‐1, herpes simplex virus‐1; RSV, respiratory syncytial virus.
Figure 2.
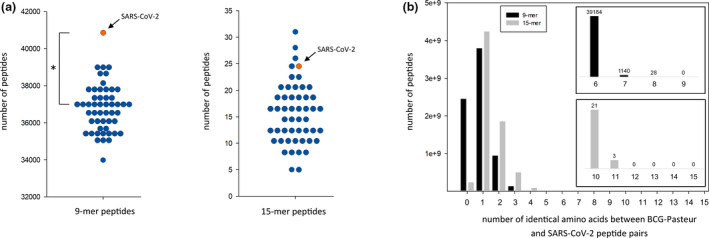
Exploration of peptide similarity between the Bacillus Calmette–Guérin (BCG)‐Pasteur proteome and the severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) proteome. (a) Comparison of the BCG‐Pasteur proteome to the SARS‐CoV‐2 proteome and randomised proteomes. 9‐mer and 15‐mer peptides of the BCG‐Pasteur proteome were mapped to the SARS‐CoV‐2 proteome and 50 random proteomes containing the same number and length proteins as the original SARS‐CoV‐2 proteome but with scrambled amino acid composition. The number of 9‐mer and 15‐mer BCG‐Pasteur peptides ≥ 67% identical to a SARS‐CoV‐2 peptide are shown. Interquartile range analysis of outliers identified values higher than 40 222 as an outlier for 9‐mer sequences and 30.25 for 15‐mer sequences. According to this analysis, the SARS‐CoV‐2 9‐mer value (40 848) was an outlier, but the SARS‐CoV‐2 15‐mer value (24) was not. Grubbs' test for outliers was applied to calculate the significance of the 40 848 value as an outlier. *P < 0.05. (b) Exploration of the identity between BCG‐Pasteur peptides and SARS‐CoV‐2 peptides. 9‐mer and 15‐mer BCG‐Pasteur peptides were mapped to the SARS‐CoV‐2 proteome. The number of BCG‐Pasteur peptides with different level of identity to a SARS‐CoV‐2 peptide is shown. Inset graphs show the occurrence of the 9‐mer and 15‐mer peptides with ≥ 67% identity.
Identification of BCG‐Pasteur epitopes that are highly similar to the putative SARS‐CoV‐2 T‐cell epitopes
Firstly, we mapped the experimentally verified SARS‐CoV‐2 T‐cell MHC‐I‐restricted epitopes to the BCG‐Pasteur proteome using an identity threshold of ≥ 67%. Further NetMHC 4.0 analysis identified 217 ‘strong binder’ BCG‐Pasteur peptides that could be presented by at least one representative allele of an HLA‐A or HLA‐B supertype. A third analysis step compared the BCG‐Pasteur peptide‐associated HLA alleles with the associated HLA allele(s) of the similar SARS‐CoV‐2 peptide. This selection resulted in 112 BCG‐Pasteur peptides that are 1. highly similar to the MHC‐I‐restricted SARS‐CoV‐2 epitope and 2. presented by an HLA allele related to the same HLA supertype (Figure 3a, Supplementary table 1). These 112 BCG‐Pasteur peptides had a similar counterpart in 16 different SARS‐CoV‐2 proteins (Figure 3b) resulting the multiple representation of SARS‐CoV‐2 epitopes by BCG‐Pasteur epitopes. SARS‐CoV‐2 epitopes such as LLSAGIFGA, FLLPSLATV, ALLADKFPV and LLLDRLNQL had 9, 8, 7 and 7 similar BCG‐Pasteur peptide pairs, respectively. An extreme example was the SARS‐CoV‐2 nsp05 protein epitope VLAWLYAAV which was the single epitope of the nsp05 protein but had 23 BCG‐Pasteur epitope counterparts with 66% identity and the 78% identical ALAWLVAAV epitope. The 24 BCG‐Pasteur epitope pairs of VLAWLYAAV represented 23 different BCG‐Pasteur proteins. The best represented SARS‐CoV‐2 proteins were the nsp05, a chymotrypsin‐like protease (M pro) and the spike glycoprotein. A similar analysis was performed for SARS‐CoV‐2 MHC‐II‐restricted T‐cell epitopes but we found no highly similar epitopes between the two microorganisms, primarily due to the longer length of most peptides presented via MHC‐II to T cells.
Figure 3.

Similarity between MHC‐I‐restricted Bacillus Calmette–Guérin (BCG)‐Pasteur and severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) T‐cell epitopes. Individual SARS‐CoV‐2 MHC‐I‐restricted T‐cell epitopes were compared with all the possible same‐length peptides of the BCG‐Pasteur proteome. BCG‐Pasteur peptides with identity of ≥ 67% were analysed with NetMHC 4.0 to identify epitopes. (a) Top‐ranked BCG‐Pasteur epitopes with the highest similarity/ NetMHC %Rank to an epitope of a SARS‐CoV‐2 protein. The NetMHC %Rank which corresponds to the predicted HLA binding affinity is shown (lower NetMHC %Rank value indicates stronger HLA binding). Note that the SARS‐CoV‐2 epitope SEETGTLIV has two identical epitope counterparts from two BCG‐Pasteur proteins. (b) Distribution of SARS‐CoV‐2 epitopes among the viral proteins. The number of SARS‐CoV‐2 epitopes and the number of their similar BCG‐Pasteur epitope counterparts are shown. SG, spike glycoprotein.
Identification of BCG‐Pasteur epitopes that are highly similar to the putative SARS‐CoV‐2 B‐cell epitopes
We applied a similar selection scheme as before to identify highly similar BCG‐Pasteur sequences with the experimentally verified and putative SARS‐CoV‐2 B‐cell epitopes. We mapped the SARS‐CoV‐2 B‐cell epitopes to the BCG‐Pasteur proteome and collected the highly similar sequences (≥ 62.5% identity). The similarity between SARS‐CoV‐2 and BCG‐Pasteur sequences ranged between 62.5% and 100%. The identified BCG‐Pasteur sequences were further analysed for potential antigenicity using the BepiPred 2.0 B‐cell epitope prediction method. 5 BepiPred analysis revealed that 690 BCG‐Pasteur sequences were located within a BCG‐Pasteur protein region that is likely to be recognised by antibodies (Supplementary table 2). These 690 BCG‐Pasteur peptides were similar to 17 putative SARS‐CoV‐2 B‐cell epitopes located in the nsp12, orf4 (envelope protein), orf5 (membrane glycoprotein), orf9 (nucleocapsid) and the spike glycoprotein (Figure 4a), indicating a high level of representation of these SARS‐CoV‐2 B‐cell epitopes. As an example of a prevalent epitope, the potential SARS‐CoV‐2 nucleocapsid epitope LLPAAD had 290 similar and likely antigenic BCG‐Pasteur counterparts, including an identical counterpart. The high similarity and the existence of several similar shared BCG‐Pasteur epitopes with SARS‐CoV‐2 B‐cell epitopes could therefore lead to the induction of cross‐specific antibodies. Interestingly, the putative SARS‐CoV‐2 spike epitope FGEVFNAT, which is located in the receptor binding domain and is likely to elicit neutralising antibodies, had five similar BCG‐Pasteur counterparts (Figure 4b and c).
Figure 4.

Similarity between Bacillus Calmette–Guérin (BCG)‐Pasteur and severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) B‐cell epitopes. (a) Individual SARS‐CoV‐2 B‐cell epitopes (putative and experimentally verified) were compared with all the possible same‐length peptides of the BCG‐Pasteur proteome. BCG‐Pasteur proteins containing the highly similar peptides (≥ 62.5% identity) were analysed with BepiPred 2.0 to identify putative B‐cell epitopes. (b) Localisation of the SARS‐CoV‐2 B‐cell epitope FGEVFNAT (red) in the receptor binding domain (turquoise) of the homotrimer SARS‐CoV‐2 spike glycoprotein (side view and top view). (c) FGEVFNAT and the similar BCG‐Pasteur B‐cell epitopes identified. SG, spike glycoprotein.
Conclusion
We systematically mapped numerous SARS‐CoV‐2 epitopes to the BCG‐Pasteur proteome to find similar epitopes that might induce adaptive cross‐immunity and explain the protective qualities of BCG vaccination against SARS‐CoV‐2. Our analysis of similar peptides of BCG‐Pasteur and other proteomes revealed that the occurrence of epitope similarity is strongly correlated to the proteome sizes of the microorganisms. As expected, the SARS‐CoV‐2 proteome behaved similarly, indicating that the coexistence of cross‐immunity with BCG‐Pasteur is likely not due to exceptional similarity between these evolutionary distant microorganisms. The fact that the SARS‐CoV‐2 proteome contained higher number of similar epitopes than all (9‐mer) or most of (15‐mer) the random proteomes further supports that the similarity does not arise by chance between the two proteomes. Rather, short conservative protein sequences exist even between these distant microorganisms. The number of highly similar peptides between the two proteomes strongly depended on the length of the peptides. Thus, the numbers of similar 9‐mer peptide pairs were 3 orders of magnitude higher than that of the 15‐mers. The immunological consequence of these findings would be that the adaptive cross‐immunity between these microorganisms has a higher chance to be induced between shorter epitopes and are therefore inherently directed towards a MHC‐I‐restricted T‐cell response. Alternatively, these data could reflect MHC‐II‐restricted T‐cell responses to shorter epitopes and B‐cell recognition of short linear epitopes or smaller conformational epitopes. Our immunological analyses of the similar peptides supported the former possibility since we identified 112 similar MHC‐I‐restricted T‐cell epitope pairs but did not identify similar MHC‐II‐restricted epitopes. However, these findings do not completely rule out MHC‐II‐restricted cross‐immunity between BCG and SARS‐CoV‐2, since epitopes with low level of full‐length sequence similarity can indeed induce cross‐immunity. 6 We found that SARS‐CoV‐2 B‐cell epitopes having BCG‐Pasteur counterparts were present mostly in structural proteins including the spike glycoprotein, a dominant target for protective antibodies. Among the 8 spike glycoprotein epitopes which had similar BCG‐Pasteur peptide counterparts, one was located in the receptor binding domain, the target of neutralising antibodies. 7
Overall, we have shown that there are shared T‐cell and B‐cell epitopes between SARS‐CoV‐2 and BCG‐Pasteur which suggests that BCG‐induced immunity could influence the adaptive immune response against SARS‐CoV‐2. Although this in silico study has not demonstrated functional cross‐reactive immunity, our work does support the further investigation of heterologous immunity between BCG and SARS‐CoV‐2. The similar BCG‐Pasteur peptides identified in this study could be used as a reference set to assist the evaluation of BCG‐induced SARS‐CoV‐2‐directed T‐cell and B‐cell responses.
Methods
Proteome comparison of BCG‐Pasteur and SARS‐CoV‐2
Severe acute respiratory syndrome‐CoV‐2 protein sequences were compared to the BCG‐Pasteur proteome (M. tuberculosis variant bovis BCG p‐1173P2 (GCF_000009445.1)). The following SARS‐CoV‐2 protein sequences were used: nsp1 (YP_009742608), nsp2 (YP_009742609), nsp3 (YP_009742610), nsp4 (YP_009742611), nsp5 (YP_009742612), nsp6 (YP_009742613), nsp7 (YP_009742614), nsp8 (YP_009742615), nsp9 (YP_009742616), nsp10 (YP_009742617), nsp11 (YP_009725312), nsp12 (YP_009725307), nsp13 (YP_009725308), nsp14 (YP_009725309), nsp15 (YP_009725310), nsp16 (YP_009725311), spike glycoprotein (YP_009724390), ORF3a (YP_009724391), ORF4 (YP_009724392), ORF5 (YP_009724393), ORF6 (YP_009724394), ORF7a (YP_009724395), ORF7b (YP_009725318), ORF8b (YP_009724396), ORF9 (YP_009724397), ORF10 (YP_009725255). For sequence comparison, the previously described sequence identity measure was used. 6 BCG‐Pasteur and SARS‐CoV‐2 peptide pairs that shared a ≥ 67% identity in their peptide sequences were collected. The same identity measure was used to compare the BCG‐Pasteur proteome to 40 viral proteomes and the C. albicans and S. aureus proteomes. The same identity measure was used to compare the BCG‐Pasteur proteome to 50 randomly generated proteomes consisting of same number of peptides with the same amino acid compositions but scrambled order the SARS‐CoV‐2 protein sequences used before. To identify whether the SARS‐CoV‐2 proteome contained more similar peptides than the random proteomes, two methods were applied: 1, Interquartile range (IQR, IQR = Q3 − Q1) based outlier detection (Q1: first quartile of the dataset , Q3: third quartile of the dataset), which marks data points as outlier, if a data point is above Q3 + 1.5*IQR; 2, Normal distribution of the data was verified using the Shapiro–Wilk normality test, and then, Grubbs' test was applied to test whether the maximum value was an outlier at the 0.05 significance level.
Mapping the experimentally verified SARS‐CoV‐2 T‐cell and B‐cell epitopes to the BCG‐Pasteur proteome
The experimentally verified SARS‐CoV‐2 T‐cell and B‐cell epitopes (linear, human, positive T‐cell assays, MHC‐I restricted and MHC‐II restricted) were downloaded from the Immune Epitope Database. Due to the low number of MHC‐II‐restricted SARS‐CoV‐2 T‐cell and B‐cell epitopes, experimentally verified SARS‐CoV T‐cell and B‐cell epitopes with identical sequence to SARS‐CoV‐2 were also included in the analysis as putative SARS‐CoV‐2 epitopes. SARS‐CoV‐2 epitopes were compared to the BCG‐Pasteur proteome using the above‐mentioned identity measure. BCG‐Pasteur peptides with identity (same amino acid at the same position) of ≥ 67% were used for further T‐cell epitope selection, because recently it was showed that if there is a ≥ 67% identity between a SARS‐CoV‐2 epitope and another coronavirus epitope than there is a 57% chance that adaptive cross‐immunity can be observed. 6 Similar threshold was observed in SARS‐CoV‐2 MHC‐1‐restricted T‐cell cross immune responses. 8 The threshold of the identification of cross binding linear B‐cell epitopes has not been described, and therefore, an identity threshold similar to the T‐cell analysis (≥ 62.5%) was used in this study. BCG‐Pasteur peptides similar to MHC‐I‐restricted epitopes were further analysed with NetMHC 4.0 software 9 using predefined alleles representative of the HLA‐A and HLA‐B supertypes. BCG‐Pasteur peptides similar to MHC‐II‐restricted SARS‐CoV‐2 peptides were analysed with NetMHCII 2.3. 10 The identified ‘strong binder’ epitopes were collected (threshold for strong binders: ≤ 0.5% Rank for NetMHC 4.0 and ≤ 2% Rank for NetMHCII 2.3). BCG‐Pasteur proteins with identity of ≥ 62.5% to a SARS‐CoV‐2 B‐cell epitopes were further analysed with the BepiPred 2.0 software. 5 Peptides that were located entirely in an epitope region by BepiPred 2.0 were collected. 3D molecular surface model of SARS‐CoV‐2 spike protein homotrimer was created with the Maestro GUI of the Schrödinger program suit (Schrödinger Inc., New York, NY, USA) using the 6X29 pdb structure from the Protein Data Bank crystallographic database (www.rcsb.org).
Author Contributions
Szabolcs Urbán: Conceptualization; Data curation; Formal analysis; Methodology; Writing‐original draft. Gábor Paragi: Conceptualization; Data curation; Formal analysis. Katalin Burián: Conceptualization; Writing‐original draft. Gary McLean: Conceptualization; Writing‐original draft.
Conflicts of Interest
The authors declare no conflict of interest.
Supporting information
Acknowledgments
Dezső P Virok was supported by the Hungarian – European Union Grant EFOP‐3.6.1‐16‐2016‐00008.
References
- 1. Shivendu S, Chakraborty S, Onuchowska A, Patidar A, Srivastava A. Is there evidence that BCG vaccination has non‐specific protective effects for COVID 19 infections or is it an illusion created by lack of testing? medRxiv 2020. 10.1101/2020.04.18.20071142 [DOI] [Google Scholar]
- 2. Moorlag SJCFM, Arts RJW, van Crevel R, Netea MG. Non‐specific effects of BCG vaccine on viral infections. Clin Microbiol Infect 2019; 25: 1473–1478. [DOI] [PubMed] [Google Scholar]
- 3. Kandasamy R, Voysey M, McQuaid F et al Non‐specific immunological effects of selected routine childhood immunisations: systematic review. BMJ 2016; 355: i5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kleinnijenhuis J, Quintin J, Preijers F et al Long‐lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J Innate Immun 2014; 6: 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred‐2.0: improving sequence‐based B‐cell epitope prediction using conformational epitopes. Nucleic Acids Res 2017; 45: W24–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mateus J, Grifoni A, Tarke A et al Selective and cross‐reactive SARS‐CoV‐2 T cell epitopes in unexposed humans. Science 2020; 370: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuan M, Liu H, Wu NC, Wilson IA. Recognition of the SARS‐CoV‐2 receptor binding domain by neutralizing antibodies. Biochem Biophys Res Commun 2020. 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelde A, Bilich T, Heitmann JS et al SARS‐CoV‐2‐derived peptides define heterologous and COVID‐19‐induced T cell recognition. Nat Immunol 2020. 10.1038/s41590-020-00808-x [DOI] [PubMed] [Google Scholar]
- 9. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016; 32: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jensen KK, Andreatta M, Marcatili P et al Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018; 154: 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


