Abstract
SARS-CoV-2is the causative agent for the ongoing COVID19 pandemic, and this virus belongs to the Coronaviridae family. The nsp14 protein of SARS-CoV-2 houses a 3′ to 5′ exoribonuclease activity responsible for removing mismatches that arise during genome duplication. A homology model of nsp10-nsp14 complex was used to carry out in silico screening to identify molecules among natural products, or FDA approved drugs that can potentially inhibit the activity of nsp14. This exercise showed that ritonavir might bind to the exoribonuclease active site of the nsp14 protein. A model of the SARS-CoV-2-nsp10-nsp14 complex bound to substrate RNA showed that the ritonavir binding site overlaps with that of the 3′ nucleotide of substrate RNA. A comparison of the calculated energies of binding for RNA and ritonavir suggested that the drug may bind to the active site of nsp14 with significant affinity. It is, therefore, possible that ritonavir may prevent association with substrate RNA and thus inhibit the exoribonuclease activity of nsp14. Overall, our computational studies suggest that ritonavir may serve as an effective inhibitor of the nsp14 protein. nsp14 is known to attenuate the inhibitory effect of drugs that function through premature termination of viral genome replication. Hence, ritonavir may potentiate the therapeutic properties of drugs such as remdesivir, favipiravir and ribavirin.
Keywords: nsp14, Exoribonuclease, SARS-CoV-2, Inhibitor, Ritonavir
1. Introduction
The SARS-CoV-2 virus is responsible for the ongoing pandemic of COVID-19, which has now spread to more than 200 countries. Presently, there are more than 3 million confirmed cases of the disease caused by this pathogen, with more than 250,000 fatalities [1]. There are no effective therapeutic or prophylactic available against COVID-19, although there are few promising drugs and vaccines currently under trial [2].
Like other members of the Coronaviridae family, SARS-CoV-2 possesses a single-stranded positive-sense RNA genome. The genome is roughly 30 kb long with a gene at the 5′ end known as orf1ab that encodes for all the polyprotein bearing the non-structural proteins [3]. The virus also possesses genes that code for structural proteins, namely spike (S), envelope (E), membrane (M), and nucleocapsid (N) [3,4]. In addition, the genome codes for nine ORF proteins namely ORF 3a, ORF 3b, ORF6, ORF 7a, ORF 7b, ORF8, ORF 9a, ORF 9b, and ORF 10 that are predicted to play accessory roles during the viral infection [3,5]. The polyprotein arising from orf1ab may undergo proteolytic processing to give rise to 16 non-structural proteins namely nsps 1–16 [6]. Among the cleaved products of the ORF1Ab polyprotein, the proteins of known function include the multi-domain nsp3 which has an adenosine diphosphate-ribose 1″-phosphatase activity [7]. The protease activity that is responsible for the cleavage of the polyprotein is present in the nsp5 protein [8]. The nsp12 protein houses the RNA-dependent RNA polymerases that is responsible for duplication of the genome [9]. The RNA helicase activity that is critical for genome duplication is present in the nsp13 protein [9]. Exoribonuclease (exoN) and N7-methyltransferase activities are present in the nsp14 protein [10]. The nsp15 protein houses a Nidoviral endoribonuclease specific for U, and the nsp16 protein has a SAM-dependent O-methyltransferase activity [3]. The structure of a number of non-structural proteins and their complexes has been determined recently. The structure of SARS-CoV-2-nsp12 in complex with nsp7 and nsp8 [11,12], nsp12-nsp7-nsp8 in complex with remdesivir [13] stalled pre-and post-translocated nsp12-nsp7-nsp8 [14], nsp12-nsp7-nsp8-RNA [15,16] nsp12-nsp7-nsp8 along with helicase and RNA [16,17] complex were determined by Cryo-EM. Similarly structures of nsp15 endoribonuclease [18] and nsp10/nsp16 in complex with sinefungin (a methyltransferase inhibitor) were also determined by X-Ray crystallography [19].
There is an urgent need for the identification of new molecules that can reduce viral titers and thus limit the severity of the disease. Towards this end, we have used a homology model of the nsp10-nsp14 complex from SARS-CoV-2 (SARS-CoV-2-nsp10-nsp14) to carry out in silico screening to identify potential inhibitors of exoribonuclease activity resident in nsp14. Our studies suggest that ritonavir may serve as an effective inhibitor of the exoribonuclease activity of nsp14 and can possibly potentiate the antiviral activity of chain terminating drugs such as remdesivir.
2. Methods
2.1. Homology model of the SARS-CoV-2-nsp10-nsp14 complex and model of the functional complex with RNA
The homology model of SARS-CoV-2-nsp10-nsp14 available at the SWISS-MODEL server [20] was used. The validation of the model was carried out by the SWISS-MODEL server [20]. To generate a computational model of the functional ternary complex (SARS-CoV-2-nsp10-nsp14:RNA), initially, DALI searches were carried with the model of apo- structure to identify structural orthologues of nsp10-nsp14 bound to RNA [21]. These searches showed that, on superimposition, MTREX-1 exhibits significant structural homology with the exoribonuclease module of nsp14 [22]. The DNA from the transformed coordinates of the binary complex of MTRX-1 (3AVX) were transferred onto the homology model of SARS-CoV-2-nsp14 [22]. The DNA was converted to RNA, and the structure prepared in this way was subjected to energy minimization using DESMOND module of the SCHRÖDINGER suite. The structure was minimized in an orthorhombic box containing single point and charge water model and subjected to steepest descent and LBGFS vectors minimization until the difference in energy converged to 0.1 kcal/mol [23].
2.2. In silico screening
The model of SARS-CoV-2- nsp10-nsp14 was assigned hydrogens using protein preparation wizard program in SCHRÖDINGER suite. Thus prepared structure was searched for potential ligand-binding sites using SITEMAP program in the PRIME module [24]. This program scores the potential sites according to its size, functionality, and extent of solvent exposure [25]. All the sites with a site score above 0.8 were carefully analyzed using PyMOL and the best site was used to generate a receptor grid using the Receptor Grid Generation program in SCHRÖDINGER.
The ligand libraries of FDA approved drugs (L1300) that contains 2698 molecules, Natural product library (L1400) that contains 2267 molecules and Antiviral Compound library that contains 347 molecules, were downloaded from selleckchem.com [26]. In total we have screened 5312 molecules with the SARS-CoV-2-nsp10-nsp14 complex. These libraries have molecules in 2D representation, and LIGPREP was used to convert them to energy minimized 3D structures. LIGPREP was also used to expand their tautomeric states, ionization states, stereoisomers, and ring conformers. The ligands prepared in this manner were then docked to the receptor grid using GLIDE program in PRIME module of SCHRÖDINGER [27]. Docking was carried out in standard-precision (SP) mode by generating multiple poses per ligand, which were scored based on the fit onto the receptor grid by ChemScore function. Poses with top scores were further minimized with respect to the receptor grid to calculate the binding energy. The ligands with high docking scores and binding energies were used for further analysis.
2.3. Minimization and molecular dynamics (MD) of SARS-CoV-2-nsp10-nsp14:ritonavir complex or SARS-CoV-2-nsp14-RNA complex
The SARS-CoV-2-nsp10-nsp14 -ligand complex or SARS-CoV-2-nsp10-nsp14-RNA complex was minimized using the minimization program in DESMOND module of SCHRÖDINGER [23]. The structure was minimized in an orthorhombic box containing single point and charge water (SPC) and Cl− ions to neutralize the system. The minimization was carried out with steepest descent (SD) and LBGFS vectors until the difference in energy between two consecutive steps was not more than 0.1 kcal/mol. For the minimization of RNA complex, a restraining force constant of 20 was applied onto RNA and coordinating metal ions.
Thus minimized complexes were subjected to molecular dynamics using the MD program in the DESMOND module of SCHRÖDINGER [23]. For the SARS-CoV-2-nsp10-nsp14:ritonavir complex, the length of the unrestrained MD run was 50 ns, and frames were written out with a time step of 50 ps. A model system similar to that of minimization with an orthorhombic box, SPC water, and Cl− ions were used for MD. The simulations were carried out at 300 K temperature and 1.0325 bar pressure. The system was equilibrated using the NVT ensemble with constant number of particles, constant volume, and constant temperature. For MD involving the SARS-CoV-2-nsp10-nsp14: RNA complex, the RNA and metal ions were restrained with force constant of 20. The length of the MD run was 100 ns, and frames were written out with a time step of 50 ps. The frames from the MD runs were further analyzed using the simulation interaction diagram program in SCHRÖDINGER as well as VMD (Visual Molecular Dynamics) program. The frames having maximum ligand /RNA interactions with nsp14 were subjected to energy minimization and used for further analysis.
2.4. Comparison of binding energies
To understand the strength of the interaction between SARS-CoV-2-nsp10-nsp14 complex and Ritonavir, the binding energy of association was calculated through Molecular Mechanics energies combined with Generalized Born and Surface Area continuum(MMGBSA) [28]. The binding energy was calculated for docked structure with the maximum number of contacts in the MD run. MMGBSA program was used to minimize the protein and ligand separately as well as in combination using VSGB 2.0 (Variable dielectric Surface Generalized Born) solvation model [29]. The energy of binding was then calculated by subtracting the energy of the optimized free receptor and optimized free ligand from the energy of the optimized complex. The binding energy was also calculated for the SARS-CoV-2-nsp14:RNA complex with RNA as ligand and the protein as a receptor. These calculations were also carried out for the frames which showed higher number of contacts during the MD run to identify the ligand pose/RNA conformation associated with maximal binding energy.
3. Results
3.1. Model of the SARS-CoV-2-nsp14 in its apo- and functional state
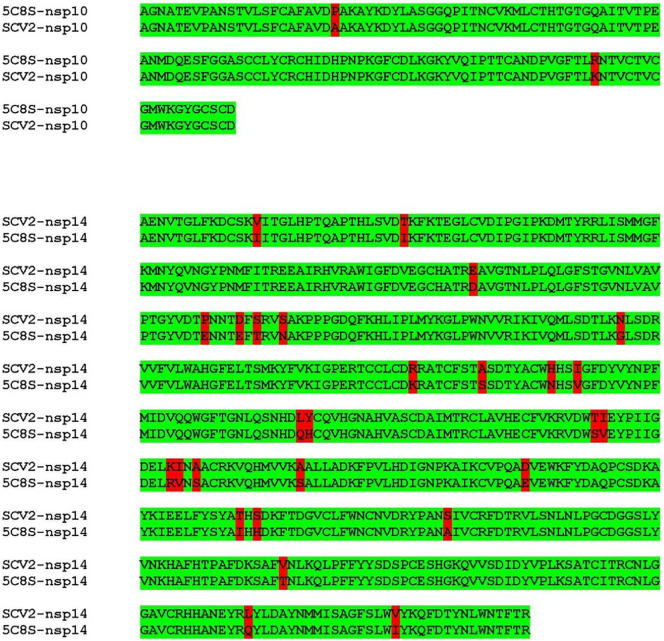
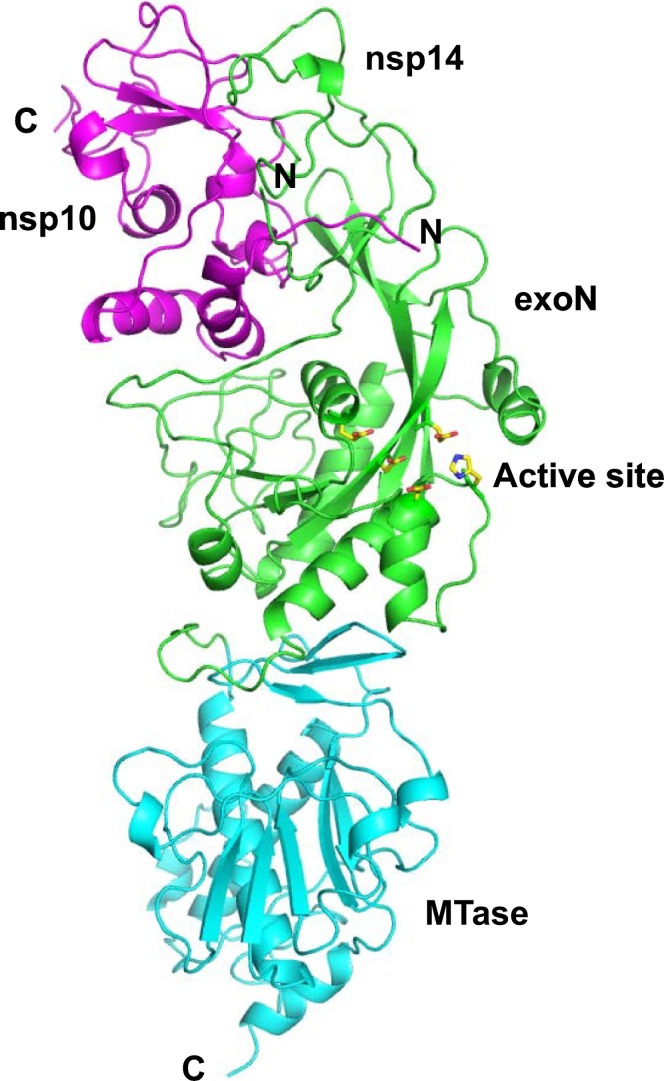
The SARS-CoV-2-nsp14 and SARS-CoV-2-nsp10 proteins show 97% identity with the corresponding protein from SARS (Fig. 1 ). The model of the SARS-CoV-2-nsp10-nsp14 complex, prepared using the structure of the corresponding complex from the SARS virus (PDB Code: 5C8S [10]) was available at the SWISS-MODEL website (Fig. 2 ). The stereochemistry of the model was good, with 98% residues in the allowed regions and less than 1% residues in the disallowed regions. The QMEAN score for this model was −2.70 which means that it can be used for further analysis [30]. The nsp14 and nsp10 models encompass residues 5925–6452 and 4239–4298 of the polyprotein translated from orf1ab of the COVID-19 genome. The exoribonuclease activity is resident in the region 1–299 and the methyltransferase activity is present in the region corresponding to residues 300–525 (Fig. 2). The active site residues for exoribonuclease activity are Asp90, Glu92, Glu191, His268 and Asp273 [10].
Fig. 1.
Alignment of the sequence of SARS-CoV-2-nsp10 and SARS-CoV-2-nsp14 with the sequence of the available structure of SARS-nsp10-nsp14 complex (5C8S). The sequence of the two proteins from SARS-CoV-2sequence exhibits about 97% identity with the corresponding sequences from SARS. The differences in the alignment are highlighted in red.
Fig. 2.
Model of SARS-CoV-2-nsp10-nsp14 complex. The homology model is displayed here and the nsp10 chain is shown in magenta. The exoribonuclease (exoN) domain of nsp14 is coloured green and the methyltransferase (MTase) region is displayed in cyan. The N- and C-termini of the two chains are highlighted. The active site residues responsible for the exoribonuclease activity are shown in stick representation and coloured according to element.
To generate the structure of the SARS-CoV-2-nsp10-nsp14 complex in its functional state, initially, a DALI search was carried out with residues 1–300 of the nsp14 enzyme to identify structural orthologs of the exoribonuclease component. The list of enzymes that showed good superimposition with the nsp14 model was analyzed to identify structures of functional ternary complexes. The structure of MTREX1 in complex with RNA (2OA8 [22]) gave a Z-score of 3.7 and an RMSD of 3.0 in the DALI search (Fig. S1). The superimposition was used to generate a model of the SARS-CoV-2-nsp14 protein in complex with RNA and two Ca2+ ions. The model was subjected to energy minimization which converged to a minimum energy of −4.96 × 105 kcal/mol. The Ramachandran plot of the minimized model showed that only 1.69% residues were in the disallowed regions.
3.2. In silico screening identifies ritonavir as a potential binder
The SITEMAP program showed three possible binding sites in the SARS-CoV-2-nsp14 protein, which were ranked according to their ability to bind to various ligands. Site 3, with a site score of 0.839 was selected as it overlapped with RNA binding site. The residues spanning site 3 were used to generate a receptor grid used for molecular docking. Molecular docking was carried out using annotated libraries of molecules that could bind to the exoribonuclease active site of SARS-CoV-2-nsp10-nsp14 complex using GLIDE docking program. The top hits from each of these libraries were ranked according to their docking score and GLIDE energy based on the interaction between the protein and ligand (Table 1 ). Based on the docking score, the top four molecules (Table 1) were Guanosine (−10.56), carfilzomib (−10.41), Inosine (−10.34) and Ritonavir (−10.144). Among these ritonavir showed the highest GLIDE energy value of −68.78 kcal/mol. Guanosine and Insoine are naturally occurring nucleotides in humans. Guanosine is a product of pentose phosphate pathway and inosine is produced by deamination of adenosine [31]. Carfilzomib on the other hand is a chemotherapy drug used in the treatment of multiple myeloma, it forms covalent bond with 20S proteasome and inhibit its activity [32]. Ritonavir is a known inhibitor of the HIV-1 protease and therefore, the possibility of adverse side effects is lower as compared to carfizomib. Also, the GLIDE energy value for ritonavir is better than that for carfilzomib (−50.11 kcal/mol) and for these reasons ritonavir was used for further analysis.
Table 1.
List of possible binder from different small molecule databases with GLIDE score better than −9.5.
| Title | Docking score | GLIDE gscore | GLIDE energy |
|---|---|---|---|
| FDA | |||
| Guanosine | −10.559 | −10.559 | −50.109 |
| Carfilzomib (PR-171) | −10.411 | −10.411 | −70.15 |
| Inosine | −10.335 | −10.335 | −45.499 |
| Ritonavir | −10.144 | −10.144 | −68.779 |
| Adrenalone hydrochloride | −10.03 | −10.03 | −40.834 |
| Paclitaxel | −9.972 | −9.972 | −62.916 |
| Danoprevir | −9.958 | −9.958 | −67.981 |
| Chlorhexidine hydrochloride | −9.59 | −9.59 | −74.705 |
| Antiviral compounds | |||
| Ritonavir | −10.144 | −10.144 | −68.779 |
| VIRA-A (vidarabine) | −10.117 | −10.117 | −47.451 |
| Ammonium glycyrrhizinate | −9.959 | −9.959 | −73.835 |
| Danoprevir (RG7227, ITMN-191, RO5190591) | −9.958 | −9.958 | −67.981 |
| Natural products | |||
| Adenosine | −10.128 | −10.128 | −47.4 |
| VIRA-A (vidarabine) | −10.117 | −10.117 | −47.451 |
| Vitamin B12 | −10.008 | −10.008 | −86.131 |
| Paclitaxel | −9.842 | −9.842 | −63.528 |
| Docetaxel | −9.81 | −9.81 | −69.759 |
| Etoposide | −9.634 | −9.634 | −57.574 |
| Tigecycline | −9.629 | −9.629 | −58.596 |
The model of the SARS-CoV-2-nsp14:ritonavir complex prepared this way was then subjected to energy minimization. The model converged to minimum energy of −5.7 × 105 kcal/mol. The final model of the complex shows the presence of one molecule of ritonavir bound in the exoribonuclease active site of the nsp14 enzyme.
3.3. Energy of binding of SARS-CoV-2-nsp10-nsp14 with natural substrates versus ritonavir
MD simulation was utilized to improve the fit between ritonavir/RNA and the SARS-CoV-2-nsp10-nsp14 complex. Based on the number of contacts, the best frame was selected from the MD run. These frames correspond to 5.9 ns and 52.5 ns for the SARS-CoV-2-nsp10-nsp14:ritonivar and SARS-CoV-2-nsp10-nsp14:RNA complexes, respectively. After energy minimization, the energy of interaction between SARS-CoV-2-nsp10-nsp14 and RNA was compared with that of between the protein complex and ritonavir. The calculated MMGBSA binding energy for the interaction of RNA and incoming nucleotide with SARS-CoV-2-nsp10-nsp14 is −98.4 kcal/mol. In comparison, the calculated MMGBSA binding energy value for the interaction of ritonavir with SARS-CoV-2-nsp10-nsp14 is −78.3 kcal/mol. These values suggest that ritonavir can bind with a significant affinity at the exoribonuclease active site of the SARS-CoV-2-nsp10-nsp14 enzyme.
Ritonavir is generally prescribed for long term use and in some patients is known to cause side effects such as hepatotoxicity, pancreatitis, retinal toxicity and allergic reactions/hypersensitivity. The oral lethal dose for this drug in mice is about 2500 mg/kg and the doses prescribed to HIV patients are much lower than this value [33]. The normal maintenance dose for adult HIV patients is 600 mg daily twice a day. Overall, the level of ritonavir induced toxicity appears to be low and adverse side effects are observed in only certain patients.
3.4. Comparison of interactions in SARS-CoV-2-nsp10-nsp14:ritonavir complex and the SARS-CoV-2-Nsp10-nsp14:RNA complex
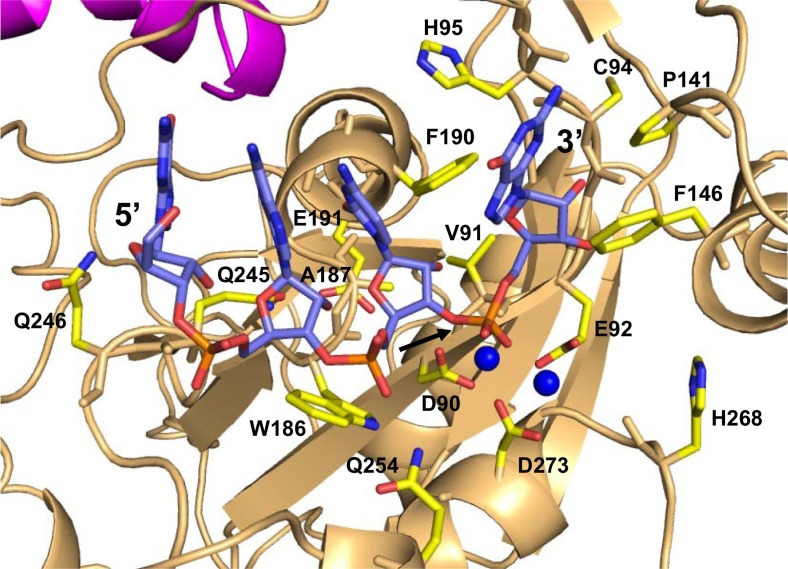
The model of the functional complex of SARS-CoV-2-nsp10-nsp14 with RNA was analyzed to identify the interacting residues (Fig. 3 ). The residues that form Van der Wall interactions with RNA are Asp90, Val91, Glu92, Gly93, Cys94, His95, Ala96, Thr97, Pro141, Phe146, Leu149, Trp186, Ala187, Gly189, Phe190, Gln245, Gln246, Gln254 and Asp27 (Fig. 3). The residues Asp90, Glu92, Gly93, His95, Gln245, Gln254, and Asp273 form polar interactions with RNA (Fig. 3).
Fig. 3.
Model of the SARS-CoV-2-nsp10-nsp14 complex bound to RNA. nsp10 and nsp14 are coloured magenta and light orange, respectively. The RNA molecule and the interacting residues from nsp14 are shown in stick representation and coloured according to element. The cofactor ions are shown in the form of blue spheres. The site of cleavage is marked by an arrow.
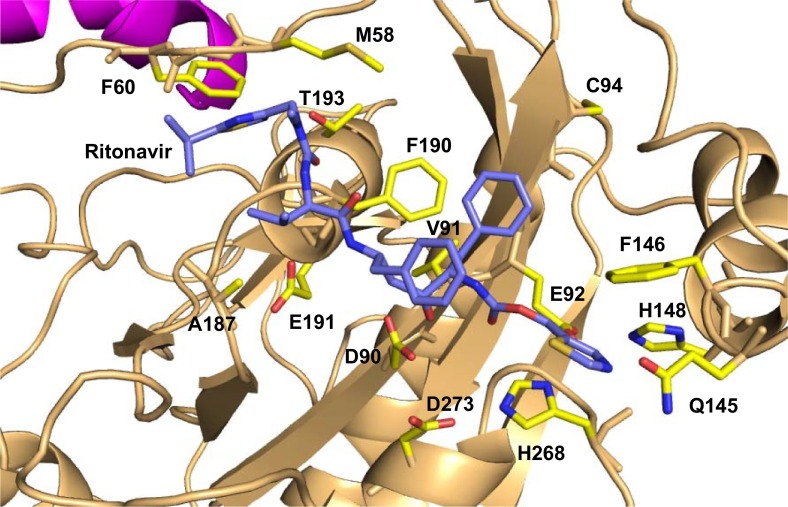
The SARS-CoV-2-nsp10-nsp14:ritonavir complex was analyzed to identify the residues that interact with the drug (Fig. 4 ). The nsp14 residues that form Van der Waal contacts with the ritonavir include Met58, Gly59, Phe60, Asp90, Val91, Glu92, Gly93, Gln145, Phe146, His148, A187, Ala187, Gly189, Phe190, Thr193, and His268. The residues Asp90, Gln145, Phe190 also form polar interactions with the ritonavir molecule. Among the five catalytic residues, Asp90, Glu92, and H268 form interactions with the Ritonavir molecule (Fig. 4).
Fig. 4.
Model of the SARS-CoV-2-nsp10-nsp14 complex bound to ritonavir. nsp10 and nsp14 are coloured magenta and light orange, respectively. The ritonavir molecule is shown in stick representation and coloured according to element (carbon = blue). The residues of nsp14 that interact with ritonavir are shown in stick representation (carbon = yellow).
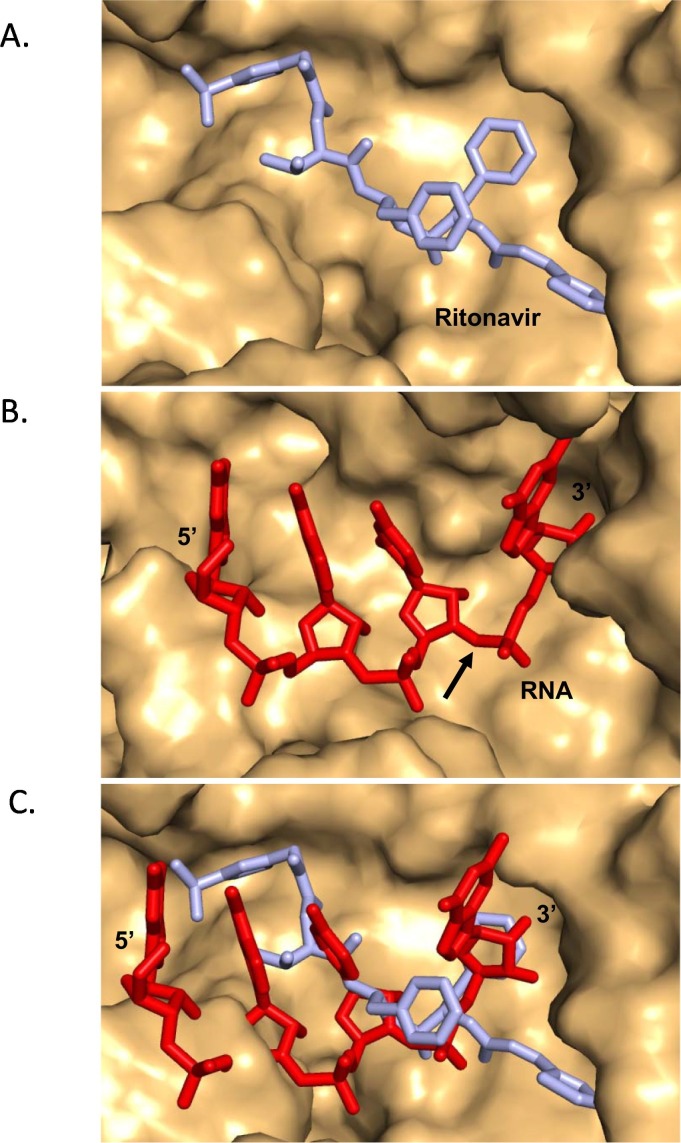
A superimposition of the model of the SARS-CoV-2-nsp10-nsp14:RNA and the SARS-CoV-2-nsp10-nsp14:ritonavir complexes were carried out to ascertain the level of overlap between the binding sites of the natural substrates of nsp14 and the ritonavir molecule. The superimposition showed that the binding site of ritonavir overlaps with that of the RNA substrate (Fig. 5 ). Therefore, the binding of ritonavir may inhibit the formation of the complex of nsp14 with RNA and thus prevent the excision of nucleotides from the 3′ end of the growing RNA strand during genome duplication.
Fig. 5.
Binding site of ritonavir overlaps with that of substrate RNA. (A) SARS-CoV-2-nsp10-nsp14:ritonavir where the surface of protein molecule is displayed in light orange and ritonavir is displayed in stick representation and coloured cyan. (B) The surface of the protein molecule is displayed and the RNA, is displayed in stick representation and coloured red. The site of cleavage on RNA is marked by an arrow. (C) Superimposition of the models of functional ternary complex (red) and that of SARS-CoV-2-nsp10-nsp14:ritonavir (cyan) is displayed. The comparison suggests that the presence of ritonavir may prevent binding of the natural RNA substrate.
Since Ritonavir is a known protease inhibitor drug we also tried to dock ritonavir into the active site of the SARS-CoV-2main protease (PDB site: 6 LU7) [34]. It was found that Ritonavir docked at the ligand binding site with a docking score of −8.527. The energy of binding as calculated using the MMGBSA method was found to be −68.66 kcal/mol. Therefore, it is possible that ritonavir may also bind to the protease active site and possibly inhibit the activity of this enzyme. However, it has been observed that ritonavir in combination with lopinavir does not provide any meaningful benefit in the treatment of COVID-19 and therefore the level of inhibition of the protease may not be significant [35].
4. Discussion
The studies presented here suggest that ritonavir may be a possible inhibitor of the 3′ to 5′ exoribonuclease activity of the SARS-CoV-2-nsp14 enzyme. The enzyme is involved in correcting mismatches that arise during genome duplication, like the proofreading exonucleases of DNA polymerases. Since this enzyme is critical for accurate replication of the viral enzyme, the inhibition of this enzyme can result in lethal mutagenesis and lower viral titres to reduce the severity of the COVID-19 disease [36,37]. In the case of Mouse hepatitis virus (MHV), a murine coronavirus, the exoribonuclease activity of nsp14 is shown to be important for evading the host innate immune response [38]. It is, therefore, possible that ritonavir treatment may render the SARS-CoV-2 virus more susceptible to detection and clearance by the human immune system.
The exoribonuclease activity of nsp14 has been implicated in the excision of drugs that are incorporated into RNA [39,40]. The inhibition of proofreading may potentiate the effect of drugs such as remdesivir and favipiravir [41,42]. These molecules are prodrugs that get converted into active triphosphate forms in the body and are incorporated into progeny RNA by the viral RNA-dependent-RNA polymerase [41,43]. The presence of the modified nucleotides corresponding to these drugs results in premature termination of viral replication [[44], [45], [46]]. It has been seen before that a mutant version of MHV without the exoribonuclease activity exhibited higher sensitivity to remdesivir [39]. The 3′ to 5′ exoribonuclease activity of nsp14 can potentially excise out the incorporated modified nucleotides and thus reduce the efficacy of the corresponding drugs. For these reasons, it has been proposed that the combination of nucleoside analogs with nsp14 inhibitors may be more effective [47]. Consequently, inhibition of ritonavir by nsp14 may potentiate the effect of drugs that function through inhibition of viral genome replication by premature chain termination. The C-terminal region of SARS-CoV-2 nsp14 houses a methyltransferase guanine-N7 methyltransferase. Recently, Selvaraj and colleagues have identified different compounds from traditional Chinese medicinal preparations that can potentially inhibit the RNA capping activity of nsp14 [48].
Ritonavir in combination with lopinavir has been found to be ineffective against COVID-19 and therefore its potential ability to inhibit the exoribonuclease or protease activity may not be enough to prevent viral replication [35]. Ritonavir may be more effective in combination with chain terminating drugs such as remdesivir, favipiravir or ribavirin. A recent clinical trial wherein the test patients exhibited significantly reduced hospitalization time on administration of a combination of ritonavir-lopinavir, ribavirin and interferon β-1B provides support for this idea [49,50]. The combination of lopinavir-ritonavir and ribavirin was also found to be helpful during the first SARS epidemic [51].
The number of patients suffering from COVID-19 are increasing daily and many of them are in serious condition. Hence, the ability of the ritonavir plus chain terminators such as remdesivir, favipiravir or ribavirin to inhibit viral replication should be tested urgently using in vivo assays and in model animals. In addition, given the urgency of the situation and the fact that separate clinical trials are already going on with ritonavir, remdesivir and favipiravir, new trials involving a combination of ritonavir plus remdesivir and ritonavir plus favipiravir may be initiated [2]. Remdesivir treatment improved the condition of about 70% of COVID-19 patients who were severely ill and treatment with a combination of ritonavir and remdesivir may significantly enhance the number of patients who recover completely from the viral infection [52]. Also, remdesivir is known to cause liver damage at the currently prescribed doses and a combination with ritonavir may permit formulation of lower doses and thus reduce the possibility of liver damage [53].
The following is the supplementary data related to this article.
Structural alignment of SARS-CoV-2-nsp14 and TREX1-RNA complex: The structural alignment of the backbone of the computational model of nsp14 (orange) with that of the structure of TREX1-RNA complex (2OA8; cyan), as obtained from the DALI server, is shown here. The alignment shows that the catalytic core of the two enzymes shows substantial overlap and all the catalytic residues superimpose except H268 (nsp14) and H195 (2OA8). The catalytic residues are shown in stick representation and the labels for residues from nsp14 are underlined. The structure of TREX1-RNA complex was therefore used to generate a model of nsp14-RNA complex.
CRediT authorship contribution statement
Prof. Deepak T. Nair: Conceptualization; Funding acquisition; Supervision; Validation; Writing – review & editing.
Mr. Naveen Narayanan: Formal analysis; Investigation; Methodology; Validation; Roles/Writing – original draft.
Acknowledgement
The study was funded by an Intramural grant from the Regional Centre for Biotechnology.
References
- 1.Johns Hopkins Coronavirus Resource Center, (n.d.).
- 2.Vanden Eynde J.J. 2020. Covid-19: a brief overview of the discovery clinical trial, Pharmaceuticals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. 2020. A new coronavirus associated with human respiratory disease in China, Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. Npj Vaccines. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michel C.J., Mayer C., Poch O., Thompson J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17:131. doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A.R. Fehr, S. Perlman, Coronaviruses: an overview of their replication and pathogenesis, in: Coronaviruses Methods Protoc., Springer New York, 2015: pp. 1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed]
- 10.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015 doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.G.M. Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Z. Chang S, Xie YC, Tian G, Jiang HW, Tao SC, Shen J, Jiang Y, Jiang H, Xu Y, X.H. S, Zhang Y, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science (80-.). (2020). doi: 10.1101/2020.02.07.937862. [DOI] [PMC free article] [PubMed]
- 12.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.W. Yin, C. Mao, X. Luan, D.-D. Shen, Q. Shen, H. Su, X. Wang, F. Zhou, W. Zhao, M. Gao, S. Chang, Y.-C. Xie, G. Tian, H.-W. Jiang, S.-C. Tao, J. Shen, Y. Jiang, H. Jiang, Y. Xu, S. Zhang, Y. Zhang, H.E. Xu, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science (80-.). (2020) eabc1560. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed]
- 14.Q. Wang, J. Wu, H. Wang, Y. Gao, Q. Liu, A. Mu, W. Ji, L. Yan, Y. Zhu, C. Zhu, X. Fang, X. Yang, Y. Huang, H. Gao, F. Liu, J. Ge, Q. Sun, X. Yang, W. Xu, Z. Liu, H. Yang, Z. Lou, B. Jiang, L.W. Guddat, P. Gong, Z. Rao, Structural basis for RNA replication by the SARS-CoV-2 polymerase, Cell. 182 (2020) 417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed]
- 15.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 16.Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., Wang Q., Rao Z., Lou Z. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. Chen, B. Malone, E. Llewellyn, M. Grasso, P.M.M. Shelton, P.D.B. Olinares, K. Maruthi, E.T. Eng, H. Vatandaslar, B.T. Chait, T.M. Kapoor, S.A. Darst, E.A. Campbell, Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex, Cell. 182 (2020) 1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed]
- 18.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P. Krafcikova, J. Silhan, R. Nencka, E. Boura, Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin, Nat. Commun. 11 (2020). doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed]
- 20.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm L., Laakso L.M. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Silva U., Choudhury S., Bailey S.L., Harvey S., Perrino F.W., Hollis T. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J. Biol. Chem. 2007 doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 23.K.J. Bowers, E. Chow, H. Xu, R.O. Dror, M.P. Eastwood, B.A. Gregersen, J.L. Klepeis, I. Kolossvary, M.A. Moraes, F.D. Sacerdoti, J.K. Salmon, Y. Shan, D.E. Shaw, Scalable algorithms for molecular dynamics simulations on commodity clusters, in: Proc. 2006 ACM/IEEE Conf. Supercomput. SC'06, ACM Press, New York, New York, USA, 2006: p. 84. doi: 10.1145/1188455.1188544. [DOI]
- 24.Halgren T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- 25.Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 26.Selleckchem.com - Inhibitor Expert (Inhibitors, Compound Libraries), (n.d.).
- 27.R.A. Friesner, J.L. Banks, R.B. Murphy, T.A. Halgren, J.J. Klicic, D.T. Mainz, M.P. Repasky, E.H. Knoll, M. Shelley, J.K. Perry, D.E. Shaw, P. Francis, P.S. Shenkin, Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J. Med. Chem. 47 (2004) 1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed]
- 28.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Li, R. Abel, K. Zhu, Y. Cao, S. Zhao, R.A. Friesner, The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling, Proteins Struct. Funct. Bioinforma. 79 (2011) 2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed]
- 30.Benkert P., Tosatto S.C.E., Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Proteins Struct. Funct. Genet. 2008 doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 31.Welihinda A.A., Kaur M., Greene K., Zhai Y., Amento E.P. The adenosine metabolite inosine is a functional agonist of the adenosine A2A receptor with a unique signaling bias. Cell. Signal. 2016;28:552–560. doi: 10.1016/j.cellsig.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J.E., Park J., Jun Y., Oh Y., Ryoo G., Jeong Y.S., Gadalla H.H., Min J.S., Jo J.H., Song M.G., Kang K.W., Bae S.K., Yeo Y., Lee W. Expanding therapeutic utility of carfilzomib for breast cancer therapy by novel albumin-coated nanocrystal formulation. J. Control. Release. 2019;302:148–159. doi: 10.1016/j.jconrel.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritonavir - DrugBank, (n.d.).
- 34.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.B. Cao, Y. Wang, D. Wen, W. Liu, J. Wang, G. Fan, L. Ruan, B. Song, Y. Cai, M. Wei, X. Li, J. Xia, N. Chen, J. Xiang, T. Yu, T. Bai, X. Xie, L. Zhang, C. Li, Y. Yuan, H. Chen, H. Li, H. Huang, S. Tu, F. Gong, Y. Liu, Y. Wei, C. Dong, F. Zhou, X. Gu, J. Xu, Z. Liu, Y. Zhang, H. Li, L. Shang, K. Wang, K. Li, X. Zhou, X. Dong, Z. Qu, S. Lu, X. Hu, S. Ruan, S. Luo, J. Wu, L. Peng, F. Cheng, L. Pan, J. Zou, C. Jia, J. Wang, X. Liu, S. Wang, X. Wu, Q. Ge, J. He, H. Zhan, F. Qiu, L. Guo, C. Huang, T. Jaki, F.G. Hayden, P.W. Horby, D. Zhang, C. Wang, A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19, N. Engl. J. Med. (2020). doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed]
- 36.Jensen J.D., Lynch M. 2020. Considering mutational meltdown as a potential SARS-CoV-2 treatment strategy, Heredity (Edinb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bull J.J., Sanjuán R., Wilke C.O. Theory of lethal mutagenesis for viruses. J. Virol. 2007 doi: 10.1128/jvi.01624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Case J.B., Li Y., Elliott R., Lu X., Graepel K.W., Sexton N.R., Smith E.C., Weiss S.R., Denison M.R. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 2017 doi: 10.1128/jvi.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. 2018. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease, MBio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferron F., Subissi L., De Morais A.T.S., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2017 doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furuta Y., Takahashi K., Fukuda Y., Kuno M., Kamiyama T., Kozaki K., Nomura N., Egawa H., Minami S., Watanabe Y., Narita H., Shiraki K. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002 doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.T.K. Warren, R. Jordan, M.K. Lo, A.S. Ray, R.L. Mackman, V. Soloveva, D. Siegel, M. Perron, R. Bannister, H.C. Hui, N. Larson, R. Strickley, J. Wells, K.S. Stuthman, S.A. Van Tongeren, N.L. Garza, G. Donnelly, A.C. Shurtleff, C.J. Retterer, D. Gharaibeh, R. Zamani, T. Kenny, B.P. Eaton, E. Grimes, L.S. Welch, L. Gomba, C.L. Wilhelmsen, D.K. Nichols, J.E. Nuss, E.R. Nagle, J.R. Kugelman, G. Palacios, E. Doerffler, S. Neville, E. Carra, M.O. Clarke, L. Zhang, W. Lew, B. Ross, Q. Wang, K. Chun, L. Wolfe, D. Babusis, Y. Park, K.M. Stray, I. Trancheva, J.Y. Feng, O. Barauskas, Y. Xu, P. Wong, M.R. Braun, M. Flint, L.K. McMullan, S.S. Chen, R. Fearns, S. Swaminathan, D.L. Mayers, C.F. Spiropoulou, W.A. Lee, S.T. Nichol, T. Cihlar, S. Bavari, Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys, Nature. (2016). doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed]
- 43.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020 doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020 doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.E.P. Tchesnokov, J.Y. Feng, D.P. Porter, M. Götte, Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir, Viruses. 11 (2019). doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed]
- 46.G.M. Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Z. Chang S, Xie YC, Tian G, Jiang HW, Tao SC, Shen J, Jiang Y, Jiang H, Xu Y, X.H. S, Zhang Y, Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science (80-.). (2020). doi: 10.1101/2020.02.07.937862. [DOI] [PMC free article] [PubMed]
- 47.Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. 2020. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites, Antiviral Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J. Biomol. Struct. Dyn. 2020;1 doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopinavir/ritonavir, ribavirin and IFN-beta combination for nCoV treatment, Case Med. Res. (2020). doi:10.31525/ct1-nct04276688.
- 50.I.F.-N. Hung, K.-C. Lung, E.Y.-K. Tso, R. Liu, T.W.-H. Chung, M.-Y. Chu, Y.-Y. Ng, J. Lo, J. Chan, A.R. Tam, H.-P. Shum, V. Chan, A.K.-L. Wu, K.-M. Sin, W.-S. Leung, W.-L. Law, D.C. Lung, S. Sin, P. Yeung, C.C.-Y. Yip, R.R. Zhang, A.Y.-F. Fung, E.Y.-W. Yan, K.-H. Leung, J.D. Ip, A.W.-H. Chu, W.-M. Chan, A.C.-K. Ng, R. Lee, K. Fung, A. Yeung, T.-C. Wu, J.W.-M. Chan, W.-W. Yan, W.-M. Chan, J.F.-W. Chan, A.K.-W. Lie, O.T.-Y. Tsang, V.C.-C. Cheng, T.-L. Que, C.-S. Lau, K.-H. Chan, K.K.-W. To, K.-Y. Yuen, Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial, Lancet. (2020). doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed]
- 51.Chu C.M., Cheng V.C.C., Hung I.F.N., Wong M.M.L., Chan K.H., Chan K.S., Kao R.Y.T., Poon L.L.M., Wong C.L.P., Guan Y., Peiris J.S.M., Yuen K.Y. 2004. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings, Thorax. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Grein, N. Ohmagari, D. Shin, G. Diaz, E. Asperges, A. Castagna, T. Feldt, G. Green, M.L. Green, F.-X. Lescure, E. Nicastri, R. Oda, K. Yo, E. Quiros-Roldan, A. Studemeister, J. Redinski, S. Ahmed, J. Bernett, D. Chelliah, D. Chen, S. Chihara, S.H. Cohen, J. Cunningham, A. D'Arminio Monforte, S. Ismail, H. Kato, G. Lapadula, E. L'Her, T. Maeno, S. Majumder, M. Massari, M. Mora-Rillo, Y. Mutoh, D. Nguyen, E. Verweij, A. Zoufaly, A.O. Osinusi, A. DeZure, Y. Zhao, L. Zhong, A. Chokkalingam, E. Elboudwarej, L. Telep, L. Timbs, I. Henne, S. Sellers, H. Cao, S.K. Tan, L. Winterbourne, P. Desai, R. Mera, A. Gaggar, R.P. Myers, D.M. Brainard, R. Childs, T. Flanigan, Compassionate use of remdesivir for patients with severe Covid-19, N. Engl. J. Med. (2020). doi: 10.1056/nejmoa2007016. [DOI] [PMC free article] [PubMed]
- 53.Zampino R., Mele F., Florio L.L., Bertolino L., Andini R., Galdo M., De Rosa R., Corcione A., Durante-Mangoni E. Liver injury in remdesivir-treated COVID-19 patients. Hepatol. Int. 2020;1:1. doi: 10.1007/s12072-020-10077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural alignment of SARS-CoV-2-nsp14 and TREX1-RNA complex: The structural alignment of the backbone of the computational model of nsp14 (orange) with that of the structure of TREX1-RNA complex (2OA8; cyan), as obtained from the DALI server, is shown here. The alignment shows that the catalytic core of the two enzymes shows substantial overlap and all the catalytic residues superimpose except H268 (nsp14) and H195 (2OA8). The catalytic residues are shown in stick representation and the labels for residues from nsp14 are underlined. The structure of TREX1-RNA complex was therefore used to generate a model of nsp14-RNA complex.