Abstract
Plasmids are autonomously replicating sequences that help cells adapt to diverse stresses. Theta plasmids are the most frequent plasmid class in enterobacteria. They co-opt two host replication mechanisms: replication at oriC, a DnaA-dependent pathway leading to replisome assembly (theta class A), and replication fork restart, a PriA-dependent pathway leading to primosome assembly through primer extension and D-loop formation (theta classes B, C, and D). To ensure autonomy from the host’s replication and to facilitate copy number regulation, theta plasmids have unique mechanisms of replication initiation at the plasmid origin of replication (ori). Tight plasmid copy number regulation is essential because of the major and direct impact plasmid gene dosage has on gene expression. The timing of plasmid replication and segregation are also critical for optimizing plasmid gene expression. Therefore, we propose that plasmid replication needs to be understood in its biological context, where complex origins of replication (redundant origins, mosaic and cointegrated replicons), plasmid segregation, and toxin-antitoxin systems are often present. Highlighting their tight functional integration with ori function, we show that both partition and toxin-antitoxin systems tend to be encoded in close physical proximity to the ori in a large collection of Escherichia coli plasmids. We also propose that adaptation of plasmids to their host optimizes their contribution to the host’s fitness while restricting access to broad genetic diversity, and we argue that this trade-off between adaptation to host and access to genetic diversity is likely a determinant factor shaping the distribution of replicons in populations of enterobacteria.
INTRODUCTION
Plasmids are autonomously replicating sequences. They are found across all kingdoms of life but are particularly abundant in prokaryotes (1). They typically carry nonessential genes that have adaptive value under specific stresses such as heavy metal exposure, antibiotics, or adapting to a host (virulence genes) (2, 3).
Here, we review the mechanism of plasmid replication known as theta, which is by far the predominant type in Enterobacteriaceae. This prokaryotic family groups a number of related genera (Escherichia, Shigella, Salmonella, Yersinia) that share the intestinal tract as their central ecological niche and that have been extensively studied due to their clinical relevance (4, 5).
Enterobacterial plasmids share similar features. Nearly all of them can be transferred between strains in one way or another, a process known as horizontal gene transfer (HGT). This allows plasmids to serve as platforms for the exchange of genetic material, for capturing new adaptive genes from the environmental microbial gene pool, and for shuffling and spreading these genes among pathogens (3, 6, 7). There are three main mechanisms of HGT, namely, cell-to-cell contact and formation of a bridge (conjugation), direct uptake from the environment (transformation), and through bacteriophages (transduction) (8). Conjugation is the predominant mechanism in Gram-negative bacteria. It involves copying the plasmid DNA, relaxing it, and moving the DNA through a type IV secretion system to a recipient cell in an ATP-dependent fashion (reviewed in references 3, 7, and 9). The majority of enterobacterial plasmids carry sets of conjugation genes, although these are not always active or complete (10). Some virulence plasmids do not have a conjugation machinery but can be transferred along with a conjugative plasmid, which acts as a helper; this process is known as plasmid mobilization (10). Transduction is also frequent in enterobacteria (11). It occurs when a bacteriophage packages bacterial genetic material along with its own genetic material and infects another bacterium (generalized transduction) or when a phage that has integrated into the bacterial chromosome is improperly excised and packages bacterial DNA (specialized transduction) (12).
Here, we present our current mechanistic understanding of theta plasmid replication and its regulation. We also present concepts that are emerging from genomic studies, which help place the replication of these plasmids in its broader biological context. Given that plasmids isolated from a variety of Enterobacteriaceae share similar features (5), we use virulence plasmids in extraintestinal pathogenic Escherichia coli (ExPEC) as representative of enterobacteria.
PLASMID REPLICATION INITIATION AT ORIGINS OF REPLICATION
Plasmid replication starts at a specific location known as the plasmid origin of replication, or ori. The origin of replication contains nucleotide sequence motifs that are recognized in trans by the replication machinery. This machinery typically involves a combination of plasmid and host elements. Since borrowing genes from the host minimizes the metabolic cost of plasmid maintenance, plasmid-encoded replicative elements are typically restricted to replication initiation, with downstream steps frequently relying exclusively on host proteins. Having plasmid-specific mechanisms of replication initiation ensures autonomy from the host and facilitates the regulation of plasmid copy number by providing specific targets for regulation. Plasmid replication initiation involves plasmid-encoded replication proteins (Reps) and/or plasmid-specific primers. Even these plasmid replication initiation factors are often assisted by host proteins (see below).
Host proteins participating in plasmid DNA synthesis are part of the machinery that replicates the chromosome at oriC (recently reviewed in references 8 and 13–15) and/or that restart replication following replication fork collapse (reviewed in references 16–19). These include proteins facilitating the melting of the DNA duplex (DnaA), helicases (PriA, DnaB), proteins that catalyze primer synthesis (DnaG) and primer extension (Pol I, Pol III holoenzyme), proteins that process primers and gaps (Pol I, ligase), proteins that control the local supercoiling status of DNA (topoisomerases), and proteins that bind and stabilize single-stranded DNA (ssDNA) (SSB). Note that replication proceeds in the 5′ to 3′ direction. The synchronous replication of leading and lagging strands involves two core Pol III enzymes, with the enzyme replicating the leading strand doing so continuously and the enzyme replicating the lagging strand doing so discontinuously, i.e., in segments. The Pol III core enzymes in the Pol III holoenzyme are joined by two τ (tau) subunits and assisted by β units that act as mobile sliding clamps, which ensures the coordination and processivity of the replication in the two strands.
MECHANISMS OF PLASMID REPLICATION
Replication initiation determines the mechanics of plasmid replication. Within theta plasmids, we can distinguish two broad mechanisms of replication initiation: oriC-like (which parallels replication initiation at oriC) and PriA-mediated (which is similar to replication restart after replication fork collapse). The mechanisms involved in these two categories of plasmid replication (as well as those of nontheta plasmids) are systematically compared in Table 1 and discussed in detail below.
Table 1.
Comparison of the different modes of plasmid replicationa
| Characteristic | Theta A | Theta Bb | Theta C | Theta D | Strand displacement | Rolling-circle |
|---|---|---|---|---|---|---|
| Directionality of replication (unidirectional vs bi-directional) | Bi/uni | Uni | Uni | Uni | Bi | Uni |
| Coupling of leading- and lagging-strand synthesis | Coupled (replisome) | Coupled (replisome) | Coupled (replisome) | Coupled (replisome) | Uncoupled | Uncoupled |
| Simultaneous leading- and lagging-strand synthesis | Yes | Yes | Yes | Yes | Yes | No, with generation of single-stranded intermediate |
| Leading strand synthesis | ||||||
| Continuous vs discontinuous | Continuous | Continuous | Continuous | Continuous | Continuous | Continuous |
| Initiation factor | Rep melts duplex and recruits DnaB helicase, often assisted by DnaA and by NAPs | NA | RepA (also primase activity) | Rep E melts duplex only and has active role in primer generation | Rep C (DnaA-like) | Rep (generates initial nick and seals it) |
| Initiation factor binding site | Multiple iterons with induced bending | NA | Iteron with two functional subregions: (I) binding and (II) initiation of DNA replication | No iterons, no bending | ssi sites (hairpins) | Dso (hairpin cruciform structure with binding domain and nicking domain) |
| Leading-strand primer | DnaG primase (host) | Transcript processed by RNaseH1 | ppApGpA produced by RepA | RepE transcript, possibly processed by RepE itself | RepB′ (interacting at the base of the hairpin) | Rep C-generated nick produces a 3′-OH |
| Helicase | DnaBb | PriA + DnaBb | PriA + DnaBb | PriA + DnaBb | RepA | DnaBb |
| Lagging-strand synthesis | ||||||
| Continuous vs discontinuous | Discontinuous | Discontinuous | Discontinuous | Discontinuous | Continuous | Continuous |
| Lagging-strand primer | DnaG | DnaG | DnaG | DnaG | RepB′ | DnaG |
| Helicase | PriA+ DnaBb | PriA+ DnaBb | PriA+ DnaBb | PriA+ DnaBb | RepA in coordination with Pol III | Not needed (it is ssDNA) |
| Additional comments | Predominant type in Gram-negative bacteria | Limits size of the plasmid to ∼40kb | Minimal size of cis-acting ori | 2 sites of replisome arrest: for Top I-like enzyme and for resolvase | More independent of host factorsLimits the size of the plasmid to ∼15 kb | Predominant type in Gram-positive bacteria, also associated with conjugation |
oriC-like plasmids correspond to class A theta plasmid replication. PriA-mediated plasmids correspond to theta classes B, C, and D. Both categories converge at the DnaB loading step and rely on the hosts’ Pol III holoenzyme to complete the replication of both strands. Thus, theta plasmids are characterized by coordinated leading- and lagging-strand replication, where lagging-strand replication is discontinuous. This leads to the generation of bubbles in the early stages of replication that resemble the Greek letter θ (theta) under the electron microscope, which gives the name to this type of plasmid replication.
oriC Chromosomal Replication Initiation
DnaA is an AAA+-family ATPase that is central for replication initiation in the chromosome. In this capacity, DnaA has three roles: melting duplex DNA at oriC, recruiting the replisome through DnaB helicase, and controlling the timing of replication initiation.
DnaA binds DnaA boxes, 9-nucleotide (nt) motifs found in the vicinity of oriC that have high affinity for DnaA. Lower-affinity motifs (with small changes to the consensus sequence) are also present, and these are only recognized by DnaA-ATP. Cooperative oligomerization of DnaA occurs when both low- and high-affinity DnaA motifs are bound and leads to the formation of a nucleoprotein helical structure, which generates torsional forces that open up the DNA in an adjacent, ATP-rich area called the DUE, for DNA unwinding element (20, 21). The opening of replication origins by DnaA-ATP occurs by a direct stretching mechanism, similar to other nucleic acid-dependent AAA+ systems (22). The continued DnaA oligomerization in the top strand of the exposed ssDNA and SSB binding to the bottom strand keeps the DNA open (23, 24). DnaA filaments on ssDNA are stabilized by an origin element composed of repeated trinucleotide motifs (25). DnaB is loaded on the bottom/lower strand first, possibly by direct interaction with DnaA; DnaC loads DnaB on the top/upper strand invaded by DnaA molecules (26, 27). The order of helicase loading is important, providing the necessary orientation for the head-to-head loading of the helicase (28). DnaB, in turn, recruits DnaG, which is the primase. Primase activity leads to the release of DnaC and to the recruitment of the β-clamp loader and β-clamp and of the Pol III core (one per each strand). Once the replisome has been loaded, replication proceeds bidirectionally until the two forks meet and resolve. The terminus region where the two forks generally meet, which is antipodal to oriC, is flanked by a series of 20-nt-long replication termination (Ter) signals that act as direction-dependent arrest sites for fork progression, where forks are allowed to enter but not leave (29, 30). This direction-dependent arrest ensures that if one of the replication forks is delayed, the two forks still meet and resolve in the terminus region of the chromosome. The mechanism conferring directionality to the fork arrest has not been completely elucidated but involves DnaB-induced strand separation and asymmetric contacts between a specific terminator protein (Tus) and its cognate Ter sequence, leading to a highly stable “locked” complex (31).
The specificity of DnaA-ATP for low-affinity DnaA sites acts as a mechanism to control the timing of replication initiation (20, 32). Following replication, DnaA-ATP moves from oriC to chromosomal titration sites such as datA (preference for DnaA-ATP) until the intrinsic ATPase activity of DnaA is stimulated by a component of the replisome (Hda) (33–35). DnaA is then recharged to DnaA-ATP through new DnaA synthesis, by relocation to DnaA reactivation sequences, or through contact with acidic membrane phospholipids (35, 36). Two nucleoid-associated proteins (NAPs), FIS and IHF, and Dam methylation also help coordinate the timing of replication initiation because DNA bending by FIS and IHF and binding of hemimethylated DNA by SeqA prevent extension of the DNA filament until the cell is ready for a new round of replication (reviewed in reference 36).
oriC-Like Plasmid Replication Initiation
Similar to chromosomal replication at oriC, oriC-like plasmid replication initiation involves recognition of a sequence motif by a replication initiation protein. In this case, however, the replication initiation protein (Rep) is plasmid-encoded. Reps are generally named with letters of the alphabet (e.g., RepA, RepB, RepE, etc.), but these names are arbitrary, i.e., two Reps with the same name being in different plasmids does not generally mean that the two Reps are phylogenetically related. Methods for typing plasmids based on sequence homology are discussed further below.
The role of Reps in class A theta replication initiation is summarized in Fig. 1. Class A theta replicons typically include a replication initiator, DnaA boxes upstream or downstream of the Rep, iterons, and an AT-rich DUE. As an example, Fig. 1 shows the arrangement found in plasmid R2K, which belongs to the IncP compatibility group. Sites for binding of NAPs (such as IHF) or for methylation by Dam methylase are frequently present as well. Reps cooperatively bind iterons until those iterons are saturated. Next, the DnaB helicase is recruited through Rep, with a variable involvement of DnaA binding to adjacent DnaA boxes, leading to open complex formation.
Figure 1.
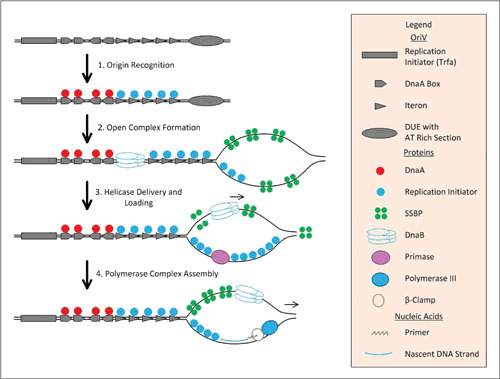
Role of replication initiation proteins in initiation in class A theta plasmid replication (based on references 43 and 97). Class A theta replicons typically include a replication initiator gene: rep (rectangle), DnaA boxes upstream or downstream of the Rep (square), iterons (arrow heads), and an AT-rich DNA unwinding element (DUE, oval). Sites for binding of nucleoid associated-proteins (such as IHF) or for methylation by Dam methylase are also frequently present as well but not shown. oriV of the RK2 plasmid (which belongs to the IncP compatibility group) is shown as an example. RK2’s replication initiator is TrfA. The trfA gene is under the control of a strong promoter. Transcription produces two products (a longer and a shorter one) that in this figure are considered largely redundant. During plasmid origin recognition (phase 1), replication initiation proteins (in their monomeric forms) cooperatively bind iterons until those iterons are saturated. The formation of an open complex (phase 2) involves unwinding of the DUE and continued binding of the replication initiator into the bottom strand of ssDNA; the rest of the bottom strand of ssDNA and the top strand are bound by SSB, which is a tetramer. In the case of RK2, the plasmid encodes its own SSB, which is under the control of the same promoter as the replication initiator gene (trfA). DnaA enhances/stabilizes the formation of the TrfA-mediated open complex and assists with the recruitment of DnaBC. Finally, the longer form of TrfA also assists in strand-specific replisome assembly in a DnaA-independent manner via direct interaction with the β clamp and through a sequence-specific interaction with one strand of the plasmid origin DUE (8, 136).
Despite clear mechanistic similarities with oriC, Rep function in these plasmids appears to be only partially redundant with that of DnaA. Some Reps have been shown to bind DnaA (37–39), and DnaA boxes are frequently found in plasmid origins of replication (40–42), suggesting some level of cooperation between the plasmid and the chromosome replication machineries. Compared to DnaA, plasmid Rep proteins in IncP plasmids (which are the ones that have been most thoroughly studied) also have important functional differences; for a side-by-side comparison, see Table 2. Both IncP Reps and DnaA cooperatively oligomerize upon binding specific repeats, which creates torsional forces leading to the melting of the DNA duplex nearby and to the formation of an open complex. However, while IncP plasmid Rep monomers bind DNA through winged helix domains, DnaA does so through a different domain (helix-turn-helix [HTH]). The cognate binding sites for Reps are unrelated to DnaA boxes: they are direct repeats of about 20 nt in length known as iterons (43–45). Iterons are intrinsically bent (8, 46), and their spacing matches the helical periodicity of the DNA double helix (47), consistent with the formation of a helical oligonucleoprotein structure, but the existence of such a structure has not been demonstrated for Reps (8). Another important difference is that Rep binding of iterons does not require ATP (48, 49); this is unlike DnaA, which has to be in the ATP-bound state to bind high-affinity sites. Finally, while DnaA is monomeric, IncP Reps can form dimers, a mechanism that is critical for the regulation of plasmid copy numbers (see below).
Table 2.
Comparison between DnaA and theta A Rep-mediated replication initiationa
| Feature | DnaA | Rep | References |
|---|---|---|---|
| Structural domains | HTH domain | Winged helix domain | 143, 144 |
| DNA cognate site | DnaA box | Iterons | 8, 43 |
| Number of nucleotides bound | 3 | 3 | 22, 144, 145 |
| DNA melting | AT-rich DUE | AT-rich DUE | 14, 20, 21, 146 |
| Can bind within single-stranded region of melted DUE | Yes | Yes | 22, 145, 147 |
| ATP binding to replication initiator required for DNA melting | Yes, used to regulate the timing | No, instead timing regulated by transcriptional autoregulation, monomer concentration and activation, and handcuffing | 48, 49, 148 |
| Presence of high- and low-affinity binding motifs in the ori | Yes, role in controlling oligomerization | No | 20, 32 |
| Melting assisted by NAPS; DnaA binding to origin of replication enhanced by the presence of NAPS IHF and HU | Yes | Sometimes | 46, 149–151 |
| Role in recruitment of replisome | Yes | Yes | 26, 28, 49, 103, 136, 144, 152 |
| Role in transcriptional autoregulation | No | Sometimes | 44, 153 |
| Formation of dimers in solution | No | Yesb | 44, 153, 154 |
PriA-Mediated Replication Fork Restart
Primosome (PriA-mediated replisome) assembly is a pathway specializing in replication fork restart (19). PriA is recruited by free 3′ ends representing the nascent leading-strand DNA close to the branch junction, which is the landmark of stalled DNA replication forks (16, 17, 19). Three-way junctions can also be found when a third strand pairs to one of the main strands in the melted double helix, displacing the other complementary main strand. If the third strand is RNA, this structure is called an R-loop; if it is DNA (for example, as a result of strand invasion during recombination or as a result of primer extension), the structure is called a D-loop. A stalled fork can also produce a large single-stranded DNA gap, in which lagging-strand DNA synthesis has proceeded past the nascent leading strand. These large ssDNA gaps appear to be the preferred substrate of a PriA homologue, PriC (50). PriA recognition is mediated by a small binding pocket at the N-terminal domain. This directs the proper positioning of the helicase domain of PriA on the double-stranded DNA (51). Primosome assembly involves two additional proteins (PriB and DnaT) that form a PriA-PriB-DnaT-DNA quaternary complex that loads the DnaB/C complex onto the lagging strand.
The PriA pathway of replication initiation was originally identified in DnaA-deficient mutants and described as stable DNA replication (SDR) (52). SDR can be achieved in at least three different ways: (i) increased R-loop formation (the processing of R-loops by RNaseE or by RNaseH leads to the formation of a free 3′-OH that can be used for replication restart, leading to constitutive SDR [cSDR] [53–55]), (ii) increased recombination, a phenomenon known as inducible SDR (iSDR) (in this case, the generation of a free 3′-OH involves exonuclease processing of the invading strand), and (iii) formation of 3′ flaps after replication fork collision in replication termination (Ter) sites [RecG SDR] [reviewed in references 30 and 53]).
R-loops have become the focus of increased attention due to their role in generating genetic instability in cancer and in neurodegenerative disease as well as their role producing genetic variation during somatic hypermutation (56–58). In prokaryotes, unscheduled R-loop formation can be highly deleterious, causing transcription blocks and inhibiting elongation (59–61). During transcription, the DNA duplex is transiently opened to allow access of the RNA polymerase (RNAP) to the template, and a short 8- to 10-bp-long RNA:DNA hybrid is generated upon initiation of RNA synthesis (62). As the RNAP moves forward, the formation of R-loops is prevented because the size of this RNA loop is maintained by the displacement of the nascent RNA via an RNA exit channel and by the re-annealing of the upstream DNA strands (53). Coupling transcription to translation and factor-dependent transcription termination are additional mechanisms preventing the hybridization of nascent transcripts with DNA in E. coli (62–64). However, increased R-loop formation can be observed in genetic backgrounds favoring negative supercoiling due to decreased topoisomerase I activity or increased gyrase activity, in strains that are deficient in RNaseH, or in strains harboring point mutations in RNAP that favor R-loop formation (reviewed in reference 53). Unscheduled R-loops may also form when the RNA polymerase falls off (a process that can be facilitated by DksA [65]) or as a result of RNaseE-mediated cleavage and facilitated rotation of the transcript (63).
Three classes of theta plasmid replication depend on PriA-dependent replisome assembly: class B, which corresponds to ColE1-like plasmids, class C, which corresponds to ColE2 and ColE3 plasmids, and class D, which corresponds to pAMβ1 and other large, low-copy streptococcal plasmids (reviewed in reference 66). All three classes recruit PriA via R-loop formation, leading to replicon assembly on the lagging strand. Despite assembling a potentially bidirectional replication fork, replication is unidirectional because these replicons have replication blocks on the 5′ end (67, 68).
Class B theta plasmid replication is illustrated in Fig. 2. These origins of replication encode an RNA transcript (RNAII) that acts as the third strand in a melted duplex DNA, generating an R-loop. The formation of this R-loop is guided by a G-rich sequence in the ori DNA that pairs with a C-rich stretch in the transcript. Two hairpin structures further downstream in the preprimer are also important functional elements for R-loop formation, preprimer processing, and extension (reviewed in reference 66). Following its complete transcription, the preprimer RNA in this R-loop is processed by RNaseH to generate a free 3′-OH terminus that is extended by DNA polymerase I (Pol I). RNaseH processing requires the transcript to be in the right conformation and orientation. Pol I extension of the RNA primer further opens the duplex and reveals a hairpin structure in the lagging strand that can serve as a primosome assembly signal (pas). Primosome assembly initiates the coordinated replication of both strands by Pol III, although there is some evidence that Pol I can functionally replace Pol III (69). The mutation footprint produced by error-prone Pol replication also suggests that when the replisome finishes replicating the leading strand, it leaves a gap of approximately 500 nucleotides on the lagging strand that is filled in by Pol I (69). The formation of the R-loop is the only essential step in this process. In the absence of both RNaseH and Pol I, replication initiation is still possible in class B theta plasmids, albeit at reduced efficiency (70).
Figure 2.
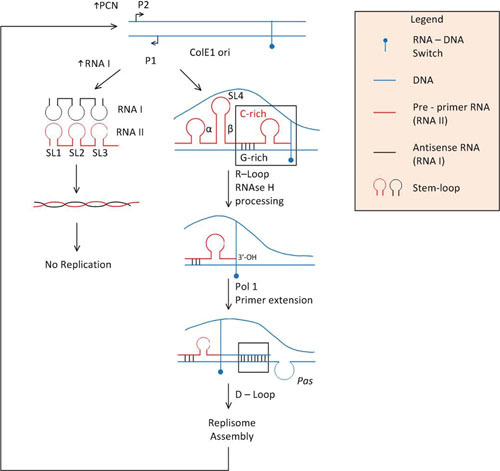
For replication initiation, the ori transcript RNAII acts as the third strand in a melted duplex DNA, generating an R-loop, guided by G-rich sequences in the ori DNA that pair with C-rich sequences in the transcript; other hairpin structures further downstream in the preprimer are also important for R-loop formation, preprimer processing, and extension (reviewed in reference 66). Processing of the R-loop terminus by RNaseH generates a free 3′-OH terminus. RNAII needs to be in the right conformation and orientation for processing by RNaseH. The 3′-OH terminus generated by RNaseH processing is extended by DNA polymerase I (Pol I), further opening the duplex and revealing a hairpin structure in the lagging strand that can serve as a primosome assembly signal (pas). PriA-mediated replisome assembly starts the coordinated replication of both strands by Pol III, although there is some evidence that Pol I can functionally replace Pol III (69). For antisense RNA regulation, as it is being transcribed from promoter P2 in the sense direction, the preprimer (RNAII) forms three symmetrical stem-loop structures (stem-loops 1, 2, and 3 [SL1, SL2, and SL3]). A small and short-lived antisense transcript (RNAI) that is transcribed from a promoter going in the opposite direction also forms these three stem-loops. The RNAII nascent transcript and the antisense RNAI contact each other through the 6- to 7-nt loop portion of their respective stem-loops. This pairing makes the preprimer incompetent for R-loop formation, thus blocking replication initiation (reviewed in references 66, 92, and 93). The half-life of RNAI is short because it contains RNase E recognition sites. Preprimer transcripts larger than 200 nt long are refractory of RNAI-induced inhibition because they form an alternate stem-loop (SL4) by pairing two sequence areas of the transcript (α and β), further reducing the effective half-life of RNAI. This short half-life ensures that RNAI-mediated suppression of replication initiation is reflective of plasmid copy number.
Class C (ColE2 and ColE3) plasmids encode a Rep (RepA) protein that has a dual function as replication initiation factor and as a primase. The primase produces a unique primer: ppApGpA (71). This primer is extended by Pol I, recruiting the replisome through the primosome assembly pathway, as seen in ColE1. Given that, in this case, the primer is produced by a plasmid-encoded Rep, the cis ori sequence required for replication initiation is minimal, only 32 to 33 nt (68).
In class D theta plasmids, replication is initiated by processing of a long transcript, but in this case, the transcript is at the same time the mRNA encoding for a replication initiator factor (RepE in the case of pAMβ1). RepE assists with duplex melting (72), but it does not appear to be involved with the recruitment of the replisome, because (similar to ColE1) replisome recruitment involves extension by Pol I of a transcript, leading to D-loop formation, and uncovering of a cryptic pas site, leading to primosome assembly (73). Two unique features of type D theta plasmid replication are that the Rep (RepE) has an active role in primer generation, either by pausing transcription or by processing the transcript (74) and that Pol I progression is arrested at two sites: ∼190 nt and ∼230 bp downstream of the RNA-DNA switch, respectively. The arrest likely assists with the switch from Pol I to Pol III and appears to be mediated by a plasmid-encoded topoisomerase I-like enzyme (first site) (75) and by a collision with the plasmid-encoded site-specific resolvase, Resβ, bound to its resolution site (second site) (76).
Nontheta Plasmid Replication
Two nontheta types of plasmid replication initiation rely on signals in the plasmid ori sequence forming higher-order secondary structures that are recognized by Reps. The two types are strand displacement and rolling-circle replication. These two alternative types of plasmid replication differ from theta plasmid replication in two additional key aspects: (i) lagging-strand synthesis is decoupled from leading-strand synthesis, and (ii) lagging-strand synthesis is continuous.
In the case of strand displacement, replication initiation is mediated by RepC, which (similar to class A theta plasmids) binds cognate iteron sequences and melts an adjacent DUE. In this case, though, DUE melting is assisted by a plasmid-encoded helicase, RepA, and exposes two initiation signals known as single-strand initiation (ssi) sites. There is one site on each strand. Each ssi forms a hairpin. The base of this hairpin is recognized by a plasmid-encoded primase, RepB, which generates a primer. Note that RepB is the only primase used here; this is unlike theta C plasmids, which have a primase that initiates replication and then switch to DnaG. This primer is extended by Pol III, forming a D-loop (reviewed in references 77 and 78). Strand-displacement plasmids are unique in the number of replication enzymes they encode in addition to replication initiation, which allows these plasmids to function in a wide range of hosts (see “Broader Biological Context: Host Range” below).
In the case of rolling-circle replication (reviewed in reference 79), replication of the two strands is completely asynchronous. Leading-strand replication starts at the double-strand origin (dso), which has a hairpin cruciform structure. The dso is recognized by a plasmid initiation factor (RepC), which nicks this structure, generating a 3′-OH terminus at the nick site. This 3′-OH is extended by Pol III. RepC also catalyzes the final trans-esterification step that joins the 5′ end generated by replication and the 3′ end generated in the cleavage site, sealing the replicated leading strand and producing a single-strand intermediate. The presence of a circular single-strand intermediate is a hallmark of this class of plasmid. Lagging-strand synthesis starts at a single-strand origin (sso) with the generation of a primer by the host primase DnaG. This primer is extended by Pol III, apparently without the need for a helicase because the template is single-stranded.
SEQUENCE-BASED CLASSIFICATION OF PLASMID REPLICONS
Replicons can also be classified according to sequence homology. The first sequence-related classification method grouped replicons based on incompatibility, i.e., on the observation that when two low-copy-number plasmids cannot be distinguished by one or more maintenance systems, the two plasmids are randomly partitioned into daughter cells, leading to the loss of one of them (80). Five compatibility groups were initially defined in E. coli according to conjugation experiments in E. coli (81). These 5 compatibility groups were later expanded to 23.
Detection of sequence homology based on primers for regions that are unique to each plasmid group was later developed; these methods are collectively known as PCR-based replicon typing, or PBRT (82). The target for the diagnostic primers in enterobacteria is listed in Table 3 (column 3). PBRT was consistent with the original conjugation-based scheme and has become the most commonly used technique for plasmid typing in Enterobacteriaceae (28 incompatibility [Inc] groups). More recently, PBRT has been extended to Pseudomonas (14 Inc groups) and Staphylococcus (18 Inc groups). Table 3 summarizes the Inc groups found in Enterobacteriaceae, listing their shared associated biological traits and some specific plasmids belonging to each of these groups. Note that there is a direct correspondence between some Pseudomonas and enterobacterium Inc groups (noted in column 1).
Table 3.
Inc plasmid classification and associated biological traits
| PBRT group (corresponding Pseudomonas PBRT in parenthesis) | Examplesa | Targets for PCR-based replicon typing (82) | Plasmid copy number (10) | Size range (kb) (10) | Host range (5, 10) | HGT (10) | Additional notes |
|---|---|---|---|---|---|---|---|
| IncP (IncP-1) | RK2 (1α)R751 (1β)pJP4pQK54pKJK5RP1RP4R68R18 | Iterons | 5–7 | 70–275 | Broad | Conjugative | Environmental: found in manure, soils, water treatment plantsExtremely stable |
| IncHI1 | R27 | Low | 75–400 | Broad | Conjugative | HI1A/B multireplicon, frequently also IncFIA/B, occasionally also IncX1 | |
| IncHI2 | R478 | Low | 75–400 | Broad | Conjugative | Usually IncF-compatible | |
| IncFIA | F | Iterons | Low | 45–200 | Enterobacteriaceae | Conjugative | Multireplicon plasmid with combinations of FIA/B, FII, and FrepBMost frequent in human (North and South America) and animal (Asia) sources |
| IncFIB | repA | ||||||
| IncFII | R1NR-1R6-5 | repA2 | |||||
| IncFrepB | RNAI/repA | ||||||
| IncN | N3pCU1R46 | repA | Low | 30–70 | Broad(relative) | Conjugative | Animal-associated environmentsOften colocalized with IncF |
| IncA/C (IncP-3) | RA1, pRMH760 | repA | Low | 18–230 | Narrow | Conjugative | |
| IncL | pK01-34R471pEL60 | repA, repB, repC | Low | 60-90 | Broad | Conjugative | Highly conserved, sharing the majority of their genes.Important role in dissemination of β-lactamases |
| IncR | pKP1780 | repB | 40–160 | Broad | Mobilizable | ||
| IncY | P1pMCR-1-P3 | repA | Low | 90–140 | Prophage | ||
| IncQ (IncP-4) | RSF1010IncP-4R1162R300B | Medium (4–12 copies/cell) | 8–14 | Broad | Mobilizable | Lacks partitioning or plasmid stability system | |
| IncW | R388R7KpSapVX2 | repA | Low | <40 | Broad | ConjugativeSmallest conjugative plasmid; can act as helper for ColE and IncQ | |
| ColE | ColE-like ori | Variable (1–20 copies/cell) | 6–40 | Narrow | Mobilizable | Produce colicins, proteins produced by certain strains of E. coli lethal for related strainsAssociated with the spread of qnrS1, qnrB19 |
Partially based on reference 97.
A set of mobility (MOB) genes, which are relaxases involved in conjugation, have been used as an alternative for replicon typing (83). Even though this method excludes nonconjugative plasmids, it produces higher granularity and is consistent with the PBRT method, as each Inc type corresponds to relaxases of a single MOB superfamily. For the remainder of the review, we are going to refer exclusively to the PBRT classification.
REGULATION OF PLASMID REPLICATION
Plasmid replication initiation is subject to tight regulation because plasmid copy number determines the plasmid gene dosage present at any given time in the cell, which in turn has a direct impact on the level of expression of all the genes encoded in the plasmid. Indeed, unlike transcription, which allows for gene- or operon-specific regulation through individual promoters, gene dosage consistently impacts the expression of all the genes encoded in the plasmid (84). Thus, tight control of steady-state plasmid copy number is critical, particularly with plasmids expressing a large number of adaptive genes. The timing of plasmid replication and plasmid segregation should also be critical for optimizing a plasmid’s adaptive value due to their impact on plasmid gene dosage.
Plasmid copy number is mainly regulated through control of replication initiation by mechanisms that are sensitive to the abundance of plasmid copy number relative to cell volume (43, 44, 85). Having multiple origins of replication and/or multiple replicons can provide additional versatility in the control of plasmid copy number (see below).
The regulation of replication initiation in class A theta plasmids is illustrated in Fig. 3. It is based on several factors: (i) Rep proteins are limiting and can be targeted by proteases, (ii) Rep proteins may need to be in an activated monomeric form to bind their target iteron sequences, a process that can be facilitated by chaperones, (iii) Rep proteins often bind iterons in their own promoter, sterically hindering transcription by the RNA polymerase, and (iv) once bound to their target iterons, Rep proteins can mediate the coupling of plasmid origins from different plasmids, blocking replication initiation (a process known as handcuffing).
Figure 3.
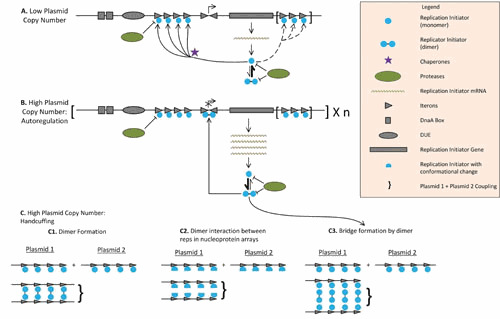
Replication initiator proteins as negative regulators of replication. The rate of replication is determined by the concentration of free (non-Rep-bound) iterons, which is determined by both plasmid copy number and cell volume. Given the cooperative interactions involved in saturating iterons, this regulatory mechanism is ultrasensitive to plasmid copy number and switch-like. Generic class A theta replicon structure: rep gene (rectangle), DnaA boxes, which can be found upstream or downstream of the Rep (squares), iterons (arrowheads), and an AT-rich DNA unwinding element (DUE, oval). (A) Replication initiator proteins (Reps) when the plasmid copy number is low. Reps (blue circles) are expressed from strong promoters and tend to be in their monomeric (active) form; in some cases, the activation of these monomers needs to be facilitated by chaperones (purple stars). Reps are also sensitive to protease activity (green ovals). Active monomers bind iterons until saturation; in some cases, additional clusters of iterons are present to decrease the level of Rep available, further tightening plasmid copy number control (86). (B) Replication initiator proteins when plasmid copy number is high. High Rep protein expression resulting from a high plasmid copy number favors the dimeric form of Rep. The symmetrical conformation of the dimeric form matches inverted iterons found in the promoter of rep genes. This results in a binding affinity for the promoter that is higher than that of the RNA polymerase, blocking rep transcription (137). Transcriptional autorepression by Reps is seen in IncFIA, IncN, and IncP plasmids and is also consistent with the structure of replicons from other groups, such as IncHI1 and IncY, although in the case of IncN and IncY, the iterons that overlap with the Rep promoter are not inverted (43, 138). Rep dimers can also bridge Rep-bound iteron arrays, one of the proposed mechanisms of handcuffing (see panel C3). (C) Different handcuffing mechanisms. Once iteron-bound Rep arrays form, they can couple two different plasmids, a reaction in trans known as plasmid handcuffing that blocks ori melting by steric hindrance (139). Three mechanisms that pair different plasmids through iteron-bound replication initiation proteins have been proposed (43). From left to right: direct dimerization of the iteron-bound initiators (proposed for the plasmid RK6 [140]), direct interaction between arrays of iteron-bound monomers, associated with a Rep conformational change induced by iteron binding (proposed for the plasmid pPS10 [141]), and bridging via dimer formation (proposed for RK2 [139]). Chaperones counteract handcuffing by facilitating the dissociation of dimers to monomers or by increasing monomer-to-dimer ratios (139, 142).
A variation on Rep autoregulation as a mechanism for control of plasmid copy number has been described for IncP plasmid RK2. The promoter for its rep protein (TrfA) is controlled by two plasmid-encoded regulators, KorA and KorB, which bind to sites that overlap with the trfA promoter sequence. Cooperatively, they suppress TrfA expression about 1,000-fold (86). So, in this case, instead of the rep itself, regulation is mediated by two other plasmid-encoded proteins.
Dimerization, illustrated in Fig. 3B, has been best studied in IncP (R2K, R6K, PS10) plasmids. The increased synthesis of Rep associated with high plasmid copy number shifts the equilibrium between the monomer and dimer Rep form toward the dimeric form, which is not competent for iteron binding, thus suppressing replication initiation (43). Chaperones such as ClpB or Dnak/DnaJ/GroE control the conversion from dimer to monomer and also the activation of monomers (44). Proteases such as ClpAP control the level of active monomers as well (87). In RK2, the activity of specific proteases against TrfA is modulated by binding to iterons, and the effects can go in either direction; i.e., iteron binding can increase or decrease initiator sensitivity to proteases depending on the specific protease (88).
High plasmid copy number can also lead to handcuffing, i.e., to the formation of bridges coupling plasmid origins from different plasmids via iteron-bound Reps. This bridge formation blocks replication initiation. In this case, replication rate is more directly linked to iteron abundance than to levels of Rep expression. Three plasmid-coupling mechanisms have been proposed and are illustrated in Fig. 3C. These are: dimerization of iteron-bound initiators (RK6), direct interaction between arrays of iteron-bound monomers (pPS10), and dimer formation bridging iteron-bound monomers (RK2) (43).
Another group of plasmids use RNAI (short and highly structured antisense transcripts) for regulation. The pairing of these antisense RNAs allows the precise targeting of specific areas of the transcript to block translation or to induce conformational changes in the transcript. These antisense transcripts have a short half-life, which is critical to enable tight regulation of replication initiation, due to the presence of RNaseE cleavage sites (89). Antisense plasmid copy number appears to be an example of parallel evolution because it is observed in three groups of plasmids with different replication mechanisms and because in each case the suppression of replication initiation occurs by different mechanisms as well. We explain them in greater detail below.
Several class A theta plasmids, including I-complex (IncI, IncK, IncBO) and IncL plasmids, have a Rep whose start codon overlaps with the stop codon of a short leader peptide immediately upstream. Translation and correct termination of the leader peptide facilitate the formation of a pseudoknot between two short (8-nt) complementary sequences of leader sequence that is necessary for Rep translation. RNAI blocks the translation of the leader peptide by sterically hindering ribosomes attempting to initiate translation of the leader peptide, therefore suppressing the translation of the Rep as well (90, 91).
In class B (ColE1-like) theta plasmids, a short-lived antisense RNAI and its complementary RNAII preprimer target sequence form three symmetrical stem-loop structures that contact each other through the 6- to 7-nt loop portion of their respective stem-loops (Fig. 2). The 5′ end of RNAI (known as antitail) nucleates the hybridization between the two RNAs to form an RNA duplex. This pairing induces conformational changes in the preprimer that make the preprimer incompetent for R-loop formation, thus blocking replication initiation (reviewed in references 66, 92, and 93). Preprimer transcripts larger than 200 nt are refractory of RNAI-induced inhibition, further reducing the effective half-life of RNAI.
In class C (ColE2) theta plasmids, RNAI targets the 5′ untranslated region of the rep transcript, possibly inducing conformational changes in the preprimer that block its translation (94).
BROADER BIOLOGICAL CONTEXT: HOST RANGE
The vast majority of plasmids found in Enterobacteriaceae are conjugative or at least mobilizable (Table 3, column 7). This means that the ability of a plasmid to replicate in a range of phylogenetically distant organisms (exhibiting broad host range, or BHR) can be beneficial in the long term because it allows a wider access to the available gene pool.
Factors conferring BHR properties have thus been the object of increased scrutiny (1, 95, 96). It turns out that plasmids have to meet a surprisingly large number of requirements to be able to replicate in different hosts. These requirements include the compatibility between plasmids and a variety of host replication factors, as well as the compatibility of plasmid replication initiation and transcriptional regulatory circuitry across hosts (84, 97).
BHR appears to be clearly advantageous under some conditions, as some plasmid replication strategies appear to have evolved to maximize host range. These include the evolution of DnaA-independent replication-initiation factors (some Reps, RNA primers, and nicks) and of plasmid-encoded replication factors (primases, helicases, topoisomerases, polymerases, resolvases), which decreases the dependency of these plasmids on adequate interaction with host-specific factors. The acquisition/evolution of plasmid-encoded replication factors is clearly the strategy adopted by IncQ plasmids, which carry their own primase and helicase (77, 78) and have an extraordinarily broad host range that includes Alpha-, Beta-, Gamma-, and Deltaproteobacteria and Cyanobacteria.
Another strategy is versatility, i.e., the availability of redundant mechanisms of replication that allow functionality in different hosts. This includes the presence of multiple origins of replication and of multiple replication initiators and decreases the plasmid’s dependence on host factor compatibility. IncN plasmids, for example, have three origins of replication, oriB, oriS, and oriV. oriB functions as the main origin of replication because it has the iterons that bind RepA; however, in the absence of these iterons, Pol I can still initiate replication at oriV (98). In the absence of oriV, oriS can still start replication (99). Another example is the IncP compatibility group R6K plasmid, which also has three origins of replication: α, β, and γ. γ appears to be the postmobilization ori (which requires rapid amplification), while α and β primarily support vegetative replication, with a lower replication rate (100). An example of redundancy in replication initiation factors is TrfA (the Rep protein in IncP plasmids), which is expressed in two different versions resulting from the alternate translation of a single transcript using separate in-frame translation start sites. In E. coli, both forms of TrfA can initiate plasmid replication, whereas in Pseudomonas, only the longer form is active because it is not dependent on DnaA (101, 102). More recently, mutants adapted to a nonnative species were shown to accumulate mutations at the N terminus of the long form of TrfA, again pointing to this form of TrfA as critical for extending host range (103).
On the other hand, the complex mechanisms of regulation of plasmid maintenance and the strong genetic evidence of plasmid adaptation to its host point to strong selective pressures restricting plasmid host range. The latter include analysis of G+C content and codon usage and even evidence of host-specific evolution of single-strand origin motifs (1, 95, 104). Thus, it seems that adaptation of plasmids to their host is generally necessary to optimize their contribution to the host’s fitness but that this adaptation restricts access to broad genetic diversity.
Figure 4 shows the distribution of plasmid replicons in 233 random ExPEC samples collected at the University of Washington hospital between 2009 and 2012 (105). We found that the three most abundant replicons (IncF, IncI-complex, and ColE) correspond to narrow-host-range (NHR) plasmids, with observed frequencies of 66%, 16%, and 5%, respectively. BHR plasmids (IncN, IncP, IncL/M, and IncQ), by contrast, were infrequent: 3%, 3%, 0.5%, and <0.5%, respectively. This snapshot is consistent with previous studies reporting IncF plasmids as one of the most prevalent incompatibility types (10, 106) and IncQ as “rarely detected” in Enterobacteriaceae (10). The observed distribution of NHR and BHR plasmids in ExPEC supports the proposed tradeoff between adaptive value for the host and access to genetic diversity. This hypothesis predicts a strong representation of NHR plasmids because they are optimally suited to their hosts and therefore tend to have a stronger positive effect on fitness. This model also predicts the retention of BHR plasmids at low frequency, driven by the benefit of broadening genetic diversity available to the population as a whole.
Figure 4.
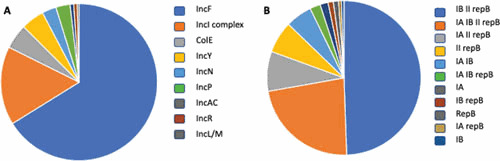
Replicon representation in ExPEC whole genomic shotgun sequences reported by the University of Washington (105). Only 233 of the reported sequences were included in this analysis because of strict quality control standards. (A) Pie chart representation of the replicons identified. The replicons are ordered clockwise from most to least abundant: IncF (n = 180), IncI complex (n = 44), ColE (n = 14), IncY (n = 13), IncN (n = 8), IncP (n = 8), IncAC (n = 2), IncR (n = 2), and incL/M (n = 1). IncF denotes the presence of at least one of the following replicons: IncFIA, IncFIB, IncFII, or IncFrepB. The Inc-I complex includes IncI1 (n = 17), IncK (n = 17), and IncBO (n = 10). Note that the number of replicons (n = 272) is greater than the number of samples included (n = 233) because, often, multiple replicons can be found in the same sample. (B) Representation of IncF replicons, which are largely mosaic combinations of IncFIA, IncFIB, IncFII, or IncFrepB. Again, these replicons are ordered clockwise from most to least abundant: IB II repB (49.4%), IA IB II repB (22.8%), IA II repB (8.3%), II repB (6.7%), IA IB (5.6%), IA IB repB (2.2%), IA (1.7%), IB repB (1.1%), repB (1.1%), IA repB (0.6%), and IB (0.6%). Note that these combinations may be found in different plasmids; we are only showing combinations present in the same cell. Note also that we are observing a rapid sequence divergence for IncFII replicons (not shown). Therefore, frequently, IncFII may not have been an exact match for our diagnostic sequence, producing false negatives.
Figure 4B shows the distribution of IncF replicons in samples that have at least one IncF replicon. Of all IncF samples, only 4% have a single replicon. The replicon mosaicism of IncF plasmids is well established (3, 107). The factors driving this mosaicism are not clear. Part of it appears to be specialization reminiscent of that of R6K, with IncFII likely involved in establishment following conjugation and IncFIA as the primary replicon supporting vegetative growth (reviewed in reference 3). The frequent linkage between IncFIA and IncFrepB may be driven by expanding the compatibility range to allow different IncF plasmids to stably coexist in host cells, as the presence of two compatible replicons overrides the incompatibility between individual replicons (107, 108). As the fraction of genetic diversity carried by one type of NHR plasmid (IncF plasmids in this case) increases, there should be increasing selective pressure for the stable acquisition of multiple variants of these plasmids in the cell. In this scenario, the availability of expanded compatibility should provide mosaic replicon plasmids such as IncF or IncHI1 with a selective advantage.
Figure 5 shows the distribution of replicons per sample, which for host cells with at least one replicon follows roughly a normal distribution centered around an average of 3.28 per sample. This high average is driven by the abundance of mosaic IncF plasmids; if we lump all IncF replicons in one category, the average goes down to 1.2, highlighting both the strong representation of IncF plasmids in our strains (Fig. 4A) and the mosaic nature of these replicons (Fig. 4B).
Figure 5.
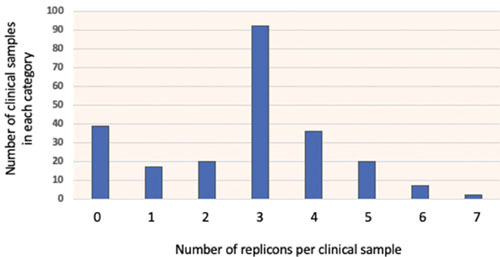
The distribution of replicon number per clinical sample is shown as a bar graph. The average number of replicons for samples that have at least one plasmid is 3.26. If all IncF replicons are considered as a single category, the average number goes down to 1.2.
BROADER BIOLOGICAL CONTEXT: PLASMID MAINTENANCE
Given that plasmid replication initiation determines the plasmid gene dosage present at any given time in the cell, which in turn has a direct impact on the level of expression of all the genes in the plasmid, controlling plasmid copy number is critical for optimizing a plasmid’s adaptive value for the host. Controlling the timing of replication initiation and plasmid segregation should be important as well, as these processes also have an impact on gene dosage. Therefore, regulation of plasmid replication needs to be understood in the broader context of plasmid maintenance, including control of plasmid segregation through partition (par) systems and post-segregational counterselection through toxin-antitoxin (TA) plasmid addiction systems.
The presence of multiple versions of these functional elements adds further complexity. This includes alternate origins of replication within a single replicon as in the cases of IncP and IncN plasmids discussed above (96, 99), mosaic replicons such as IncF and IncHI1 (3, 109), and examples of cointegrated replicons (110–112) and of multiple par (113, 114) and/or TA systems (115, 116) coexisting in the same plasmid. We believe that all these systems are tightly interconnected, maintaining the delicate balance between optimizing the adaptive value of the plasmids and access to genetic diversity.
TA systems counterselect plasmid-free cells through post-segregational killing or growth inhibition of plasmid-free cells. According to their mechanism of action and regulation, TA systems can be classified into six types (types I through VI). They typically consist of two elements: a stable protein that targets essential cellular processes (the toxin) and short-lived protein or an untranslated antisense RNA species inhibiting the toxin activity (the antitoxin). Antitoxins are more labile than toxins because they are degraded more efficiently by host enzymes. Loss of the antitoxin when the plasmid is no longer present allows the toxin to reach or act on its target, causing death or growth restriction in the cell (for comprehensive reviews, see references 117 and 118). More recently, additional roles for TA systems have been recognized, including adaptation to intracellular environments, stress response (induction of persistent state and of programmed cell death), and postrecombinational killing (117, 119).
Most TA systems found in plasmids correspond to types II and III, with type II being the most abundant in virulence plasmids (5). Type II TA systems have been categorized into eight superfamilies: RelBE, MazEF, VapBC, CcdAB, ParDE, HigAB, HipBA, and Phd-Doc (120). In these TA systems, both the toxin and the antitoxin are proteins. The toxin usually consists of two distinct domains: a DNA-binding domain at the amino-terminus and a toxin-interacting domain at the carboxy-terminus. The antitoxin binds the toxin, forming a protein complex that results in toxin sequestration and inactivation.
Partition (par) systems are found in replicons of above 25 kb and ensure that all daughter cells receive at least one plasmid following segregation. Par systems may also be needed to counteract plasmid eviction toward nucleoid edges (121). These systems have three components: a motor protein, a centromere-like specific DNA sequence, and a DNA-binding protein that oligomerizes and serves as an adaptor to connect the motor with the centromere (reviewed in reference 122). The two main types found in plasmids differ in the mode of action of their motor proteins. Type I par systems, such as parABS and sopAB, have Walker-type ATPases as motor proteins that form dynamic gradients in the cell that pull plasmids along. In contrast, type II systems, such as parMRC and stbAB, have actin-like motor proteins that push plasmids apart as they oligomerize. Most plasmids of above 180 kb carry ParAB systems, so type I par systems appear to be better suited for large plasmids (121).
CLUSTERING OF PLASMID MAINTENANCE ELEMENTS
Functionally related elements tend to cluster together as a way to facilitate their stable coregulation/cross-talk (2, 123, 124). This is particularly important in plasmids, which undergo a very high rate of recombination and could then lose elements that are needed (125). With this in mind, we looked for protein functional domains (using the PFAM database [126]) that are consistently found within a 10-kb window centered on the PCR amplicon used for PBRT classification in 761 fully assembled E. coli genomic sequences available in NCBI as of April 2019. We defined “consistently” as seen in >75% of the isolates for one or more of the Inc groups. The results are shown in Fig. 6, which shows protein family (PFAM) representation compiled using color-coding, with darker colors representing a higher prevalence of protein families within each Inc group. Inc groups were arranged by similarity using a hierarchical clustering algorithm and were grouped into five functional categories: replication initiation, plasmid maintenance, environmental stress protection, conjugation/mobilization, and other factors.
Figure 6.
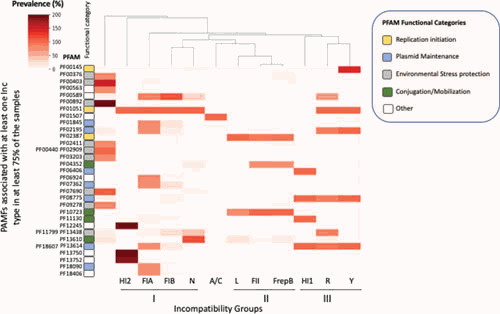
PFAM representation in the vicinity of plasmid replicons. Completely assembled E. coli plasmid sequences obtained from NCBI as of April 2019 were grouped by Inc type based on the diagnostic PCR amplicons for the PBRT system (82). Within each incompatibility group, we looked for PFAM matches within a 10-kb window centered around the diagnostic PCR amplicons. PFAM protein families present in at least 75% of the samples in at least one Inc group were identified and mapped to individual open reading frames. These PFAMs were also identified in other Inc groups. These PFAM representation data were compiled into a matrix and plotted using color-coding, with darker colors representing a higher prevalence of protein families within each Inc group. Inc groups are arranged by similarity using a hierarchical clustering algorithm for PFAMs. Note that the percentage can be higher than 100% in the case of duplications or if two domains are found in the same ORF. PFAMs are listed in the y axis, grouped in the following color-coded functional categories. Replication initiation-related PFAMs (orange): m5C methylase (PF00145), initiator replication protein (PF01051), and IncFII RepA protein family (PF02387). Plasmid maintenance PFAMs (light blue): PemK endoribonuclease toxin (PF01845) ParB-like nuclease domain (PF02195), StnA protein (PF06406), post-segregation TA CcdA (PF07362), ParB family (PF08775), ParA AAA+ and HT domains (PF13614 PF18607), centromere-binding protein HTH domain (PF18090). Environmental stress protection PFAMs (light gray): MerR family regulatory protein (PF00376), heavy metal-associated domain (PF00403), EamA-like transporter family (PF00892), mercuric transport protein (PF02411), tetracycline repressor family (PF02909 PF00440), MerC mercury resistance protein (PF03203), major facilitator superfamily (PF07690), MerR (PF09278), impB/mucB/samB Y family polymerase (PF13438, PF11799). Conjugation/mobilization PFAMs (green): phage integrase family (PF00589), ProQ/FINO family (PF04352), replication regulatory protein repB (PF10723), TraC_F_IV F pilus assembly (PF11130), DDE domain found in transposases (PF13610). Other PFAMs (white): EAL domain (PF00563), phosphoadenosine phosphosulfate reductase family (PF01507), DUF1281 (PF06924), bacterial IgE domain (PF12245), bacterial IgE-like domain (group3) (PF13750), DUF4165 (PF13752), ferredoxin-like domain in Api92-like protein (PF18406).
IncFIA replicons stand out for the presence of plasmid maintenance systems associated with the ori. We find a consistent association between the IncFIA ori and genes encoding ParB, ParBc, and another segregation-related PFAM (centromere-binding protein HTH domain). The association of IncF with plasmid par genes is well established (3, 127). IncFIA is also consistently associated with the CcdA/CcdB TA (type II) system. F-like plasmids have been reported to carry at least one representative of the various subfamilies of type II TA systems (3). The location of ccdAB genes adjacent to the origin of replication of the F plasmids has been previously reported and thought to enhance IncFIA replicon stability by coupling plasmid replication to host cell division (128).
Notably, our clustering of consistently represented PFAM groups in Fig. 6 identified three clusters that include Inc replicons that share functional commonalities, highlighting the functional connection between replicons and TA and segregation systems that are found in close physical proximity.
Cluster I includes IncFIA, IncFIB, and IncN. These three replicons share a homologous Rep, although their regulatory mechanisms show considerable divergence (95). They also share the presence of a phage integrase in the vicinity of ori. Y-family polymerase UmuDC is close to the ori at some frequency in these three incompatibility types (as well as in IncP and IncHI1). This polymerase may provide increased genetic diversity (129) and/or protection from DNA damage (130, 131), although how its association with ori is relevant to its functionality is not known.
Cluster II comprises IncL, IncFII, and IncFrepB replicons. These replicons are all regulated by RNAI suppression of leader peptide translation, with the presence of a pseudoknot (85, 90, 91). IncFII, IncFrepB, and IncL share rolling-circle replication regulatory protein repB, which is likely associated with conjugation.
Cluster III includes IncHI1A, IncR, and incY, which share a homologous Rep (95). ParAB (type I partition system) is consistently associated with the ori in these three incompatibility groups. In addition, IncR plasmids have ParBc close to their plasmid ori, and IncHI1A plasmids have a type II par gene. ParB-type proteins have recently been recognized as a new class of CTP hydrolases that act as molecular switches. During plasmid segregation, these proteins work in concert with ParA and the prokaryotic SMC/condensin complex, mediating centromere condensation into kinetochore-like structures (132, 133).
In contrast, IncP, IncHI2, and IncA/C have unique PFAM ori profiles. IncA/C stands out for the low density of PFAMs consistently found in the vicinity of the ori. IncP is very enriched for genes involved in protection from environmental stress (8 of the 9 consistent ori-proximal PFAMs). This is consistent with the association of this plasmid compatibility group with bacteria isolated from manure, soils, and water treatment plants (134, 135).
CONCLUDING REMARKS
The plasmid gene content, level and timing of gene expression, and DNA sequence (G+C content, codon usage, etc.) need to be adjusted to the host for a given plasmid to have maximal adaptive impact. This selective pressure favoring adaptation tends to narrow plasmid host range and conflicts with the need for access to a broader pool of genetic diversity. Here, we argue that the trade-off between these two selective forces likely shapes the distribution of replicons in populations of enterobacteria based on two considerations: (i) a predominance of NHR plasmids in the population of ExPEC, supporting the adaptive value of these plasmids, coupled with a low but consistent prevalence of BHR plasmids, suggesting that expanded access to genetic diversity is important, and (ii) the success of NHR mosaic replicon plasmids such as IncF or IncHI1, which maximizes adaptive value while moderately increasing access to genetic diversity by making these plasmids compatible with other plasmids of the same Inc group.
We also argue that, given its critical importance for modulation of plasmid gene expression, the regulation of plasmid replication needs to be understood broadly. This includes the presence of redundant origins of replication, of mosaic and cointegrated replicons, of plasmid segregation systems, and of TA systems. The reason is that all these elements control not only plasmid copy number at steady state but also variations in plasmid copy number relative to the host’s cell cycle, the presence of additional plasmids, and the physiological state of the host cell. Highlighting their tight functional integration with ori function, all these elements are frequently found in close proximity to the plasmid ori.
ACKNOWLEDGEMENTS
We acknowledge Steve Salipante’s help accessing and processing the data from his study, Giselle Migglioranza Ricci’s help with the initial literature search, and Marc Drolet (Université de Montréal) and Igor Konieczny (University of Gdansk) for critical reading of the manuscript ahead of publication.
This work was funded in part by CITRIS Seed Funding proposal 2015-324 and by NIAID award 1R41AI122740.
Contributor Information
Jay W. Kim, Department of Microbiology and Environmental Toxicology, University of California at Santa Cruz, Santa Cruz, CA, 95064
Vega Bugata, Department of Microbiology and Environmental Toxicology, University of California at Santa Cruz, Santa Cruz, CA, 95064.
Gerardo Cortés-Cortés, Department of Microbiology and Environmental Toxicology, University of California at Santa Cruz, Santa Cruz, CA, 95064.
Giselle Quevedo-Martínez, Department of Microbiology and Environmental Toxicology, University of California at Santa Cruz, Santa Cruz, CA, 95064.
Manel Camps, Department of Microbiology and Environmental Toxicology, University of California at Santa Cruz, Santa Cruz, CA, 95064.
James M. Slauch, The School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL
Gregory Phillips, College of Veterinary Medicine, Iowa State University, Ames, IA.
REFERENCES
- 1.Shintani M, Suzuki H. 2019. Plasmids and their hosts, p 109–132. In Nishida H, Oshima T (ed), DNA Traffic in the Environment. Springer, Singapore. 10.1007/978-981-13-3411-5_6 [DOI] [Google Scholar]
- 2.Thomas CM. 2000. Paradigms of plasmid organization. Mol Microbiol 37:485–491 10.1046/j.1365-2958.2000.02006.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Koraimann G. 2018. Spread and persistence of virulence and antibiotic resistance genes: a ride on the F plasmid conjugation module. Ecosal Plus 2-18 10.1128/ecosalplus.ESP-0003-2018. 10.1128/ecosalplus.ESP-0003-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitout JD. 2013. Enterobacteriaceae that produce extended-spectrum β-lactamases and AmpC β-lactamases in the community: the tip of the iceberg? Curr Pharm Des 19:257–263 10.2174/138161213804070348. [DOI] [PubMed] [Google Scholar]
- 5.Pilla G, Tang CM. 2018. Going around in circles: virulence plasmids in enteric pathogens. Nat Rev Microbiol 16:484–495 10.1038/s41579-018-0031-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Norris V, Merieau A. 2013. Plasmids as scribbling pads for operon formation and propagation. Res Microbiol 164:779–787 10.1016/j.resmic.2013.04.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Norman A, Hansen LH, Sørensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci 364:2275–2289 10.1098/rstb.2009.0037. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wegrzyn KE, Gross M, Uciechowska U, Konieczny I. 2016. Replisome assembly at bacterial chromosomes and iteron plasmids. Front Mol Biosci 3:39 10.3389/fmolb.2016.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. 2015. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 39:81–95. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137 10.1093/jac/dkx488. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Colavecchio A, Cadieux B, Lo A, Goodridge LD. 2017. Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family: a review. Front Microbiol 8:1108 10.3389/fmicb.2017.01108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–650 10.1038/nrmicro3527. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Lewis JS, Jergic S, Dixon NE. 2016. The E. coli DNA replication fork. Enzymes 39:31–88 10.1016/bs.enz.2016.04.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Leonard AC, Grimwade JE. 2015. The orisome: structure and function. Front Microbiol 6:545 10.3389/fmicb.2015.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakley AJ. 2019. A structural view of bacterial DNA replication. Protein Sci 28:990–1004 10.1002/pro.3615. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabbai CB, Marians KJ. 2010. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst) 9:202–209 10.1016/j.dnarep.2009.12.009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masai H, Tanaka T, Kohda D. 2010. Stalled replication forks: making ends meet for recognition and stabilization. BioEssays 32:687–697 10.1002/bies.200900196. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Michel B, Sinha AK, Leach DRF. 2018. Replication fork breakage and restart in Escherichia coli. Microbiol Mol Biol Rev 82:82 10.1128/MMBR.00013-18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Windgassen TA, Wessel SR, Bhattacharyya B, Keck JL. 2018. Mechanisms of bacterial DNA replication restart. Nucleic Acids Res 46:504–519 10.1093/nar/gkx1203. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. 2004. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci USA 101:2811–2816 10.1073/pnas.0400340101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leonard AC, Grimwade JE. 2011. Regulation of DnaA assembly and activity: taking directions from the genome. Annu Rev Microbiol 65:19–35 10.1146/annurev-micro-090110-102934. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duderstadt KE, Chuang K, Berger JM. 2011. DNA stretching by bacterial initiators promotes replication origin opening. Nature 478:209–213 10.1038/nature10455. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. 2008. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem 283:8351–8362 10.1074/jbc.M708684200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. 2010. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem 285:28229–28239 10.1074/jbc.M110.147975. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson TT, Harran O, Murray H. 2016. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534:412–416 10.1038/nature17962. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soultanas P. 2012. Loading mechanisms of ring helicases at replication origins. Mol Microbiol 84:6–16 10.1111/j.1365-2958.2012.08012.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mott ML, Erzberger JP, Coons MM, Berger JM. 2008. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135:623–634 10.1016/j.cell.2008.09.058. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weigel C, Seitz H. 2002. Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol Microbiol 46:1149–1156 10.1046/j.1365-2958.2002.03232.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Bastia D, Zaman S. 2014. Mechanism and physiological significance of programmed replication termination. Semin Cell Dev Biol 30:165–173 10.1016/j.semcdb.2014.04.030. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gowrishankar J. 2015. End of the beginning: elongation and termination features of alternative modes of chromosomal replication initiation in bacteria. PLoS Genet 11:e1004909 10.1371/journal.pgen.1004909. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berghuis BA, Raducanu VS, Elshenawy MM, Jergic S, Depken M, Dixon NE, Hamdan SM, Dekker NH. 2018. What is all this fuss about Tus? Comparison of recent findings from biophysical and biochemical experiments. Crit Rev Biochem Mol Biol 53:49–63 10.1080/10409238.2017.1394264. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Kawakami H, Keyamura K, Katayama T. 2005. Formation of an ATP-DnaA-specific initiation complex requires DnaA arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem 280:27420–27430 10.1074/jbc.M502764200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Kim JS, Nanfara MT, Chodavarapu S, Jin KS, Babu VMP, Ghazy MA, Chung S, Kaguni JM, Sutton MD, Cho Y. 2017. Dynamic assembly of Hda and the sliding clamp in the regulation of replication licensing. Nucleic Acids Res 45:3888–3905 10.1093/nar/gkx081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su’etsugu M, Harada Y, Keyamura K, Matsunaga C, Kasho K, Abe Y, Ueda T, Katayama T. 2013. The DnaA N-terminal domain interacts with Hda to facilitate replicase clamp-mediated inactivation of DnaA. Environ Microbiol 15:3183–3195 10.1111/1462-2920.12147. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Skarstad K, Katayama T. 2013. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol 5:a012922 10.1101/cshperspect.a012922. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama T, Kasho K, Kawakami H. 2017. The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front Microbiol 8:2496 10.3389/fmicb.2017.02496. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abeles AL, Reaves LD, Austin SJ. 1990. A single DnaA box is sufficient for initiation from the P1 plasmid origin. J Bacteriol 172:4386–4391 10.1128/JB.172.8.4386-4391.1990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawasaki Y, Matsunaga F, Kano Y, Yura T, Wada C. 1996. The localized melting of mini-F origin by the combined action of the mini-F initiator protein (RepE) and HU and DnaA of Escherichia coli. Mol Gen Genet 253:42–49 10.1007/s004380050294. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Caspi R, Helinski DR, Pacek M, Konieczny I. 2000. Interactions of DnaA proteins from distantly related bacteria with the replication origin of the broad host range plasmid RK2. J Biol Chem 275:18454–18461 10.1074/jbc.M000552200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Rajewska M, Wegrzyn K, Konieczny I. 2012. AT-rich region and repeated sequences: the essential elements of replication origins of bacterial replicons. FEMS Microbiol Rev 36:408–434 10.1111/j.1574-6976.2011.00300.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Park K, Chattoraj DK. 2001. DnaA boxes in the P1 plasmid origin: the effect of their position on the directionality of replication and plasmid copy number. J Mol Biol 310:69–81 10.1006/jmbi.2001.4741. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Doran KS, Helinski DR, Konieczny I. 1999. Host-dependent requirement for specific DnaA boxes for plasmid RK2 replication. Mol Microbiol 33:490–498 10.1046/j.1365-2958.1999.01491.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Konieczny I, Bury K, Wawrzycka A, Wegrzyn K. 2014. Iteron plasmids. Microbiol Spectr 2:PLAS-0026-2014 10.1128/microbiolspec.PLAS-0026-2014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Chattoraj DK. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol 37:467–476 10.1046/j.1365-2958.2000.01986.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Nordström K. 1990. Control of plasmid replication: how do DNA iterons set the replication frequency? Cell 63:1121–1124 10.1016/0092-8674(90)90405-4. [DOI] [PubMed] [Google Scholar]
- 46.Fekete RA, Venkova-Canova T, Park K, Chattoraj DK. 2006. IHF-dependent activation of P1 plasmid origin by DnaA. Mol Microbiol 62:1739–1751 10.1111/j.1365-2958.2006.05479.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Bowers LM, Krüger R, Filutowicz M. 2007. Mechanism of origin activation by monomers of R6K-encoded pi protein. J Mol Biol 368:928–938 10.1016/j.jmb.2007.02.074. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Konieczny I, Doran KS, Helinski DR, Blasina A. 1997. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J Biol Chem 272:20173–20178 10.1074/jbc.272.32.20173. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Lu YB, Datta HJ, Bastia D. 1998. Mechanistic studies of initiator-initiator interaction and replication initiation. EMBO J 17:5192–5200 10.1093/emboj/17.17.5192. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wessel SR, Marceau AH, Massoni SC, Zhou R, Ha T, Sandler SJ, Keck JL. 2013. PriC-mediated DNA replication restart requires PriC complex formation with the single-stranded DNA-binding protein. J Biol Chem 288:17569–17578 10.1074/jbc.M113.478156. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Windgassen TA, Leroux M, Satyshur KA, Sandler SJ, Keck JL. 2018. Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase. Proc Natl Acad Sci USA 115:E9075–E9084 10.1073/pnas.1809842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kogoma T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61:212–238 10.1128/.61.2.212-238.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drolet M, Brochu J. 2019. R-loop-dependent replication and genomic instability in bacteria. DNA Repair (Amst) 84:102693 10.1016/j.dnarep.2019.102693. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Usongo V, Martel M, Balleydier A, Drolet M. 2016. Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair (Amst) 40:1–17 10.1016/j.dnarep.2016.02.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Usongo V, Drolet M. 2014. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10:e1004543 10.1371/journal.pgen.1004543. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groh M, Gromak N. 2014. Out of balance: r-loops in human disease. PLoS Genet 10:e1004630 10.1371/journal.pgen.1004630. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hegazy YA, Fernando CM, Tran EJ. 2020. The balancing act of R-loop biology: the good, the bad, and the ugly. J Biol Chem 295:905–913 10.1074/jbc.REV119.011353. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiedemann EM, Peycheva M, Pavri R. 2016. DNA replication origins in immunoglobulin switch regions regulate class switch recombination in an R-loop-dependent manner. Cell Rep 17:2927–2942 10.1016/j.celrep.2016.11.041. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.Baaklini I, Usongo V, Nolent F, Sanscartier P, Hraiky C, Drlica K, Drolet M. 2008. Hypernegative supercoiling inhibits growth by causing RNA degradation. J Bacteriol 190:7346–7356 10.1128/JB.00680-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drolet M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59:723–730 10.1111/j.1365-2958.2005.05006.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 61.Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Massé E, Baaklini I. 2003. The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci 8:d210–d221 10.2741/970. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Ray-Soni A, Bellecourt MJ, Landick R. 2016. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem 85:319–347 10.1146/annurev-biochem-060815-014844. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Gowrishankar J, Leela JK, Anupama K. 2013. R-loops in bacterial transcription: their causes and consequences. Transcription 4:153–157 10.4161/trns.25101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts JW. 2019. Mechanisms of bacterial transcription termination. J Mol Biol 431:4030–4039 10.1016/j.jmb.2019.04.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Myka KK, Küsters K, Washburn R, Gottesman ME. 2019. DksA-RNA polymerase interactions support new origin formation and DNA repair in Escherichia coli. Mol Microbiol 111:1382–1397 10.1111/mmi.14227. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lilly J, Camps M. 2015. Mechanisms of theta plasmid replication. Microbiol Spectr 3:PLAS-0029-2014. [DOI] [PubMed] [Google Scholar]
- 67.Nakasu S, Tomizawa J. 1992. Structure of the ColE1 DNA molecule before segregation to daughter molecules. Proc Natl Acad Sci USA 89:10139–10143 10.1073/pnas.89.21.10139. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takechi S, Itoh T. 1995. Initiation of unidirectional ColE2 DNA replication by a unique priming mechanism. Nucleic Acids Res 23:4196–4201 10.1093/nar/23.20.4196. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Troll C, Yoder J, Alexander D, Hernández J, Loh Y, Camps M. 2014. The mutagenic footprint of low-fidelity Pol I ColE1 plasmid replication in E. coli reveals an extensive interplay between Pol I and Pol III. Curr Genet 60:123–134 10.1007/s00294-013-0415-9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cesareni G, Helmer-Citterich M, Castagnoli L. 1991. Control of ColE1 plasmid replication by antisense RNA. Trends Genet 7:230–235 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 71.Takechi S, Matsui H, Itoh T. 1995. Primer RNA synthesis by plasmid-specified Rep protein for initiation of ColE2 DNA replication. EMBO J 14:5141–5147 10.1002/j.1460-2075.1995.tb00196.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Chatelier E, Jannière L, Ehrlich SD, Canceill D. 2001. The RepE initiator is a double-stranded and single-stranded DNA-binding protein that forms an atypical open complex at the onset of replication of plasmid pAMbeta 1 from Gram-positive bacteria. J Biol Chem 276:10234–10246 10.1074/jbc.M010118200. [DOI] [PubMed] [Google Scholar]
- 73.Bruand C, Le Chatelier E, Ehrlich SD, Jannière L. 1993. A fourth class of theta-replicating plasmids: the pAM beta 1 family from Gram-positive bacteria. Proc Natl Acad Sci USA 90:11668–11672 10.1073/pnas.90.24.11668. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bruand C, Ehrlich SD. 1998. Transcription-driven DNA replication of plasmid pAMbeta1 in Bacillus subtilis. Mol Microbiol 30:135–145 10.1046/j.1365-2958.1998.01044.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Bidnenko V, Ehrlich SD, Jannière L. 1998. In vivo relations between pAMbeta1-encoded type I topoisomerase and plasmid replication. Mol Microbiol 28:1005–1016 10.1046/j.1365-2958.1998.00862.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Jannière L, Bidnenko V, McGovern S, Ehrlich SD, Petit MA. 1997. Replication terminus for DNA polymerase I during initiation of pAM beta 1 replication: role of the plasmid-encoded resolution system. Mol Microbiol 23:525–535 10.1046/j.1365-2958.1997.d01-1874.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Loftie-Eaton W, Rawlings DE. 2012. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67:15–34 10.1016/j.plasmid.2011.10.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Meyer R. 2009. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62:57–70 10.1016/j.plasmid.2009.05.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruiz-Maso JA, Macho NC, Bordanaba-Ruiseco L, Espinosa M, Coll M, Del Solar G. 2015. Plasmid rolling-circle replication. Microbiol Spectr 3:PLAS-0035-2014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Austin S, Nordström K. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351–354 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 81.Datta N, Hedges RW. 1971. Compatibility groups among fi–R factors. Nature 234:222–223 10.1038/234222a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228 10.1016/j.mimet.2005.03.018. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Garcillán-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687 10.1111/j.1574-6976.2009.00168.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.San Millan A, MacLean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr 5:MTBP-0016-2017. 10.1128/microbiolspec.MTBP-0016-2017. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brantl S. 2014. Plasmid replication control by antisense RNAs. Microbiol Spectr 2:PLAS-0001-2013. 10.1128/microbiolspec.PLAS-0001-2013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Thomas CM, Hussain AA. 1984. The korB gene of broad host range plasmid RK2 is a major copy number control element which may act together with trfB by limiting trfA expression. EMBO J 3:1513–1519 10.1002/j.1460-2075.1984.tb02004.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bury K, Wegrzyn K, Konieczny I. 2017. Handcuffing reversal is facilitated by proteases and replication initiator monomers. Nucleic Acids Res 45:3953–3966 10.1093/nar/gkx166. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kubik S, Wegrzyn K, Pierechod M, Konieczny I. 2012. Opposing effects of DNA on proteolysis of a replication initiator. Nucleic Acids Res 40:1148–1159 10.1093/nar/gkr813. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nishio SY, Itoh T. 2009. Arginine-rich RNA binding domain and protein scaffold domain of RNase E are important for degradation of RNAI but not for that of the Rep mRNA of the ColE2 plasmid. Plasmid 62:83–87 10.1016/j.plasmid.2009.04.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Praszkier J, Pittard AJ. 2005. Control of replication in I-complex plasmids. Plasmid 53:97–112 10.1016/j.plasmid.2004.12.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Athanasopoulos V, Praszkier J, Pittard AJ. 1999. Analysis of elements involved in pseudoknot-dependent expression and regulation of the repA gene of an IncL/M plasmid. J Bacteriol 181:1811–1819 10.1128/JB.181.6.1811-1819.1999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Polisky B. 1988. ColE1 replication control circuitry: sense from antisense. Cell 55:929–932 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 93.Camps M. 2010. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat DNA Gene Seq 4:58–73 10.2174/187221510790410822. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hiraga S, Sugiyama T, Itoh T. 1994. Comparative analysis of the replicon regions of eleven ColE2-related plasmids. J Bacteriol 176:7233–7243 10.1128/JB.176.23.7233-7243.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 192:6045–6055 10.1128/JB.00277-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.del Solar G, Alonso JC, Espinosa M, Díaz-Orejas R. 1996. Broad-host-range plasmid replication: an open question. Mol Microbiol 21:661–666 10.1046/j.1365-2958.1996.6611376.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Yano H, Shintani M, Tomita M, Suzuki H, Oshima T. 2018. Reconsidering plasmid maintenance factors for computational plasmid design. Comput Struct Biotechnol J 17:70–81 10.1016/j.csbj.2018.12.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Banerjee SK, Luck BT, Kim HY, Iyer VN. 1992. Three clustered origins of replication in a promiscuous-plasmid replicon and their differential use in a PolA+ strain and a delta PolA strain of Escherichia coli K-12. J Bacteriol 174:8139–8143 10.1128/JB.174.24.8139-8143.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim HY, Banerjee SK, Iyer VN. 1994. The incN plasmid replicon: two pathways of DNA polymerase I-independent replication. J Bacteriol 176:7735–7739 10.1128/JB.176.24.7735-7739.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Filutowicz M, Dellis S, Levchenko I, Urh M, Wu F, York D. 1994. Regulation of replication of an iteron-containing DNA molecule. Prog Nucleic Acid Res Mol Biol 48:239–273 10.1016/S0079-6603(08)60857-0. [DOI] [PubMed] [Google Scholar]
- 101.Jiang Y, Pacek M, Helinski DR, Konieczny I, Toukdarian A. 2003. A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin. Proc Natl Acad Sci USA 100:8692–8697 10.1073/pnas.1532393100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caspi R, Pacek M, Consiglieri G, Helinski DR, Toukdarian A, Konieczny I. 2001. A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J 20:3262–3271 10.1093/emboj/20.12.3262. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yano H, Wegrzyn K, Loftie-Eaton W, Johnson J, Deckert GE, Rogers LM, Konieczny I, Top EM. 2016. Evolved plasmid-host interactions reduce plasmid interference cost. Mol Microbiol 101:743–756 10.1111/mmi.13407. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nishida H. 2012. Comparative analyses of base compositions, DNA sizes, and dinucleotide frequency profiles in archaeal and bacterial chromosomes and plasmids. Int J Evol Biol 2012:342482 10.1155/2012/342482. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. 2015. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128 10.1101/gr.180190.114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983 10.1128/AEM.02171-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529 10.1093/jac/dkq347. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Newnham PJ, Taylor DE. 1994. Molecular analysis of RepHI1A, a minimal replicon of the IncHI1 plasmid R27. Mol Microbiol 11:757–768 10.1111/j.1365-2958.1994.tb00353.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Phan MD, Wain J. 2008. IncHI plasmids, a dynamic link between resistance and pathogenicity. J Infect Dev Ctries 2:272–278. [DOI] [PubMed] [Google Scholar]
- 110.Papagiannitsis CC, Miriagou V, Giakkoupi P, Tzouvelekis LS, Vatopoulos AC. 2013. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-β-lactamase. J Antimicrob Chemother 68:2259–2262 10.1093/jac/dkt196. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Froehlich B, Parkhill J, Sanders M, Quail MA, Scott JR. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J Bacteriol 187:6509–6516 10.1128/JB.187.18.6509-6516.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deng Y, He L, Chen S, Zheng H, Zeng Z, Liu Y, Sun Y, Ma J, Chen Z, Liu JH. 2011. F33:A-:B- and F2:A-:B- plasmids mediate dissemination of rmtB-blaCTX-M-9 group genes and rmtB-qepA in Enterobacteriaceae isolates from pets in China. Antimicrob Agents Chemother 55:4926–4929 10.1128/AAC.00133-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ebersbach G, Gerdes K. 2001. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc Natl Acad Sci USA 98:15078–15083 10.1073/pnas.261569598. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cerin H, Hackett J. 1993. The parVP region of the Salmonella typhimurium virulence plasmid pSLT contains four loci required for incompatibility and partition. Plasmid 30:30–38 10.1006/plas.1993.1031. [PubMed] [DOI] [PubMed] [Google Scholar]
- 115.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801 10.1371/journal.pone.0008801. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Christensen SK, Gerdes K. 2003. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol 48:1389–1400 10.1046/j.1365-2958.2003.03512.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Harms A, Brodersen DE, Mitarai N, Gerdes K. 2018. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell 70:768–784 10.1016/j.molcel.2018.01.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 118.Goeders N, Van Melderen L. 2014. Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 6:304–324 10.3390/toxins6010304. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kędzierska B, Hayes F. 2016. Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules 21:790 10.3390/molecules21060790. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee KY, Lee BJ. 2016. Structure, biology, and therapeutic application of toxin-antitoxin systems in pathogenic bacteria. Toxins (Basel) 8:305 10.3390/toxins8100305. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Planchenault C, Pons MC, Schiavon C, Siguier P, Rech J, Guynet C, Dauverd-Girault J, Cury J, Rocha EPC, Junier I, Cornet F, Espéli O. 2020. Intracellular positioning systems limit the entropic eviction of secondary replicons toward the nucleoid edges in bacterial cells. J Mol Biol 432:745–761 10.1016/j.jmb.2019.11.027. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Bouet JY, Funnell BE. 2019. Plasmid localization and partition in Enterobacteriaceae. Ecosal Plus 8:10.1128/ecosalplus.ESP-0003-2019 10.1128/ecosalplus.ESP-0003-2019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moreno-Hagelsieb G, Santoyo G. 2015. Predicting functional interactions among genes in prokaryotes by genomic context. Adv Exp Med Biol 883:97–106 10.1007/978-3-319-23603-2_5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Lawrence JG, Roth JR. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li JJ, Spychala CN, Hu F, Sheng JF, Doi Y. 2015. Complete nucleotide sequences of bla(CTX-M)-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob Agents Chemother 59:3002–3007 10.1128/AAC.04772-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47(D1):D427–D432 10.1093/nar/gky995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Phan MD, Forde BM, Peters KM, Sarkar S, Hancock S, Stanton-Cook M, Ben Zakour NL, Upton M, Beatson SA, Schembri MA. 2015. Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS One 10:e0122369 10.1371/journal.pone.0122369. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ogura T, Hiraga S. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci USA 80:4784–4788 10.1073/pnas.80.15.4784. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol 3:e176 10.1371/journal.pbio.0030176. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Murli S, Opperman T, Smith BT, Walker GC. 2000. A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J Bacteriol 182:1127–1135 10.1128/JB.182.4.1127-1135.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Munoz-Najar U, Vijayakumar MN. 1999. An operon that confers UV resistance by evoking the SOS mutagenic response in streptococcal conjugative transposon Tn5252. J Bacteriol 181:2782–2788 10.1128/JB.181.9.2782-2788.1999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Osorio-Valeriano M, Altegoer F, Steinchen W, Urban S, Liu Y, Bange G, Thanbichler M. 2019. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell 179:1512-1524.e15. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Soh YM, Davidson IF, Zamuner S, Basquin J, Bock FP, Taschner M, Veening JW, De Los Rios P, Peters JM, Gruber S. 2019. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 366:1129–1133 10.1126/science.aay3965. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Popowska M, Krawczyk-Balska A. 2013. Broad-host-range IncP-1 plasmids and their resistance potential. Front Microbiol 4:44 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Adamczyk M, Jagura-Burdzy G. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol 50:425–453 10.18388/abp.2003_3696. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Wawrzycka A, Gross M, Wasaznik A, Konieczny I. 2015. Plasmid replication initiator interactions with origin 13-mers and polymerase subunits contribute to strand-specific replisome assembly. Proc Natl Acad Sci USA 112:E4188–E4196 10.1073/pnas.1504926112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kelley W, Bastia D. 1985. Replication initiator protein of plasmid R6K autoregulates its own synthesis at the transcriptional step. Proc Natl Acad Sci USA 82:2574–2578 10.1073/pnas.82.9.2574. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Papp PP, Iyer VN. 1995. Determination of the binding sites of RepA, a replication initiator protein of the basic replicon of the IncN group plasmid pCU1. J Mol Biol 246:595–608 10.1016/S0022-2836(05)80109-3. [DOI] [PubMed] [Google Scholar]
- 139.Zzaman S, Bastia D. 2005. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol Cell 20:833–843 10.1016/j.molcel.2005.10.037. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Kunnimalaiyaan S, Inman RB, Rakowski SA, Filutowicz M. 2005. Role of pi dimers in coupling (“handcuffing”) of plasmid R6K’s gamma ori iterons. J Bacteriol 187:3779–3785 10.1128/JB.187.11.3779-3785.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gasset-Rosa F, Maté MJ, Dávila-Fajardo C, Bravo J, Giraldo R. 2008. Binding of sulphonated indigo derivatives to RepA-WH1 inhibits DNA-induced protein amyloidogenesis. Nucleic Acids Res 36:2249–2256 10.1093/nar/gkn067. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Das N, Chattoraj DK. 2004. Origin pairing (‘handcuffing’) and unpairing in the control of P1 plasmid replication. Mol Microbiol 54:836–849 10.1111/j.1365-2958.2004.04322.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Pierechod M, Nowak A, Saari A, Purta E, Bujnicki JM, Konieczny I. 2009. Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system. Protein Sci 18:637–649 10.1002/pro.68. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Swan MK, Bastia D, Davies C. 2006. Crystal structure of pi initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication. Proc Natl Acad Sci USA 103:18481–18486 10.1073/pnas.0609046103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cheng HM, Gröger P, Hartmann A, Schlierf M. 2015. Bacterial initiators form dynamic filaments on single-stranded DNA monomer by monomer. Nucleic Acids Res 43:396–405 10.1093/nar/gku1284. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kaur G, Vora MP, Czerwonka CA, Rozgaja TA, Grimwade JE, Leonard AC. 2014. Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol Microbiol 91:1148–1163 10.1111/mmi.12525. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wegrzyn K, Fuentes-Perez ME, Bury K, Rajewska M, Moreno-Herrero F, Konieczny I. 2014. Sequence-specific interactions of Rep proteins with ssDNA in the AT-rich region of the plasmid replication origin. Nucleic Acids Res 42:7807–7818 10.1093/nar/gku453. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Erzberger JP, Mott ML, Berger JM. 2006. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 13:676–683 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 149.Hwang DS, Kornberg A. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem 267:23083–23086. [PubMed] [PubMed] [Google Scholar]
- 150.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. 2002. IHF and HU stimulate assembly of pre-replication complexes at Escherichia colioriC by two different mechanisms. Mol Microbiol 46:113–124 10.1046/j.1365-2958.2002.03129.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Shah DS, Cross MA, Porter D, Thomas CM. 1995. Dissection of the core and auxiliary sequences in the vegetative replication origin of promiscuous plasmid RK2. J Mol Biol 254:608–622 10.1006/jmbi.1995.0642. [PubMed] [DOI] [PubMed] [Google Scholar]
- 152.Maestro B, Sanz JM, Díaz-Orejas R, Fernández-Tresguerres E. 2003. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J Bacteriol 185:1367–1375 10.1128/JB.185.4.1367-1375.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Filutowicz M, McEachern MJ, Mukhopadhyay P, Greener A, Yang SL, Helinski DR. 1987. DNA and protein interactions in the regulation of plasmid replication. J Cell Sci Suppl 7(Suppl 7):15–31 10.1242/jcs.1987.Supplement_7.2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 154.Ingmer H, Fong EL, Cohen SN. 1995. Monomer-dimer equilibrium of the pSC101 RepA protein. J Mol Biol 250:309–314 10.1006/jmbi.1995.0378. [PubMed] [DOI] [PubMed] [Google Scholar]


