Abstract
Recent scientific evidence suggests a link between epigenetic changes (DNA methylation) and tumorigenesis. Moreover, a potential carcinogenic mechanism of cadmium was associated with changes in DNA methylation. In this study we investigated the impact of CdCl2 and CuSO4 aqueous solutions on DNA methylation in HT-29 cells by quantifying DNA methyltransferase (DNMT1, DNMT3A and DNMT3B) mRNA expression. Furthermore, we also studied the cytotoxic and anti-migratory potential of these substances. The results showed a dose-dependent decrease of viable cell percentage following 24 h of exposure (at concentrations of 0.05; 0.2; 1; 10 and 100 µg/ml), and an inhibitory effect on HT-29 cell migration capacity. In addition, RT-qPCR results showed that cadmium acts as a hypomethylating agent by suppressing DNMT expression, whereas copper acts as a hypermethylating compound by increasing DNMT expression. These findings suggest a cytotoxic potential of both cadmium and copper on HT-29 cells and their capacity to induce epigenetic changes.
Keywords: cadmium, copper, HT-29 cells, DNA methylation, cytotoxicity
Introduction
Cadmium (Cd) is a heavy metal with a wide occurrence in the environment, classified as group 1 human carcinogen since 1993 by the International Agency for Research on Cancer (IARC) (1,2). Cadmium was described as a cumulative toxin due to its long half-life (up to 1-3 decades) and its very low rate of excretion (3), parameters that are directly linked to the noxious effects on key organs such as kidneys, liver, heart, thyroid, bones and reproductive system (1,4-6). In addition, long-term exposure to Cd was associated with an increased risk for breast, lung, genitourinary, prostate, hepatic and colon cancer (1,7,8). Although important progress has been made towards the elucidation of mechanisms involved in cadmium toxicity (such as oxidative stress, endocrine disruption, interference with sulfhydryl groups, impairment of zinc-linked cellular processes, weak mutagenicity and poor DNA binding capacity), the molecular mechanisms underlying Cd carcinogenic properties are not fully known and understood (3,6,9,10). It was fairly recently found that Cd has the ability to trigger genomic instability through epigenetic mechanisms, involving DNA methylation machinery (DNMT-DNA methyltransferases) (9-12).
Colorectal cancer is the fourth cause of cancer-related mortality globally (13-15). This disease presents a heterogeneous profile characterized by chromosomal instability (most cases-over 85%) and high-frequency microsatellite instability (15% of the cases). Its occurrence is prevalent in persons with no apparent genetic predisposition or hereditary colorectal pathologies (65%). The remaining cases are associated with familial inherited susceptibility (13). Besides the genetic risk factors, age and health status, environmental factors also play an important role in the pathogenesis of colorectal cancer (14). Previous studies supported a possible link between exposure to heavy metals such as cadmium, chromium, mercury and arsenic and the risk for colorectal cancer (16,17). A recent study asserted a potential molecular mechanism for the implication of cadmium in colorectal tumor pathogenesis (1).
Copper is one of the essential metals involved in the activity of different enzymes, either as a structural component, or as a cofactor. Nevertheless, at high concentrations it is a toxic compound (18). Copper also interferes with key processes related to tumorigenesis (progression, metastasis and reluctance to conventional treatments) by direct or indirect mechanisms such as activation of the oncogenic MAPK pathway (promotion), impairment of cancer cell redox status, angiogenic activity (19), and aberrant DNA methylation and changes in expression patterns of DNMT genes (20,21). Moreover, elevated copper concentrations in serum of patients represent a reliable marker for the diagnosis of a malignancy (including colorectal cancer) in advanced stage, with a low response to therapy. The link between copper and colon cancer is even stronger, since copper modulates BRAF signaling and BRAFV600E mutation, which represents 90% of all mutations encountered in colon cancer patients (18,19).
Epigenetic changes can be defined as inherited modifications in DNA with no alterations within the sequence. They include DNA methylation, miRNA regulation, and histone deacetylation (22). DNA methylation is fundamental for embryogenesis and maintenance of the specificity of cell-lineage gene expression for life (9). Dysregulated DNA methylation induces improper organism development, chronic pathologies and even tumorigenesis (9,20-22). A feature of colorectal cancer and other types of malignancies in terms of epigenetic changes, is the decreased global DNA methylation and hypermethylation of locus specific gene promoters (23-25).
In light of the data mentioned above, the present study aimed at characterizing the impact of CdCl2 and CuSO4 aqueous solutions on DNA methylation in human colorectal carcinoma HT-29 cells by quantifying DNA methyltransferase (DNMT1, DNMT3A and DNMT3B) mRNA expression. In addition, studies on cell viability and morphology, and migratory capacity were performed.
Materials and methods
Reagents
CuCl2 and CuSO4 were acquired from Sigma- Aldrich; Merck KGaA, as powders of analytical grade purity. The cell culture media: McCoy's 5a Medium Modified and supplements-fetal bovine serum (FBS), penicillin/streptomycin antibiotic mixture were from ATCC (American Type Cell Collection), Thermo Fisher Scientific, Inc., and Sigma-Aldrich; Merck KGaA. The other reagents used in the present experimental design: Phosphate saline buffer (PBS), Trypan blue, and Alamar blue were acquired from Sigma-Aldrich; Merck KGaA, and applied as recommended by the manufacturers.
Cell line
The in vitro experimental part of the present study was conducted on a human colorectal carcinoma cell line-HT-29 (ATCC® HTB-38™), that was acquired as frozen vial from ATCC.
Cell culture protocol
HT-29 cells were grown in specific cell culture medium, McCoy's 5a modified medium (ATCC® 30-2007™) supplemented with 10% fetal bovine serum (FBS) purchased from Thermo Fisher Scientific, Inc., and 1% solution of antibiotic mixture (Penicillin and Streptomycin; Sigma-Aldrich; Merck KGaA) added to minimize the risk of contamination in the culture.
Cell viability assessment-Alamar blue test
In order to assess the impact of the test compounds (CdCl2 and CuSO4) on HT-29 cell viability, the Alamar blue assay was performed. HT-29 cells were seeded in 96-well plates, 1x104 cells/well/200 µl culture medium and were allowed to adhere to the plate for 24 h. When the cells reached the optimal confluence, the old culture medium was replaced with fresh culture medium 200 µl/well that contained different concentrations of the test solutions (CdCl2 or CuSO4) 0.05; 0.2; 1; 10 and 100 µg/ml for 24 h. The 24 h exposure period was followed by addition of 20 µl/well of Alamar blue reagent, incubation for 3 h at 37˚C and reading of the absorbance at 570 and 600 nm by a xMark™ Microplate spectrophotometer (Bio-rad). The percentage of viable cells (%) was calculated according to the formula presented in a previous study (26), as follows:
Percentage of viable cells (%)={[(εOX)λ2 Aλ1-(εOX)λ1 Aλ2 of test agent dilution]/[(εOX)λ2 A°λ1-(εOX)λ1 A°λ2 of untreated positive growth control]} x100,
Where: εOX=molar extinction coefficient of Alamar blue oxidized form (BLUE)
A=absorbance of test wells
A°=absorbance of positive growth control well (cells without tested compounds)
λ1=570 nm and λ2=600 nm.
Cell morphology microscopic evaluation
Changes in cell morphology and shape induced by different compounds represent specific signs of cytotoxicity. Therefore, the effect of the tested compounds on HT-29 cell morphology was evaluated as a part of the toxicological profile. The cells were photographed under bright field illumination of the Olympus IX73 inverted microscope (Olympus) at 24 h post-stimulation with the test compounds (CdCl2 or CuSO4, 0.05; 0.2; 1; 10 and 100 µg/ml) and the photos were analyzed by the cellSens Dimensions v.1.8. software (Olympus).
Migration rate evaluated by the Scratch assay
To evaluate the impact of the test compounds (CdCl2 and CuSO4) on migratory capacity of HT-29 cells, the scratch assay (also known as wound healing assay) was performed. The protocol applied was similar to the one described in our previous articles (27,28) and was adapted to the present experimental conditions. A number of 2x105 HT-29 cells/well/1.5 ml culture medium were cultured in 12-well plates. When confluence reached 90-95%, the old medium was removed, a scratch was drawn manually using a pipette tip (2-200 µl) and the detached cells were washed with PBS. Fresh culture medium containing the test compounds (0.05, 0.2 and 1 µg/ml) was added to cells for a period of 24 h. Pictures were taken at 0 and 24 h following addition of the medium containing the test compounds, using an Olympus IX73 inverted microscope equipped with a DP74 camera (Olympus). The scratch widths were measured at 0 and 24 h by the means of CellSense Dimension 1.17. software (Olympus). The scratch closure/migration rate (%) was calculated with the following formula:
Scratch closure %=[(At=0 h-At=24 h)/At=0 h] X100,
Where: At=0 h is the width of the wound measured immediately after scratching at 0 h.
At=24 h is the width of the wound measured after 24 h (29,30).
RNA extraction and reverse-transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from HT-29 cells using the Trizol reagent (Thermo Fisher Scientific, Inc.) and the Quick-RNA™ purification kit (Zymo Research). cDNA was generated by reverse transcription with the Maxima® First Strand cDNA Synthesis Kit (Fermentas). Quantitative real-time PCR analysis was performed in 20 µl reactions containing Power SYBR-Green PCR Master Mix (Thermo Fisher Scientific, Inc.)
Cycling conditions in a Quant Studio 5 real-time PCR system (Thermo Fisher Scientific, Inc.) were: 95˚C for 10 sec followed by 40 cycles of denaturing at 95˚C for 15 sec and annealing and extension at 55˚C for 1 min. The following primer pairs (Eurogentec) were used:
18S (as housekeeping gene): F: 5'GTAC-CCGT-TGAA-CCCC-ATT3', R: 5'CCAT-CCAA-TCGG-TAGT-AGCG3', DNMT1: F: 5'ACCG-CTTC-TACT-TCCT-CGAG-GCCTA3' R: 5'GTTG-CAGT-CCTC-GTGA-ACAC-TGTGG3', DNMT3A: F: 5'CACA-CAGA-AGCA-TATC-CAGG-AGTG3' R: 5'AGTG-GACT-GGGA-AACC-AAAT-ACCC3', DNMT3B: F: 5'AATG-TGAA-TCCA-GTCA-GGAA-AGGC3' R: 5'ACTG-GATT-ACAC-TCCA-GGAA-CCGT3'.
Statistical analysis
Graph Pad Prism 8 was used for the statistical interpretation of the data. The results were expressed as the mean ± standard deviation (SD). One-way ANOVA was applied to determine the statistical differences followed by Dunnett's post-test (*P<0.05; ****P<0.0001).
Results
CdCl2 and CuSO4 reduced viability of HT-29 cells in a concentration-dependent manner
The effect of CdCl2 on the viability of human colorectal HT-29 cells was evaluated after stimulation of cells with different concentrations (0.05; 0.2; 1; 10 and 100 µg/ml) of the tested compound for 24 h. Our results showed that the lowest concentration tested, 0.05 µg/ml, had a stimulatory effect on cell viability. On the contrary, a dose-dependent cytotoxicity was observed when the other concentrations were tested. The effect was statistically significant only at the two highest concentrations, 10 and 100 µg/ml, the calculated percentages of viable cells were 83 and 13%, respectively (Fig. 1). The calculated IC50 value was 16.21 µg/ml.
Figure 1.
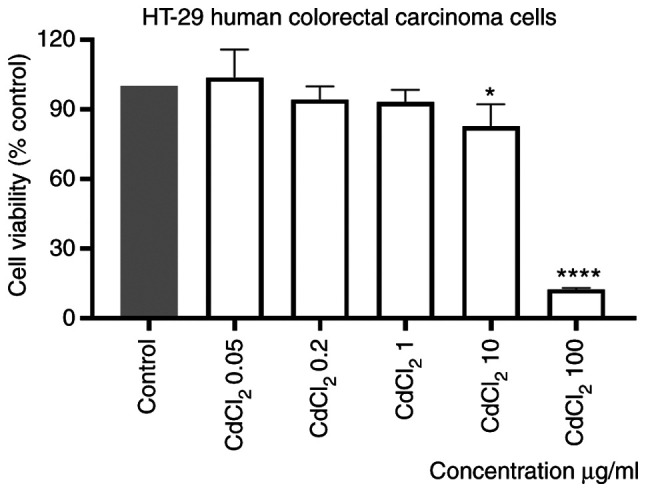
In vitro evaluation of the CdCl2 effect (0.05; 0.2; 1; 10 and 100 µg/ml) on viability of HT-29 cells at 24 h post-stimulation by the Alamar blue assay. The results are expressed as cell viability percentage (%) normalized to control (unstimulated) cells. The data represent the mean values ± SD of three independent experiments performed in triplicate. One-way ANOVA analysis was applied to determine the statistical differences compared with the control group, followed by Dunnett's multiple comparisons post-test (*P<0.05 and ****P<0.0001).
Stimulation of the cells with the same volume of sterile water did not influence the viability of the cells as compared with control cells (unstimulated cells).
Stimulation of HT-29 cells with a CuSO4 aqueous solution (prepared at the same concentrations as for CdCl2), induced a dose-dependent decrease of human colorectal carcinoma cell viability percentage, compared with control (unstimulated) cells. The effect was statistically significant only at the two highest concentrations tested, 10 and 100 µg/ml (Fig. 2). The calculated percentages of viable cells were 68.83 and 41.02%, respectively. Moreover, the smallest concentrations appeared to have a stimulatory effect (Fig. 2). The IC50 value obtained was 4.4 µg/ml.
Figure 2.
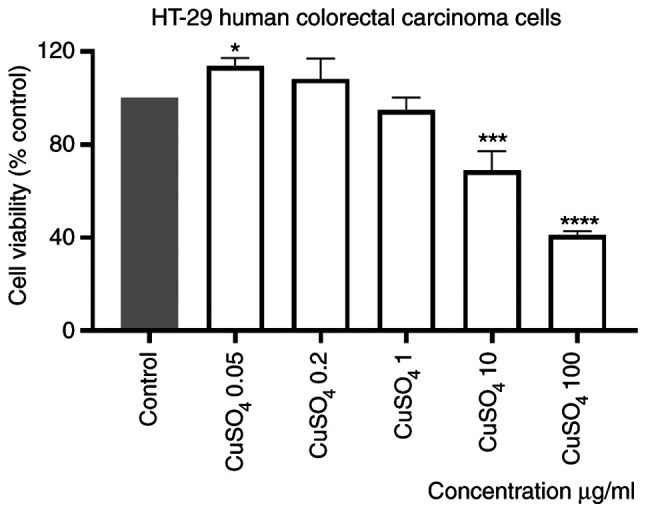
In vitro evaluation of the CuSO4 effect (0.05; 0.2; 1; 10 and 100 µg/ml) on HT-29 cells viability at 24 h post-stimulation by the Alamar blue assay. The results are expressed as cell viability percentage (%) normalized to control (unstimulated) cells. The data represent the mean values ± SD of three independent experiments performed in triplicate. One-way ANOVA analysis was applied to determine the statistical differences compared with the control group, followed by Dunnett's multiple comparisons post-test (*P<0.05, ***P<0.001 and ****P<0.0001).
Effect of test compounds (CdCl2 and CuSO4) on morphology of HT-29 cells
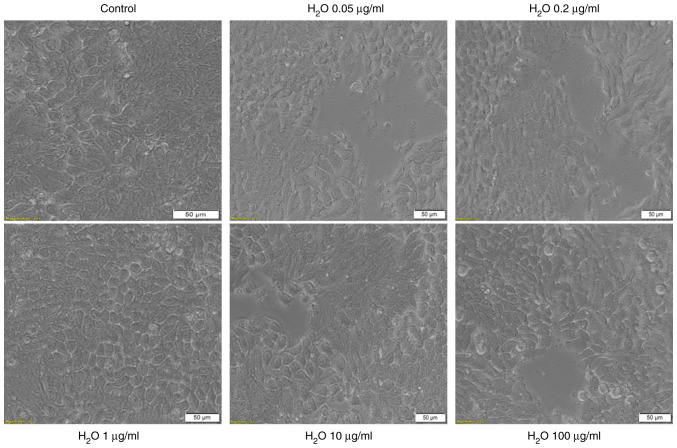
As shown in Fig. 3, the solvent used for the two test solutions, sterile water, had no impact on HT-29 cell morphology as compared with control cells (unstimulated cells): The cells stimulated with sterile water present the same shape as control cells, are well-attached to the culture plate and their confluence is not affected (Fig. 3). These data are in agreement with the results obtained for viability experiments.
Figure 3.
Microscopic image of HT-29 human colorectal carcinoma cells in culture: Control cells (non-stimulated) and cells stimulated with different concentrations (0.05; 0.2; 1; 10 and 100 µg/ml) of sterile H2O for 24 h. H2O is the solvent used for the CdCl2 and CuSO4 solutions. The pictures were taken following 24 h of stimulation and the scale bars represent 50 µm.
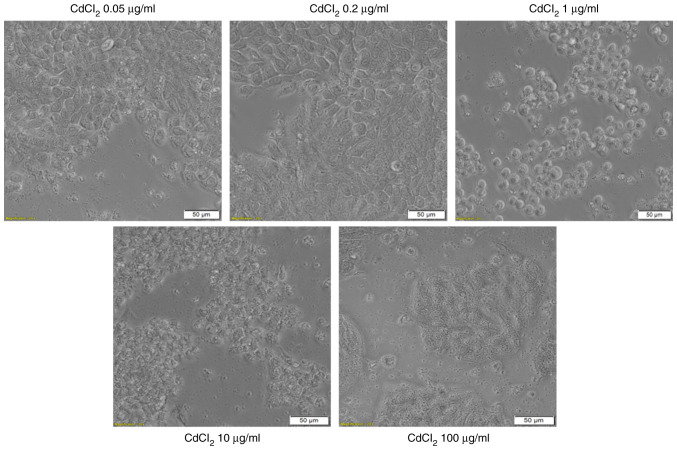
The addition of CdCl2 into the medium of HT-29 cells and exposure of the cells to this compound for 24 h was associated with several changes in cell morphology, compared with control (unstimulated) cells and solvent (sterile water) treated cells. The lowest concentrations, 0.05 and 0.2 µg/ml had no impact on the cell shape, confluence or adherence to the culture plate. At the concentration of 1 µg/ml the cells become round and were floating in the medium. At the highest concentration tested, 100 µg/ml, the cells appeared disintegrated (Fig. 4-lower panel). These results reinforce the viability data and indicate a cytotoxic effect.
Figure 4.
Microscopic appearance of HT-29 human colorectal carcinoma cells stimulated with different concentrations (0.05; 0.2; 1; 10 and 100 µg/ml) of CdCl2 for 24 h. The pictures were taken following 24 h of stimulation and the scale bars represent 50 µm.
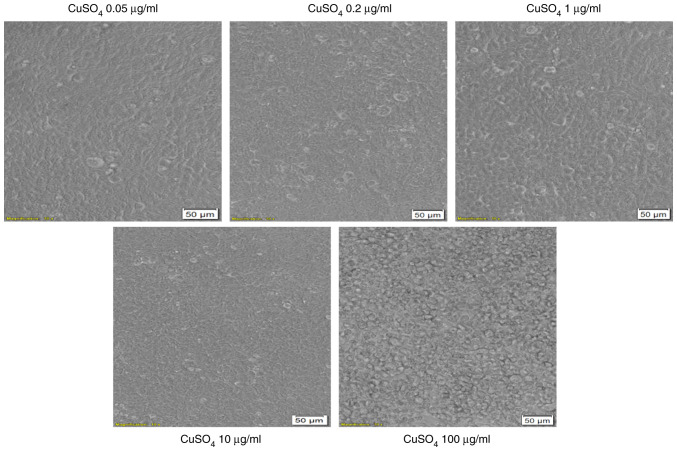
CuSO4 stimulation (at concentrations of 0.05; 0.2 and 1 µg/ml) for 24 h had no impact on HT-29 cells morphology in terms of shape, adherence or confluence as compared with control cells or solvent-stimulated cells (Fig. 5). Several changes were observed in the cells exposed to the highest concentrations of 10 and 100 µg/ml. These cells presented different morphology related to the other groups of cells, round cells that floated (Fig. 5).
Figure 5.
Microscopic appearance of HT-29 human colorectal carcinoma cells stimulated with different concentrations (0.05; 0.2; 1; 10 and 100 µg/ml) of CuSO4 for 24 h. The pictures were taken following 24 h of stimulation and the scale bars represent 50 µm.
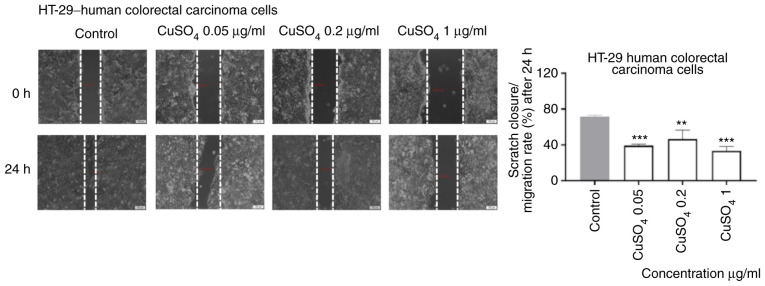
CdCl2 and CuSO4 stimulation interferes with the migratory capacity of HT-29 cells
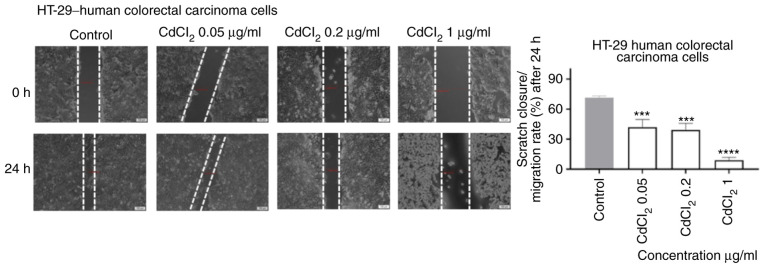
As shown in Fig. 6, 1 µg/ml of CdCl2 induced a significant inhibition of cell migration following 24 h of exposure. Furthermore, the shape of the cells was also different (round) as compared with control cells. An inhibitory effect, less pronounced, was also recorded for the other concentrations tested (Fig. 6).
Figure 6.
The impact of CdCl2 (0.05, 0.2 and 1 µg/ml) on HT-29 human colorectal carcinoma cells migratory capacity following 24 h of stimulation. Scratch widths were initially recorded by bright field microscopy, 0 and 24 h post exposure, respectively. Scale bars, 100 µm. The results are expressed as scratch closure/migration rate (%) following 24 h of stimulation. The data represent the mean values ± SD of three independent experiments performed in triplicate. One-way ANOVA analysis was used to determine the statistical differences from the control cells, followed by Dunnett's multiple comparisons post-test (***P<0.001 and ****P<0.0001).
Exposure to CuSO4 (0.05, 0.2 and 1 µg/ml) for 24 h induced a similar effect on the migratory capacity of HT-29 cells as the one described for CdCl2, namely an inhibitory effect even at the lowest concentration tested (Fig. 7).
Figure 7.
The impact of CuSO4 (0.05, 0.2 and 1 µg/ml) on HT-29 human colorectal carcinoma cells migratory capacity following 24 h of stimulation. Scratch widths were initially recorded by bright field microscopy, 0 and 24 h post exposure. Scale bars represent 100 µm. The results are expressed as scratch closure/migration rate (%) following 24 h of stimulation. The data represent the mean values ± SD of three independent experiments performed in triplicate. One-way ANOVA analysis was applied to determine the statistical differences from the control cells, followed by Dunnett's multiple comparisons post-test (**P<0.01, ***P<0.001).
CdCl2 and CuSO4 stimulation interferes with DNMT1, DNMT3A and DMNT3B gene expression in HT-29 human colorectal carcinoma cells Cadmium chloride
There was a significant effect on the expression of DNMT1, DNMT3A and DNMT3B for all cadmium concentrations tested in the present experiment (ANOVA, P<0.05).
The DNMT3A expression was significantly increased at the two lowest concentrations tested, as compared with the expression of DNMT1 and DNMT3B (Newman-Keuls tests, P<0.05). The increase in CdCl2 concentration resulted in significant changes in the expression of DNMT1, DNMT3A and DNMT3B (ANOVA, P<0.05). Expression of DNMT1 and DNMT3B showed a similar pattern of evolution as the concentration of CdCl2 increased. The determined values decreased from the lowest cadmium concentration to the concentration of 10 µg/ml and increased at the concentration of 100 µg/ml (Table I).
Table I.
Relative expression of DNMT1, DNMT3A and DNMT3B in human colorectal carcinoma cells (HT-29) exposed to CdCl2 and CuSO4 normalized to control (unstimulated cells).
| CdCl2 (µg/ml) | CuSO4 (µg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | 0.05 | 1 | 10 | 100 | 0.05 | 1 | 10 | 100 |
| DNMT1 | -3.72 | -5.67 | -8.69 | -1.03 | 18.58 | 16.81 | 17.3 | 11.69 |
| (-0.24) | (-0.64) | (-0.54) | (-0.23) | (3.83) | (3.20) | (2.93) | (1.25) | |
| DNMT3A | 5.72 | 1.03 | -17.12 | -7.19 | 0 | 0 | 0 | 0 |
| (0.45) | (0.13) | (-1.98) | (-0.87) | - | - | - | - | |
| DNMT3B | -2.85 | -5.82 | -13.7 | -8.7 | 22.19 | 12.64 | 25.53 | 21.02 |
| (-0.45) | (-0.38) | (-1.67) | (-0.63) | (2.54) | (1.45) | (2.86) | (3.45) | |
Copper sulphate
In the case of copper, the expression of DNMT1 remained relatively constant up to the concentration of 10 µg/ml but showed a significant decrease for the highest dose treatment (ANOVA P<0.05). There was no detectable expression of DNMT3A. However, for DNMT3B there were significant differences (ANOVA, P<0.05). The expression decreased significantly at the second lowest dose, compared with the lowest dose (Table I).
Discussion
Colorectal cancer presents lower mortality rates compared with the past, due to the efficient measures implemented (screening methods, early detection and intervention, optimized treatments) (14). Nevertheless, it is characterized by a poor survival rate of 13% if metastasis occurs, with an overall median survival period of 24 months (19,31). Several innovative methods were developed in recent years, to gather insights into the cancer development, diagnostic, and finding novel therapeutic approaches, such as: Cellomics (32), proteomics (33), and QSAR machine learning-based models (34). The various risk factors for colorectal cancer include heredity, age, health status (different pathologies, obesity, inflammatory Bowel disease and ulcerative colitis), as well as lifestyle. Similarly, risk factors such as lifestyle (nutrition, smoking, alcohol consumption), long-term use of contraceptives, exposure to environmental toxicants were described in the tumorigenesis related to human papilloma virus infection (35,36). A novel identified risk factor for colorectal cancer is microbial dysbiosis (biofilms) in gut microbiome (37), biofilms being also responsible for most types of infections (~65% nosocomial infections and ~80% of all microbial infections) (38). Furthermore, environmental toxicants such as heavy metals, including cadmium, play an asserted role in the pathogenesis of the disease, according to recent studies (14,39).
The wide use of Cd in industry (refining industries, metal mining, construction, Cd-containing batteries, and shipyard) and its environmental dissemination (7,40-42) led to the ranking of this metal among the most toxic ones (the seventh most toxic heavy metal) (16). It is responsible for multiple noxious effects such as nephrotoxicity, cardiotoxicity, bone diseases, reproductive toxicity, inflammatory disorders and tumorigenesis. To reduce the toxicity of the metals used for biomedical applications, there were proposed multiple coatings, diamond-like carbon films being a promising tool (43). The underlying molecular pathways involved in the carcinogenic properties of cadmium are not fully understood. The possible mechanisms include: i) Induction of reactive oxygen species (ROS) via activation of the mitogen activated protein kinases (MAPKs) pathway with oxidative impairment of proteins, lipids, and DNA; ii) the estrogen-like properties that were linked to the development of endometrial, breast and prostate tumors; iii) initiation of inflammation via upregulation of COX-2 (cyclooxygenase-2) expression, a key player in colorectal cancer via its major metabolite PGE2-prostaglandin E2, iv) triggering of malignant transformation in normal cells (human bronchial epithelial cells) by inducing apoptosis followed by DNA impairment, reduced DNA repair capacity and genomic instability through accumulation of mutations in DNA repair genes, creating a suitable ground for cancer development, v) epigenetic changes by interfering in DNA methylation, and vi) mitochondrial toxicity both in healthy (HPNE) and tumor (AsPC-1) pancreatic cells by altering mitochondrial function (1,2,7,9,10,44-46).
Tumor cell migration represents an essential condition for invasion and metastasis, and numerous studies have focused lately to identify the factors that regulate the migration process that is directly connected to signal biomolecules of the immune and neuroendocrine systems (47).
In a recent experimental study, Naji and colleagues showed that exposure to low levels of CdCl2 (100 and 1,000 nM equivalent to 0.01833 and 0.1833 µg/ml, respectively) for 9 and 12 h induced an increased migratory effect on HT-29 cells by activating several signaling pathways, such as the ROS-p38-COX-2-PGE2 and the ROS-Akt (1). These data are in accordance with our results that showed an increased percentage of viable cells at the lowest concentrations tested (0.05 and 0.2 µg/ml), compared with control cells (Fig. 1). Regarding the effect of Cd on the migratory capacity of HT-29 cells in our study, the longer exposure time (24 h), as compared with 9 and 12 h reported by Naji and colleagues, induced an inhibitory effect on HT-29 cell migration rate as compared with control cells (Fig. 6). These results are also confirmed by the study of Naji and colleagues who found that no further increase of wound closure was observed after 12 h (1).
An interesting finding of this work is the cytotoxic effect of CdCl2 on human colorectal carcinoma HT-29 cells which was observed at concentrations ≥1 µg/ml. This effect was characterized by a decrease in the viable cell percentage (Fig. 1) and changes in cell morphology (Fig. 4).
This novel cytotoxic property of Cd on cancer cells was also reported by Hajrezaie et al in a study where colon cancer HT-29 cells were exposed for 72 h to a complex of Cd, a Schiff based complex, CdCl2(C14H21N3O2) that induced apoptosis via activation of mitochondrial pathway (31). Another study that supports our data on the cytotoxic effect of Cd on cancer cells, was published by Guo et al who showed the antiproliferative and proapoptotic effects of CdCl2 as a single agent, as well as in combination with hSmac, in hepatocellular carcinoma cells (48).
Zhou et al showed that Cd exerted its cytotoxic effects on multiple epithelial-like cells, both healthy, as well as of tumor origin (hepatoma cell line FLC-4, breast carcinoma MCF7, gastric adenocarcinoma AGS, colon carcinoma HCT116, esophageal carcinoma TE4 and embryonic kidney cell line HEK293) in a cell-type and dose-dependent manner (49). A dose- and time-dependent cytotoxic effect of CdCl2 was shown on MDA-MB468 breast carcinoma cells (50), data that are in agreement with our results.
The connection between colorectal cancer and copper could be considered stronger, compared with cadmium, since elevated serum concentrations of copper in patients diagnosed with colorectal cancer indicate an advanced disease with a high risk of mortality (19).
The role of copper in the modulation of tumorigenesis via direct or indirect mechanisms (angiogenic cofactor, a redox-active metal and activation of MAPK pathway) is established. Moreover, it was shown that depletion of copper determines apoptosis of cancer cells (19). To highlight the function of Cu in colon cancer, different copper chelators were tested as potential anticancer therapies (18,19).
A strong connection also lies between Cd and Cu, since Cd is responsible for a disrupted balance of essential metals (Zn, Cu, Ca) leading to depletion of these metals and inducing noxious effects on human health including cancer of the intestine (39). In our previous study, conducted on land snails-Cantareus aspersus, we showed that low doses of dietary cadmium interfere with hepatopancreas copper deposition (51).
Our results on the effect of Cu on HT-29 cell viability showed a dose-dependent cytotoxic effect and are supported by the findings of another study that tested a CuCl2 solution (1-1,000 µM) on MDA-MB458 breast carcinoma cells (50). The cytotoxic effect of a CuSO4 solution was also studied on a human glioma cell line-U87-MG (52) and HeLa cells (53). The findings of both studies are consistent with our results. Moreover, our experimental data indicate an anti-migratory effect of copper on HT-29 cells (Fig. 7).
Among the multiple mechanisms of toxicity linked to cadmium and copper exposure, the interference with DNA methylation has also been described (3,9-11,20,21). DNA methylation is prevalent in the mammalian genome and is responsible for genomic stability, chromatin structure modulation, and transcriptional regulation of specific genes (22,25). This process occurs at the cytosines in specific regions of CpG dinucleotides and is catalyzed by DNA methyltransferases (DNMTs), DNMT1, DNMT3A and DNMT3B (22,25,54). DNMT1 is responsible for maintaining the patterns of DNA methylation during cell division and shows an affinity for hemi-methylated DNA strands, whereas DNMT3A and DNMT3B are described as de novo methyltransferases with different target sites from DNMT1 (22,25,54). Impaired DNA methylation has been associated with carcinogenesis (including colorectal cancer) and with the carcinogenic potential of cadmium (3,24).
The expression of DNMT1 and DNMT3B, key players in establishing and maintaining the DNA methylation patterns, are known to be disrupted in the HT-29 cell line. It was found that these genes are generally silenced in different colorectal cancer cell lines, including the HT-29 cell line, and the consequent disruptions in the methylation pattern contribute directly to the process of carcinogenesis (55). Other studies indicate that DNMT1, DNMT3A and DNMT3B are highly expressed in colon cancer (56,57), and the inhibition of their expression determines a reduction of tumorigenesis process (58). With respect to copper, scientific evidence on humans does not support an effect of this trace metal on genomic or gene-specific DNA methylation levels (59). However, our results show that copper has an effect on the expression of DNMT1 and DNMT3B, and subsequently on DNA methylation in the HT-29 cell line. Our data show that the expression of these genes is increased in response to copper exposure over a broad range of concentrations. This is most probably related to the dysregulated methylation control cycle in these tumor cells, which favors their uncontrolled growth and spreading (60). The relationship between the concentrations of CuSO4 and changes in the expression of the three enzymes were strikingly different; DNMT1 expression in copper exposed cells showed a significant decrease and approached the expression levels in control (unstimulated cells) only at the highest CuSO4 concentration (100 µg/ml). DNMT3B exhibited the same trend at a significantly lower CuSO4 concentration (1 µg/ml). These patterns might be related to their different role in establishing and maintaining the DNA methylation pattern.
The effect of cadmium on DNMT1 and DNMT3B expression showed a similar pattern. The expression of both enzymes was suppressed by cadmium. This finding is consistent with the results of a previous study that showed an association between acute cadmium exposure and decreased DNA methylation in the TRL1215 rat liver cells. It showed that the decrease in genomic DNA methylation levels is caused by noncompetitive inhibition of DNMT activity, a mechanism which could also explain our results (61). In this context, it is noteworthy to highlight the differential effect of Cu and Cd on these two genes in terms of direction and magnitude in the changes in DNMT1 and DNMT3B expression. Based on our results, it appears that for acute exposure 24 h of Cu increases expression and acts as a hypermethylating agent, while Cd suppresses expression and acts as a hypomethylating agent in the HT-29 cells.
We have currently shown that both CdCl2 and CuSO4 solutions induced cytotoxicity on HT-29 cells in a dose-dependent manner. The toxic effect included a decrease of cell viability, changes in cell morphology, and anti-migratory effects even at the lowest concentrations tested (0.05; 0.2 and 1 µg/ml). Moreover, cadmium acts as a hypomethylating agent by suppressing DNMT expression, whereas, copper acts as a hypermethylating compound by increasing DNMT expression. A limitation of this study could be considered the selection of a single time point of 24 h, but further studies are in progress to determine the mechanistic insights of a possible connection between the cytotoxic effects and the epigenetic changes observed.
Acknowledgements
The in vitro experiments were conducted within the Center of Pharmaco-toxicological evaluations from the Faculty of Pharmacy, ‘Victor Babes’ University of Medicine and Pharmacy (Timisoara, Romania).
Funding
The present study was supported by the Executive Agency for Higher Education, Research, Development and Innovation Funding Institution [grant no. PN-III-P1-1.1-PD-2016-1982] and the grant was awarded to DEC.
Availability of data and materials
All data generated or analyzed during the study are included in this published article.
Authors' contributions
GAD, AI, DEC, CD, AMT and OC conceived and designed the study. IoM, IaM, DEC, GAD and AT acquired the data and drafted the manuscript. DEC, GAD, RD and LK analysed the data. DEC, AT, AI and GAD wrote the manuscript. LK, AMT, OC and CD revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Naji S, Issa K, Eid A, Iratni R, Eid AH. Cadmium induces migration of colon cancer cells: Roles of reactive oxygen species, P38 and cyclooxygenase-2. Cell Physiol Biochem. 2019;52:1517–1534. doi: 10.33594/000000106. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Z, Wang C, Liu H, Huang Q, Wang M, Lei Y. Cadmium induced cell apoptosis, DNA damage, decreased DNA repair capacity, and genomic instability during malignant transformation of human bronchial epithelial cells. Int J Med Sci. 2013;10:1485–1496. doi: 10.7150/ijms.6308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6:820–827. doi: 10.4161/epi.6.7.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes-Hinojosa D, Lozada-Pérez CA, Zamudio Cuevas Y, López-Reyes A, Martínez-Nava G, Fernández-Torres J, Olivos-Meza A, Landa-Solis C, Gutiérrez-Ruiz MC, Rojas Del Castillo E, Martínez-Flores K. Toxicity of cadmium in musculoskeletal diseases. Environ Toxicol Pharmacol. 2019;72(103219) doi: 10.1016/j.etap.2019.103219. [DOI] [PubMed] [Google Scholar]
- 5.Geng HX, Wang L. Cadmium: Toxic effects on placental and embryonic development. Environ Toxicol Pharmacol. 2019;67:102–107. doi: 10.1016/j.etap.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Buha A, Matovic V, Antonijevic B, Bulat Z, Curcic M, Renieri EA, Tsatsakis AM, Schweitzer A, Wallace D. Overview of cadmium thyroid disrupting effects and mechanisms. Int J Mol Sci. 2018;19(1501) doi: 10.3390/ijms19051501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luevano J, Damodaran C. A review of molecular events of cadmium-induced carcinogenesis. J Environ Pathol Toxicol Oncol. 2014;33:183–194. doi: 10.1615/jenvironpatholtoxicoloncol.2014011075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal D, Suman S, Kolluru V, Sears S, Das TP, Alatassi H, Ankem MK, Freedman JH, Damodaran C. Inhibition of autophagy prevents cadmium-induced prostate carcinogenesis. Br J Cancer. 2017;117:56–64. doi: 10.1038/bjc.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hossain MB, Vahter M, Concha G, Broberg K. Low-level environmental cadmium exposure is associated with DNA hypomethylation in Argentinean women. Environ Health Perspect. 2012;120:879–884. doi: 10.1289/ehp.1104600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou ZH, Lei YX, Wang CX. Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium. Toxicol Sci. 2012;125:412–417. doi: 10.1093/toxsci/kfr320. [DOI] [PubMed] [Google Scholar]
- 11.Šrut M, Drechsel V, Höckner M. Low levels of Cd induce persisting epigenetic modifications and acclimation mechanisms in the earthworm Lumbricus terrestris. PLoS One. 2017;12(e0176047) doi: 10.1371/journal.pone.0176047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura T, Hosaka T, Nakanishi T, Aozasa O. Long-term cadmium exposure enhances metallothionein-1 induction after subsequent exposure to high concentrations of cadmium in P1798 mouse lymphosarcoma cells. J Toxicol Sci. 2019;44:309–316. doi: 10.2131/jts.44.309. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol Lett. 2018;16:9–18. doi: 10.3892/ol.2018.8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11(164) doi: 10.3390/nu11010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krasanakis T, Nikolouzakis TK, Sgantzos M, Mariolis-Sapsakos T, Souglakos J, Spandidos DA, Tsitsimpikou C, Tsatsakis A, Tsiaoussis J. Role of anabolic agents in colorectal carcinogenesis: Myths and realities (Review) Oncol Rep. 2019;42:2228–2244. doi: 10.3892/or.2019.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaishankar M, Tseten T, Anbalagan N, Mathew BB, Beeregowda KN. Toxicity, mechanism and health effects of some heavy metals. Interdiscip Toxicol. 2014;7:60–72. doi: 10.2478/intox-2014-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metals toxicity and the environment. Exp Suppl. 2012;101:133–164. doi: 10.1007/978-3-7643-8340-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fatfat M, Merhi RA, Rahal O, Stoyanovsky DA, Zaki A, Haidar H, Kagan VE, Gali-Muhtasib H, Machaca K. Copper chelation selectively kills colon cancer cells through redox cycling and generation of reactive oxygen species. BMC Cancer. 2014;14(527) doi: 10.1186/1471-2407-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldari S, Di Rocco G, Heffern MC, Su TA, Chang CJ, Toietta G. Effects of copper chelation on BRAFV600E positive colon carcinoma cells. Cancers (Basel) 2019;11(659) doi: 10.3390/cancers11050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Liu C, Liu Y, Hosokawa T, Saito T, Kurasaki M. Changes in the expression of epigenetic factors during copper-induced apoptosis in PC12 cells. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2014;49:1023–1028. doi: 10.1080/10934529.2014.894847. [DOI] [PubMed] [Google Scholar]
- 21.Dorts J, Falisse E, Schoofs E, Flamion E, Kestemont P, Silvestre F. DNA methyltransferases and stress-related genes expression in zebrafish larvae after exposure to heat and copper during reprogramming of DNA methylation. Sci Rep. 2016;6(34254) doi: 10.1038/srep34254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong KK, Lawrie CH, Green TM. Oncogenic roles and inhibitors of DNMT1, DNMT3A, and DNMT3B in acute myeloid leukaemia. Biomark Insights. 2019;14(1177271919846454) doi: 10.1177/1177271919846454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarabi MM, Naghibalhossaini F. Association of DNA methyltransferases expression with global and gene-specific DNA methylation in colorectal cancer cells. Cell Biochem Funct. 2015;33:427–433. doi: 10.1002/cbf.3126. [DOI] [PubMed] [Google Scholar]
- 24.Wu W, Ye S, Tan W, Zhou Y, Quan J. Analysis of promoter methylation and epigenetic regulation of miR-32 in colorectal cancer cells. Exp Ther Med. 2019;17:3209–3214. doi: 10.3892/etm.2019.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honeywell RJ, Sarkisjan D, Kristensen MH, de Klerk DJ, Peters GJ. DNA methyltransferases expression in normal tissues and various human cancer cell lines, xenografts and tumors. Nucleosides Nucleotides Nucleic Acids. 2018;37:696–708. doi: 10.1080/15257770.2018.1498516. [DOI] [PubMed] [Google Scholar]
- 26.Soica C, Oprean C, Borcan F, Danciu C, Trandafirescu C, Coricovac D, Crăiniceanu Z, Dehelean CA, Munteanu M. The synergistic biologic activity of oleanolic and ursolic acids in complex with hydroxypropyl-γ-cyclodextrin. Molecules. 2014;19:4924–4940. doi: 10.3390/molecules19044924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gheorgheosu D, Jung M, Ören B, Schmid T, Dehelean C, Muntean D, Bruene B. Betulinic acid suppresses NGAL-induced epithelial-to-mesenchymal transition in melanoma. Biol Chem. 2013;394:773–781. doi: 10.1515/hsz-2013-0106. [DOI] [PubMed] [Google Scholar]
- 28.Coricovac D, Farcas C, Nica C, Pinzaru I, Simu S, Stoian D, Soica C, Proks M, Avram S, Navolan D, et al. Ethinylestradiol and Levonorgestrel as active agents in normal skin, and pathological conditions induced by UVB exposure: In vitro and in ovo assessments. Int J Mol Sci. 2018;19(3600) doi: 10.3390/ijms19113600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felice F, Zambito Y, Belardinelli E, Fabiano A, Santoni T, Di Stefano R. Effect of different chitosan derivatives on in vitro scratch wound assay: A comparative study. Int J Biol Macromol. 2015;76:236–241. doi: 10.1016/j.ijbiomac.2015.02.041. [DOI] [PubMed] [Google Scholar]
- 30.Grada A, Otero-Vinas M, Prieto-Castrillo F, Obagi Z, Falanga V. Research techniques made simple: Analysis of collective cell migration using the wound healing assay. J Invest Dermatol. 2017;137:e11–e16. doi: 10.1016/j.jid.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Hajrezaie M, Paydar M, Looi CY, Moghadamtousi SZ, Hassandarvish P, Salga MS, Karimian H, Shams K, Zahedifard M, Majid NA, et al. Apoptotic effect of novel Schiff based CdCl2(C14H21N3O2) complex is mediated via activation of the mitochondrial pathway in colon cancer cells. Sci Rep. 2015;5(9097) doi: 10.1038/srep09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boda D. Cellomics as integrative omics for cancer. Current Proteomics. 2013;10:237–245. [Google Scholar]
- 33.Ion A, Popa IM, Papagheorghe LM, Lisievici C, Lupu M, Voiculescu V, Caruntu C, Boda D. Proteomic approaches to biomarker discovery in cutaneous T-cell lymphoma. Dis Markers. 2016;2016(9602472) doi: 10.1155/2016/9602472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ancuceanu R, Dinu M, Neaga I, Laszlo FG, Boda D. Development of QSAR machine learning-based models to forecast the effect of substances on malignant melanoma cells. Oncol Lett. 2019;17:4188–4196. doi: 10.3892/ol.2019.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boda D, Docea AO, Calina D, Ilie MA, Caruntu C, Zurac S, Neagu M, Constantin C, Branisteanu DE, Voiculescu V, et al. Human papilloma virus: Apprehending the link with carcinogenesis and unveiling new research avenues (Review) Int J Oncol. 2018;52:637–655. doi: 10.3892/ijo.2018.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boda D, Neagu M, Constantin C, Voinescu RN, Caruntu C, Zurac S, Spandidos DA, Drakoulis N, Tsoukalas D, Tsatsakis AM. HPV strain distribution in patients with genital warts in a female population sample. Oncol Lett. 2016;12:1779–1782. doi: 10.3892/ol.2016.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, Roslani AC, Wick EC, Mongodin EF, Loke MF, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017;3(34) doi: 10.1038/s41522-017-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andor B, Tucuina AAT, Berceanu-Vaduva D, Lazureanu V, Cheveresan A, Poenaru M. Antimicrobial activity and cytotoxic effect on gingival cells of silver nanoparticles obtained by biosynthesis. Rev Chim (Bucharest) 2019;70:781–783. [Google Scholar]
- 39.Klimczak M, Dziki A, Kilanowicz A, Sapota A, Duda-Szymańska J, Daragó A. Concentrations of cadmium and selected essential elements in malignant large intestine tissue. Prz Gastroenterol. 2016;11:24–29. doi: 10.5114/pg.2015.52563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nedelescu M, Baconi D, Neagoe A, Iordache V, Stan M, Constantinescu P, Ciobanu AM, Vardas AI, Vinceti M, Tsatsakis AM. Environmental metal contamination and health impact assessment in two industrial regions of Romania. Sci Total Environ. 2017;580:984–995. doi: 10.1016/j.scitotenv.2016.12.053. [DOI] [PubMed] [Google Scholar]
- 41.Renieri EA, Alegakis AK, Kiriakakis M, Vinceti M, Ozcagli E, Wilks MF, Tsatsakis AM. Cd, Pb and Hg Biomonitoring in fish of the Mediterranean region and risk estimations on fish consumption. Toxics. 2014;2:417–442. [Google Scholar]
- 42.Renieri EA, Safenkova IV, Alegakis AΚ, Slutskaya ES, Kokaraki V, Kentouri M, Dzantiev BD, Tsatsakis AM. Cadmium, lead and mercury in muscle tissue of gilthead seabream and seabass: Risk evaluation for consumers. Food Chem Toxicol. 2019;124:439–449. doi: 10.1016/j.fct.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Calenic B, Greabu M, Caruntu C, Nicolescu MI, Moraru L, Surdu-Bob CC, Badulescu M, Anghel A, Logofatu C, Boda D. Oral keratinocyte stem cells behavior on diamond like carbon films. Romanian Biotechnological Lett. 2016;21:11914–11922. [Google Scholar]
- 44.Nair AR, Degheselle O, Smeets K, Van Kerkhove E, Cuypers A. Cadmium-induced pathologies: Where is the oxidative balance lost (or Not)? Int J Mol Sci. 2013;14:6116–6143. doi: 10.3390/ijms14036116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace DR, Spandidos DA, Tsatsakis A, Schweitzer A, Djordjevic V, Djordjevic AB. Potential interaction of cadmium chloride with pancreatic mitochondria: Implications for pancreatic cancer. Int J Mol Med. 2019;44:145–156. doi: 10.3892/ijmm.2019.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao M, Kargman S, Lam EC, Kelly CR, Zheng Y, Luk P, Kwong E, Evans JF, Wolfe MM. Inhibition of Cyclooxygenase-2 by Rofecoxib attenuates the growth and metastatic potential of colorectal carcinoma in mice. Cancer Res. 2003;63:586–592. [PubMed] [Google Scholar]
- 47.Solomon I, Voiculescu VM, Caruntu C, Lupu M, Popa A, Ilie MA, Albulescu R, Caruntu A, Tanase C, Constantin C, et al. Neuroendocrine factors and head and neck squamous cell carcinoma: An affair to remember. Dis Markers. 2018;2018(9787831) doi: 10.1155/2018/9787831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo C, Li Y, Zhang H, Wang Z, Jin M, Zhang L, An L, Hu G, Liu X, Liu Y, et al. Enhancement of antiproliferative and proapoptotic effects of cadmium chloride combined with hSmac in hepatocellular carcinoma cells. Chemotherapy. 2011;57:27–34. doi: 10.1159/000321031. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X, Koizumi Y, Zhang M, Natsui M, Koyota S, Yamada M, Kondo Y, Hamada F, Sugiyama T. Cadmium-coordinated supramolecule suppresses tumor growth of T-cell leukemia in mice. Cancer Sci. 2015;106:635–641. doi: 10.1111/cas.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panjehpour M, Taher MA, Bayesteh M. The growth inhibitory effects of cadmium and copper on the MDA-MB468 human breast cancer cells. J Res Med Sci. 2010;15:279–286. [PMC free article] [PubMed] [Google Scholar]
- 51.Nica DV, Draghici GA, Andrica FM, Popescu S, Coricovac DE, Dehelean CA, Gergen II, Kovatsi L, Coleman MD, Tsatsakis AM. Short-term effects of very low dose cadmium feeding on copper, manganese and iron homeostasis: A gastropod perspective. Environ Toxicol Pharmacol. 2019;65:9–13. doi: 10.1016/j.etap.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Hu J, Guan F, Song L, Fan R, Zhu H, Hu X, Shen E, Yang B. Copper induces cellular senescence in human glioblastoma multiforme cells through downregulation of Bmi-1. Oncol Rep. 2013;29:1805–1810. doi: 10.3892/or.2013.2333. [DOI] [PubMed] [Google Scholar]
- 53.Chen SY, Liu ST, Lin WR, Lin CK, Huang SM. The mechanisms underlying the cytotoxic effects of copper via differentiated embryonic chondrocyte gene 1. Int J Mol Sci. 2019;20(5225) doi: 10.3390/ijms20205225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fragou D, Fragou A, Kouidou S, Njau S, Kovatsi L. Epigenetic mechanisms in metal toxicity. Toxicol Mech Methods. 2011;21:343–352. doi: 10.3109/15376516.2011.557878. [DOI] [PubMed] [Google Scholar]
- 55.Jin B, Yao B, Li JL, Fields CR, Delmas AL, Liu C, Robertson D. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: A novel target for prevention and therapy. Front Oncol. 2014;4(80) doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fouad MA, Salem SE, Hussein MM, Zekri ARN, Hafez HF, El Desouky ED, Shouman SA. Impact of global DNA methylation in treatment outcome of colorectal cancer patients. Front Pharmacol. 2018;9(1173) doi: 10.3389/fphar.2018.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li S, Han Z, Zhao N, Zhu B, Zhang Q, Yang X, Sheng D, Hou J, Guo S, Wei L, Zhang L. Inhibition of DNMT suppresses the stemness of colorectal cancer cells through down-regulating Wnt signaling pathway. Cell Signal. 2018;47:79–87. doi: 10.1016/j.cellsig.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 59.Ryu HW, Lee DH, Won HR, Kim KH, Seong YJ, Kwon SH. Influence of toxicologically relevant metals on human epigenetic regulation. Toxicol Res. 2015;31:1–9. doi: 10.5487/TR.2015.31.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogoshi K, Hashimoto S, Nakatani Y, Qu W, Oshima K, Tokunaga K, Sugano S, Hattori M, Morishita S, Matsushima K. Genome-wide profiling of DNA methylation in human cancer cells. Genomics. 2011;98:280–287. doi: 10.1016/j.ygeno.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 61.Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res. 2003;286:355–365. doi: 10.1016/s0014-4827(03)00062-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the study are included in this published article.