Abstract
Background & Aims
Gastrointestinal (GI) manifestations have been increasingly reported in patients with coronavirus disease 2019 (COVID-19). However, the roles of the GI tract in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are not fully understood. We investigated how the GI tract is involved in SARS-CoV-2 infection to elucidate the pathogenesis of COVID-19.
Methods
Our previously established nonhuman primate (NHP) model of COVID-19 was modified in this study to test our hypothesis. Rhesus monkeys were infected with an intragastric or intranasal challenge with SARS-CoV-2. Clinical signs were recorded after infection. Viral genomic RNA was quantified by quantitative reverse transcription polymerase chain reaction. Host responses to SARS-CoV-2 infection were evaluated by examining inflammatory cytokines, macrophages, histopathology, and mucin barrier integrity.
Results
Intranasal inoculation with SARS-CoV-2 led to infections and pathologic changes not only in respiratory tissues but also in digestive tissues. Expectedly, intragastric inoculation with SARS-CoV-2 resulted in the productive infection of digestive tissues and inflammation in both the lung and digestive tissues. Inflammatory cytokines were induced by both types of inoculation with SARS-CoV-2, consistent with the increased expression of CD68. Immunohistochemistry and Alcian blue/periodic acid–Schiff staining showed decreased Ki67, increased cleaved caspase 3, and decreased numbers of mucin-containing goblet cells, suggesting that the inflammation induced by these 2 types of inoculation with SARS-CoV-2 impaired the GI barrier and caused severe infections.
Conclusions
Both intranasal and intragastric inoculation with SARS-CoV-2 caused pneumonia and GI dysfunction in our rhesus monkey model. Inflammatory cytokines are possible connections for the pathogenesis of SARS-CoV-2 between the respiratory and digestive systems.
Keywords: COVID-19, Viral Infection, Inflammatory Cytokines, Fecal-Oral Route
Abbreviations used in this paper: AB-PAS, Alcian blue and periodic acid–Schiff; ACE2, angiotensin converting enzyme 2; COVID-19, coronavirus disease 2019; dpi, days postinfection; GI, gastrointestinal; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IEC, intestinal epithelial cells; IFNγ, interferon gamma; IHC, immunohistochemistry; NHP, non-human primate; PBS, phosphate-buffered saline; PFU, plaque-forming units; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome CoV-2; TGF, transforming growth factor; TMPRSS2, type II transmembrane serine protease; TNF, tumor necrosis factor; VEGF, vascular epithelial growth factor
Graphical abstract
See Covering the Cover synopsis on page 1435; See editorial on page 1467.
What You Need to Know.
Background and Context
Gastrointestinal manifestations have been increasingly reported in COVID-19 patients. However, the roles of the GI tract in SARS-CoV-2 pathogenesis are not fully understood.
New Findings
Single route of intragastric or intranasal inoculation with SARS-CoV-2 leads to dysfunctions in both respiratory and digestive systems.
Limitations
Further investigation of the microbiome and immune cells in respiratory and digestive systems is necessary to determine the mechanisms by which SARS-COV-2 infection leads to pneumonia and GI dysfunction.
Impact
This study indicates GI tract plays an unneglectable role in SARS-CoV-2 pathogenesis and transmission, which is an important guideline for treatment and prevention of COVID-19.
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported in December 2019 and has spread globally.1 , 2 Clinical syndromes caused by SARS-CoV-2 infection include acute respiratory distress, severe lung injury, multiple organ failure, and even death.2 , 3 Although respiratory symptoms are predominant among clinical syndromes of COVID-19, 20%–50% of patients with COVID-19 experience gastrointestinal (GI) manifestations such as abdominal pain, vomiting, nausea, and diarrhea.4, 5, 6 Patients in the intensive care unit have a higher frequency of abdominal pain than patients in the general care unit.7 The genome of SARS-CoV-2 is approximately 80% identical to that of severe acute respiratory syndrome coronavirus (SARS-CoV),4 but the GI symptoms of SARS-CoV-2 are more common and severe than those of SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV).8 , 9 Moreover, SARS-CoV-2 RNA has been detected in patient stool10, 11, 12 and rectal swabs, even after nasal and throat swab results became negative.8 , 12 , 13 Recently, infectious SARS-CoV-2 was isolated from the stool specimen of a patient with COVID-19 with diarrhea.14 These observations raise concerns that the fecal-oral route may be another potential transmission mode of SARS-CoV-2.
SARS-CoV-2 enters human host cells using the same receptor, angiotensin-converting enzyme 2 (ACE2), as SARS-CoV, and the spike glycoprotein (S) is processed by type II transmembrane serine protease (TMPRSS2) to enter the cytoplasm of host cells.15, 16, 17, 18, 19 ACE2 and TMPRSS2 are highly expressed in pulmonary alveolar epithelial type II and ciliated cells.20 Therefore, SARS-CoV-2 can actively replicate in upper respiratory tract tissues, suggesting that the respiratory tract is the key route of transmission.15 However, both of these proteins are also highly expressed in the digestive system, including in intestinal epithelial cells (IECs) and cholangiocytes.21 In fact, an increasing number of reports have shown that SARS-CoV-2 productively replicates in human and bat small intestinal enteroids,14 , 22 , 23 implying that the GI tract plays an important role in SARS-CoV-2 transmission. Clinical signs are the results of SARS-CoV-2 infection. However, it is very difficult to determine the initial infection routes of patients with COVID-19, which is critical for the elucidation of the pathogenesis of COVID-19. Even though a couple of animal models are established to recapitulate COVID-19, the mechanisms of SARS-CoV-2 infection are not completely clear because of the variation of viral inoculation routes.
Here, using a single route of viral challenge in a nonhuman primate (NHP) model of COVID-19, we sought to elucidate how the GI tract is involved in SARS-CoV-2 infection and causes COVID-19. To our knowledge, we show direct evidence, for the first time, that intranasal inoculation with SARS-CoV-2 caused pneumonia and GI manifestations on the basis of our established rhesus monkey model. Notably, intragastric challenge with SARS-CoV-2 also led to respiratory and digestive dysfunction. These results indicate that the GI tract plays a central role in SARS-CoV-2 pathogenesis.
Materials and Methods
Ethics and Biosafety Statement
All animal procedures in this article were approved by the Institutional Animal Care and Use Committee of Institute of Medical Biology, Chinese Academy of Medical Science (ethics number DWSP202002001). All experimental animals were male rhesus macaques that were provided by our Experimental Animal Department. All animal experiments were performed in the high-level biosafety facility of National Kunming High-level Biosafety Primate Research Center. All animals were housed with 1 animal per negative-pressure cage in a controlled environment (12-hour daylight cycle and lights off at 8 PM) with free access to food and water.
Virus Amplification and Identification
SARS-CoV-2 was obtained from the Center of Disease Control of Guangdong Province, China. Viruses were amplified in Vero E6 cells, purified, and concentrated with an ultrafilter system with a 300-kDa module (Millipore US). SARS-CoV-2 was confirmed via reverse-transcription polymerase chain reaction (RT-PCR), sequencing, and transmission electronic microscopy, titrated via a plaque assay (107 plaque-forming units [PFU]/mL).
Animal Experimental Procedures
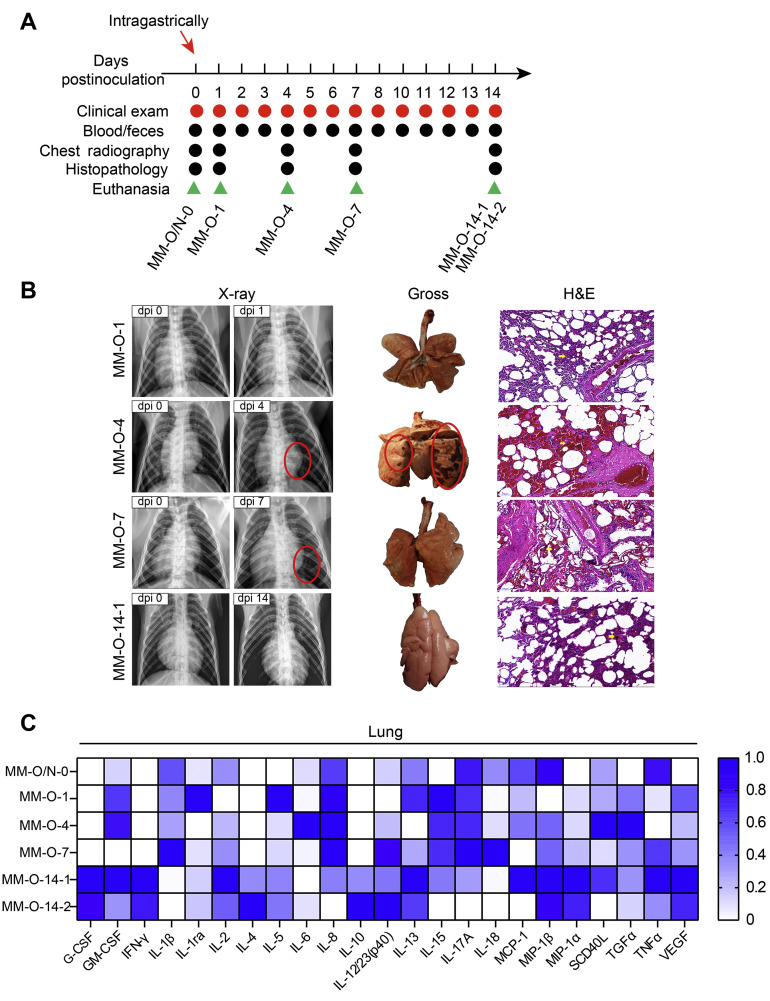
Animal information is detailed in Supplementary Table 1. Eleven rhesus monkeys were used for 2 infection experiments in this study. Animals were randomly divided into 3 groups: phosphate-buffered saline (PBS) treatment (MM-O/N-0), intragastric inoculation with SARS-CoV-2 (MM-O-1/4/7/14), and intranasal inoculation with SARS-CoV-2 (MM-N-1/4/7/14). The numbers 0, 1, 4, 7, and 14 represent the day of necropsy postviral inoculation. To reduce the number of experimental animals, PBS treatment was used as a shared control for both the intragastric and intranasal inoculation groups. Rhesus monkeys were intragastrically or intranasally challenged with 1 mL of 1 × 107 PFU SARS-CoV-2 or with PBS via an intragastric or intranasal route. We performed a gavage for intragastric inoculation. Briefly, the animals were anesthetized and laid supine on the operating table. An approximately 20-cm length of silicone rubber gastric tube (3.3 mm/10F) was slowly inserted into the esophagus. A syringe at the end of the gastric tube was drawn back to test whether the tube reached the stomach. If no air was drawn out, the gastric tube had reached the stomach via the esophagus. At this time, 107 PFU SARS-CoV-2 in another syringe was pushed into the stomach through the gastric tube, and then at least 20 mL of air in another syringe was pushed into the tube to ensure that all SARS-CoV-2 reached the stomach. For the intranasal route, animals were challenged with 500 μL of SARS-CoV-2 (107 PFU/mL) for each nostril. As outlined in Figures 1 A and 2 A , clinical signs (Supplementary Table 2), weight, and rectal temperature were recorded after virus inoculation. At the same time, we collected nasal, throat, and anal swabs and feces for further analysis. At the indicated timepoints, animals from each group were anesthetized with ketamine, and chest radiographs were recorded; animals were then killed, and necropsy was performed. At each timepoint, monkeys with severe chest radiography signs were chosen for necropsy and evaluation of the gross and histologic changes of tissues. Blood and GI contents were collected for further analysis.
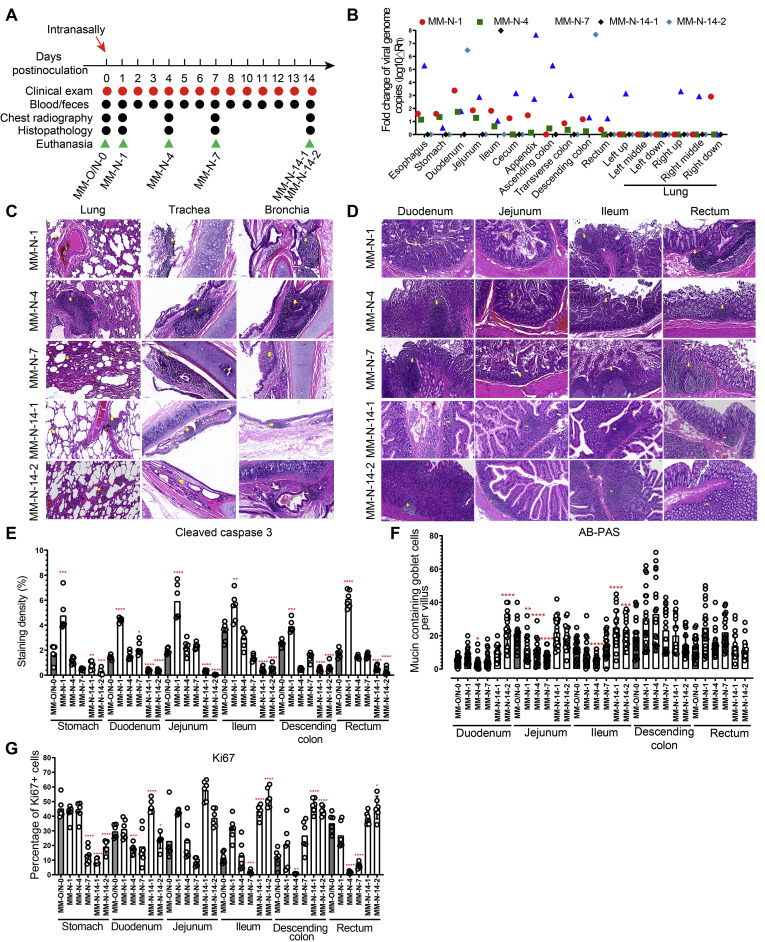
Figure 1.
Intranasal challenge with SARS-CoV-2 caused both pulmonary and enteric infections in rhesus monkeys. (A) Experimental design. Six rhesus monkeys were used for this experiment. Five animals were intranasally challenged with 1 mL of 107 PFU SARS-CoV-2, followed by clinical examinations, sampling, chest radiography, necropsy, and histopathologic analysis at the timepoints indicated. At 0 dpi, the animal MM-O/N-0 was necropsied as a control for both the intranasal and intragastric routes to reduce the number of animals. The animals MM-N-1, MM-N-4, MM-N-7, MM-N-14-1, and MM-N-14-2 were dissected on 1, 4, 7, 14, and 14 dpi, respectively. (B) Virus loads in tissue samples from the respiratory and digestive systems were measured by qRT-PCR. (C) Histopathology of respiratory tissues was analyzed after H&E staining after intranasal inoculation. (D) Histopathology of digestive tissues was analyzed after H&E staining after intranasal inoculation. (E) Cleaved caspase-3, as a marker for apoptosis, was evaluated by the IHC staining of digestive tissues. (F) The integrity of the digestive barrier was evaluated by AB-PAS staining after intranasal inoculation. (G) The marker for cell proliferation Ki67 was analyzed by the IHC staining of digestive tissues.
Figure 2.
Intragastric challenge with SARS-CoV-2 caused pneumonia in rhesus monkeys. (A) Experimental scheme. Rhesus monkeys were intragastrically challenged with 1 mL of 1 × 107 pfu SARS-CoV-2. Clinical signs were recorded every day after virus inoculation. At the same time, blood and fecal samples were collected for further analysis. The animals MM-O-1, MM-O-4, MM-O-7, MM-O-14-1/2 were necropsied on 1, 4, 7, and 14 dpi, respectively. (B) Pulmonary changes after infection. At left, chest radiographs (red circles indicate areas of nodular lesions caused by pulmonary infiltrates); in the middle, gross lesions of the lungs (lesions are circled in red); and at right, histopathology of the lung. One representative animal (MM-O-14-1) on 14 dpi is shown here because 2 animals had similarly mitigatory manifestations. (C) Twenty-three inflammatory cytokines were analyzed in lung tissue lysates after intragastric challenge with SARS-CoV-2. The highest level of cytokine, set as 1, is used to normalize other samples within 1 column, where the color density represents the relative level of the cytokines.
Viral RNA Extraction and Quantitative Reverse-Transcription Polymerase Chain Reaction
A total of 400 μL of a TRIzol suspension of swab samples or 20–50 mg of tissue samples were used for RNA extraction by using a Direct-zol RNA Miniprep Extraction Kit (Zymo Research, catalog no. R2052) according to the manufacturer’s instructions. Then, 50 μL of DNase/RNase-free water were used to elute RNA. Quantitative RT-PCR (qRT-PCR) was used to quantify the viral genome using TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific), and SARS-CoV-2 RNA was used as a standard curve. RT-PCR was performed on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) with the following conditions: 25°C for 2 minutes, 50°C for 15 minutes, 95°C for 2 minutes, and 40 cycles at 95°C for 5 seconds and 60°C for 31 seconds. Primers and probes specific for the NP gene were synthesized according to sequences reported by the Chinese Center for Disease Control and Prevention: target-2-F, GGGGAACTTCTCCTGCTAGAAT; target-2-R, CAGACATTTTGCTCTCAAGCTG; and target-2-P, 5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′. The copy number of the NP gene was calculated and expressed as log10 according to the standard curve of viral RNA to quantitate the genomic RNA of SARS-CoV-2 in the GI contents. For GI tissues, the genomic RNA of SARS-CoV-2 was also determined via the NP gene; however, the copy number was calculated via the comparative cycle threshold ΔΔCt) method, normalized to the housekeeping gene GAPDH, and then expressed as log10.
Virus Isolation and Transmission Electron Microscopy
Vero E6 cells were cultured in Dulbecco’s modified Eagle medium with 10% fetal bovine serum. Viral RNA–positive fecal specimens were used to make a PBS suspension, followed by filtration with a 0.22-μm syringe filter and dilution (1:10) with Dulbecco’s modified Eagle medium containing 2% fetal bovine serum and antibiotics. Cells were infected at 37°C for 1 hour. The inoculum was removed and replaced with fresh culture medium. The cells were incubated at 37°C, and cytopathic effects were observed daily. If there was no obvious cytopathic effect until 6 days postinfection (dpi), the cells and supernatant were scraped up and freeze-thawed once before the second-round passage. The culture supernatant was analyzed for viral RNA by qRT-PCR and transmission electron microscopy.
Histopathologic Analysis
Tissues of a proper size were fixed in 10% neutral-buffered formalin for 3–7 days. Then, 5-μm sections were prepared for H&E staining and histopathologic analysis.
Immunohistochemical Analysis
For immunohistochemical (IHC) staining, tissue sections were incubated with the following antibodies: cleaved caspase-3 (1:500; catalog no. GB13436, Servicebio), Ki67 (1:200; catalog no. GB13030-M, Servicebio), and CD68 (1:300; catalog no. GB13067-M-2, Servicebio). The stained sections were scanned and analyzed on a 3-dimensional (3D) HISTECH system.
For the analysis of Ki67, the brown-yellow nuclei were used as the standard to identify the Ki-67–positive cells via Image Pro Plus 6.0 software. The blue nuclei were identified as negative cells. The percentage of Ki67-positive cells is expressed as the ratio of the number of positive cells to the total number of cells (positive cells plus negative cells) multiplied by 100%.
Areal density was used for analysis of CD68 and cleaved caspase-3 staining. Six images were randomly taken for analysis in 1 section of each animal. When taking photos, we tried to make the tissue fill the whole field of vision and ensured that the background of each photo was consistent. The brown yellow was used as the standard to define the positive staining via Image Pro Plus 6.0 software. The integrated optical density and the pixel area were measured in each photo. The areal density was calculated as integrated optical density divided by pixel area.
In Situ Hybridization of Virus RNA
The hybridization of viral RNA was performed on formalin-fixed, paraffin-embedded sections according to the manufacturer’s recommended procedures (RNAscope Multiplex Fluorescent v2 [Advanced Cell Diagnostics]) with some modifications. The probes were specific for analysis of the S gene of SARS-CoV-2 in NHPs (RNAscope Probe-V-nCoV2019-S, ACD, catalog no. 848561, targeting the region of 21631–23303 [NC_045512.2]). The stained sections were scanned and analyzed on a 3D HISTECH system.
Alcian Blue and Periodic Acid–Schiff Staining
Formalin-fixed, paraffin-embedded sections were stained according to the manufacturer’s recommended procedure for an Alcian blue and periodic acid–Schiff (AB-PAS) staining kit (catalog no. G1285, Solarbio). The stained sections were scanned and analyzed on a 3D HISTECH system. Twenty crypts in 1 section of each animal were randomly selected for counting goblet cells in 1 crypt.
Electron Microcopy
Cells infected with SARS-CoV-2 were scraped off culture dishes and harvested by centrifugation at 500 g for 5 minutes. The supernatant was removed without disrupting the pellet, followed by the gentle and slow addition of 2.5% glutaraldehyde without resuspending the pellet. Fixation was performed at 4°C overnight. The next day, cells were embedded in epoxy resin, and ultrathin sections were made and stained with 2% uranium acetate and lead citrate. Slides were imaged under an electron microscope.
Multiplex Assay of Inflammatory Cytokines
A MILLIPLEX MAP Non-Human Primate Cytokine Magnetic Bead Panel–Immunology Multiplex Assay (Millipore) was performed according to the manufacturer’s protocol on a Bio-Plex machine (Bio-Rad). The inflammatory cytokines in this panel were interleukin (IL) 1β, IL4, IL5, IL6, IL8/CXCL8, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon gamma (IFNγ), IL1ra, IL2, IL10, I-12/23 (p40), IL13, IL15, IL17A/CTLA8, MCP-1/CCL2, MIP-1β/CCL4, MIP-1α/CCL3, sCD40L, transforming growth factor (TGF) α, tumor necrosis factor (TNF) α, vascular endothelial growth factor (VEGF) A, and IL18.
Statistical Analysis
A Student t test was performed to compare differences between the 2 groups. A P value of <.05 was considered statistically significant, and significant values are denoted with asterisks.
Results
Intranasal Inoculation With Severe Acute Respiratory Syndrome Coronavirus 2 Leads to Pathologic Damages in Both the Respiratory and Digestive Systems
GI manifestations are frequently reported in patients with COVID-19. However, there is no direct evidence that shows the relationship between digestive dysfunction and pneumonia caused by SARS-CoV-2 infection. The single route of intranasal challenge with SARS-CoV-2 in rhesus monkeys used in this study is outlined in Figure 1 A. On dpi 1, 4, 7, and 14, no significant changes of body weight and body temperature were observed (Supplementary Table 3). During the 2-week period of the experiment, we could not observe diarrhea and other manifestations (Supplementary Tables 4 and 5). Viral RNA was detectable in both respiratory and digestive tissues of rhesus monkeys after the intranasal inoculation of SARS-CoV-2 (Figure 1 B).
SARS-CoV-2 infections in the respiratory system caused histopathologic changes in the lung, trachea, and bronchia, including the infiltration of inflammatory cells, congestion and edema of the mucous membrane, exfoliation of epithelial cells, and vascular wall thickening (Figure 1 C). Histopathologic damage in the digestive system included the infiltration of inflammatory cells and exfoliation of mucosal epithelium in the stomach, duodenum, jejunum, ileum, descending colon, and rectum (Figure 1 D). Immunohistochemical staining of cleaved caspase-3 and Ki67 showed that intranasal infection with SARS-CoV-2 significantly induced the expression of cleaved caspase 3 on 1 dpi (Figure 1 E and Supplementary Figure 1 A) and inhibited the expression of Ki67 in the GI tract on 4 and 7 dpi (Figure 1 G and Supplementary Figure 2 A). Further AB-PAS staining for mucin showed that the number of mucin-containing goblet cells was significantly lower in the duodenum, jejunum, and ileum of infected animals at the earlier stage of infection than in uninfected animals (Figure 1 F and Supplementary Figure 3). However, on 14 dpi, most of the infected GI segments tended to recover, which was supported by less severe pathologic lesions, decreased cleavage of caspase-3, increased expression of Ki67, and elevated number of goblet cells (Figure 1 C–G). These results indicated that intranasal inoculation with SARS-CoV-2 caused infection and pathology in both the respiratory and digestive tracts, including gut barrier impairment.
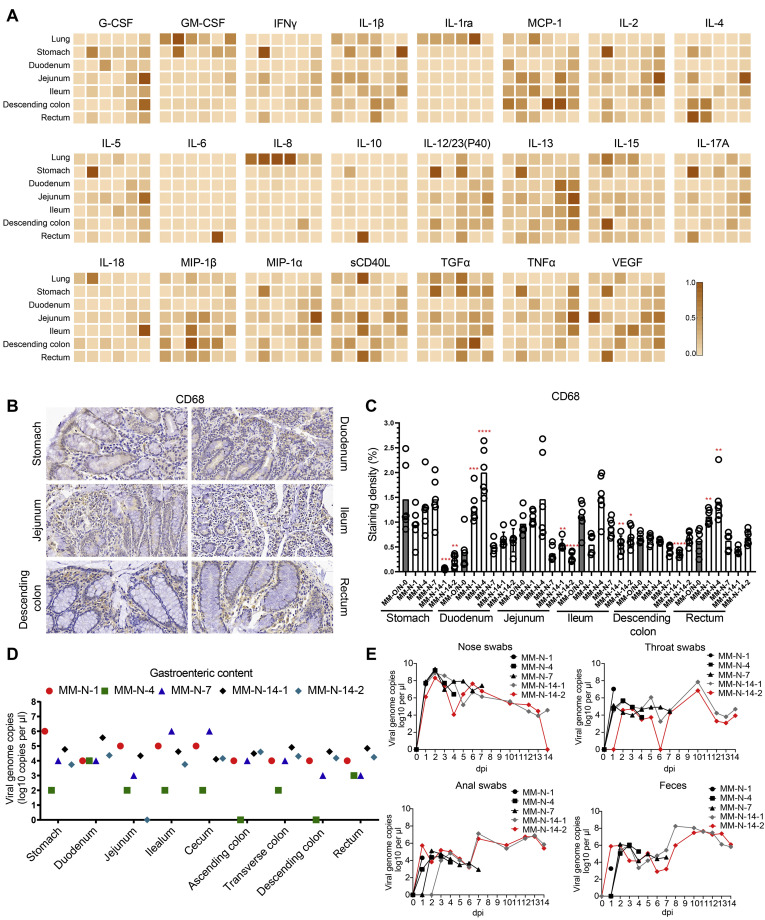
Inflammatory cytokines were further evaluated in lung and GI tissues after intranasal infection. Among the 23 inflammatory cytokines analyzed, 10 cytokines (GM-CSF, IL1ra, MCP-1, IL8, IL12/23 (p40), IL15, IL18, sCD40L, TGFα, and VEGF) were induced in respiratory tissues by SARS-CoV-2 infection. In digestive tissues infected by SARS-CoV-2, 21 cytokines (all analyzed cytokines except IL1ra and IL8) were up-regulated after infection (Figure 3 A). As the infection progressed, inflammatory cytokines were induced in more GI segments. On 14 dpi, some anti-inflammatory or protective cytokines increasingly expressed in the GI segments, such as G-CSF, IL4, IL6, IL13, IL18, MIP-1β and TNF-α. IHC staining showed that CD68 expression significantly increased in the duodenum and rectum at the earlier stage of infection and came back to the normal level on 14 dpi (Figure 3 B and C). Together, these results indicated that intranasal inoculation with SARS-CoV-2 activated macrophages in the digestive system to secrete inflammatory cytokines that induced tissue damages.
Figure 3.
Inflammatory responses in the digestive system and virus shedding in rhesus monkeys intranasally challenged with SARS-CoV-2. (A) Multiplex assay of inflammatory cytokines in tissue lysates. For each panel, MM-O/N-0, MM-N-1, MM-N-4, MM-N-7, and MM-N-14 are shown in 1 column from left to right. The highest level of cytokine, set as 1, is used to normalize other samples within 1 panel, where the color density represents the relative levels of cytokines. (B) The macrophage surface marker CD68 was analyzed by the IHC staining of digestive tissues. Representative images from 1 intranasally inoculated animal are shown here. (C) This plot shows the quantitative analysis of macrophages in panel B. Compared with the control MM-O/N-0, significant differences in the digestive tissues of the other animals are indicated by asterisks (∗P < .05, ∗∗P < .01, ∗∗∗P < .005, ∗∗∗∗P < .001). (D) Viral loads in the digestive tract contents were determined by qRT-PCR after intranasal inoculation. (E) Virus shedding was monitored by qRT-PCR every day after intranasal inoculation.
Finally, we have detected viral RNA in the contents of the GI tract, nasal swabs, throat swabs, anal swabs and fecal samples after intranasal inoculation (Figure 3 D and E). The results in the TCID50 assay suggested that viruses in tissues and GI contents are infectious (Supplementary Figure 4 A, B, D, E).
Together, these results suggested that intranasal inoculation of SARS-CoV-2 results in pathologic damage to both the respiratory system and digestive system and that viruses were released from the infected tissues, which caused virus shedding.
Intragastric Inoculation With Severe Acute Respiratory Syndrome Coronavirus 2 Causes Respiratory Manifestations in Rhesus Monkeys
Then, we hypothesized that GI inoculation with SARS-CoV-2 could result in infections in digestive tissues or even in respiratory tissues. To test this hypothesis, single-route intragastric inoculation with SARS-CoV-2 was performed to infect 5 rhesus monkeys, as shown in Figure 2 A. One milliliter of 107 PFU SARS-CoV-2 was delivered directly into the stomach, followed by the observation of clinical signs and sampling. Chest radiographs showed nodular pulmonary infiltrations in the lung on 4 and 7 dpi but not on 14 dpi. To characterize the early events after intragastric SARS-CoV-2 inoculation, rhesus monkeys were killed on days 1, 4, and 7 after challenge, respectively. Gross lesions of the lungs showed mild hyperemia on 1 dpi, massively focal hyperemia on 4 dpi, and consolidation on 7 dpi. Histologically, vascular congestion and wall thickening, interstitial hyperplasia, and inflammatory infiltration were observed in the lungs of rhesus monkeys on 1 dpi. More severe lesions, including alveolar hemorrhage, alveolar dilation, vasodilation, hyperemia, infiltration, and aggregation of peripheral lymphocytes, were detected on 4 dpi, followed by alveolar edema, interstitial edema, lymphocyte aggregation, and alveolar cavity expansion on 7 dpi. These pathologic changes were slightly mitigated on 14 dpi (Figure 2 B). Twenty-three inflammatory cytokines were analyzed in lung tissue lysates after intragastric challenge with SARS-CoV-2. Compared with the lung from the uninfected monkey, 15 inflammatory cytokines (GM-CSF, IL1β, IL1ra, IL5, IL6, IL12, IL13, IL15, IL17A, IL18, MIP-1α, sCD40L, TGFα, TNF-α, and VEGF) were induced by the early infection (on 1, 4, and 7 dpi) (Figure 2 C). Then, some anti-inflammatory or protective cytokines increased (G-CSF, IFNγ, IL2, IL4, IL10, and MIP-1α), and some inflammatory cytokines decreased (IL1β, IL1ra, IL5, IL6, IL8, IL15, IL17A) on 14 dpi. These results suggested that intragastric inoculation with SARS-CoV-2 led to pneumonia, possibly via the induction of inflammatory cytokines.
The Fecal-Oral Route Could Be a Potential Transmission for Severe Acute Respiratory Syndrome Coronavirus 2 Infection
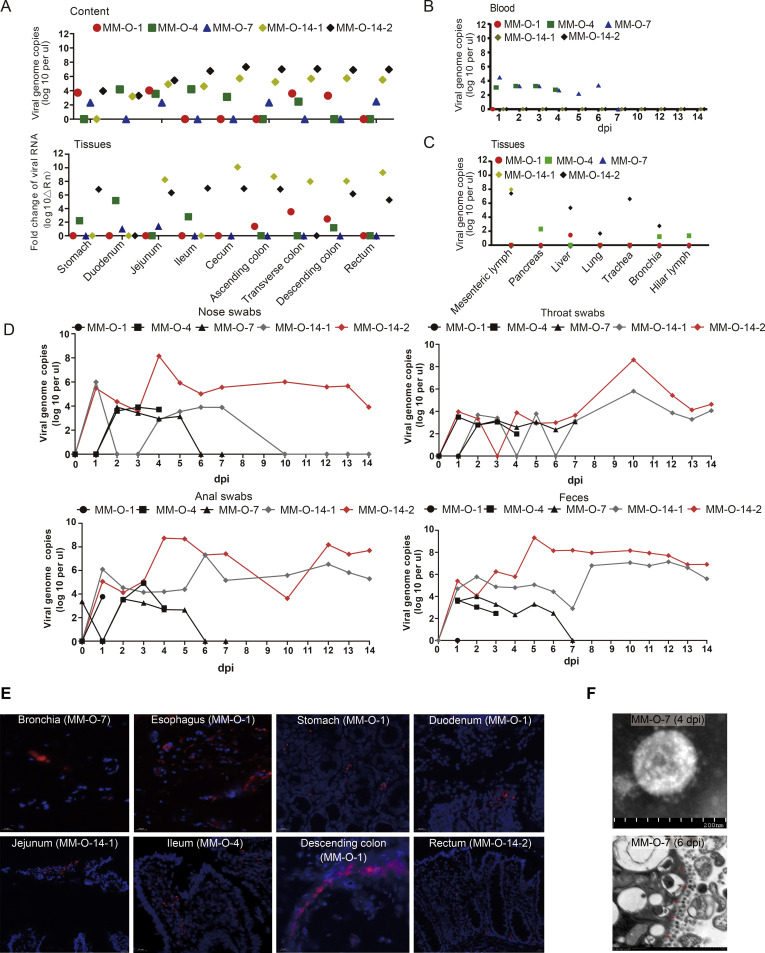
To examine the dynamics of SARS-CoV-2 after intragastric inoculation, we quantitated viral genomic RNA in samples from respiratory tissues, digestive tissues, GI contents, and blood. We had detected viral RNA in digestive tissues and their contents (Figure 4 A), some pulmonary tissues (but not lung tissue), mesenteric lymph, and pancreatic and hepatic tissues (Figure 4 C). The TCID50 assay suggested that the majority of viruses detected in these tissues and the gastroenteric contents were infectious (Supplementary Figure 4 C–E). In situ hybridization confirmed the existence of viral RNA in some tissues (Figure 4 E). Furthermore, viral RNA was also detectable in blood on 1, 4, and 7 dpi but not 14 dpi (Figure 4 B). More importantly, viruses in the alimentary tract were discharged from the nose, throat, and anus, which was evidenced by the presence of viral RNA in these swab samples (Figure 4 D). In fact, we reisolated viruses from 1 fecal sample of MM-O-7, which was confirmed by electronic microscopy (Figure 4 F) and TCID50 assay (105/mL). In vitro, the human epithelial cells Caco-2 are susceptible to SARS-CoV-2 (Supplementary Figure 5). These results indicated that the fecal-oral route could be a potential transmission for SARS-CoV-2 infection.
Figure 4.
Dynamic analysis of viruses in rhesus monkeys after intragastric inoculation with SARS-CoV-2. (A) Viral genomic RNA was detected with qRT-PCR in digestive tissues (top) and their contents (bottom) from several parts of the alimentary tract and (B) blood after intragastric inoculation. (C) Viral loads were measured via qRT-PCR in other tissues and pulmonary tissues after intragastric inoculation. (D) Virus shedding from the nose, throat, and feces was monitored by qRT-PCR. (E) Viral genomic RNA was confirmed in situ by RNAscope technology, and representative pictures are shown. The RNAscope results are summarized in Supplementary Table 6. (F) SARS-CoV-2 was isolated from feces and identified by electron microscopy.
Intragastric Severe Acute Respiratory Syndrome Coronavirus 2 Infection Results in Inflammation and Intestinal Barrier Damage
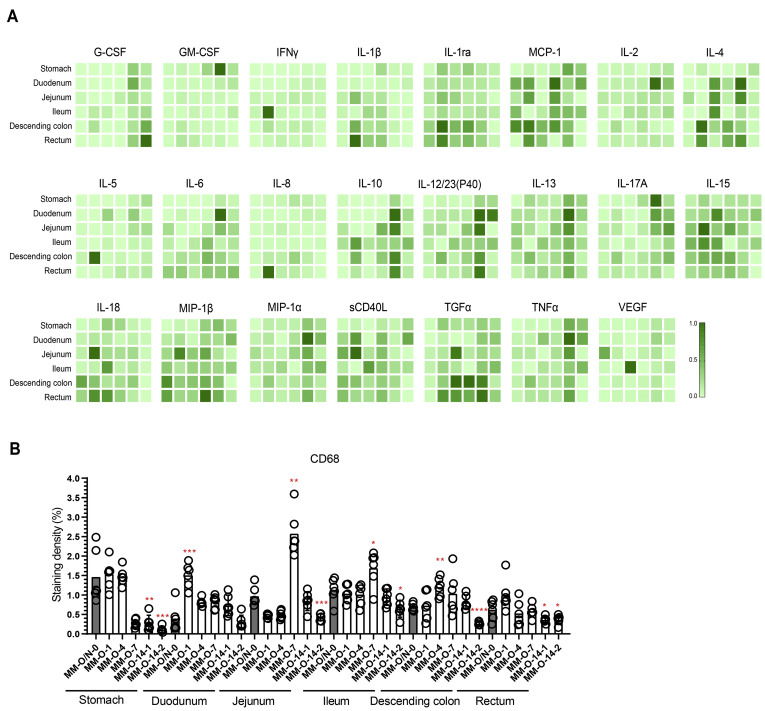
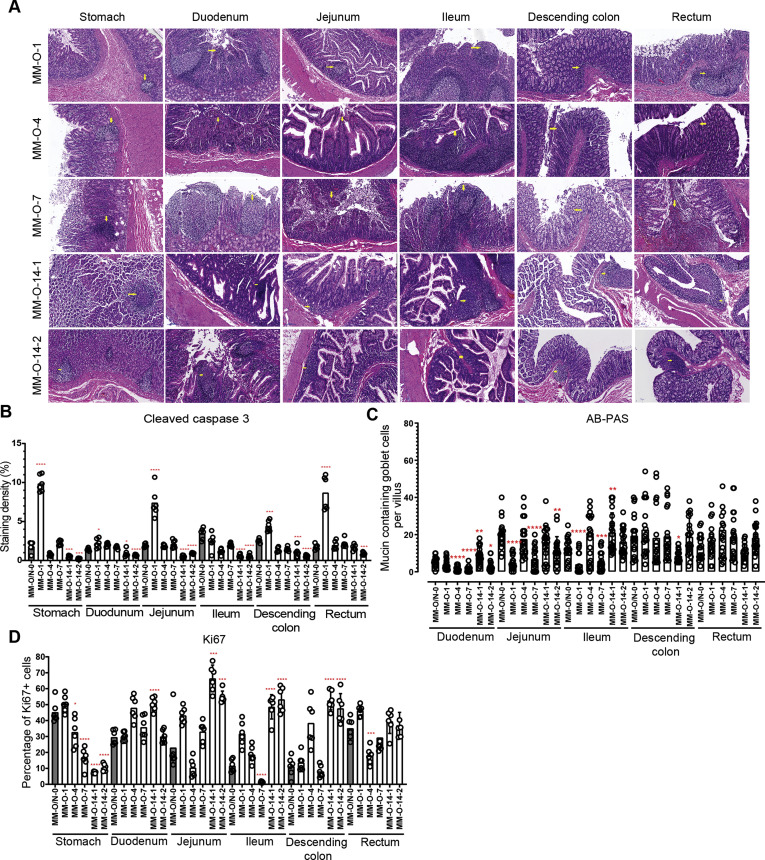
To examine the host response to intragastric SARS-CoV-2 infection, inflammatory cytokines were analyzed in GI tissues. Compared with the uninfected animal, the 23 inflammatory cytokines assayed in each segment of the GI tract increased to varying degrees, especially in the middle and terminal segments of the digestive tract from SARS-CoV-2–infected animals (Figure 5 A). On 14 dpi, some anti-inflammatory cytokines (G-CSF, IL2, IL10, IL13 and MIP-1α) were induced and inflammatory cytokines (IFNγ, IL1β) were inhibited. Next, we evaluated the expression of CD68, which is a marker for macrophages. Increased CD68 expression was observed in the duodenum, jejunum, ileum, and descending colon (Figure 5 B), consistent with the expression of inflammatory cytokines (Figure 5 A). To further investigate the immunopathologic effects of these inflammatory responses, we necropsied 5 monkeys on 1, 4, 7, and 14 dpi, as outlined in Figure 4 A. Histopathologic analysis showed various degrees of pathologic changes in the stomach, duodenum, jejunum, ileum, descending colon, and rectum. These pathologic changes included the infiltration of inflammatory cells and loss of mucosal epithelium (Figure 6 A), which was similar to the changes induced by intranasal inoculation (Figure 1 D). The IHC staining of cleaved caspase-3 indicated that apoptosis was observed in the stomach, jejunum, descending colon, and rectum on 1 dpi and then was gradually mitigated (Figure 6 B and Supplementary Figure 1 B). As intestinal goblet cells secrete mucins to form a mucus layer for the host defense barrier, we determined the number of mucin-containing goblet cells by the AB-PAS staining of acidic and neutral mucins. The decreased number of mucin-containing goblet cells was observed in the GI segments of duodenum, jejunum, and ileum from infected monkeys (Figure 6 C and Supplementary Figure 6). Thus, intragastric SARS-CoV-2 infection caused dysfunction of the mucus barrier by affecting the goblet cells. The cell proliferation marker Ki67 also significantly inhibited in the stomach, ileum, and rectum in response to SARS-CoV-2 infection (Figure 6 D and Supplementary Figure 2 B). On 14 dpi, most of the GI segments showed the normal levels of cleaved caspase-3, Ki67, and goblet cells (Figure 6 B–D). These results indicated that intragastric inoculation with SARS-CoV-2 impaired the GI barrier at the early stage of infection.
Figure 5.
Inflammatory responses in the digestive system were induced by SARS-CoV-2 infection. (A) Multiplex assay of inflammatory cytokines in digestive tissues. For each panel, MM-O/N-0, MM-O-1, MM-O-4, MM-O-7, and MM-O-14 are shown in 1 column from left to right. The highest level of cytokine, set as 1, is used to normalize the other samples within 1 panel, where the color density represents the relative levels of cytokines. (B) The macrophage marker CD68 was analyzed by IHC staining. This plot shows the quantitative analysis of macrophages. Compared with the control MM-O/N-0, significant differences in the digestive tissues of the other animals are indicated by asterisks (∗P < .05, ∗∗P < .01, ∗∗∗P < .005, ∗∗∗∗P < .001).
Figure 6.
Intragastric infection with SARS-CoV-2 resulted in damage to the alimentary tract. (A) Histopathologic analysis after H&E staining was performed on every part of the digestive system after intragastric inoculation. (B) Cleaved caspase-3, as a marker for apoptosis, was evaluated by the IHC staining of digestive tissues. (C) The integrity of the digestive barrier was evaluated by AB-PAS staining. (D) IHC staining using the marker Ki67 was used to evaluate the proliferation of epithelial cells of the alimentary tract after SARS-CoV-2 infection. For B–D, compared with the control MM-O/N-0, significant differences in each digestive tissue of other animals are indicated by asterisks (∗P < .05, ∗∗P < .01, ∗∗∗P < .005, and ∗∗∗∗P < .001).
Discussion
The COVID-19 pandemic is still ongoing since the first report of SARS-CoV-2 infection in December 2019. However, important issues related to public health, such as transmission and infection routes, remain to be fully elucidated. Because clinical manifestations of SARS-CoV-2 are mainly respiratory symptoms, airway exposure is assumed to be one of the major infection routes that contributes to human SARS-CoV-2 infections.24 Expectedly, epidemiologic studies, viral biological assessments, and intestinal organoid infection models all indicate that humans may also be infected with SARS-CoV-2 through the GI tract.4 , 6 , 13 , 22 , 25, 26, 27, 28 However, direct evidence for the involvement of the GI tract in the pathogenesis of COVID-19 is insufficient. On the basis of our established NHP model of COVID-19, a single route of viral inoculation (intranasal or intragastric) was used to investigate the roles of the GI tract in SARS-CoV-2 infection. We found that a single route of intranasal or intragastric inoculation with SARS-CoV-2 caused infection and pathologic changes in both respiratory and alimentary tissues, as well as virus shedding from the digestive tract. Inflammatory cytokines induced by SARS-CoV-2 infection may be the connection for viral pathogenesis between the respiratory and digestive systems. To our knowledge, this is the first direct evidence that the GI tract plays important roles in SARS-CoV-2 infection and transmission.
SARS-CoV-2 intestinal infection was suggested in several studies of patients with COVID-19, NHPs, and hACE2 transgenic mouse models, in which an increasing viral load was observed in stomachs, intestines, colons, and rectums.29, 30, 31, 32, 33 However, the infection route in these cases was either intratracheal, intranasal, a combination of multiple inoculations in animals, or unclear in patients. Thus, these studies could not provide direct evidence of GI infection in vivo. Here, we characterized the GI infection of SARS-CoV-2 in rhesus monkeys after intranasal or intragastric inoculation. Clinically, respiratory symptoms may not be the initial presentations of patients with SARS-CoV-2.5 , 6 Respiratory infection was documented as a subsequent manifestation in some patients with SARS-CoV-2 whose initial symptoms were diarrhea and fever.34 However, in this study, we still could not determine which system was first infected after intranasal administration of SARS-CoV-2. These viruses, after intranasal inoculation, traveled to the GI tissues, possibly via the lymphatic system or bloodstream35 and bound to the essential receptor ACE2. Although the 2 routes of SARS-CoV-2 inoculation in this study induced both pneumonia and GI tract dysfunction, there were at least 2 major differences between them. Intranasal inoculation with SARS-CoV-2 led to a higher viral load in respiratory and digestive tissues than intragastric inoculation. A possible explanation for this observation is that most SARS-CoV-2 that is intragastrically inoculated could be killed by gastric acid or other factors in the harsh GI environment.9 Nevertheless, viruses protected by mucus or GI contents remained alive, and their replication led to infection, inflammation, and pathologic damage in the digestive system. In addition, inflammatory cytokines in serum, particularly sCD40L, TGFα, IL15, and IL1ra, were differentially induced by intranasal and intragastric inoculation with SARS-CoV-2 (Supplementary Figure 7). sCD40L, IL-15, and IL-1ra were induced by intranasal inoculation with SARS-CoV-2. TGFα was likely induced by intragastric inoculation. TGFα has been reported to inhibit the secretion of gastric acid.36 It will be interesting to investigate whether intragastric inoculation with SARS-CoV-2 induces TGFα to allow viruses to escape from gastric acid. Therefore, the intranasal or intragastric inoculation with SARS-CoV-2 may lead to 2 distinct forms of pathogenesis.
Cytokine storms, but not the virus itself, are commonly recognized as a major pathogenic factor in SARS-CoV-2–induced pneumonia.37 In this study, intragastric inoculation with SARS-CoV-2 led to pneumonia, but we could not assay viral RNA in the lung. SARS-CoV-2 that was intragastrically delivered to the digestive tract induced a large amount of inflammatory cytokines (Figure 5 A) that overflowed to pulmonary tissues via the bloodstream or lymphatic system and caused inflammation,38 which might further expel viruses from the lung. This may explain why viruses are sometimes not detectable in the lung.
The GI barrier, which consists of different types of IECs, plays important roles in preventing the entry of pathogens. The rapid turnover of IECs due to the vigorous proliferation of epithelial progenitors is thought to be an important defensive mechanism that works by expelling pathogens, confining bacterial spreading, and localizing inflammation.39 The IHC staining of Ki67 and cleaved caspase-3 in this study indicated that SARS-COV-2 infection inhibited cellular proliferation and induced apoptosis in the GI tract. Mucin secreted by goblet cells, the first defense layer, is important for preventing pathogen invasion. The absence of mucin makes the host more susceptible to pathogens.40 Both routes of viral inoculation in this study decreased the number of mucin-containing goblet cells. These results indicated that SARS-CoV-2 infection inhibits the turnover of IECs and mucin and induces apoptosis to disrupt the intestinal barrier, causing further infection in multiple tissues. However, further investigation of the microbiome and immune cells underneath the epithelial layer are necessary to determine the mechanisms by which SARS-COV-2 infection leads to pneumonia and GI dysfunction.
Coronaviruses can infect human and animals. There are at least 17 species of coronaviruses found in nature. Diseases caused by coronavirus infection involve the respiratory, GI, and neurologic systems. The GI manifestations play important roles in the pathogenesis, transmission, and epidemiology of coronaviruses. It is known that there are 11 species of coronaviruses causing GI manifestations, including SARS-CoV and MERS-CoV.41 Clinical studies and animal models of COVID-19 indicate that SARS-CoV-2 could be an enteric coronavirus.30 , 33 , 42 , 43 In this study, the first evidence from the NHP model of COVID-19 showed the replication and pathogenesis of SARS-CoV-2 in the gastrointestinal tract. Viral receptor expressed in the gastrointestinal tract is the essential host factor for coronavirus pathogenesis, such as ACE2 for SARS-CoV and SARS-CoV-2 and DPP4 for MERS-CoV. The protein amino-peptidase N (APN) is the receptor for several enteric coronaviruses, including human coronavirus 229E, feline coronavirus serotype II, transmissible gastroenteritis virus, porcine epidemic diarrhea virus, and canine coronavirus genotype II.44 Enteric coronaviruses attach and enter the host cells via these receptors. Gastroenteritis is caused by virus direct damages,9 , 45 viremia, or systemic inflammation. Here, we found that SARS-CoV-2 infection leads to apoptosis and inflammation in the GI tract. Inflammatory factors induced by SARS-CoV-2 are a potential bridge for viral pathogenesis between the respiratory and GI systems. Interestingly, the results of this study indicate a possible recovery mechanism at the late stage of SARS-CoV-2 infection. In our monkey model, whether with intranasal or intragastric challenge, viral infection induced inflammatory cytokines and histopathologic lesions in the digestive system at the early stage (on 1, 4, and 7 dpi). After 2 weeks (on 14 dpi), anti-inflammatory or protective factors were increasingly expressed and resulted in recovery signs, such as the decreased cleavage of caspase-3, increases of cell proliferation and goblet cells, and improvement of pathologic lesions. Whether there is such a self-recovery mechanism in patients with COVID-19 needs further research.
COVID-19 is first well-known as a respiratory infectious disease. Therefore, wearing surgical masks is strongly recommended in effectively preventing SARS-CoV-2 transmission. However, SARS-CoV-2 is still reported to transmit in the population wearing masks, indicating the presence of other transmission routes. Environmental factors, in particular wastewater or sewage, play critical roles in the prevalence of some important diseases, such as polio. SARS-CoV-2 RNA fragments have been detected from patients’ feces and wastewater in several countries, including Italy,46 Spain,47 Australia,48 the Netherlands,49 the United States,50 , 51 France,52 and Pakistan.53 Studies in some countries indicate that viral RNA concentrations may correlate with and even predict COVID-19 cases.49, 50, 51, 52 These published data indicate that contact transmission and gastrointestinal infection are not to be ignored in controlling SARS-CoV-2 infection. Routine monitoring of wastewater or sewage is scheduled as part of the national surveillance of SARS-CoV-2 in the Netherlands,54 Germany,55 Australia, and New Zealand.56 The NHP model of COVID-19 in this study and others30 , 33 , 42 , 43 showed that infectious SARS-CoV-2 was excreted from feces after intranasal and intragastric inoculation, which provides experimental evidence that GI diseases and environmental factors play critical roles in controlling SARS-CoV-2 transmission.
Although we show the direct evidence of GI SARS-CoV-2 infection in an NHP model of COVID-19, there are 2 major limitations in the current study. Because of the limited NHP source, only 1 animal was used for each timepoint. Therefore, more animals (different sexes and ages included) are necessary for future study to fully explore the mechanism of GI involvement in SARS-CoV-2 infection. The second limitation of this study is that we could only speculate about the possible mechanism of respiratory and digestive involvement in SARS-CoV-2 infection based on our current studies of viral load, inflammatory cytokines, apoptosis, cell proliferation, and histopathologic lesions. Deep understanding of a comprehensive mechanism of SARS-CoV-2 infection needs further integrative studies via multiomics approaches, including transcriptomics, proteomics, metabolomics, microbiomics, and so on.
Acknowledgments
CRediT Authorship Contributions
Li Jiao, PhD (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Visualization: Equal; Writing – original draft: Lead); Haiyan Li, MSc (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Methodology: Equal); Jingwen Xu, MSc (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal); Mengli Yang, MSc (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Methodology: Equal); Chunxia Ma, MSc (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Methodology: Equal); Jingmei Li, MSc (Conceptualization: Equal; Data curation: Equal; Investigation: Equal; Methodology: Equal); Siwen Zhao, MSc (Investigation: Supporting; Methodology: Supporting); Haixuan Wang, MSc (Investigation: Supporting; Methodology: Supporting); Yun Yang, MSc (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal); Wenhai Yu, MSc (Investigation: Equal; Methodology: Equal); Junbin Wang, MSc (Investigation: Equal; Methodology: Equal); Jing Yang, MSc (Data curation: Equal); Haiting Long, BSc (Investigation: Equal; Methodology: Equal); Jiahong Gao, MSc (Investigation: Equal; Methodology: Equal); Kaiyun Ding, BSc (Investigation: Equal; Methodology: Equal)
Daoju Wu, BSc (Methodology: Supporting); Dexuan Kuang, PhD (Investigation: Equal; Methodology: Equal); Yuan Zhao, MSc (Investigation: Equal; Methodology: Equal); Jiansheng Liu, MSc (Methodology: Equal); Shuaiyao Lu, MSc (Conceptualization: Equal; Investigation: Equal; Methodology: Equal; Project administration: Lead; Supervision: Lead; Writing – review & editing: Lead); Hongqi Liu, PhD (Conceptualization: Equal; Investigation: Equal; Supervision: Equal; Writing – review & editing: Equal); Xiaozhong Peng, PhD (Conceptualization: Lead; Investigation: Lead; Project administration: Lead; Supervision: Lead; Writing – review & editing: Lead).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the National Key R&D Project of China (2020YFA0707602, 2020YFC0846400, 2020YFC0841100), CAMS Innovation Fund for Medical Sciences (2016-I2M-2-001, 2016-I2M-2-006, and 2020-I2M-CoV19-012), and Yunnan Key R&D Project (202003AC100003).
Author names in bold designate shared co-first authorship.
Note: To access the supplementary mater’ial accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.12.001.
Supplementary Material
References
- 1.Wang C., Horby P.W., Hayden F.G. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W.J., Ni Z.Y., Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L., Jiang X., Zhang Z. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 5.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan L., Mu M., Yang P. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holshue M.L., DeBolt C., Lindquist S. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J., Li C., Zhao G. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv. 2017;3(11) doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Openshaw P.J. Crossing barriers: infections of the lung and the gut. Mucosal Immunol. 2009;2:100–102. doi: 10.1038/mi.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y., Guo C., Tang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao F., Tang M., Zheng X. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Xu Y., Gao R. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J., Li C., Liu X. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 15.Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls A.C., Park Y.J., Tortorici M.A. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukassen S., Chua R.L., Trefzer T. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39(10) doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B., Ni C., Gao R. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771–775. doi: 10.1007/s13238-020-00718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamers M.M., Beumer J., van der Vaart J. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zang R., Castro M.F.G., McCune B.T. TMPRSS2 and TMPRSS4 mediate SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nardell E.A., Nathavitharana R.R. Airborne spread of SARS-CoV-2 and a potential role for air disinfection. JAMA. 2020;324:141–142. doi: 10.1001/jama.2020.7603. [DOI] [PubMed] [Google Scholar]
- 25.Aroniadis O.C., DiMaio C.J., Dixon R.E. Current knowledge and research priorities in the digestive manifestations of COVID-19. Clin Gastroenterol Hepatol. 2020;18:1682–1684. doi: 10.1016/j.cgh.2020.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du M., Cai G., Chen F. Multiomics evaluation of gastrointestinal and other clinical characteristics of COVID-19. Gastroenterology. 2020;158:2298–2301. doi: 10.1053/j.gastro.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gul F., Lo K.B., Peterson J. Meta-analysis of outcomes of patients with COVID-19 infection with versus without gastrointestinal symptoms. Proc (Bayl Univ Med Cent) 2020;33:366–369. doi: 10.1080/08998280.2020.1771164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X., Lian J.S., Hu J.H. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao L., Deng W., Gao H. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. Nat Rev Immunol. 2020;20:351. [Google Scholar]
- 30.Munster V.J., Feldmann F., Williamson B.N. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockx B., Kuiken T., Herfst S. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6494):1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao L., Deng W., Huang B. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 33.Sun S.H., Chen Q., Gu H.J. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han C., Duan C., Zhang S. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115:916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenner F., Bachmann P.A., Gibbs E.P.J. Pathogenesis: infection and the spread of viruses in the body. Vet Virol. 1987:133–152. [Google Scholar]
- 36.Rhodes J.A., Tam J.P., Finke U. Transforming growth factor alpha inhibits secretion of gastric acid. Proc Natl Acad Sci U S A. 1986;83:3844–3846. doi: 10.1073/pnas.83.11.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the ‘cytokine storm’ in COVID-19. J Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mjösberg J., Rao A. Lung inflammation originating in the gut. Science. 2018;359(6371):36–37. doi: 10.1126/science.aar4301. [DOI] [PubMed] [Google Scholar]
- 39.Ramanan D., Cadwell K. Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe. 2016;19:434–441. doi: 10.1016/j.chom.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarepour M., Bhullar K., Montero M. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2013;81:3672–3683. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding S., Liang T.J. Is SARS-CoV-2 also an enteric pathogen with potential fecal–oral transmission? A COVID-19 virological and clinical review. Gastroenterology. 2020;159:53–61. doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartman AL, Nambulli S, McMillen CM, et al. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and prolonged shedding of infectious virus from both respiratory and gastrointestinal tracts. Preprint. Posted online June 21, 2020. bioRxiv 2020:2020.06.20.137687. doi:10.1101/2020.06.20.137687 [DOI] [PMC free article] [PubMed]
- 43.Lu S., Zhao Y., Yu W. Comparison of nonhuman primates identified the suitable model for COVID-19. Sig Transduct Target Ther. 2020;5(1):157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung W.K., To K.F., Chan P.K. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rimoldi SG, Stefani F, Gigantiello A, et al. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. Preprint. Posted online May 5, 2020. medRxiv 2020:2020.05.01.20086009. https://doi:10.1101/2020.05.01.20086009
- 47.Randazzo W., Truchado P., Cuevas-Ferrando E. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed W., Angel N., Edson J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medema G., Heijnen L., Elsinga G. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 50.Peccia J, Zulli A, Brackney DE, et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. Preprint. Posted online June 12, 2020. medRxiv 2020:2020.05.19.20105999. https://doi:10.1101/2020.05.19.20105999
- 51.Wu F, Xiao A, Zhang J, et al. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Preprint. Posted online July 6, 2020. medRxiv 2020.06.15.20117747v1. https://doi:10.1101/2020.06.15.20117747
- 52.Wurtzer S, Marechal V, Mouchel JM, et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. Preprint. Posted online May 6, 2020. medRxiv 2020.04.12.20062679. https://doi:10.1101/2020.04.12.20062679
- 53.Sharif S, Ikram A, Khurshid A, et al. Detection of SARS-Coronavirus-2 in wastewater, using the existing environmental surveillance network: an epidemiological gateway to an early warning for COVID-19 in communities. Preprint. Posted online June 24, 2020. medRxiv 2020.06.03.20121426v2. https://doi:10.1101/2020.06.03.20121426
- 54.Dutch Water Sector Sewer surveillance part of Dutch national Covid-19 dashboard. https://www.dutchwatersector.com/news/sewer-surveillance-part-of-dutch-national-covid-19-dashboard
- 55.CNN Sewage could hold the key to stopping new coronavirus outbreaks. https://edition.cnn.com/2020/06/01/europe/germany-sewage-coronavirus-detection-intl/index.html Updated June 1, 2020.
- 56.Deere D., Sobsey M., Sinclair M. Historical context and initial expectations on sewage surveillance to inform the control of COVID-19. HealthStream, Water Research Australia. https://www.waterra.com.au/_r9779/media/system/attrib/file/2272/ HealthStream_Newsletter-97_FINAL.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.