Abstract
Aberrant activation of Wnt/β-catenin axis occurs in several gastrointestinal malignancies due to inactivating mutations of adenomatous polyposis coli (in colorectal cancer) or activating mutations of β-catenin itself [in hepatocellular carcinoma (HCC)]. These lead to β-catenin stabilization, increase in β-catenin/T-cell factor (TCF)–mediated transcriptional activation, and target gene expression, many of which are involved in tumor progression. While studying pharmaceutical agents that can target β-catenin in cancer cells, we observed that the plant compound berberine (BBR), a potent activator of AMP-activated protein kinase (AMPK), can reduce β-catenin expression and downstream signaling in HCC cells in a dose-dependent manner. More in-depth analyses to understand the mechanism revealed that BBR-induced reduction of β-catenin occurs independently of AMPK activation and does not involve transcriptional or post-translational mechanisms. Pretreatment with protein synthesis inhibitor cycloheximide antagonized BBR-induced β-catenin reduction, suggesting that BBR affects β-catenin translation. BBR treatment also antagonized mammalian target of rapamycin (mTOR) activity and was associated with increased recruitment of eukaryotic translation initiation factor 4E–binding protein (4E-BP) 1 in the translational complex, which was revealed by 7-methyl-cap–binding assays, suggesting inhibition of cap-dependent translation. Interestingly, knocking down 4E-BP1 and 4E-BP2 significantly attenuated BBR-induced reduction of β-catenin levels and expression of its downstream target genes. Moreover, cells with 4E-BP knockdown were resistant to BBR-induced cell death and were resensitized to BBR after pharmacological inhibition of β-catenin. Our findings indicate that BBR antagonizes β-catenin pathway by inhibiting β-catenin translation and mTOR activity and thereby reduces HCC cell survival. These also suggest that BBR could be used for targeting HCCs that express mutated/activated β-catenin variants that are currently undruggable.
SIGNIFICANCE STATEMENT
β-catenin signaling is aberrantly activated in different gastrointestinal cancers, including hepatocellular carcinoma, which is currently undruggable. In this study we describe a novel mechanism of targeting β-catenin translation via utilizing a plant compound, berberine. Our findings provide a new avenue of targeting β-catenin axis in cancer, which can be utilized toward the designing of effective therapeutic strategies to combat β-catenin–dependent cancers.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common forms of gastrointestinal cancers and a major cause of cancer-related death worldwide (Torre et al., 2015). A vast majority of HCCs develop in the setting of chronic liver diseases and cirrhosis. The first-line Food and Drug Administration–approved therapy available for treating advanced, unresectable HCC is the multikinase inhibitor sorafenib, which, despite some promising results (Llovet et al., 2008), is only effective for a few months. More recently, immune checkpoint inhibitors (nivolumab and pembrolizumab), several other multikinase inhibitors (lenvatinib, cabozantinib, regorafenib), and human monoclonal antibodies (ramucirumab) have been approved as first- or second-line therapies for advanced HCC (Pinyol et al., 2019; Caruso et al., 2020). Despite this, the overall survival of patients is still not significantly improved because of resistance. Newer and more effective therapeutic approaches are necessary for combating this deadly malignancy (Llovet and Bruix, 2008; Porta and Paglino, 2010).
Among various pro-oncogenic pathways that are aberrantly activated in HCC, Wnt/β-catenin signaling cascade is specifically important. About 18.5% of HCCs harbor oncogenic mutations of β-catenin gene (CTNNB1) (de La Coste et al., 1998; Miyoshi et al., 1998; Russell and Monga, 2018; Perugorria et al., 2019) leading to its activation (Nhieu et al., 1999). This scenario is much higher in hepatoblastomas, which harbor >50% CTNNB1 mutations. Mutations of adenomatous polyposis coli (APC) or β-catenin itself (Morin et al., 1997; Nhieu et al., 1999) or activation of Wnt signaling result in stabilization of β-catenin (Korinek et al., 1997; de La Coste et al., 1998) with increased nuclear translocation, interaction with transcription factors of the TCF/lymphoid enhancer factor family, and induction of target gene transcription (Bienz and Clevers, 2000; Cadoret et al., 2002), which include cyclin D1, c-myc, matrix metalloproteinase-7, vascular endothelial growth factor, B-cell lymphoma–extra large, survivin, and glutamine synthetase (Vlad et al., 2008; Perugorria et al., 2019). Several inhibitors that can antagonize key steps of β-catenin/TCF axis have been developed, but none became successful clinically, although some clinical trials are currently ongoing to test the efficacy of Wnt antagonists on various liver diseases (Perugorria et al., 2019) and cancers (Zhong and Virshup, 2020).
Berberine (BBR), an alkaloid extracted from herbal plants, has been used in ancient Chinese medicine for a long time to treat microbial infections and diarrhea, and BBR also possesses antidiabetic (Yin et al., 2008; Zhang et al., 2010) and cholesterol-lowering effects (Kong et al., 2004; Krishan et al., 2015). In addition, various studies have shown antineoplastic properties of BBR in various cancers, such as colon cancer, gastric cancer, breast cancer (Eom et al., 2008; Tillhon et al., 2012; Kim et al., 2013; Wang et al., 2013; Zhang et al., 2013), and others, including HCC (Tsang et al., 2015; Huang et al., 2018). Despite these, the detailed in-depth mechanism by which BBR antagonizes pro-oncogenic pathways in various cancers is still unclear. BBR is also an activator of AMP-activated protein kinase (AMPK) (Hawley et al., 2010), and some reports suggest that the antineoplastic effects of BBR are linked with AMPK activation (Kim et al., 2012; Park et al., 2012; Yu et al., 2014; Li et al., 2015). Other studies, however, have shown antagonism of β-catenin pathway as a potential mechanism (Wu et al., 2012; Albring et al., 2013). A more recent study has shown that BBR can antagonize Wnt/β-catenin axis in colon cancer via inducing β-catenin proteasomal degradation involving retinoid X receptor α (Ruan et al., 2017).
Since β-catenin is a major pro-oncogenic axis in HCC and there are no effective pharmaceutical options available yet, the current studies were undertaken to determine whether BBR antagonizes β-catenin signaling in HCC and to elucidate the underlying mechanism. Our studies revealed that BBR can antagonize β-catenin and its downstream signaling in HCC in an AMPK-independent manner. Instead, this involved a novel, translational regulation via eukaryotic translation initiation factor 4E (e1F4E) –binding protein (4E-BP) 1 and 4E-BP2, which are known inhibitors of cap-dependent translation. Treatment with BBR antagonized mammalian target of rapamycin (mTOR) activity associated with reduced phosphorylated 4E-BP1 at Thr 37, 46 (p4E-BP1Thr37/46) levels and promoted interaction between eIF4E with 4E-BP1. Knocking down 4E-BP1 and 4E-BP2 significantly attenuated BBR-induced reduction of β-catenin expression and downstream signaling. In addition, cells with 4E-BP1 and 4E-BP2 knockdown were resistant toward BBR-induced cell death and were resensitized by pharmacological targeting of β-catenin with inhibitor of β-catenin responsive transcription (iCRT-14). Taken together, these studies reveal a novel translational regulation of β-catenin, which contributes toward BBR-induced HCC cell death.
Materials and Methods
Reagents and Antibodies.
Dulbecco’s modified Eagle’s medium (DMEM), DMEM/F12, minimum essential medium (MEM) and Opti-MEM media, TRIzol, RNase solution, and LipofectAMINE 2000 were purchased from Invitrogen (Carlsbad, CA); berberine was from Sigma (St. Louis, MO); luciferase assay reagent was from Promega (Madison, WI); actinomycin D and iCRT-14 were from Tocris (Minneapolis, MN); Lactacystin, MG-132, and cycloheximide were from Millipore Sigma (Burlington, MA); 4EGI-1 was from Selleckchem (Houston, TX); immobilized r-aminophenyl-m7GTP (C10-spacer) agarose beads were from Jena Science (Jenna, Germany); propidium iodide (PI) and FITC Annexin V Apoptosis Detection Kit were from BD Biosciences (San Jose, CA); and JC-1 dye and rhodamine phalloidin were from Thermo-Fisher Scientific (Waltham, MA). The antibodies used were obtained from the following sources: poly (ADP-ribose) polymerase, caspase-3, caspase 8, caspase 9, phosphorylated AMPK at Thr 172 (pAMPKT172), total AMPK, AMPKα1 and AMPKα2, phosphorylated β-catenin at Ser 33, 37/Thr 41, cyclin D1, Axin2, c-Myc, p4E-BP1Thr37/46, 4E-BP1, 4E-BP2, phosphorylated p70S6K at Thr 389 (p-p70S6KThr389), the 70-kDa ribosomal protein S6 kinase, pAKTSer473, AKT, phosphorylated GSK3β at Ser 9 (pGSKβSer9), GSKβ, phosphorylated eIF4E at Ser 209, eIF4E, eIF4G1, eIF4G, cytochrome c, phosphorylated ACC at Ser 79, and Acetyl-CoA-carboxylase were from Cell Signaling Technologies (Danvers, MA); β-catenin was from BD Transduction Laboratories; mitochondrial cytochrome c oxidase subunit IV was from abcam (Cambridge, MA); and GAPDH was from Ambion Inc. (Austin, TX).
Cell Culture.
HCC cells (Hep3B, HepG2) and HEK293 were obtained from ATCC, and Huh7 cells were obtained as described (Sureau et al., 1992; Senthivinayagam et al., 2009). Hep3B and HepG2 cells were maintained in MEM media supplemented with 10% FBS, 1% Pen/Strep, 1% HEPES, 1% sodium pyruvate, and 1% nonessential amino acids; Huh7 cells were maintained in DMEM/F12 media with 10% FBS and 1% Pen/Strep; and HEK293 cells were maintained in DMEM media with 10% FBS and 1% Pen/Strep. In BBR-related experiments, cells were treated with 50 µM BBR (unless indicated otherwise) for 16–24 hours followed by Western blot or quantitative polymerase chain reaction (qPCR) analyses.
Preparation of Wnt3a-Conditioned Medium.
L-Wnt3A (ATCC CRL2647) cells and L cells (ATCC CRL2648) were obtained from ATCC to prepare Wnt3a- and control-conditioned media, respectively, as per manufacturer’s instructions. Cells were cultured in DMEM media supplemented with 10% FBS and 0.4 mg/ml G-418 for L-Wnt3a cells and 1% Pen/Strep for L cells. For Wnt3a-conditioned medium (CM), cells were split in the ratio of 1:10 in 10 ml of DMEM media without G-418 in 10-cm dishes and allowed to grow. After 4 days, the first batch of CM was collected, and another 10 ml of fresh media was added to the cells. The second batch of CM was collected after 3 days, and cells were discarded thereafter. CM from two batches was mixed at 1:1 ratio, filtered using 0.22-μm filter, and stored at −20°C. Similarly, control CM was prepared by culturing L cells in DMEM with 10% FBS and 1% Pen/Strep.
RNA Isolation and qPCR Analysis.
qPCR analysis was performed as described earlier (Viswakarma et al., 2017; Ke et al., 2018). Briefly, total RNA was extracted from HCC cells treated with vehicle (DMSO) or BBR using TRIzol reagent, and the integrity of 18S and 28S ribosomal RNA was assessed by gel electrophoresis. cDNA synthesis was then performed using Superscript III First-Strand Synthesis System kit (Invitrogen) and amplified using SYBR Green Polymerase Chain Reaction (PCR) Master Mix (Applied Biosystems) in ABI StepOnePlus detection system (Applied Biosystems). The PCR cycling condition was set as follows: an initial denaturation step at 95°C for 2 minutes, 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute, finally subjecting to melting temperature to check amplification curve. The relative changes in gene expression were estimated using the 2–ΔΔCt method using 18S ribosomal RNA as a housekeeping gene. The lists of primers used are included in Supplemental Table 1.
Transient Transfection and Luciferase Assays.
Subconfluent populations of cells were transiently transfected using LipofectAMINE-2000 with β-catenin/TCF-responsive luciferase-reporter construct (pGL3-OT) or the corresponding mutant construct (pGL3-OF) along with β-galactosidase (β-gal) vector in the presence of empty vector or β-catenin–expressing vector as reported earlier (Thylur et al., 2011). β-catenin/TCF transcriptional activity was assessed using luciferase reporters containing TCF sites linked to luciferase reporter (pGL3-OT) and compared with a control reporter (pGL3-OF), in which the TCF sites were mutated. These reporter constructs were derived from the TOPFLASH and FOPFLASH vectors, respectively, and obtained from Dr. Bert Vogelstein (Morin et al., 1996; Korinek et al., 1997; Shih et al., 2000). Treatment with vehicle or BBR was initiated after 48 hours of transfection and luciferase and β-gal assays were performed using a luminometer (Berthold Technologies, TriStar2 LB 942) and a microplate reader (EPOCH2; BioTek), respectively. Each transfection was performed in triplicate, and each experiment was repeated at least twice. The results obtained were calculated as the ratio of relative light units (RLUs) to β-gal values. For the luciferase assays with AMPKα1 or AMPKα2 knockdown, cells were cotransfected as above in the presence of control–small interference RNA (siRNA), AMPKα1-siRNA, AMPKα2-siRNA, or AMPKα1+2-siRNA and analyzed as above.
siRNA.
siRNA smart pool against human AMPKα1 (L-005027-00) and human AMPKα2 (L-005361-00) was purchased from Dharmacon (Lafayette, CO). A negative control siRNA from Ambion Inc. was used as control siRNA. siRNA transfection was performed using LipofectAMINE 2000 as per the manufacturer’s instructions and as described previously (Santha et al., 2015). Briefly, subconfluent cells plated in 35-mm plates were transfected with 50 nM of either control siRNA or target siRNA for 24 hours followed by recovery in serum-containing medium. The transfected cells were treated after 48–72 hours of transfection with either DMSO or BBR for an additional 16 hours followed by Western blot analysis.
Stable Cell Line Creation.
Lentiviral particles containing human AMPKα1/2-shRNA (sc-45312-V) obtained from Santa Cruz Biotechnology was used to knock down endogenous AMPKα1/2 expressions following manufacturer’s protocol as reported (Laderoute et al., 2014). A control lentiviral preparation encoding a scrambled shRNA sequence (sc-108080) was used as negative control. Briefly, subconfluent HCC cells plated in 12-well plates were transduced overnight with lentiviral particles (∼40,000–80,000 infectious units) in complete medium containing polybrene (5 µg/ml), and this was followed by selection in puromycin-containing medium. Puromycin-resistant colonies were propagated and stored, and the degree of knockdown was determined by Western blot analysis.
For creating the eIF4E-BP1 and eIF4E-BP2 knockdown stable cells, we used pLKO.1-CMV-puro-eIF4E-BP1-shRNA (TRCN0000040203) and pLKO.1-CMV-neo-eIF4E-BP2-shRNA (TRCN0000117814) lentiviral constructs from Sigma. Lentiviral particles were produced as described earlier (Das et al., 2019) by cotransfecting HEK293 FT cells (Life Technologies) with either one of the lentiviral plasmids along with psPAX2 and pMD2G packaging plasmids using LipofectAMINE 2000. Titrations were performed first to achieve a multiplicity of infection of 0.3–0.5, and the infection efficiency was confirmed by Western blots. Huh7 cells were then transduced with the lentiviral particles as described above, and stable cell lines were selected using both puromycin (for eIF4E-BP1 shRNA) and neomycin (for eIF4E-BP2 shRNA).
Cap-Binding Affinity Assay.
Cap-binding affinity assay was performed as described (Zhan et al., 2015) with modifications. Briefly, cells treated with vehicle or BBR were lysed in NP40 lysis buffer (20 mM Tris-HCl, pH 8.0; 150 mM NaCl; 2 mM EGTA, pH 8.0; 10% glycerol; 1% NP40; 50 mM β-glycero phosphate; 1 mM Na-orthovanadate; 1 mM dithiothreitol; 1 mM phenylmethylsulfonyl fluoride containing a mixture of proteinase inhibitors). Equal amounts of protein extracts were then added to the m7GTP agarose beads (30 µl) and incubated on a rotator for 3 hours at 4°, which was followed by washing with NP40 lysis buffer four times. m7GTP-bound proteins were then analyzed by Western blots.
Apoptosis Assay.
Apoptosis assay was performed using FITC Annexin V Apoptosis Detection Kit (BD Biosciences) as per manufacturer’s instructions. Cells seeded at a density of 1.5 × 106 cells/plate in 35-mm plates were treated with BBR, iCRT alone, or both in combination for various lengths of time. At the time of harvest, they were trypsinized and distributed equally into two parts: one part was used for apoptosis assay, and the other part was used for JC-1 assay (described below). Cells for apoptosis assay were centrifuged at 2000 rpm for 5 minutes, washed once with ice-cold PBS, and washed once with 1X binding buffer, and this was followed by resuspension in 100 µl 1X binding buffer and incubation with 5 µl each of Annexin V and PI at room temperature in dark for 15 minutes. 1X binding buffer (400 μl) was then added to each sample, and apoptosis assays were performed using Gallios flow cytometer (Beckman Coulter). The data were analyzed using FlowJo software.
JC-1 Assay.
To determine changes in mitochondrial membrane potential, a distinctive feature of early apoptosis, JC-1 assay was performed as per manufacturer’s instructions. Cells harvested as above were suspended in 1 ml of warm growth media, which was followed by incubation in 2 µM JC-1 dye at 37°C in 5% CO2 incubator for 15 minutes. Cells were then washed twice with PBS and resuspended in 500 µl of PBS, and readings were acquired on Gallios flow cytometer (Beckman Coulter) using 488-nm laser. JC-1 red emission of healthy mitochondria was obtained at 590 nm, and the green emission of apoptotic mitochondria (with reduced membrane potential) was obtained at 530 nm. The data were analyzed using FlowJo software.
Cell-Cycle Analysis.
Cells seeded at a density of 0.6 × 106 cells/plate in 35-mm plates were treated with BBR for different lengths of time. At the time of harvest, they were washed in PBS, fixed in ice-cold 70% ethanol, and stored at −20°C overnight. Next day, the fixed cells were washed twice with PBS and treated with 50 µl of RNase at room temperature (stock 100 µg/ml). After 15 minutes, 200 µl of PI (stock 50 µg/ml) was added, and cell-cycle analysis was performed using Gallios flow cytometer (Beckman Coulter). The data were analyzed using FlowJo software.
Migration Assay.
The effect of BBR on the cell migration was determined using the ORIS cell migration assay kit (Platypus, NJ). The ORIS cell migration assay was performed using a 96-well plate, in which a “stopper” barrier was used to create a central cell-free detection zone for cells to migrate. Briefly, cells at a density of 10,000 cells/well (in quadruplicate) were added to each well through wedge of the stopper and allowed to grow overnight. Next day, the stoppers were removed, and cells were washed with media and treated with DMSO or BBR. The readings were taken by Celligo Imaging Cytometer (Nexcelom Bioscience, MA) after 48 hours of BBR treatment. Amount of cells migrated into the central migration zone was used to calculate percent increase in migration.
Detection of Cytochrome c Release.
To detect cytochrome c release from mitochondria to cytoplasm, cells were separated into cytoplasmic and mitochondrial fractions as described with modifications (Chandra et al., 2004). For cytoplasmic and mitochondrial fractionation, cells washed with PBS were resuspended in homogenization buffer (20 mM HEPES, pH 7.5; 10 mM KCl; 1.5 mM MgCl2; 1 mM EDTA; 1 mM EGTA; 1 mM dithiothreitol; 250 mM sucrose) and incubated on ice for 30 minutes with intermittent mixing. Cells were then homogenized (with 10–12 strokes) in cold, and the homogenate was centrifuged at 1000 rpm for 5 minutes at 4°C to remove nuclei, cellular debris, and intact cells. The resultant supernatant was collected and centrifuged at 12,000 rpm for 20 minutes at 4°C to separate cytoplasmic fraction (as supernatant) and mitochondrial fraction (as pellet). The cytoplasmic protein extracts were preserved, and mitochondrial pellet was washed with homogenizing buffer twice at 12,000 rpm for 10 minutes each at 4°C. The mitochondrial pellets were resuspended in mitochondrial protein extraction buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 2 mM EDTA; 2 mM EGTA; 0.2% Triton X-100; 0.3% NP40; 2.5 mM phenylmethylsulfonyl fluoride; and protease inhibitor cocktail). After incubation on ice for 10 minutes, they were centrifuged at 12,000 rpm for 20 minutes at 4°C, and mitochondrial proteins were collected as supernatant and preserved. Both cytoplasmic and mitochondrial proteins were boiled and denatured using Laemmli buffer and used for Western blotting.
Western Blot Analysis.
Western blot analysis was performed following procedures described previously (Pradeep et al., 2004; Mishra et al., 2010). Briefly, equal amounts of total cell extracts were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and subjected to Western blot analysis utilizing various antibodies. The bar graphs for most proteins represent the ratio of the respective protein/GAPDH, and those for pAMPK represent the ratio of pAMPK/total AMPK.
Statistical Analyses.
All data were presented as mean with ±S.D. For determining significance between control (vehicle) and BBR-treated samples, Student’s t test was performed and expressed as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001, with ns: P > 0.05 not significant. The data of Supplemental Table 2 were presented either as mean of the group with 95% confidence interval (CI) or, for the key comparisons, the difference of means between pairwise groups together with 95% CI of the difference using the pooled S.E. Two or three-way ANOVA was employed to test the significance of treatment group, vehicle type, or time. To control the overall type I error at 0.05 for multiple tests, the Tukey’s follow-up tests were used for the pairwise comparisons, and Dunnett’s tests were used instead to compare each mean with the control mean. The results of ANOVA, 95% CI of difference, Tukey’s test, and Dunnett’s test have been summarized in Supplemental Table 2.
Results
BBR Antagonizes β-Catenin Pathway in HCC Cells.
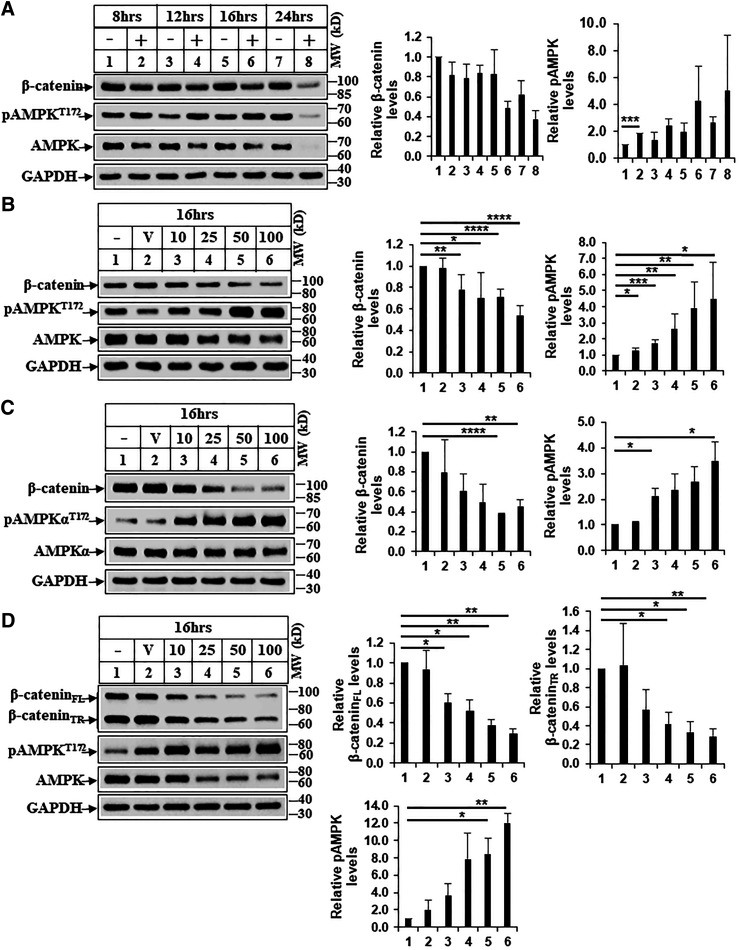
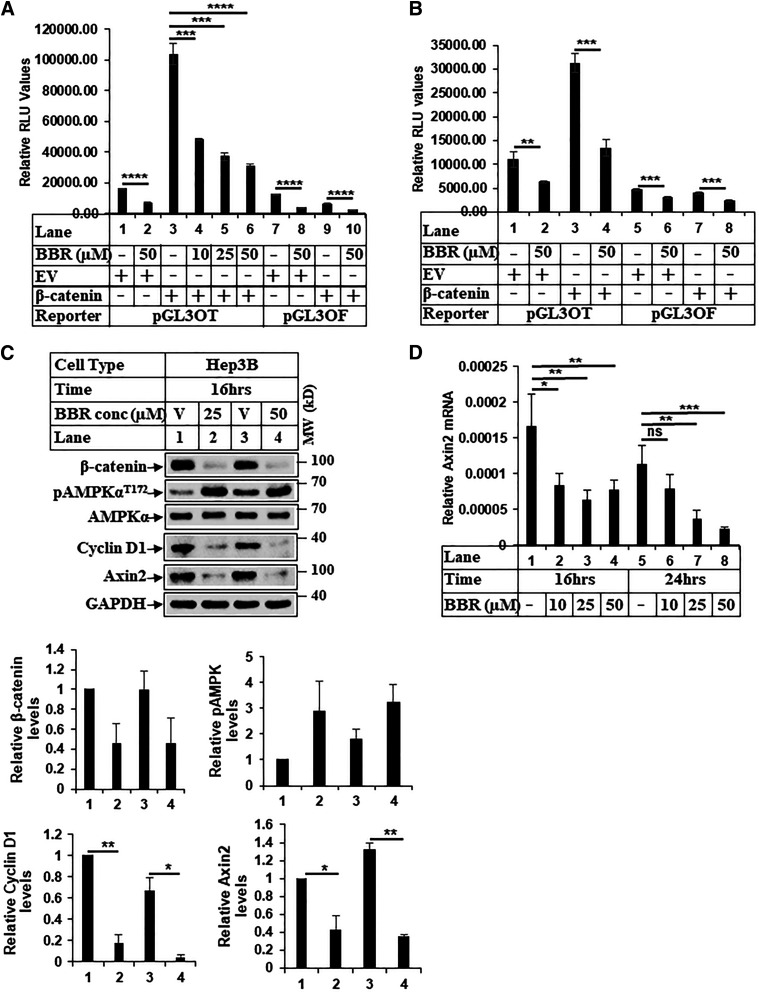
In an attempt to identify pharmacological agents that can effectively target β-catenin, we determined the effect of BBR on β-catenin axis in HCC. Treatment of HCC cells (Huh7 and Hep3B) with BBR showed a dose- and time-dependent reduction of β-catenin (Fig. 1, A–C), suggesting an antagonism of this pathway. Interestingly, BBR treatment also reduced the expression of mutated β-catenin in the HepG2 cells (Fig. 1D), which is resistant to degradation via conventional APC/GSK3β pathway. Treatment of HEK293 cells with BBR also showed a dose-dependent reduction of β-catenin expression (Supplemental Fig. 1A). To determine whether BBR can antagonize β-catenin/TCF transcriptional activity and downstream target gene expression, luciferase assays were performed with β-catenin/TCF–responsive reporter (pGL3-OT) and the corresponding mutant (pGL3-OF) as reported (Thylur et al., 2011). BBR suppressed OT-reporter activity in a dose-dependent manner (Fig. 2, A and B) and antagonized β-catenin target gene (Fig. 2D) and protein expression (Fig. 2C; Supplemental Fig. 1B). These observations suggested that BBR might be an effective therapeutic approach to target β-catenin in HCCs.
Fig. 1.
Treatment with BBR reduces β-catenin expression in HCC cells. (A) Huh7 HCC cells were treated with DMSO (−) or BBR 50 µM (+) for the indicated time points and then harvested. Equal amounts of protein extracts were analyzed by Western blots against the antibodies indicated. (B) Huh7, (C) Hep3B, and (D) HepG2 cells were treated with DMSO (V) or increasing concentrations of BBR (10, 25, 50, 100 µM) for 16 hours and analyzed by Western blots. β-catenin-FL and β-catenin-TR indicate full-length (FL) and truncated (TR) forms, respectively. The bar graphs represent the ratio of various proteins/controls observed in the Western blots. The data represent the mean ± S.D. of two to five independent experiments. Statistical analysis was performed using Student’s t test and indicated as follows: ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001. MW, molecular weight.
Fig. 2.
Treatment with BBR antagonizes β-catenin/TCF transcriptional activity and expression of target genes. (A) HEK-293 and (B) Huh7 cells were transiently transfected with β-catenin/TCF–responsive reporter (pGL3-OT) or the corresponding mutant (pGL3-OF) along with empty vector (EV) or β-catenin–expressing vector and treated with DMSO (−) or indicated concentrations of BBR for 16 hours. Luciferase and β-gal assays were performed next, and the results were expressed as RLU/β-gal values. Each transfection was performed in triplicate, and each experiment was repeated at least two times. The data represent the mean ± S.D. of three independent transfections. (C) Equal amounts of protein from Hep3B cells treated with DMSO (V) or BBR for 16 hours were analyzed by Western blots with the antibodies indicated. The bar graphs represent the ratio of various proteins/controls observed in the Western blots. The data represent the mean ± S.D. of two independent experiments. Statistical analysis was performed using Student’s t test and indicated as: *P ≤ 0.05; **P ≤ 0.01; . (D) Total RNA extracted from Huh7 cells treated with DMSO or increasing concentrations of BBR for 16 hours and 24 hours was subjected to qPCR analysis for determining changes in Axin2 gene expression. The experiment was repeated at least three times, and data represent the mean ± S.D. of four independent PCR reactions. Significant differences for (A, B, and D) were determined by t test and indicated as follows: ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001. MW, molecular weight ns, not significant.
BBR Regulates β-Catenin Pathway Independent of AMPK.
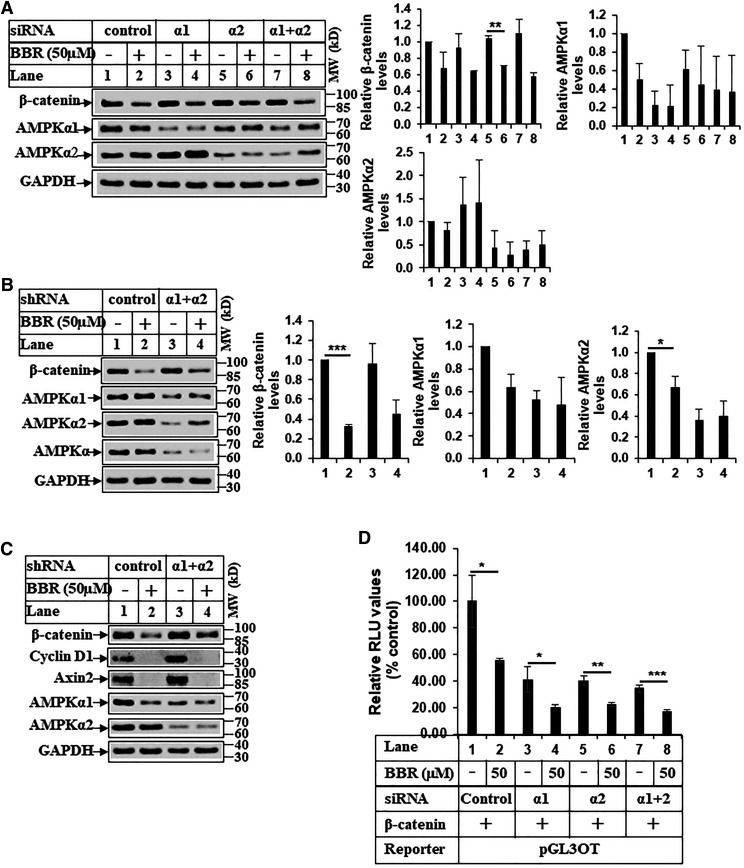
Our earlier studies showed that BBR and TRAIL combination–induced apoptosis is mediated via AMPK activation (Ke et al., 2018). BBR-induced activation of AMPK has been reported by others as well (Lee et al., 2006). Treatment with BBR also showed an increase in pAMPKT172 levels—suggesting its activation mostly—during the time of β-catenin reduction (Fig. 1; Supplemental Fig. 1, A and B). The reduction of pAMPT172 observed with BBR at 24 hours is most likely due to a reduction of total AMPK expression, as suggested by the bar graphs of pAMPK/AMPK (Fig. 1A, lanes 7, 8). Results from this study on BBR-induced activation of AMPK are consistent with previous findings (Hawley et al., 2010; Li et al., 2015). To determine whether AMPK was involved in BBR-induced antagonism of β-catenin, AMPKα1 and AMPKα2 levels were transiently knocked down using corresponding siRNAs. Surprisingly, knocking down AMPKα was unable to restore β-catenin expression in the presence of BBR (Fig. 3A; Supplemental Fig. 2A). To confirm this further, HCC cells with stable AMPKα1 and AMPKα2 knockdown were generated, which also showed BBR-induced reduction of β-catenin in the absence of AMPKα1/α2 (Fig. 3, B and C). Similarly, BBR reduced expression of β-catenin target proteins (cyclin D1 and Axin2) even with AMPKα1/α2 knockdown (Fig. 3C). Furthermore, luciferase assays showed that BBR was capable of reducing OT-luciferase activity in the absence of AMPKα1 and AMPKα2 (Fig. 3D; Supplemental Fig. 2B). In addition, treatment with various AMPK agonists, despite activating AMPK pathway (Lin et al., 2017), was unable to reduce β-catenin expression in each case (Supplemental Fig. 2C, compare β-catenin and phosphorylated ACC at Ser 79 in DMSO, BBR, salicylate, A769662). These suggested that BBR can reduce β-catenin expression and antagonize its downstream signaling in an AMPK-independent manner.
Fig. 3.
BBR regulates β-catenin pathway independent of AMPK. (A) Subconfluent populations of Hep3B cells were transiently transfected with control siRNA or AMPKα1-siRNA or AMPKα2-siRNA alone or in combination followed by treatment with DMSO or BBR for 16 hours. Western blot analyses were performed with the antibodies indicated. (B) Hep3B or (C) Huh-7 cells stably overexpressing control-shRNA or AMPKα1- and AMPKα2-shRNA were treated with DMSO or BBR for 16 hours and analyzed by Western blots. The bar graphs in (A and B) represent the ratio of various proteins/controls observed in the Western blots. The data represent the mean ± S.D. of two independent experiments. Statistical analysis was performed using Student’s t test and indicated as follows: *P ≤ 0.05; **P ≤ 0.01; and ***P ≤ 0.001. (D) Huh7 cells were cotransfected with β-catenin/TCF–responsive reporter (pGL3-OT) and β-catenin–expressing vector along with either control siRNA, AMPKα1-siRNA, AMPKα2-siRNA, or AMPKα1+α2-siRNA and treated with DMSO or BBR for 16 hours. Luciferase and β-gal assays were performed as in Fig. 2A, and RLU/β-gal values were expressed as % control. Each transfection was performed in triplicate, and each experiment was repeated at least two times. The data represent the mean ± S.D. of three independent transfections. Significant differences were determined by t test and indicated as follows: *P ≤ 0.05; **P ≤ 0.01; and ***P ≤ 0.001. MW, molecular weight.
BBR Regulates β-Catenin Expression at the Level of Translation.
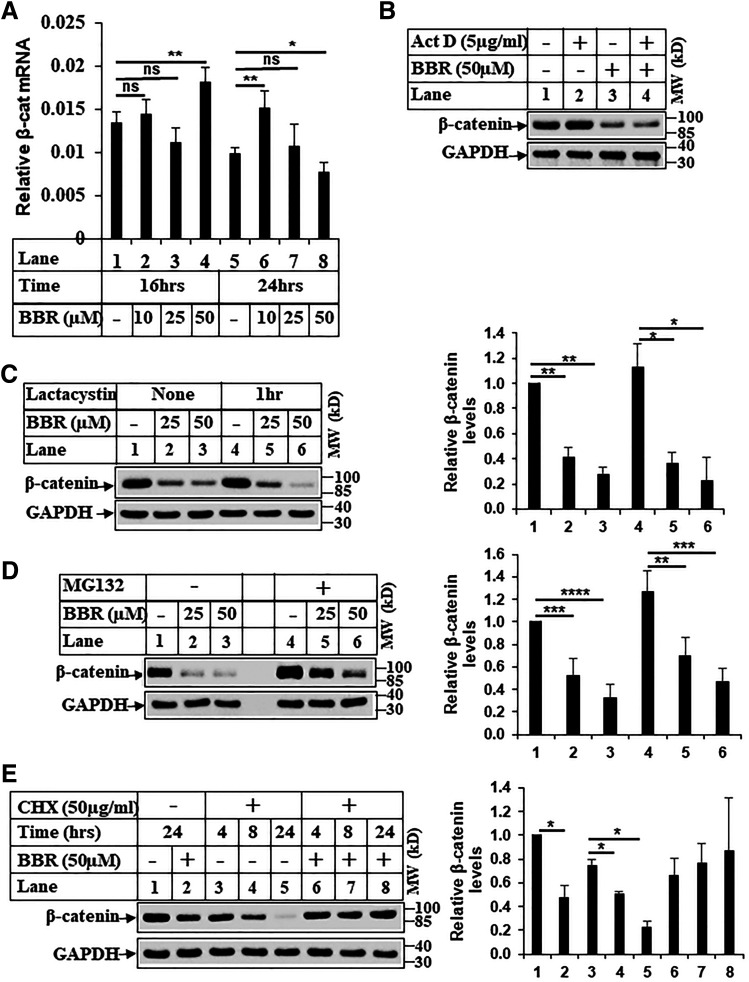
We next focused on determining whether BBR-induced reduction of β-catenin was at the level of transcription, translation, or post-translation. Recently, BBR was shown to target β-catenin post-translationally via a proteasome-dependent pathway in colon cancer cells (Ruan et al., 2017). Estimation of β-catenin mRNA expression with and without BBR treatment showed no significant difference (Fig. 4A) at a time when protein expression was reduced significantly (Fig. 1A). In addition, BBR was able to reduce β-catenin expression even in the presence of an inhibitor of transcription, actinomycin D (Fig. 4B), suggesting this was regulated independent of transcription. To determine the possibility of a post-translational regulation mediated via proteasomes, cells were pretreated with two different proteasomal inhibitors, lactacystin and MG132 (Alao et al., 2006), which were unable to rescue β-catenin expression after BBR treatment (Fig. 4, C and D). Furthermore, the levels of phosphorylated β-catenin at Ser 33, 37/Thr 41 were also reduced with BBR treatment (Supplemental Fig. 3, A and B) and correlated with an increase in pGSK3βSer9 levels (indicating inhibition) under these conditions (Fig. 5, A and B), suggesting BBR does not significantly regulate post-translational modification of β-catenin. Taken together, these suggested the possibility that BBR might target β-catenin at the level of translation. To explore this possibility, cells were pretreated with translational inhibitor cycloheximide (CHX) prior to BBR treatment. Interestingly, BBR was unable to reduce β-catenin expression in the presence of CHX pretreatment (Fig. 4E, compares lanes 4 and 5 with 7 and 8), suggesting BBR-induced reduction involves a translational regulation. It is unclear at this time why treatment with CHX and BBR did not show the regular decay of β-catenin observed under CHX-DMSO conditions (lanes 3–5), thus indicating additional mechanisms. The mostly likely explanation is that CHX also inhibited translation of those that mediate β-catenin degradation, including those involved in proteasomal pathway. To rule out the proteasomal pathway involvement, we have performed studies with two different proteasomal inhibitors (lactacystin and MG132), which were unable to reverse BBR-induced β-catenin reduction (Fig. 4, C and D).
Fig. 4.
BBR regulates β-catenin expression at the level of translation. (A) Total RNA extracted from Huh7 cells as in Fig. 2D was subjected to qPCR analysis for determining changes in CTNNB1 (β-catenin) gene expression. The experiment was repeated at least three times, and data represent the mean ± S.D. of four independent PCR reactions. Significant differences were determined by t test and indicated as follows: ns, P > 0.05; *P ≤ 0.05; and **P ≤ 0.01. (B) Huh7 cells were treated with either DMSO or actinomycin D (5 µg/ml) or BBR (50 µM) or a combination of actinomycin D and BBR for 24 hours and analyzed by Western blots. (C) Huh7 cells were treated with DMSO (−) or with indicated concentrations of BBR for 24 hours after a pretreatment in the absence or presence of 10 µM lactacystin for 1 hour. Equal amounts of lysates were analyzed by Western blots. (D) Huh7 cells were treated with DMSO (−) or with indicated concentrations of BBR for 16 hours after a 2-hour pretreatment in the absence (−) or presence (+) of 20 µM MG132 and analyzed by Western blots. (E) Huh7 cells were pretreated with 50 µg/ml CHX for 24 hours followed by treatment with DMSO (−) or BBR 50 µM (+) in combination with CHX for the indicated periods of time. Lanes 1 and 2 were only treated with DMSO or BBR. The samples were analyzed by Western blots with the antibodies indicated. The bar graphs in (C–E) represent the ratio of various proteins/controls observed in the Western blots. The data represent the mean ± S.D. of two to four independent experiments. Statistical analysis was performed using Student’s t test and indicated as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001. Act D, actinomycin D; β-cat, β-catenin; MW, molecular weight; ns, not significant.
Fig. 5.
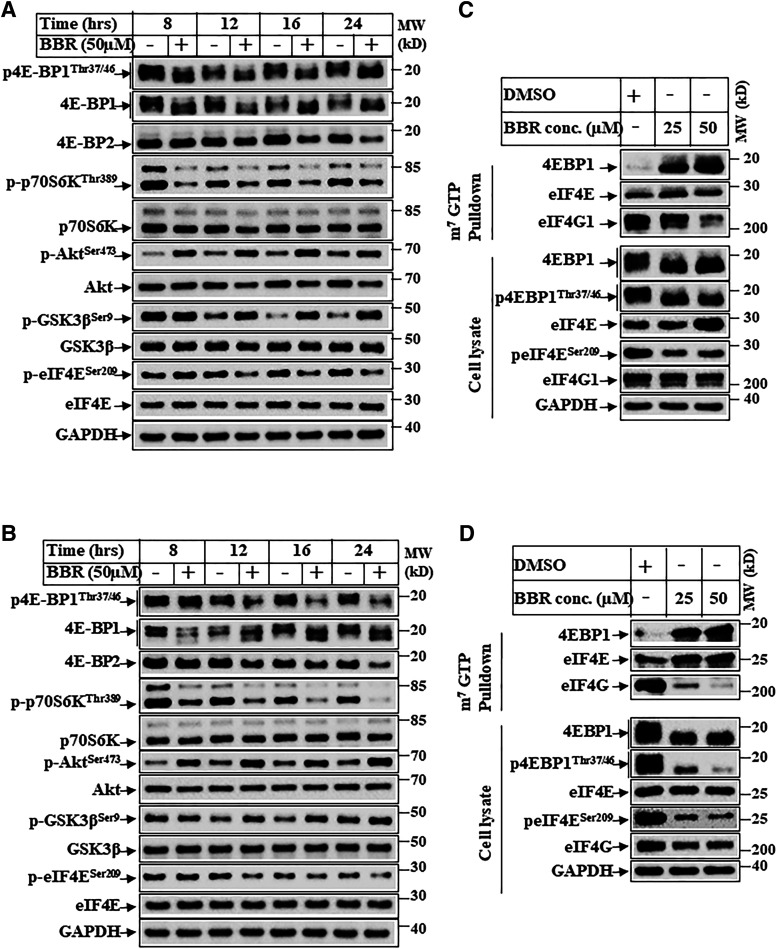
BBR inhibits mTOR pathway and cap-dependent translation. (A) Hep3B and (B) Huh7 cells were treated with DMSO (−) or BBR 50 µM (+) for various lengths of time and analyzed by Western blots. (C) Huh7 and (D) Hep3B cells were treated with DMSO or with the indicated concentrations of BBR for 16 hours and harvested. Equal amounts of cell extracts were incubated with m7GTP agarose beads for 3 hours at 4oC . The top panels show m7GTP-bound proteins analyzed by Western blots, and the bottom panels show expression of the corresponding proteins in whole-cell lysates. MW, molecular weight.
BBR Inhibits mTOR Pathway and Cap-Dependent Translation.
Regulation of β-catenin expression via cap-dependent translation has been reported earlier. The proto-oncogene c-Src was shown to induce β-catenin levels via cap-dependent translation (Karni et al., 2005), and mitogen-activated protein kinase–interacting serine/threonine kinase (MNK)-eIF4E axis was shown to regulate increased β-catenin translation and activity (Lim et al., 2013). Since mTOR is a positive regulator of cap-dependent translation (Ma and Blenis, 2009), we first detected the effect of BBR on mTOR activation. HCC cells treated with BBR showed that BBR can inhibit p-p70S6KThr389 and p4E-BP1Thr37/46 levels in a time-dependent manner (Fig. 5, A and B), suggesting inhibition of mTORC1 axis. In addition, BBR also showed increase in pAKTSer473 and its downstream pGSK3βSer9 levels, which was likely due to a corresponding activation of mTORC2. The levels of p-eIF4ESer209 also were reduced with BBR treatment (Fig. 5, A and B), which is known to be phosphorylated by MNKs. Taken together, these suggested a potential inhibition of cap-dependent translation when treated with BBR because of inhibition of mTORC1 and eIF4E phosphorylation. In fact, m7GTP pulldown assays performed with BBR-treated extracts showed increased recruitment of 4E-BP1 and a corresponding reduction of eIF4G in the translation complex when treated with increasing dose of BBR (Fig. 5, C and D).
BBR Antagonizes β-Catenin Pathway via Targeting Cap-Dependent Translation.
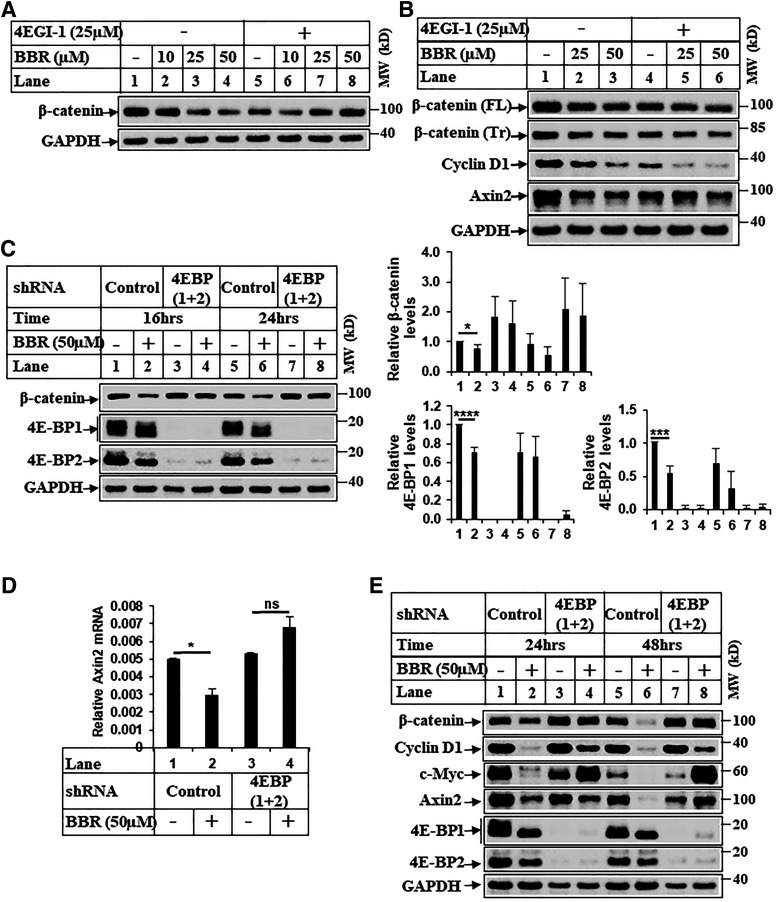
Since BBR antagonized cap-dependent translation, we determined next whether BBR reduced β-catenin expression via targeting cap-dependent translation. Treatment with 4EGI-1, a small molecule that inhibits interaction between initiation factors eIF4E and eIF4G (Moerke et al., 2007), reduced β-catenin expression to very low levels almost similar to those of BBR (Fig. 6A, compares lanes 1 and 5). Interestingly, using HepG2 cells (which express a degradation-resistant, truncated form of β-catenin), we determined that BBR as well as 4EGI-1 reduced expression of full-length and truncated β-catenin and its downstream targets (Fig. 6B). This suggested that in HCC cells, mutated β-catenin expression can be targeted by 4EGI-1. To validate this further, stable HCC cells overexpressing either scrambled shRNA (control-shRNA) or 4E-BP1 and 4E-BP2 shRNA (4E-BP1+2-shRNA) were generated, which showed a significant reduction of 4E-BPs in the 4E-BP1+2 shRNA cells (Fig. 6C). Interestingly, treatment with BBR showed reduction of β-catenin expression in the control-shRNA cells, whereas BBR was unable to reduce it in the 4E-BP1+2-shRNA cells (Fig. 6C), thus suggesting that BBR-induced reduction of β-catenin expression involves antagonism of cap-dependent translation via 4E-BPs. BBR treatment was also unable to reduce Wnt3a-induced β-catenin/TCF transcriptional activity in the 4E-BP1+2-shRNA cells, whereas it effectively antagonized this in control-shRNA cells (Supplemental Fig. 4, A and B). Further analysis performed to estimate changes in β-catenin downstream target gene expressions showed that BBR was unable to reduce these levels in 4E-BP1+2-shRNA cells (Fig. 6, D and E; Supplemental Fig. 5, A and B). Taken together, these confirmed that BBR can antagonize β-catenin expression and downstream signaling via targeting cap-dependent translation.
Fig. 6.
BBR-induced reduction of β-catenin involves cap-dependent translation. (A) Huh7 cells were treated with DMSO (−) or increasing concentrations of BBR for 16 hours after a 2-hour pretreatment with or without 4EGI-1 (25 µM) and analyzed by Western blots. (B) Western blot analysis of extracts from HepG2 cells treated with DMSO (−) or BBR for 24 hours after a 2-hour pretreatment with or without 4EGI-1. (C) Stable Huh7 cells overexpressing either a control-shRNA (Huh7-control-shRNA) or eIF4E-BP1– and eIF4E-BP2-shRNA (Huh7-4E-BP-1+2-shRNA) were treated with DMSO (−) or BBR (+) for the indicated periods of time. Equal amounts of protein were analyzed by Western blots with the antibodies indicated. The bar graphs represent the ratio of various proteins/controls observed in the Western blots. The data represent the mean ± S.D. of four independent experiments. Statistical analysis was performed using Student’s t test and indicated as follows: *P ≤ 0.05; ***P ≤ 0.001; and ****P ≤ 0.0001. (D) Total RNA extracted from stable Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells treated with DMSO (−) or BBR (+) for 48 hours were analyzed by qPCR for Axin2 gene expression. The experiment was repeated twice, and data represent the mean ± S.D. of two independent PCR reactions. Significant differences were determined by t test and indicated as follows: ns, P > 0.05 and *P ≤ 0.05. (E) Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells treated with DMSO (−) or BBR (+) for the indicated periods of time were analyzed by Western blots. FL, full-length; ns, not significant; Tr, truncated. MW, molecular weight.
BBR Induces HCC Cell Apoptosis via Antagonizing Cap-Dependent Translation and β-Catenin Axis.
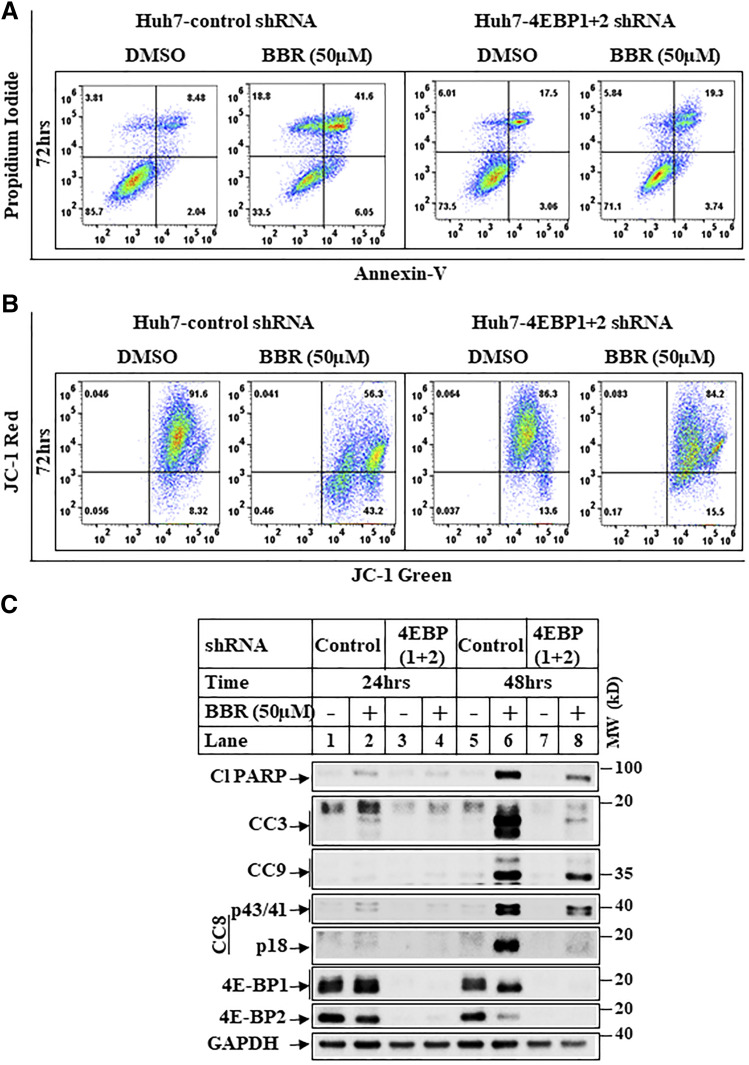
To determine which of the biologic effects of BBR in HCC are mediated via inhibition of cap-dependent translation, we focused on the effects of BBR on cell proliferation, migration, and apoptosis. As shown in Supplemental Fig. 6, A and B, although BBR can induce cell-cycle arrest in these cells (increase in G2/M stage), knocking down 4E-BP1/2 was unable to reverse this. Similarly, BBR reduction of migration was not rescued with 4E-BP1/2 knockdown (Supplemental Fig. 6C), suggesting BBR-induced cell-cycle arrest and inhibition of migration are likely to be independent of cap-dependent translation. To determine the effects on cell death, apoptosis assays were designed first after treatment of HCC cells with BBR. These showed an increase in cellular apoptosis and mitochondrial damage by BBR in a time- and dose-dependent manner in Huh-7 (Supplemental Fig. 7, A–C) and Hep3B cells (Supplemental Fig. 8, A–C). Interestingly, similar apoptosis assays carried out with BBR showed that knocking down 4E-BP1 and 4E-BP2 significantly attenuates BBR-induced apoptosis and mitochondrial damage (Fig. 7, A and B), whereas high-level apoptosis was observed in the control-shRNA cells. Similarly, Western blot analysis revealed that BBR-induced cleavages of poly (ADP-ribose) polymerase and caspase 3, 8, 9 involve antagonism of cap-dependent translation (Fig. 7C).
Fig. 7.
BBR induces apoptosis via antagonizing cap-dependent translation. (A) Representative pictures showing flowcytometric detection of apoptosis assays performed in Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells treated with DMSO or BBR (50 µM) for 72 hours. Cells were then harvested, and apoptosis assays were performed using the FITC Annexin V Apoptosis Detection Kit. The quadrant lines were adjusted to divide the cells in four distinct populations with respect to control. Lower left quadrant represents cells negative for both Annexin V and PI, upper left quadrant represents necrotic cells (only positive for PI), lower right quadrant represents cells in early apoptosis (single positive for Annexin V), and upper right quadrant represents cells in late apoptosis (positive for both Annexin V and PI). (B) Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells treated as in Fig. 7A were analyzed by JC-1 assay to detect changes in mitochondrial membrane potential. The quadrant lines were adjusted to highlight changes in the fluorescent intensity of cell populations with respect to control. Upper right quadrant represents cells containing healthy mitochondria with higher mitochondrial potential emitting a red and a green fluorescence at 590 and 530 nm, respectively. Lower right quadrant represents apoptotic cells with reduced mitochondrial membrane potential emitting green fluorescence at 530 nm. (C) Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells treated with DMSO (−) or BBR 50 µM (+) for the indicated periods of time were analyzed by Western blots with the antibodies indicated. CC, cleaved caspase; PARP, poly (ADP-ribose) polymerase. MW, molecular weight.
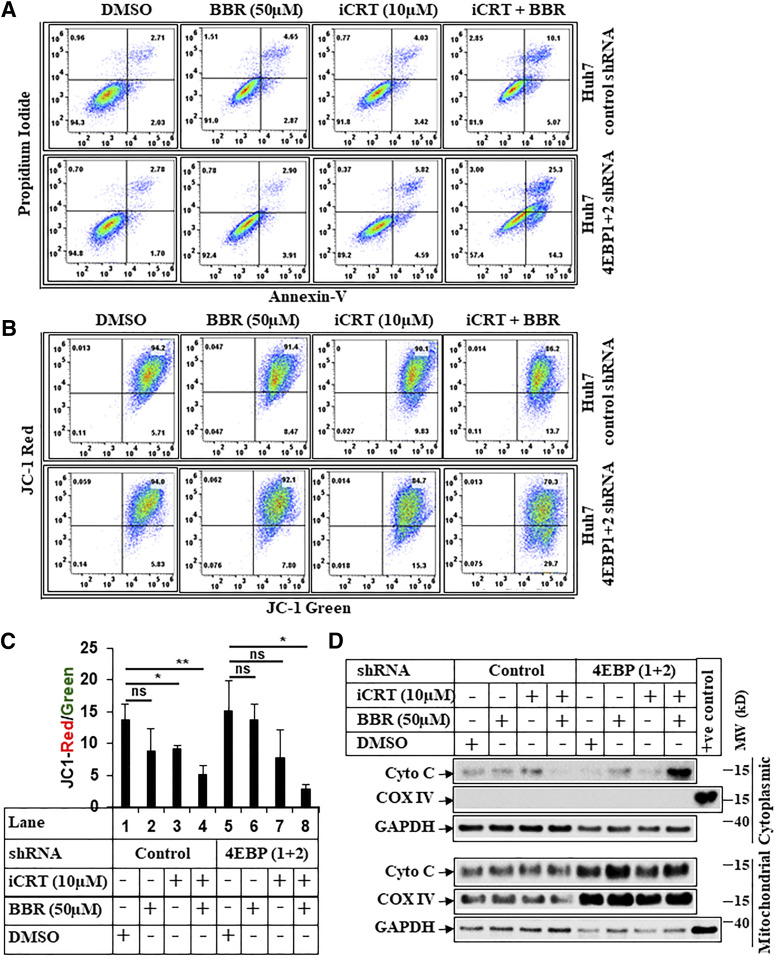
Since BBR-induced apoptosis and antagonism of β-catenin pathway were both reversed with 4E-BP1/2 knockdown, we hypothesized that inhibiting β-catenin might resensitize these cells. To address this, we used iCRT-14 (iCRT), which disrupts β-catenin signaling by antagonizing β-catenin–TCF interaction (Gonsalves et al., 2011; Watanabe and Dai, 2011). In fact, iCRT at a lower dose (10 µM) inhibited β-catenin/TCF–responsive OT activity (Supplemental Fig. 9A) but not OF activity (Supplemental Fig. 9B). Interestingly, treating 4E-BP1+2-shRNA cells with low dose of iCRT resulted in increased apoptosis with or without BBR, which seemed more pronounced than in the control-shRNA cells (Fig. 8, A–C). These suggested that in the absence of 4E-BP1/2, the cells become too addicted to β-catenin for survival, and so antagonizing β-catenin by iCRT increases their susceptibility to BBR-induced cell death. Despite increased apoptosis, iCRT was unable to induce caspase 3 cleavage in the 4E-BP1+2 shRNA cells (Supplemental Fig. 9C). Since iCRT-BBR combination increased mitochondrial damage (Fig. 8, B and C), it suggested the possibility of mitochondrial apoptosis cascade. In fact, treatment with iCRT and BBR promoted a significant increase in cytoplasmic cytochrome c release in the 4E-BP1+2 shRNA cells (Fig. 8D). Combined, these studies reveal a novel translational control of β-catenin by BBR, which can be used to promote cell death in β-catenin–mutated HCCs involving intrinsic apoptotic pathway.
Fig. 8.
BBR induces intrinsic apoptosis via targeting β-catenin. (A) Representative picture showing flowcytometric analysis of apoptosis assays performed in Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells pretreated with iCRT-14 (10 µM) for 24 hours, which was followed by treatment with iCRT-14 (10 µM) alone or in combination with BBR (50 µM) for an additional 24 hours. The cells that received DMSO and BBR (50 µM) were treated with these for 24 hours. At the end of treatment, cells were harvested and subjected to apoptosis assays and analyzed similarly as described under Fig. 7A. (B) Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells treated as in Fig. 8A were analyzed by JC-1 assay, following procedures described under Fig. 7B. (C) The bar graphs represent the ratio of JC-1 Red/JC-1 green. The data represent ±S.D. of two independent experiments. Significant differences were determined by t test and indicated as follows: ns, P > 0.05; *P ≤ 0.05; and **P ≤ 0.01. (D) Huh7-control-shRNA or Huh7-4E-BP 1+2-shRNA cells were pretreated with iCRT-14 (10 µM) for 24 hours, which was followed by treatment with iCRT-14 (10 µM) alone or in combination with BBR (50 µM) for an additional 16 hours. The cells that received DMSO and BBR (50 µM) were treated with these for 16 hours. At the end of treatment, cells were harvested and fractionated into mitochondrial and cytoplasmic fractions and analyzed by Western blots. COX IV was used a positive control to show the purity of mitochondrial fraction, and GAPDH was used as a positive control for cytoplasmic fraction. COX IV, mitochondrial cytochrome c oxidase subunit IV; Cyto C, cytochrome c; MW, molecular weight; ns, not significant.
Discussion
Identification of more effective therapeutic approaches that can antagonize β-catenin axis is critical for targeting aberrant β-catenin activation in HCCs. In an attempt to identify specific signaling pathways that can ameliorate TRAIL resistance, we reported earlier that combination of TRAIL and troglitazone can sensitize TRAIL-resistant cells toward apoptosis that required the activation of AMPK (Senthivinayagam et al., 2009; Santha et al., 2015). Interestingly, treatment with troglitazone alone also inhibited β-catenin and its downstream axis by an APC-independent mechanism (Sharma et al., 2004). More recently we observed that treatment with the natural compound BBR can significantly reduce HCC cell viability and induce apoptosis in combination with TRAIL in an AMPK-dependent manner (Ke et al., 2018). In addition, BBR also showed antagonistic effects on β-catenin axis involving a novel mechanism, which is the focus of the current studies.
Earlier studies have shown that BBR can antagonize cancer progression, and this anticancer activity is linked with an antagonism of Wnt/β-catenin signaling (Wu et al., 2012; Ruan et al., 2017). Despite this, the mechanism involved in β-catenin antagonism is largely unknown. Since β-catenin is often mutated and its downstream signaling is activated in HCC, our goal here was to elucidate in detail the mechanism by which BBR antagonized β-catenin pathway in HCC. Our results demonstrate that BBR can inhibit β-catenin expression in various HCC cells in a dose- and time-dependent manner. Interestingly, BBR also suppressed β-catenin levels in HepG2 cells, which express a truncated/mutated form of β-catenin that is resistant to conventional GSK3β/APC/Axin-mediated proteasomal degradation pathway. β-Catenin is known to interact with TCF or lymphoid enhancer factor transcription factors, which in turn activate target gene transcription (Behrens et al., 1996; Huber et al., 1996; Molenaar et al., 1996). Thus, reduction of β-catenin expression is expected to reduce β-catenin/TCF–mediated transcriptional activity. In fact, BBR-induced reduction of β-catenin expression also antagonized β-catenin/TCF–mediated transcriptional activity and downstream signaling. This suggests that BBR (or its derivatives) might have the potential to be developed as therapeutic agents to target β-catenin–mutated HCCs. BBR is a known agonist of AMPK (Hawley et al., 2010), and earlier studies have established a link between BBR’s antineoplastic effects with AMPK signaling (Kim et al., 2012; Park et al., 2012; Yu et al., 2014; Li et al., 2015). A crosstalk of AMPK and β-catenin has also been reported (Zhao et al., 2010). BBR treatment in our studies showed an increase in pAMPKT172 levels, which followed an inverse correlation with β-catenin reduction, suggesting the possibility of AMPK involvement. However, AMPKα1 and AMPKα2 transient knockdown was unable to reverse BBR-induced reduction of β-catenin or downstream signaling. These were further validated by stable knockdown of AMPKα1 and AMPKα2, thus confirming that BBR-induced antagonism of β-catenin signaling is independent of AMPK. These are consistent with earlier studies that showed that BBR could antagonize nuclear factor κB signaling in colon cancer via AMPK-independent mechanism (Li et al., 2015).
A recent study in colon cancer cells have demonstrated that BBR antagonizes β-catenin via promoting its proteasomal degradation involving Retinoid X Receptor α (Ruan et al., 2017). However, in our studies, pretreatment with two different inhibitors of proteasomal degradation (lactacystin and MG132) was unable to fully rescue β-catenin expression in the HCC cells. These data, together with the observation that BBR can reduce mutant β-catenin expression in the HepG2 cells (this mutant form is resistant toward degradation by proteasomal pathway), suggested the possibility that BBR might target β-catenin via additional mechanisms. The likely explanation behind these discrepancies is that BBR-induced reduction of β-catenin in colon and liver cancer cells might be different. It is important to note that although APC mutations are found in colorectal cancer, β-catenin mutations are prevalent in liver cancer (Miyoshi et al., 1998), the majority of which enable β-catenin to evade GSK3β/APC-induced proteasomal degradation. To elucidate this pathway further, we also determined that this antagonism of β-catenin is not at the level of mRNA expression since it was reduced in the presence of actinomycin D, and β-catenin mRNA levels did not show significant inhibition with BBR when protein levels were reduced. Interestingly, pretreatment with protein synthesis inhibitor cycloheximide almost completely antagonized BBR-induced β-catenin reduction. This suggested a very novel translational mechanism involved in BBR-induced reduction of β-catenin in HCC.
Translational regulation of β-catenin involving cap-dependent translation has been shown before. Activation of Src pathway induced β-catenin expression and its downstream transcriptional activity by promoting cap-dependent translation (Karni et al., 2005). Activation of eIF4E (an essential component of cap-dependent mRNA translation) can increase β-catenin mRNA translation and promote stem cell function in blast crisis chronic myeloid leukemia (Lim et al., 2013). Cap-dependent translation is promoted by mTORC1 via phosphorylation and inhibition of 4E-BPs (4E-BP1, 4E-BP2, 4E-BP3), leading to the association of eIF4E and eIF4G, formation of eIF4F, and initiation of translation (Pause et al., 1994; Haghighat et al., 1995). Inactivation of mTORC1 signaling will thus lead to activation of 4E-BPs and repression of cap-dependent translation. In addition to mTORC1, GSK3β (Shin et al., 2014b) and casein kinase 1ε (Shin et al., 2014a) have also been shown to phosphorylate and inhibit 4E-BPs and promote cap-dependent translation. Our results with BBR show a time-dependent inhibition of mTORC1, as indicated by reduced levels of p-p70S6KThr389 and p4E-BP1Thr37/46, two downstream targets of mTORC1 pathway. Our findings support earlier observations, which showed BBR-mediated antagonism of mTOR pathway (Wang et al., 2010). In addition, and likely due to activation of mTORC2 axis, BBR also increased pAKTS473 and pGSK3βSer9 (indicating inhibition of GSK3β signaling) levels. Taken together, this suggests that BBR can reduce phosphorylation and activate 4E-BPs via two different mechanisms: one by inhibiting mTORC1 and the second by inactivating GSK3β, thus setting the stage for inhibition of cap-dependent translation. In fact, m7GTP pulldown assays confirmed this by showing that BBR treatment reduces the association of eIF4E with eIF4G while increasing its association with 4E-BP1. To determine whether BBR reduced β-catenin via antagonizing cap-dependent translation, HCC cells with stable knockdown of 4E-BP1 and 4E-BP2 (4E-BP1+2 shRNA) were created. Interestingly, although treatment with BBR reduced β-catenin expression in the control-shRNA cells, it was completely unable to reduce this in the 4E-BP1+2 shRNA cells. Similarly, BBR was unable to reduce the levels of β-catenin downstream targets in the 4E-BP1+2 shRNA cells, confirming that BBR antagonizes β-catenin expression and downstream signaling via targeting cap-dependent translation. This is further validated by the fact that 4EGI-1 (inhibitor of translation) (Moerke et al., 2007) can decrease β-catenin expression and downstream signaling significantly even in the absence of BBR. The exact reason behind the increase of β-catenin with 4EGI-1 in the Huh7 cells (Fig. 6A, lanes 7, 8) and not in other HCC cells (Fig. 6B) is unclear and might indicate cell-specific effects. Similar cancer cell–specific effects of 4EGI-1 on c-myc translation were also reported in malignant pleural mesothelioma (De et al., 2018). In addition, under certain stress conditions, translation of oncogenes can switch from cap-dependent translation to internal ribosome entry segment (IRES)-dependent translation. This was observed with c-myc, whose translation is normally induced by cap-dependent mechanism downstream of mTOR pathway (West et al., 1998) but was induced by IRES-dependent pathway when cells underwent apoptosis (Stoneley et al., 2000). Similarly, translation of vascular endothelial growth factor is induced by IRES during hypoxia, when cap-dependent translation is inhibited (Stein et al., 1998). It is thus possible that in certain cells, inhibition of β-catenin cap-dependent translation (by 4EGI-1) results in a switch toward cap-independent translation. To our knowledge, this is the first report to show that BBR, in fact, antagonizes β-catenin axis in HCC via targeting cap-dependent translation. Further studies to determine the biologic effects showed that BBR promoted cell death via antagonism of cap-dependent translation, since this process was significantly attenuated in 4E-BP1+2 shRNA cells. Interestingly, pharmacological targeting of β-catenin/TCF transcriptional activity by iCRT-14 (Gonsalves et al., 2011; Watanabe and Dai, 2011) resensitized these cells to cell death with or without BBR via increasing cytoplasmic cytochrome c release. Based on our current data and published results, we have proposed a model by which BBR regulates β-catenin translation and its impact on HCC cell survival/death (Fig. 9).
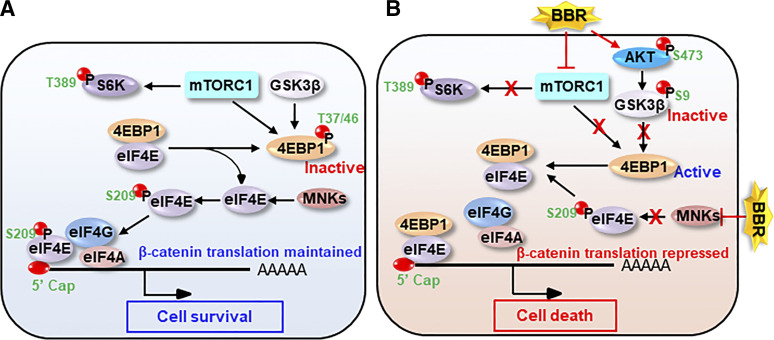
Fig. 9.
Model illustrating translational regulation of β-catenin by BBR: (A) In the absence of BBR, an active mTORC1 pathway phosphorylates and inactivates 4E-BP1, thus releasing eIF4E from 4E-BP1. MNKs then phosphorylate and activate eIF4E. eIF4E interacts with other components of the translational machinery to form eIF4F, which is recruited to 5′-mRNA cap to initiate cap-dependent translation of β-catenin. This leads to increased cell survival. (B) BBR inactivates mTORC1, leading to dephosphorylation and activation of 4E-BP1 and its interaction with eIF4E. BBR also inhibits MNK-induced eIF4E phosphorylation. This leads to disassembly of eIF4F and repression of β-catenin translation. Repression of β-catenin sensitizes the cells toward increased cell death.
In conclusion, our studies demonstrate a novel mechanism by which BBR inhibits β-catenin and its downstream signaling axis in HCC and which involves inhibition of cap-dependent translation. This pathway also seems to mediate BBR-induced cell death involving cytoplasmic c release. Some limitations of the current study include the in vitro cellular approaches used, which need to be validated in vivo utilizing animal models of HCC. However, BBR has been used in earlier in vivo studies without any reported toxicity, suggesting its potential to be developed as a cancer therapeutic agent (Ruan et al., 2017). Since β-catenin is often mutated in patients with HCC, most of which are resistant to degradation by conventional proteasomal pathway, BBR (or its derivatives) and other translation inhibitors (e.g., 4EGI-1) might provide a novel/alternate therapeutic axis by which these can be targeted. In addition, because of its antagonistic effects on GSK3β activity, BBR seems to provide a therapeutic opportunity for those cancers that are dependent on aberrant GSK3β activation (Kotliarova et al., 2008; Wilson and Baldwin, 2008). Although the pharmacological effects of BBR on β-catenin pathway antagonism are quite remarkable, the oral bioavailability and hence druggability of BBR is currently low because of several reasons (Wang et al., 2017). Based on the current studies, combination of BBR with antagonists of β-catenin or inhibitors of cap-dependent translation might help overcoming the difficulty of high dose administration. In fact, a current phase I trial with BBR combination showed promising results in patients with ulcerative colitis (Xu et al., 2020).
Acknowledgments
We are grateful to Dr. Bert Vogelstein for providing the pGL3-OT and pGL3-OF luciferase reporters [(Morin et al., 1996), (Korinek et al., 1997), (Shih et al., 2000)]. Research reported in this publication was supported in part by the University of Illinois Cancer Center Biostatistics Shared Resource Core (BSRC).
Abbreviations
- AMPK
AMP-activated protein kinase
- APC
adenomatous polyposis coli
- ATCC
American Type Culture Collection
- BBR
berberine
- BP
binding protein
- CHX
cycloheximide
- CI
confidence interval
- CM
conditioned medium
- CTNNB1
β-catenin
- DMEM
Dulbecco’s modified Eagle’s medium
- 4E-BP
eIF4E-BP
- eIF4E
eukaryotic translation initiation factor 4
- β-gal
β-galactosidase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GSK
glycogen synthase kinase
- HCC
hepatocellular carcinoma
- HEK293
human embryonic kidney 293
- iCRT
β-catenin responsive transcription (iCRT-14)
- IRES
internal ribosome entry segment
- m7
7-methyl
- MEM
minimum essential medium
- MNK
mitogen-activated protein kinase–interacting serine/threonine kinase
- mTOR
mammalian target of rapamycin
- p-p70S6K
phosphorylated p70S6K at Thr 389 (p-p70S6KThr389)
- p4E-BP1
phosphorylated 4E-BP1 at Thr 37, 46 (p4E-BP1Thr37/46)
- pAMPK
phosphorylated AMPK at Thr 172 (pAMPKT172)
- PCR
polymerase chain reaction
- Pen/Strep
penicillin/streptomycin
- pGSK
phosphorylated GSK3β at Ser 9 (pGSK3βSer9)
- PI
propidium iodide
- qPCR
quantitative PCR
- RLU
relative light unit
- shRNA
short hairpin RNA
- siRNA
small interference RNA
- TCF
T-cell factor
- TRAIL
tumor necrosis factor–related apoptosis-inducing ligand
Authorship Contributions
Participated in research design: Vishnoi, Ke, Saini, Viswakarma, Nair, Das, A. Rana, B. Rana.
Conducted experiments: Vishnoi, Ke, Saini, Viswakarma, Nair, Das.
Performed data analysis: Vishnoi, Ke, Saini, Viswakarma, Nair, Das, Chen, A. Rana, B. Rana.
Wrote or contributed to the writing of the manuscript: Vishnoi, Ke, A. Rana, B. Rana.
Footnotes
This work was supported by National Institutes of Health National Cancer Institute [Grants R01 CA178063 and R03 CA219764] (to B.R.) and [Grants R01 CA176846 and R01 CA216410] (to A.R.) as well as the Veterans Affairs Merit Award [Grant I01 BX003296] (to B.R.) and [Grants I01 BX002703 and I01 BX002355] (to A.R).
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Alao JP, Stavropoulou AV, Lam EW, Coombes RC, Vigushin DM. (2006) Histone deacetylase inhibitor, trichostatin A induces ubiquitin-dependent cyclin D1 degradation in MCF-7 breast cancer cells. Mol Cancer 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albring KF, Weidemüller J, Mittag S, Weiske J, Friedrich K, Geroni MC, Lombardi P, Huber O. (2013) Berberine acts as a natural inhibitor of Wnt/β-catenin signaling--identification of more active 13-arylalkyl derivatives. Biofactors 39:652–662. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. (1996) Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638–642. [DOI] [PubMed] [Google Scholar]
- Bienz M, Clevers H. (2000) Linking colorectal cancer to Wnt signaling. Cell 103:311–320. [DOI] [PubMed] [Google Scholar]
- Cadoret A, Ovejero C, Terris B, Souil E, Lévy L, Lamers WH, Kitajewski J, Kahn A, Perret C. (2002) New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene 21:8293–8301. [DOI] [PubMed] [Google Scholar]
- Caruso S, O’Brien DR, Cleary SP, Roberts LR, Zucman-Rossi J. (2020) Genetics of HCC: novel approaches to explore molecular diversity. Hepatology DOI: 10.1002/hep.31394 [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Chandra D, Choy G, Deng X, Bhatia B, Daniel P, Tang DG. (2004) Association of active caspase 8 with the mitochondrial membrane during apoptosis: potential roles in cleaving BAP31 and caspase 3 and mediating mitochondrion-endoplasmic reticulum cross talk in etoposide-induced cell death. Mol Cell Biol 24:6592–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Nair RS, Mishra R, Sondarva G, Viswakarma N, Abdelkarim H, Gaponenko V, Rana B, Rana A. (2019) Mixed lineage kinase 3 promotes breast tumorigenesis via phosphorylation and activation of p21-activated kinase 1. Oncogene 38:3569–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, Jacobson BA, Peterson MS, Jay-Dixon J, Kratzke MG, Sadiq AA, Patel MR, Kratzke RA. (2018) 4EGI-1 represses cap-dependent translation and regulates genome-wide translation in malignant pleural mesothelioma. Invest New Drugs 36:217–229. [DOI] [PubMed] [Google Scholar]
- de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, et al. (1998) Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA 95:8847–8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom KS, Hong JM, Youn MJ, So HS, Park R, Kim JM, Kim TY. (2008) Berberine induces G1 arrest and apoptosis in human glioblastoma T98G cells through mitochondrial/caspases pathway. Biol Pharm Bull 31:558–562. [DOI] [PubMed] [Google Scholar]
- Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S, Nagourney R, Cardozo T, Brown AM, DasGupta R. (2011) An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci USA 108:5954–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N. (1995) Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J 14:5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, Towler MC, Brown LJ, Ogunbayo OA, Evans AM, et al. (2010) Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab 11:554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang K, Gu C, Yu G, Zhao D, Mai W, Zhong Y, Liu S, Nie Y, Yang H. (2018) Berberine, a natural plant alkaloid, synergistically sensitizes human liver cancer cells to sorafenib. Oncol Rep 40:1525–1532. [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev 59:3–10. [DOI] [PubMed] [Google Scholar]
- Karni R, Gus Y, Dor Y, Meyuhas O, Levitzki A. (2005) Active Src elevates the expression of beta-catenin by enhancement of cap-dependent translation. Mol Cell Biol 25:5031–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Vishnoi K, Viswakarma N, Santha S, Das S, Rana A, Rana B. (2018) Involvement of AMP-activated protein kinase and Death Receptor 5 in TRAIL-Berberine-induced apoptosis of cancer cells. Sci Rep 8:5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim MJ, Kim EJ, Yang Y, Lee MS, Lim JS. (2012) Berberine-induced AMPK activation inhibits the metastatic potential of melanoma cells via reduction of ERK activity and COX-2 protein expression. Biochem Pharmacol 83:385–394. [DOI] [PubMed] [Google Scholar]
- Kim S, Oh SJ, Lee J, Han J, Jeon M, Jung T, Lee SK, Bae SY, Kim J, Gil WH, et al. (2013) Berberine suppresses TPA-induced fibronectin expression through the inhibition of VEGF secretion in breast cancer cells. Cell Physiol Biochem 32:1541–1550. [DOI] [PubMed] [Google Scholar]
- Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, Wang Y, Wang Z, Si S, Pan H, et al. (2004) Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med 10:1344–1351. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 275:1784–1787. [DOI] [PubMed] [Google Scholar]
- Kotliarova S, Pastorino S, Kovell LC, Kotliarov Y, Song H, Zhang W, Bailey R, Maric D, Zenklusen JC, Lee J, et al. (2008) Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res 68:6643–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishan S, Richardson DR, Sahni S. (2015) Adenosine monophosphate-activated kinase and its key role in catabolism: structure, regulation, biological activity, and pharmacological activation. Mol Pharmacol 87:363–377. [DOI] [PubMed] [Google Scholar]
- Laderoute KR, Calaoagan JM, Chao WR, Dinh D, Denko N, Duellman S, Kalra J, Liu X, Papandreou I, Sambucetti L, et al. (2014) 5′-AMP-activated protein kinase (AMPK) supports the growth of aggressive experimental human breast cancer tumors. J Biol Chem 289:22850–22864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Kim WS, Kim KH, Yoon MJ, Cho HJ, Shen Y, Ye JM, Lee CH, Oh WK, Kim CT, et al. (2006) Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes 55:2256–2264. [DOI] [PubMed] [Google Scholar]
- Li W, Hua B, Saud SM, Lin H, Hou W, Matter MS, Jia L, Colburn NH, Young MR. (2015) Berberine regulates AMP-activated protein kinase signaling pathways and inhibits colon tumorigenesis in mice. Mol Carcinog 54:1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J, Lim AQ, Chang CT, Fruman DA, Rizzieri DA, et al. (2013) Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc Natl Acad Sci USA 110:E2298–E2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Ying Y, Wang YY, Wang G, Jiang SS, Huang D, Luo L, Chen YG, Gerstenfeld LC, Luo Z. (2017) AMPK downregulates ALK2 via increasing the interaction between Smurf1 and Smad6, leading to inhibition of osteogenic differentiation. Biochim Biophys Acta Mol Cell Res 1864:2369–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Bruix J. (2008) Molecular targeted therapies in hepatocellular carcinoma. Hepatology 48:1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. SHARP Investigators Study Group (2008) Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359:378–390. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10:307–318. [DOI] [PubMed] [Google Scholar]
- Mishra P, Paramasivam SK, Thylur RP, Rana A, Rana B. (2010) Peroxisome proliferator-activated receptor gamma ligand-mediated apoptosis of hepatocellular carcinoma cells depends upon modulation of PI3Kinase pathway independent of Akt. J Mol Signal 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, Murata M, Shimano T, Nakamura Y. (1998) Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res 58:2524–2527. [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. (2007) Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128:257–267. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. (1996) XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86:391–399. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275:1787–1790. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Vogelstein B, Kinzler KW. (1996) Apoptosis and APC in colorectal tumorigenesis. Proc Natl Acad Sci USA 93:7950–7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhieu JT, Renard CA, Wei Y, Cherqui D, Zafrani ES, Buendia MA. (1999) Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol 155:703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Seo SM, Kim EJ, Lee YJ, Ko YG, Ha J, Lee M. (2012) Berberine inhibits human colon cancer cell migration via AMP-activated protein kinase-mediated downregulation of integrin β1 signaling. Biochem Biophys Res Commun 426:461–467. [DOI] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donzé O, Lin TA, Lawrence JC, Jr, Sonenberg N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762–767. [DOI] [PubMed] [Google Scholar]
- Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L, Banales JM. (2019) Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol 16:121–136. [DOI] [PubMed] [Google Scholar]
- Pinyol R, Sia D, Llovet JM. (2019) Immune exclusion-wnt/CTNNB1 class predicts resistance to immunotherapies in HCC. Clin Cancer Res 25:2021–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta C, Paglino C. (2010) Medical treatment of unresectable hepatocellular carcinoma: going beyond sorafenib. World J Hepatol 2:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep A, Sharma C, Sathyanarayana P, Albanese C, Fleming JV, Wang TC, Wolfe MM, Baker KM, Pestell RG, Rana B. (2004) Gastrin-mediated activation of cyclin D1 transcription involves beta-catenin and CREB pathways in gastric cancer cells. Oncogene 23:3689–3699. [DOI] [PubMed] [Google Scholar]
- Ruan H, Zhan YY, Hou J, Xu B, Chen B, Tian Y, Wu D, Zhao Y, Zhang Y, Chen X, et al. (2017) Berberine binds RXRα to suppress β-catenin signaling in colon cancer cells. Oncogene 36:6906–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JO, Monga SP. (2018) Wnt/β-catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol 13:351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santha S, Viswakarma N, Das S, Rana A, Rana B. (2015) Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-troglitazone-induced apoptosis in prostate cancer cells involve AMP-activated protein kinase. J Biol Chem 290:21865–21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthivinayagam S, Mishra P, Paramasivam SK, Yallapragada S, Chatterjee M, Wong L, Rana A, Rana B. (2009) Caspase-mediated cleavage of beta-catenin precedes drug-induced apoptosis in resistant cancer cells. J Biol Chem 284:13577–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C, Pradeep A, Wong L, Rana A, Rana B. (2004) Peroxisome proliferator-activated receptor gamma activation can regulate beta-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J Biol Chem 279:35583–35594. [DOI] [PubMed] [Google Scholar]
- Shih IM, Yu J, He TC, Vogelstein B, Kinzler KW. (2000) The beta-catenin binding domain of adenomatous polyposis coli is sufficient for tumor suppression. Cancer Res 60:1671–1676. [PubMed] [Google Scholar]
- Shin S, Wolgamott L, Roux PP, Yoon SO. (2014a) Casein kinase 1ε promotes cell proliferation by regulating mRNA translation. Cancer Res 74:201–211. [DOI] [PubMed] [Google Scholar]
- Shin S, Wolgamott L, Tcherkezian J, Vallabhapurapu S, Yu Y, Roux PP, Yoon SO. (2014b) Glycogen synthase kinase-3β positively regulates protein synthesis and cell proliferation through the regulation of translation initiation factor 4E-binding protein 1. Oncogene 33:1690–1699. [DOI] [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol 18:3112–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. (2000) c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol 20:1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau C, Moriarty AM, Thornton GB, Lanford RE. (1992) Production of infectious hepatitis delta virus in vitro and neutralization with antibodies directed against hepatitis B virus pre-S antigens. J Virol 66:1241–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thylur RP, Senthivinayagam S, Campbell EM, Rangasamy V, Thorenoor N, Sondarva G, Mehrotra S, Mishra P, Zook E, Le PT, et al. (2011) Mixed lineage kinase 3 modulates β-catenin signaling in cancer cells. J Biol Chem 286:37470–37482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillhon M, Guamán Ortiz LM, Lombardi P, Scovassi AI. (2012) Berberine: new perspectives for old remedies. Biochem Pharmacol 84:1260–1267. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65:87–108. [DOI] [PubMed] [Google Scholar]
- Tsang CM, Cheung KC, Cheung YC, Man K, Lui VW, Tsao SW, Feng Y. (2015) Berberine suppresses Id-1 expression and inhibits the growth and development of lung metastases in hepatocellular carcinoma. Biochim Biophys Acta 1852:541–551. [DOI] [PubMed] [Google Scholar]
- Viswakarma N, Nair RS, Sondarva G, Das S, Ibrahimi L, Chen Z, Sinha S, Rana B, Rana A. (2017) Transcriptional regulation of mixed lineage kinase 3 by estrogen and its implication in ER-positive breast cancer pathogenesis. Oncotarget 8:33172–33184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad A, Röhrs S, Klein-Hitpass L, Müller O. (2008) The first five years of the Wnt targetome. Cell Signal 20:795–802. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhu C, Ying Y, Luo L, Huang D, Luo Z. (2017) Metformin and berberine, two versatile drugs in treatment of common metabolic diseases. Oncotarget 9:10135–10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cao H, Lu N, Liu L, Wang B, Hu T, Israel DA, Peek RM, Jr, Polk DB, Yan F. (2013) Berberine inhibits proliferation and down-regulates epidermal growth factor receptor through activation of Cbl in colon tumor cells. PLoS One 8:e56666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Feng Y, Zhu M, Tsang CM, Man K, Tong Y, Tsao SW. (2010) Berberine induces autophagic cell death and mitochondrial apoptosis in liver cancer cells: the cellular mechanism. J Cell Biochem 111:1426–1436. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Dai X. (2011) Winning WNT: race to Wnt signaling inhibitors. Proc Natl Acad Sci USA 108:5929–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Stoneley M, Willis AE. (1998) Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17:769–780. [DOI] [PubMed] [Google Scholar]
- Wilson W, III, Baldwin AS. (2008) Maintenance of constitutive IkappaB kinase activity by glycogen synthase kinase-3alpha/beta in pancreatic cancer. Cancer Res 68:8156–8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Yang Q, Mu Y, Zhou L, Liu Y, Zhou Q, He B. (2012) Berberine inhibits the proliferation of colon cancer cells by inactivating Wnt/β-catenin signaling. Int J Oncol 41:292–298. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhang Y, Xue X, Liu J, Li ZS, Yang GY, Song Y, Pan Y, Ma Y, Hu S, et al. (2020) A Phase I trial of berberine in Chinese with ulcerative colitis. Cancer Prev Res (Phila) 13:117–126. [DOI] [PubMed] [Google Scholar]
- Yin J, Xing H, Ye J. (2008) Efficacy of berberine in patients with type 2 diabetes mellitus. Metabolism 57:712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Zhang ZQ, Wang B, Jiang HX, Cheng L, Shen LM. (2014) Berberine-induced apoptotic and autophagic death of HepG2 cells requires AMPK activation. Cancer Cell Int 14:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Dahabieh MS, Rajakumar A, Dobocan MC, M’Boutchou MN, Goncalves C, Lucy SL, Pettersson F, Topisirovic I, van Kempen L, et al. (2015) The role of eIF4E in response and acquired resistance to vemurafenib in melanoma. J Invest Dermatol 135:1368–1376. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wei J, Xue R, Wu JD, Zhao W, Wang ZZ, Wang SK, Zhou ZX, Song DQ, Wang YM, et al. (2010) Berberine lowers blood glucose in type 2 diabetes mellitus patients through increasing insulin receptor expression. Metabolism 59:285–292. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cao H, Zhang B, Cao H, Xu X, Ruan H, Yi T, Tan L, Qu R, Song G, et al. (2013) Berberine potently attenuates intestinal polyps growth in ApcMin mice and familial adenomatous polyposis patients through inhibition of Wnt signalling. J Cell Mol Med 17:1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yue W, Zhu MJ, Sreejayan N, Du M. (2010) AMP-activated protein kinase (AMPK) cross-talks with canonical Wnt signaling via phosphorylation of beta-catenin at Ser 552. Biochem Biophys Res Commun 395:146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Zheng, Virshup David M. (2020) Wnt Signaling and Drug Resistance in Cancer. Mol Pharmacol 97:72–89. [DOI] [PubMed] [Google Scholar]