SUMMARY
Precisely deciphering the molecular mechanisms of age-related memory loss is crucial to create appropriate therapeutic interventions. We have previously shown that the histone-binding protein RbAp48/Rbbp4 is a molecular determinant of Age-Related Memory Loss. By exploring how this protein regulates the genomic landscape of the hippocampal circuit, we find that RbAp48 controls the expression of BDNF and GPR158 proteins, both critical components of osteocalcin (OCN) signaling in the mouse hippocampus. We show that inhibition of RbAp48 in the hippocampal formation inhibits OCN’s beneficial functions in cognition and causes deficits in discrimination memory. In turn, disruption of OCN/GPR158 signaling leads to the downregulation of RbAp48 protein, mimicking the discrimination memory deficits observed in the aged hippocampus. We also show that activation of the OCN/GPR158 pathway increases the expression of RbAp48 in the aged dentate gyrus and rescues age-related memory loss.
Graphical Abstract
In Brief
Kosmidis et al. show how osteocalcin, a hormone produced by the bones, can ameliorate symptoms of age-related memory loss by upregulating the histone binding protein RbAp48.
INTRODUCTION
Normal aging produces significant alterations in hippocampal circuitry, thereby leading to deficits in key components of episodic memory (Leal and Yassa, 2015). Studies in human and in mouse model systems have established that aging may lead to spatial memory impairment (Bach et al., 1999; Pavlopoulos et al., 2013; Small et al., 2004) and to deficits in pattern separation/completion processes (Clark et al., 2017; McAvoy et al., 2016; Stark et al., 2013). Imbalances between pattern separation and pattern completion, which are mediated by the dentate gyrus (DG) and the CA3 hippocampal sub-regions, ultimately contribute to memory deficits and to age-related memory loss (Leal and Yassa, 2015).
Learning and memory formation and maintenance are complex processes that are tightly controlled at the cellular level by regulation of gene expression, which in turn can be affected by DNA modifications such as histone acetylation and DNA methylation (Alarcón et al., 2004; Guan et al., 2002). Cellular aging has also been associated with DNA changes and remodeling of chromatin architecture, leading to deregulation of gene transcription programs and perturbations to cellular homeostasis (Pegoraro and Misteli, 2009); transcription of several hippocampal plasticity-dependent genes, including ARC, ZIF268, and BDNF, is also governed by similar modifications, such as DNA methylation or histone acetylation (Penner et al., 2011). Deregulation of plasticity-dependent genes in brain regions susceptible to the aging process may therefore contribute to age-related cognitive deficits.
Decrease in histone-binding protein Rbbp4/RbAp48 expression is a molecular hallmark of normal age-related memory loss in humans and in wild-type mice. Inhibition of RbAp48 function interferes with its capacity for histone interaction and recapitulates age-related memory loss in young mice. Overexpression of RbAp48 in the DG of older mice alleviates the cognitive deficits of aging and restores histone acetylation (Pavlopoulos et al., 2013). Despite its unequivocal importance, the signaling cascades responsible for the dysregulation of RbAp48 in age-related memory loss remain unknown.
Here, we explore the molecular mechanisms of RbAp48 function in the brain and its role in age-related memory loss. RbAp48 was found to control the expression of GPR158, a G-protein-coupled receptor (GPCR) protein involved in osteocalcin (OCN) signaling in the brain (Khrimian et al., 2017). In turn, GPR158/OCN signaling regulates RbAp48 expression in the hippocampal formation. We show that the OCN/GPR158/RbAp48 signaling pathway acts on the DG/CA3c and CA3a brain regions to modulate contextual fear discrimination and completion, respectively. Finally, we find that activation of this signaling pathway in the aged DG alleviates the cognitive deficits associated with aging.
RESULTS
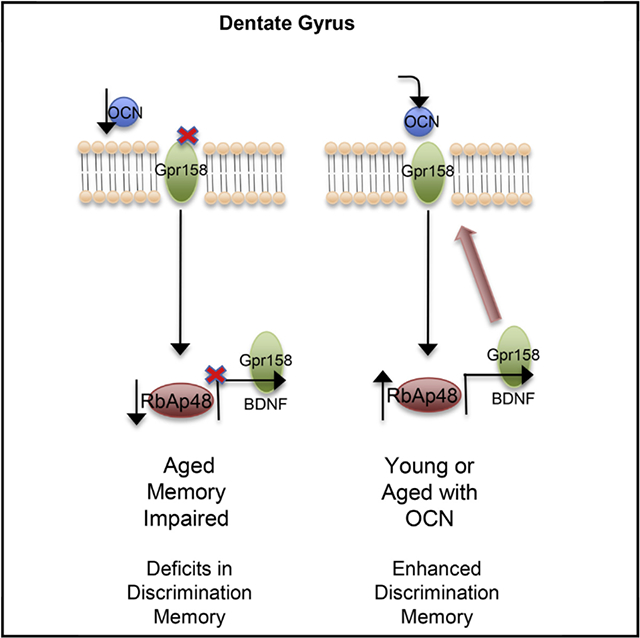
RbAp48 Is Involved in OCN Signaling and Regulates the Levels of GPR158
We first aimed to identify the genomic loci of RbAp48 binding in the mouse DG. We performed chromatin immunoprecipitation experiments coupled with deep sequencing. Analysis of two biological replicates revealed that RbAp48 binds mainly to heterochromatin regions and particularly to olfactory receptor genes, consistent with the well-described role of RbAp48 in repression complexes (Loyola and Almouzni, 2004; Yamada et al., 2014). Since other biological functions of RbAp48 protein could have been masked by its olfactory receptor genes interactions, we excluded olfactory receptor genes from our analysis and merged the two biological replicates to increase the number of the potential RbAp48 binding sites. We found that RbAp48 protein binds to 7,179 sites (Table S1, RbAp48 binding sites related to Figure 1). 14% of these sites were around gene promoters (−/+ 5 kb from transcription start sites [TSSs]), 18% were within gene coding sequences, and 68% were intergenic–enhancer binding sites (Figure 1A). Gene ontology analysis revealed neuronal pathways such as perception of taste, nervous system development, and learning (Figure 1B and Table S2, gene ontology and pathways analysis of RbAp48 chromatin immunoprecipitation sequencing [ChlP-seq] related to Figure 1). Since RbAp48 participates in several transcriptional complexes (NuRD, PRC2, etc.), we surveyed our results for RbAp48 binding sites in genes already implicated in age-related memory loss. We focused on the BDNF locus, because its expression decreases with aging (Rowe et al., 2007), and on the GPCR GPR158, since it mediates the beneficial actions of the bone-derived hormone OCN in age-related memory loss by controlling BDNF protein levels in the mouse hippocampus (Khrimian et al., 2017) (Figure 1C). Using ChlP-qPCR, we found that RbAp48 occupies the P5 BDNF promoter and the GPR158 promoter and within the gene (intron) (Figures 1D–1F). To verify our results, we used a transgenic mouse model of inhibition of RbAp48 function, where a truncated form of RbAp48 lacking the first 54 aa responsible for binding to histones, is overexpressed in forebrain neurons under the Camk2-tTA transgene (referred to as dominant-negative mice [DT]). Expression of this transgenic form of RbAp48 is suppressed in presence of doxycycline (Pavlopoulos et al., 2013). Western blot and immunohistochemistry experiments revealed a reduction in the protein levels of GPR158 in the hippocampus of DT mice compared with single transgenic (ST) controls (Figures 1G–1I). (DT mice bear two transgenes required for RbAp48 overexpression, whereas ST mice only one). We next wondered whether RbAp48 is part of the OCN signaling cascade, which controls GPR158 and BDNF protein levels. Therefore, we measured GPR158 and BDNF mRNA levels in DT and ST mice 24 hr after PBS or OCN injection in the DG. GPR158 mRNA levels were significantly decreased in the DG of the DT mice compared to ST controls (Figure 1J).
Figure 1. RbAp48 Controls GPR158 and OCN Signaling.
(A) Distribution of RbAp48 binding sites in the mouse genome.
(B) Top ten biological processes for genes that RbAp48 binds to.
(C) Peaks in the mouse genome for RbAp48 binding compared with the input DNA for BDNF and GPR158 loci.
(D-F) Quantitative real-time PCR of ChIP assays from dentate gyrus (DG) lysates for the GPR158 promoter (D), the GPR158 intron (E), and P5 BDNF promoter (F) compared to input DNA (n = 6 mice per group). Fold enrichment of RbAp48 binding in the respective genomic regions (one-way ANOVA treatment between columns post hoc Bonferroni; p = 0.0129, p = 0.0275, p = 0.0036, respectively).
(G) Western blot images of GPR158, transgenic Flag-tagged DN-RbAp48, and tubulin proteins from hippocampal lysates from control (ST) and dominant-negative RbAp48 overexpression mice (DT).
(H) Quantification of GPR158 protein levels (unpaired t test, p = 0.0038; n = 6 mice per group).
(I) Representative images of the DG of DT and ST mice stained for GPR158 and RbAp48 (scale bar, 20 μm).
(J) Quantitative real-time PCR of GPR158 mRNA from the DG/CA3c of ST or DT mice injected with PBS or OCN. GPR158 mRNA is downregulated in the DG of DT mice compared to ST controls injected with PBS (one-way ANOVA treatment between columns post hoc Bonferroni; n = 4–5 mice per treatment, respectively; p = 0.0113).
All data represent means ± SEM. *p < 0.05, **p < 0.01.
In order to further correlate our molecular findings with OCN signaling and RbAp48, we examined whether inhibition of RbAp48 blocks BDNF expression in the DG (Khrimian et al., 2017; Liu et al., 2004). We detected BDNF upregulation both in the mRNA and protein levels only in the DG of ST animals injected with OCN (Figures S1A and S1B), but not in the DG of DT animals. We also measured the RbAp48 mRNA levels in the same animals and found a significant increase upon OCN injection in the DG (Figure S1C) in control ST mice, which was completely abolished in the DT RbAp48 mice, further demonstrating that RbAp48 participates in OCN signaling.
OCN Increases RbAp48 in the DG and Enhances Fear Expression during Context Re-exposure in a Contextual Fear Conditioning Paradigm
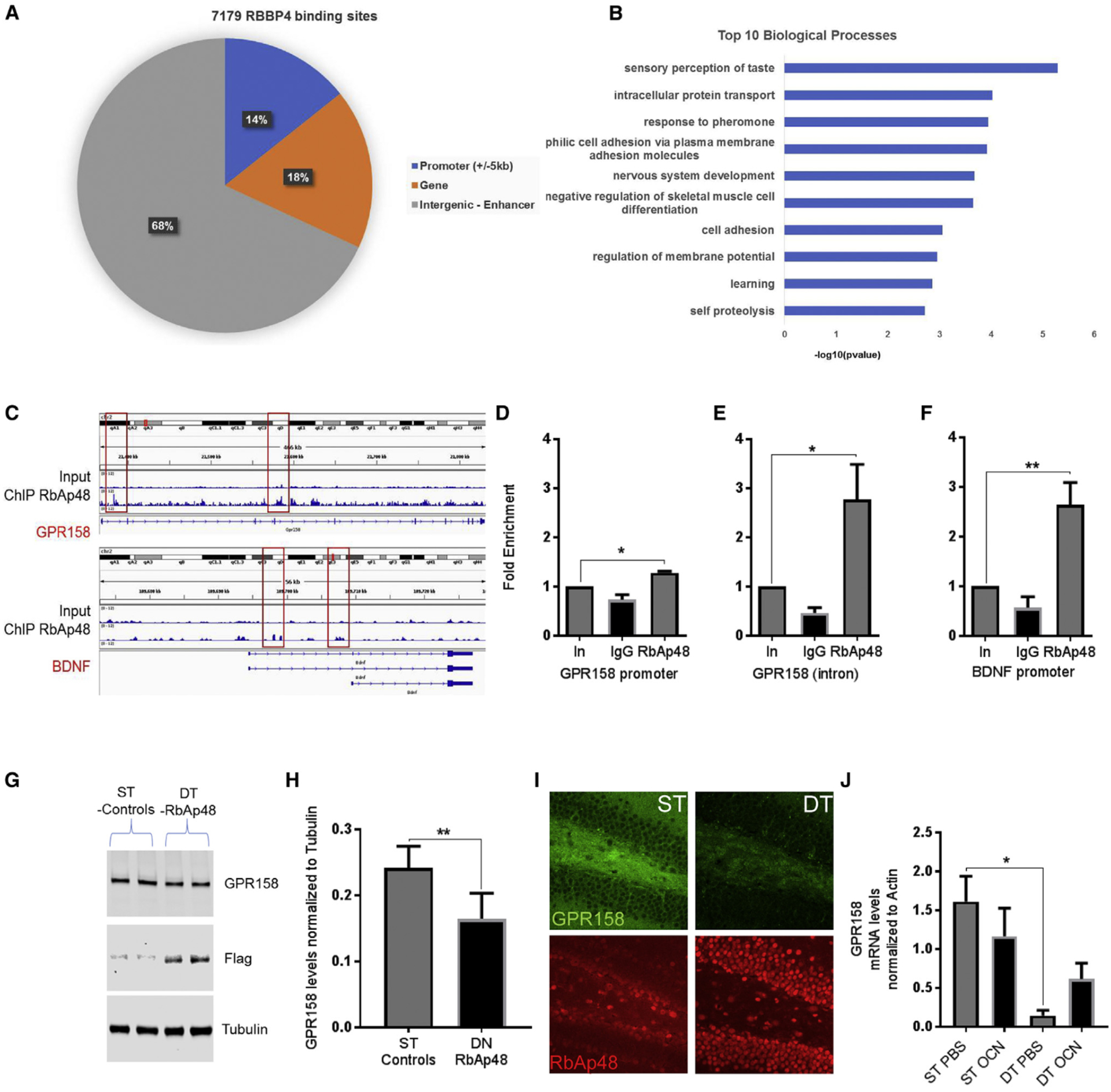
Since OCN injection increased RbAp48 mRNA levels in the DG, we tested whether the presence of OCN in the DG can also affect the expression of RbAp48 protein. We injected OCN directly into the DG of 3-month-old mice, and we observed a modest increase in RbAp48 in the DG compared with PBS-injected controls within 24 hr (Figures 2A and 2B).
Figure 2. OCN Increases RbAp48 in the DG and Enhances Fear Expression during Context Re-exposure (Contextual Fear Conditioning).
(A) Images of the DG stained for RbAp48 (red) 24 hr after administration of PBS (top) or OCN (bottom) in the DG (scale bar, 100 μm).
(B) OCN injection increases RbAp48 24 hr after an injection (unpaired t tests; n = 8 mice per group; p = 0.0103).
(C) Schematic representation of contextual fear conditioning (CFC) in c-FosCreERT2 × tdTomato mice.
(D) Images from DG of c-FosCreERT2/tdTomato mice stained for mCherry (red) PBS (left) or OCN (right) (scale bar, 100 μm).
(E) c-FosCreERT2 × tdTomato mice were administered a three-shock CFC paradigm. OCN administration did not affect fear acquisition behavior, but it resulted in increased fear expression during context re-exposure (unpaired t tests; n = 6 mice per group; p = 0.016).
(F) Images from c-Fos+ dendrites in the outer molecular layer (OML), OCN (right) and PBS (left) (scale bar, 10 μm).
(G) Quantification of spines in c-Fos-+ dendrites in the OML, OCN (right) and PBS (left). OCN increased spine density in c-Fos+ neurons (unpaired t tests; n = 3 mice per group; p = 0.0146; <35 dendrites per animal).
All data represent means ± SEM. *p < 0.05.
Similarly, 24-hr treatment with OCN in hippocampal neuronal cultures (17 days in vitro [DIV]) produced a nuclear accumulation of RbAp48 compared with control-treated neurons and upregulation of PSD95 protein (Figures S2A–S2C) consistent with the notion that OCN enhances long-term potentiation (LTP) in pyramidal cells of the CA3 region via a synaptic specific mechanism (Khrimian et al., 2017).
We next wondered whether upregulation of RbAp48/PSD95 upon OCN administration in the DG can affect memory. We used a transgenic c-FosCreERT2/tdTomato reporter mouse line to label activity-dependent neurons (c-Fos+) and their processes with tdTomato red fluorescent protein (Guenthner et al., 2013). We argued that since OCN can regulate cFos expression and LTP in the CA3a region through binding to the GPR158 receptor (Khrimian et al., 2017), the use of this transgenic mouse would be a suitable tool to visualize any synaptic effects of OCN administration. We subjected the c-FosCreERT2/tdTomato line to an experimental setting illustrated in Figure 2C. OCN administration in the DG increased the percentage of freezing compared to control (Figures 2D and 2E). The numbers of c-Fos+ cells were not significantly different between the two groups (Figure S2D), showing that our manipulation had a similar number of activation of cFos throughout the experimental setting. Since the number of labeled neurons was the same and OCN administration was directed into the whole DG/CA3a region, we measured the number of spines in the tdTomato-positive neurons to examine any plasticity-depended effects of OCN. OCN-treated mice had more dendritic spines than the PBS-treated controls (Figures 2F and 2G). Taken together, these results suggest that (1) OCN regulates the RbAp48 protein, which in turn controls the expression of GPR158; (2) OCN administration in the DG/CA3c hippocampal region can lead to enhanced 7-day fear-memory recall; and (3) OCN administration coupled with a fear memory recall behavioral task can lead to increased spine formation in neurons of the outer molecular layer of the DG.
OCN Signaling Modulates Learning and Memory through an RbAp48-Dependent Mechanism in the DG/CA3
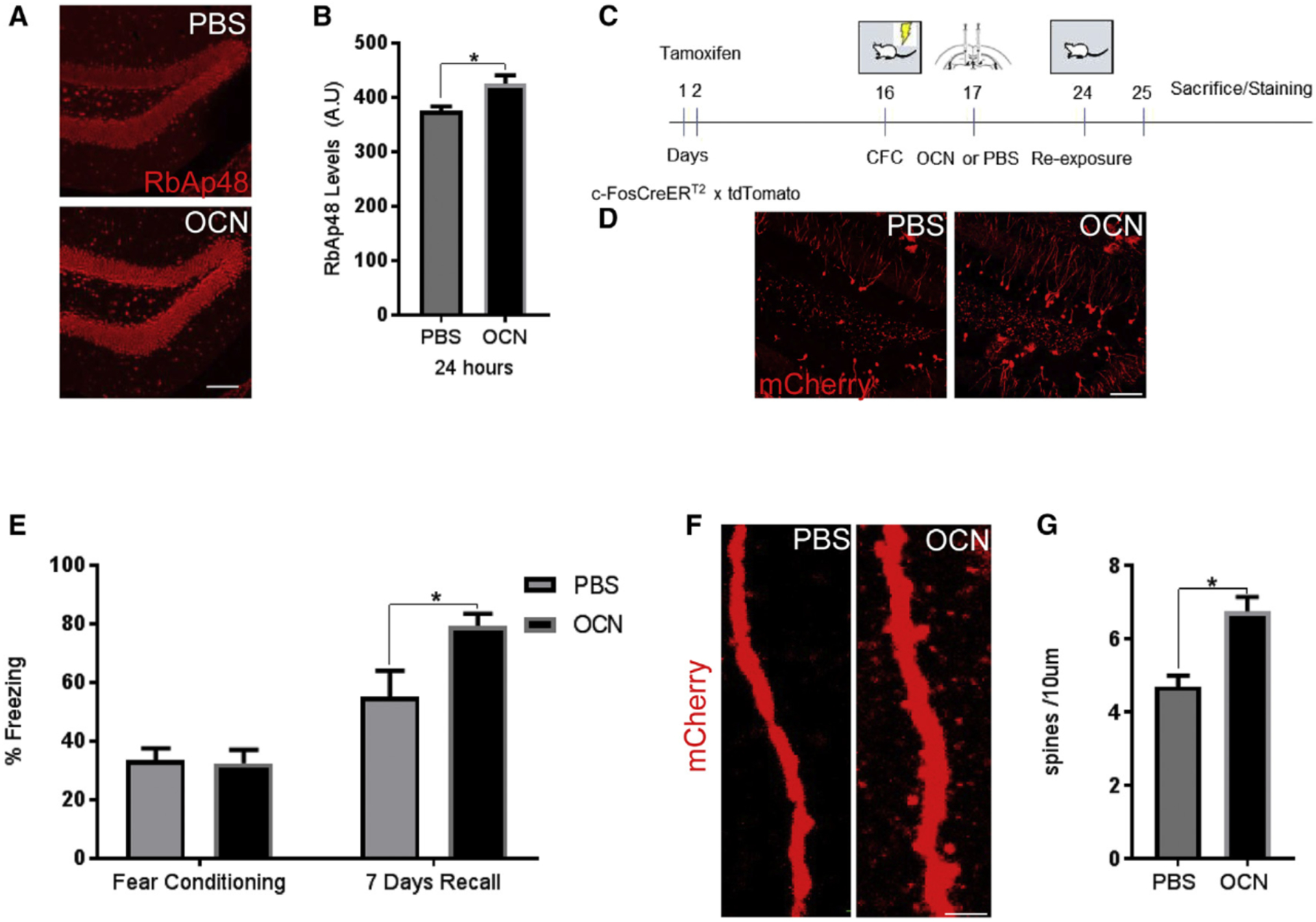
Having found that RbAp48 participates in GPR158/OCN signaling, and that RbAp48 regulates GPR158 expression, we asked whether histone binding by RbAp48 is essential to OCN’s role in memory. OCN seems to act both in the DG, a region required to separate two similar memories with very distinct differences, and in the CA3, which helps to retrieve a memory when partial cues are presented (Besnard and Sahay, 2016). We therefore first subjected cohorts of ST and DT mice to a novel object recognition (NOR) task, which is a hippocampal-dependent memory task (Clark et al., 2000; Pavlopoulos et al., 2013). Administration of OCN (Figures 3A and S3A, bottom) or PBS (Figure 3A and S3A, top) in the DG (Figure 3A) or in CA3a (Figure S3A) had no effect on the total exploration time between the familiar and the novel phase in both DT and ST mice (Figures 3B and S3B). However, the discrimination index of ST animals injected with OCN in either hippocampal region was increased in the novel phase of the task (Figures 3C and S3C). OCN administration either in the DG or in CA3a of DT animals failed to rescue the deficit caused by inhibition of RbAp48 (Figures 3C and S3C), demonstrating once again that inhibiting RbAp48 function in young animals is important for spatial memory, and that OCN signaling is blocked upon RbAp48 inhibition.
Figure 3. OCN Function Is Blocked by RbAp48 Inhibition.
(A) Images of the DG showing accurate targeting of OCN (100 ng) 15 min after injection of mStrawberry alone (top) or fusion mStrawberry-OCN (bottom). Total OCN (green) staining was performed as control (scale bar, 100 μm).
(B and C) Novel object recognition (NOR) test after injection of PBS or OCN in the DG of ST and DT mice (n = 16 mice per group). No differences were detected in total exploration time between the familiarization and the testing phase (three-way ANOVA, post hoc Bonferroni) (B), and discrimination index for novel object (C). OCN had an effect only on ST mice compared to DT OCN-treated mice (two-way repeated-measures [RM] ANOVA comparisons between OCN or PBS and ST versus DT; post hoc Bonferroni, p < 0.0001).
(D) Schematic contextual fear discrimination task.
(E-H) Contextual fear discrimination in ST or DT mice injected with OCN in the DG/CA3c (F and H). Both genotypes were able to discriminate between the two contexts after PBS injection (E and G), but only ST were able to discriminate the two contexts after OCN injection (F) (two-way RM ANOVA, post hoc Bonferroni; three-way RM ANOVA, post hoc Bonferroni for treatment × contexts in ST mice, p = 0.0003, and treatment x contexts in DT mice; n = 16 mice per group).
(I) Discrimination index of ST and DT mice injected in the DG with OCN or PBS 10 days after the end of contextual fear discrimination. OCN-injected ST mice were still able to discriminate better than the ST PBS or DT OCN mice (one-way ANOVA, post hoc Bonferroni, p = 0.0029 and p = 0.0194).
All data represent means ± SEM. *p < 0.05, **p<0.01, ***p < 0.001.
Since memory deficits can be associated with impairments in pattern separation (Dillon et al., 2017; Holden and Gilbert, 2012), the same groups of animals were further tested in a contextual fear discrimination paradigm commonly used to assess pattern separation (Jinde et al., 2012; Sahay et al., 2011) (Figure 3D). OCN injection in the DG greatly increased discrimination in ST animals but had no effect in DT animals (Figures 3E–3H). In addition, we found that OCN had no effect in the same contextual fear discrimination task when delivered into the CA3a region (Figures S3D–S3G). ST animals injected with OCN in the DG were able to discriminate better than those injected with PBS or compared to DT animals injected either with PBS or OCN, even 10 days after the end of the behavioral assay (Figure 3I). In contrast, cohorts injected in CA3a did not display significant differences (Figure S3H), demonstrating that OCN can mediate differential effects in spatial and discrimination memory tasks depending on the hippocampal region that it binds to.
Inability of OCN to produce a significant improvement in the NOR task and in contextual fear discrimination paradigms when injected in the DG or CA3 of DT mice further suggests that RbAp48 protein is necessary for OCN signaling, and that the memory deficits observed in DT mice can be attributed to the deregulation of the OCN/GPR158/BDNF pathway.
Knockdown of GPR158 in the DG Affects the Levels of RbAp48 Protein and Discrimination Memory
We next wondered whether OCN/GPR158 signaling mediates a loop that controls RbAp48 protein, which then regulates GPR158 expression. We confirmed that endogenous OCN protein co-localizes with GPR158 in the DG and CA3 (Figure S4A) and further verified the interaction between GPR158 and OCN in vitro. We tagged several variants of OCN with mStrawberry (mStrawberry-OCN), and GPR158 with GFP (GFP-GPR158). Overexpression of GFP-GPR158 in HEK293FT cells co-localized with exogenously added fluorescent-tagged wild-type (WT) mStrawberry-OCN (Figure S4B) and produced a dose-dependent response with fluorescence resonance energy transfer (FRET) (Figure S4C). GFP alone or mStrawberry alone were not able to produce significant FRET (data not shown). In addition, we performed a competitive binding experiment based on the displacement of the interaction between His-tagged unlabeled OCN and GFP-GPR158 with the mStrawberry-OCN. To produce significant FRET signal, mStrawberry-OCN was required in 40-fold higher amounts (10 ng versus 0.25 ng), presumably to displace the unlabeled bound OCN (Figure S4D). To further explore the specificity of OCN for GPR158, we used fluorescent forms of OCN with point mutations at the glutamate residue 13, which partially mimics the carboxylation states (calcium binding sites) of OCN (E13Q and E13F). Mutant OCN produced significantly less FRET, compared with WT OCN (Figure S4E), indicating that the observed response was specific to GPR158-GFP/mStrawberry-OCN.
We then injected knockdown of GPR158 (shGPR158) and control (shGFP) lentiviral particles into the DG and observed a ~50% reduction in the protein levels of GPR158 expressed in this region. Immunostaining for GPR158 revealed a similar decrease in the protein levels of GPR158 and OCN (Figure S4G) particularly in the granule cell layer but not in the hilus of the DG. We also found a concomitant reduction in the protein levels of RbAp48 (Figures 4A–4C), corroborating our finding that OCN/GPR158 signaling mediates the upregulation of RbAp48.
Figure 4. GPR158 Knockdown in the DG Decreases RbAp48 Levels and Impairs Discrimination Memory.
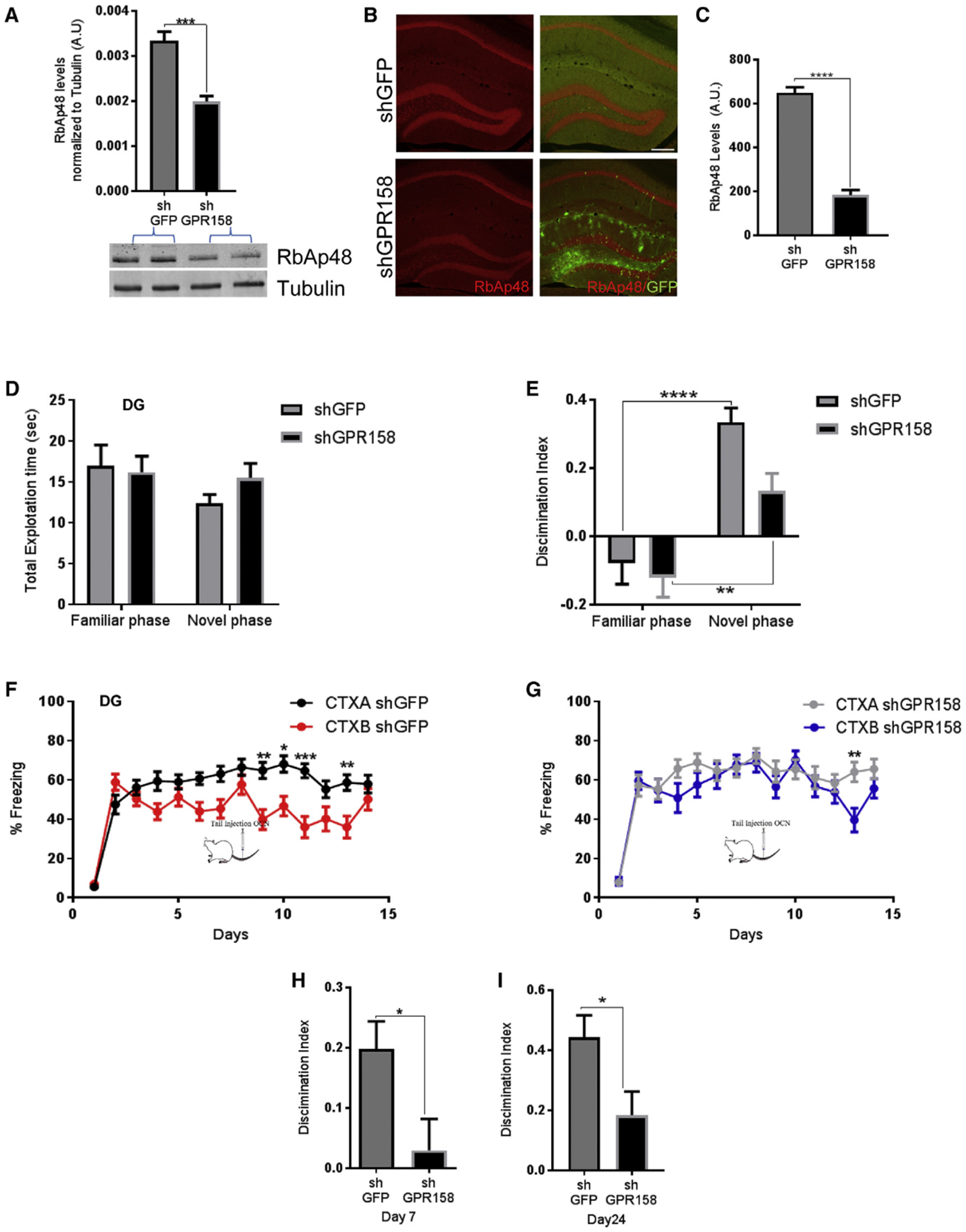
(A) Quantification of RbAp48 protein from dissected DG after injection of shGFP (control) or shGPRI 58 (knockdown) (unpaired t test, p = 0.0005; n = 4–5 mice per group).
(B) Images of shGFP-injected (control, top) or shGPRI 58-injected (knockdown, bottom) DG stained for RbAp48 (red) and GFP (green) (scale bar, 200 μm).
(C) Knockdown with shGPRI 58 results in decreased RbAp48 expression in the DG when compared with shGFP control mice (unpaired t test, p < 0.0001) (n = 4 mice per group).
(D) No differences were detected in total exploration time between the familiarization and the testing phase after injections of shGFP or shGPRI 58 viruses in the DG.
(E) Discrimination index for novel object during a NOR test. shGFP mice were able to discriminate the novel object better than shGPRI 58-injected mice (two-way RM ANOVA comparisons, post hoc Bonferroni; n = 20 mice; p = 0.0193) (Fam versus Nov shGFP, p < 0.0001, and shGPRI 58, p = 0.0061).
(F and G) Contextual fear discrimination in mice injected with shGFP (F) or shGPRI 58 (G) in the DG (three-way RM ANOVA comparisons of treatment × contexts in shGFP versus shGPRI 58 mice, post hoc Bonferroni, p = 0.0158).
(H and I) Discrimination indexes between shGFP and shGPR158 at day 7 (H) and day 24 (I) (unpaired t test, p = 0.0224, day 7; and unpaired t test, p = 0.0232, day 24; n = 16 mice).
All data represent means ± SEM. *p < 0.05, **p<0.01, ***p < 0.001.
We next investigated whether knockdown of GPR158 in the DG could exert a behavioral change similar to the one observed upon inhibition of RbAp48 in forebrain neurons. We therefore performed the same behavioral tests as we did above (Figures 3 and S3). We found that knockdown of GPR158 in the DG produced a lower discrimination ability in a NOR assay, compared with the functional control group (shGFP) (Figures 4D and 4E).
Similarly, mice with reduced levels of GPR158 in the DG (shGPR158) showed a reduced discrimination ability by day 7 of a contextual fear discrimination behavioral task compared to control animals (Figures 4F–4H). At day 7, we administered OCN systemically in both groups via tail injection to exclude the possibility that OCN signaling in other brain regions, and particularly in CA3, is not compensating for the deficits observed in the discrimination memory task. Control (shGFP) animals showed an increase in their discrimination ability until day 14, whereas GPR158 knockdown animals (shGPR158) displayed a significant deficit even with OCN administration (Figures 4F and 4G). Ten days after the end of the experiment (day 24), mice were re-exposed to the same two contexts and their discrimination ability was assessed. Although shGPR158-injected animals were able to discriminate between the two contexts to some extent, they underperformed compared to controls (Figure 4I). The overall reduced discrimination ability upon knockdown of GPR158 is likely to be due to the reduction in the protein levels of RbAp48 and to the failure of OCN binding in the DG. These results are similar to what was observed in DT mice upon OCN administration particularly in the DG.
OCN/GPR158 Signaling Controls Pre-exposure-Mediated Contextual Fear Conditioning
Although tail vein administration of recombinant OCN was ineffective during contextual fear discrimination when GPR158 expression was knocked down in the DG/CA3c regions (Figure 4G), the discrimination ability of this control group (Figure 4F) was different than that of the shGFP group, which received direct administration of OCN in the DG (Figure 3F). We therefore wondered whether OCN binding in the CA3, a sub-region responsible for pattern completion, may explain the reduced discrimination ability of the tail-injected control mice.
To test whether tail vein administration of OCN can bind to the DG and to CA3, we injected purified, recombinant His-tagged, OCN to WT mice via the tail vein and found that in the hippocampus the recombinant hormone, along with the endogenous form, binds mostly to CA3 and some to the DG (Figure S5A).
Using a three-foot-shock contextual fear conditioning paradigm (Figure S5B), we sought to determine whether tail vein administration of OCN was able to influence freezing levels upon exposure to a shock-associated context versus exposure to a novel context not associated with shock. OCN-and PBS-injected control mice showed similar freezing levels when re-exposed to the same context (A) 24 or 48 hr after fear conditioning (Figures S5C–S5E). However, OCN-injected mice froze significantly less than the controls when introduced to a similar context (B) that was novel (Figure S5F), suggesting that OCN administration might be able to either decrease generalization or increase discrimination between two similar but different contexts processes, both of which involve the hippocampal formation.
Because CA3 region is responsible for pattern completion (McHugh et al., 2007; Nakashiba et al., 2009), we investigated the potential role of OCN binding in this region (Khrimian et al., 2017) using a pre-exposure contextual fear conditioning task (Nakashiba et al., 2012) (Figure 5A). We hypothesized that binding of OCN in CA3 might account for the reduced performance of control mice with OCN tail administration. Different cohorts of young WT mice were pre-exposed to a specific context D for 10 min per day for 5 consecutive days (Figure S5G). On the sixth day, they received a strong foot-shock for 2 s and remained in the chamber for an additional 30 s (Figure 5B). 24 hr later, they received injections of either OCN or PBS into the CA3a region, and 48 hr after administration they were re-exposed to the same context (D). OCN-injected mice showed significantly higher freezing than the PBS-injected group, indicating enhanced pre-exposure-mediated contextual fear conditioning (Figure 5C).
Figure 5. OCN/GPR158 Signaling in the CA3a Affects Pre-exposure-Mediated Contextual Fear Conditioning.
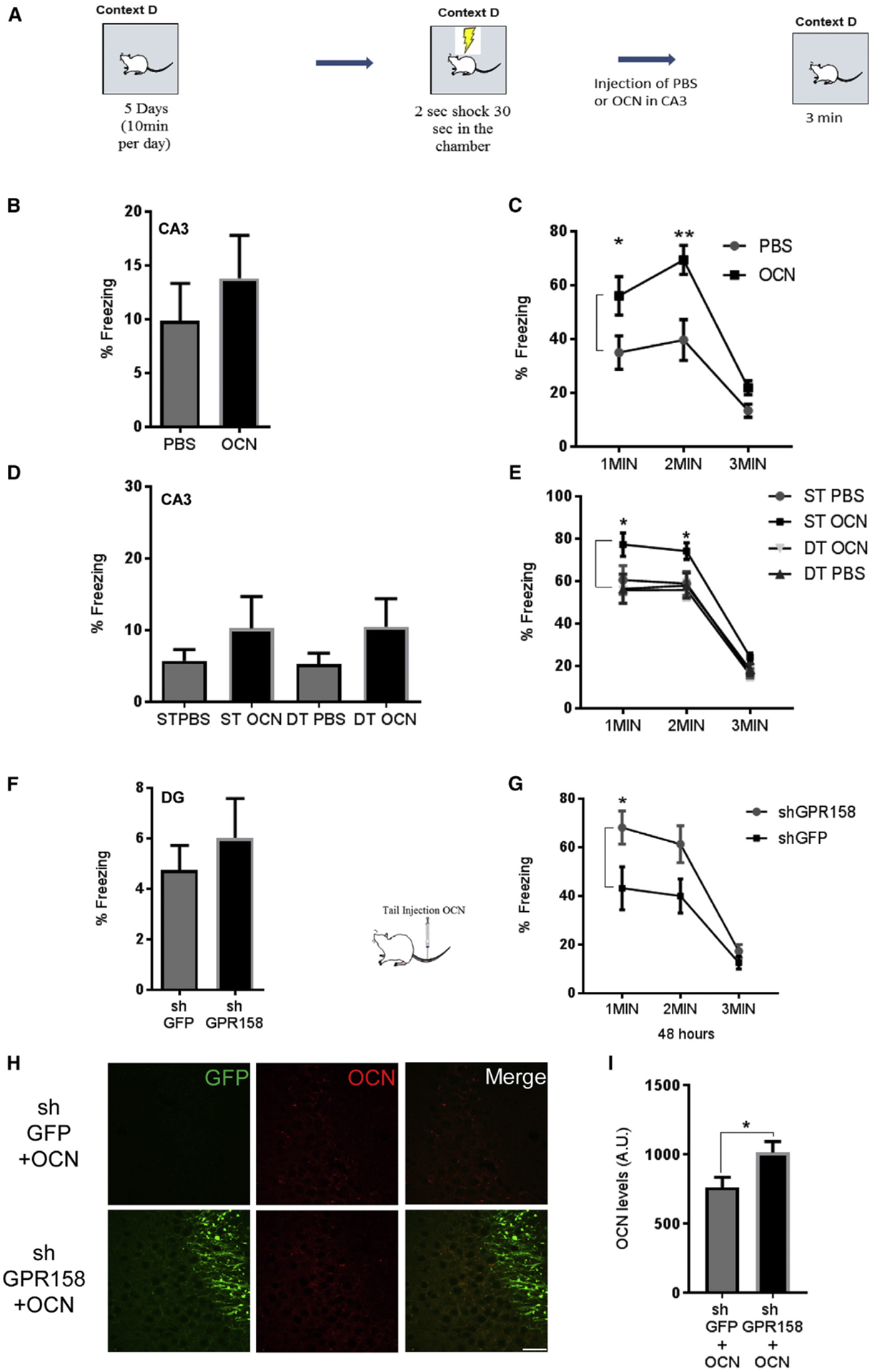
(A) Schematic of pre-exposure contextual fear conditioning.
(B) Freezing levels of mice injected with PBS or OCN 30 s after one 1.5-mA foot-shock.
(C) Freezing levels during 1–3 min of exposure to context D 48 hr after injection of OCN or PBS in CA3a. OCN enhances freezing after injection in CA3a (two-way RM ANOVA, post hoc Bonferroni; n = 10 mice per group; p = 0.0311 and p = 0.0013).
(D) Freezing levels of ST and DT mice injected with OCN or PBS in CA3a 30 s after one 1.5-mA foot-shock.
(E) Freezing levels of ST and DT mice 1–3 min during exposure to context D 48 hr after injection of OCN or PBS in CA3. OCN had an effect only in ST mice (two-way RM ANOVA for OCN versus PBS, Bonferroni post hoc, p = 0.0136 and p = 0.0295 for first minute and second minute comparison between ST OCN and ST PBS mice; three-way ANOVA for genotype x treatment effect, p = 0.0094; n = 12 mice).
(F) Freezing levels of shGFP and shGPR158 mice injected in DG/CA3c 30 s after one 1.5-mA foot-shock.
(G) Freezing levels of shGFP and shGPRI 58 mice in DG/CA3c during exposure to context D, 48 hr after tail injection of OCN. Tail injection of OCN in shGPRI 58 mice had an increase in freezing (two-way RM ANOVA, post hoc Bonferroni; treatment, p = 0.0409; n = 11 mice per group; p = 0.0228 for first minute).
(H) Images from CA3a regions of shGFP (control, top) or shGPRI 58 (knockdown, bottom) animals injected in the DG after peripheral administration of OCN and pre-exposure-mediated contextual fear conditioning, stained for GFP (green) and OCN (red) (scale bar, 20 μm).
(I) Quantification of OCN protein accumulation in the CA3a region 7 days after the end of the behavioral task (unpaired t test, p = 0.0397; n = 6 mice per group). All data represent means ± SEM.
In contrast, when OCN or PBS was injected into the CA3a region of DT or ST mice, there was an increase in the freezing levels on pre-exposure-mediated contextual fear conditioning only in the ST animals injected with OCN (Figures 5D and 5E). No significant difference was observed upon OCN administration in DT animals or in PBS injected ST or DT animals (Figures 5D and 5E). We also found no significant difference between the groups upon the 5-day pre-exposure (Figure S5H). These results confirm that OCN signaling is dispensable upon inhibition of RbAp48 also in the CA3a hippocampal region and can potentially explain the inability of OCN to affect contextual fear discrimination memory when injected in the CA3a hippocampal sub-region.
Finally, to investigate whether OCN binding in the CA3a region interferes with contextual fear discrimination memory, we knocked down GPR158 in the DG and subjected the two cohorts (shGPR158 and shGFP) in a pre-exposure-mediated contextual fear conditioning paradigm. We found no significant differences between the two groups upon the 5-day pre-exposure (Figure S5I) or during the shock phase (Figure 5F). We then administered OCN via the tail vein in both groups and re-exposed them 48 hr later to context D. Mice with GPR158 knockdown in the DG showed increased freezing levels compared to shGFP controls (Figure 5G), presumably because of preferential binding of OCN to CA3a rather than the DG/CA3c hippocampal regions. Seven days after the end of the behavioral task, both groups were sacrificed and stained for total OCN. Mice injected with shGPR158 in the DG (Figure S5J) had more OCN accumulated in the CA3a region than mice injected with shGFP (Figures 5H, 5I, and S5K). Taken together, these data show that OCN/GPR158/RbAp48 signaling pathway can differentially affect fear discrimination memory in the hippocampal formation.
Disruption of GPR158 Protein in Different Cell Types of the DG Affects Discrimination Memory
To date, several cell types of the hippocampus have been implicated in contextual fear discrimination, with particular emphasis on neuronal progenitor cells and their lineage (Drew et al., 2010; Jinde et al., 2012; Sahay et al., 2011). So far, we have demonstrated that forebrain-specific inhibition of RbAp48 and decrease of RbAp48 protein levels through viral knockdown of GPR158 in the DG can disrupt contextual fear discrimination. To better understand the relationship of OCN, GPR158, and contextual fear discrimination, we used a transgenic mouse line NestinCreERT2/Rosa26/E-YFP (Dranovsky et al., 2011) in combination with pSico/pSicoR CRE/LoxP lentiviral vector system (Ventura et al., 2004). Using this viral system (Figure 6A), we were able to restrict GPR158 deletion to a subset of neural progenitor cells when injecting pSico-shGPR158 (termed Nestin+), or we could restrict GPR158 knockdown to non-Nestin-positive cells (termed Nestin−), by injecting pSicoR-shGPR158 virus (Figure 6A).
Figure 6. Disruption of OCN/GPR158 Signaling in Nestin+ and Nestin Cells Affect Contextual Fear Discrimination.
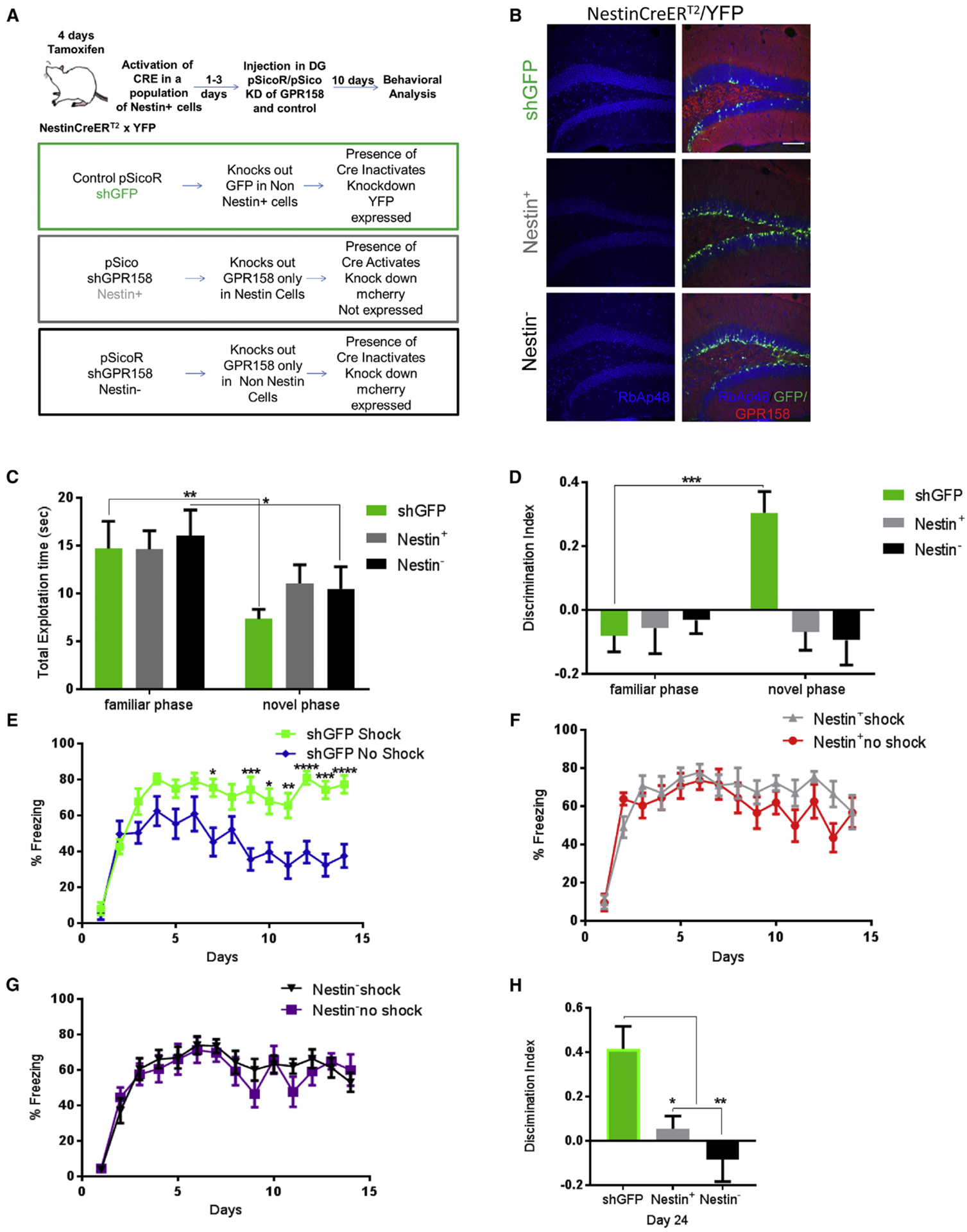
(A) Schematic of viral strategy to disrupt GPR158/OCN signaling in Nestin+ and Nestin− cells.
(B) Images of DG from control and GPR158 knockdown mice, in Nestin+ or Nestin− cells showing RbAp48 (blue) co-localization with Nestin YFP (green) and GPR158 (red) (scale bar, 100 μm).
(C) shGFP and Nestin− mice spent less time exploring both objects in the novel phase than the familiar phase (two-way RM ANOVA comparisons, post hoc Bonferroni; n = 9 mice per genotype; p = 0.001, Fam versus Nov, p = 0.001 for shGFP and p = 0.0126 for Nestin−. No differences were observed between the genotypes in total exploration time in the familiarization or the testing phase.
(D) Discrimination index for novel object during a NOR test in control (shGFP) and Nestin+ or Nestin− GPR158 knockdown mice. Only shGFP mice were able to discriminate the novel object (two-way RM ANOVA comparisons, post hoc Bonferroni; n = 9 mice per group; p = 0.0008).
(E-G) Contextual fear discrimination in control (shGFP) (E) and Nestin+ (F) or Nestin− (G) GPR158 knockdown mice. shGFP mice were able to discriminate between contexts (two-way RM ANOVA comparisons of treatment × contexts between control shGFP and GPR158 knockdown in Nestin+ and Nestin− cells; post hoc Bonferroni, p < 0.0001).
(H) Discrimination index of control (shGFP) and Nestin+ or Nestin− GPR158 knockdown in the DG, 10 days after the end of contextual fear discrimination. Only shGFP mice were able to discriminate between the two contexts (one-way ANOVA, post hoc Bonferroni, p = 0.0016 and p = 0.025 and p = 0.0016).
All data represent means ± SEM. *p < 0.05, **p<0.01, ***p < 0.001.
We showed that the GPR158 knockdown reduced RbAp48 levels in Nestin+ and Nestin− cells (Figures 6B and S6A–S6C), and that this effect was dependent on the tamoxifen treatment, particularly in Nestin+ cells (Figures S6D and S6E). Ten days after virus injection, we subjected the three groups of mice to a NOR task. Although control and Nestin− mice displayed reduced exploration times between the familiar and novel phases, there were no significant differences between the control (shGFP) mice and the Nestin+ or Nestin− groups in the total exploration times during the familiar or the novel phase (Figure 6C). However, control mice discriminated the novel object better compared to the groups of mice in which GPR158 was knocked down in Nestin+ or Nestin− cells (Figure 6D).
We then subjected the same groups of mice to a contextual fear discrimination behavioral task. Control mice (shGFP) were able to discriminate between the contexts A (shock) versus context B (no shock) compared to GPR158 knockdown in the Nestin+ or Nestin− (Figures 6E–6G). This deficit in GPR158 knockdown in Nestin+ or Nestin− cells was still detectable even 10 days after the end of the task (Figure 6H). Taken together, these data indicate that knockdown of GPR158 directly affects the function of RbAp48 also in non-neuronal progenitor cell types, leading to discrimination memory deficits similar to those observed in DT mice with or without OCN administration.
OCN Administration Ameliorates Age-Related Memory Loss and Increases RbAp48
Our data were indicative that OCN/GPR158 signaling controls RbAp48 protein and affects discrimination memory. Since we have previously found an involvement of both RbAp48 and OCN proteins in age-related memory loss (Khrimian et al., 2017; Pavlopoulos et al., 2013), we wondered whether activation of OCN/GPR158 pathway can elevate RbAp48 levels in middle-aged animals and rescue any age-associated deficits in discrimination memory. We administered OCN or PBS into the DG of 16-month-old mice and found that OCN upregulated the protein levels of RbAp48 along with CBP and BDNF protein levels within 48-hr administration (Figures 7A–7E and S7A). Furthermore, OCN administration in the aged DG was able to improve NOR memory (Figures 7F and 7G) and restore the discrimination ability in 16-month-old mice, compared to PBS-injected controls (Figures 7H and 7I). Ten days after the end of these tasks, 16-month-old, OCN-injected mice displayed improved contextual fear discrimination when compared with age-matched control PBS-injected mice (Figure 7J).
Figure 7. OCN Signaling in the DG Upregulates RbAp48 and Ameliorates Age-Related Memory Loss.
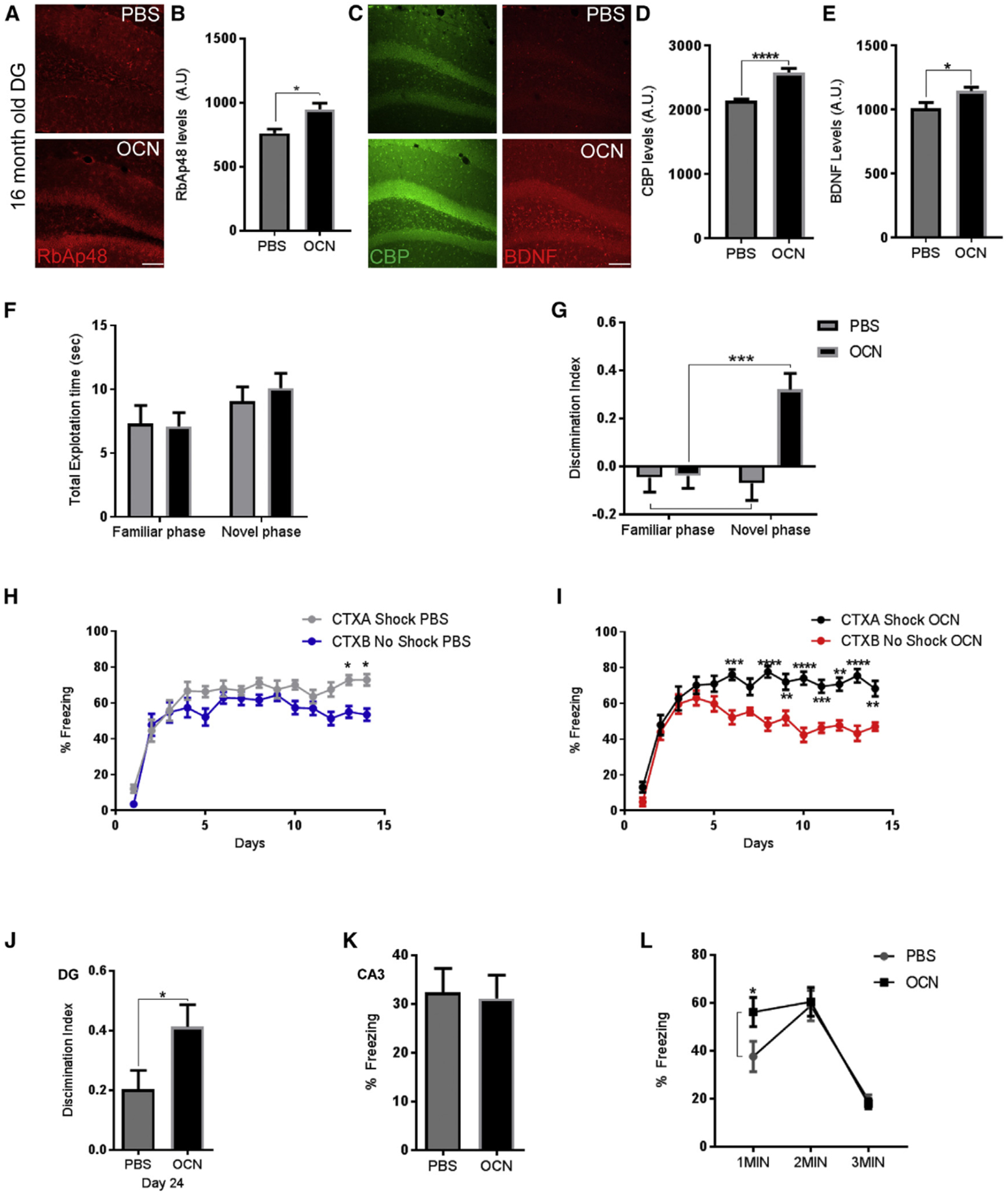
(A) Images of the DG from 16-month-old wild-type (WT) mice stained for RbAp48 (red) 48 hr after injection of PBS (20 ng) (top) or OCN (bottom) (scale bar, 100 μm).
(B) Quantification of RbAp48 protein after injection of OCN or PBS in the DG of 16-month-old WT mice. OCN increases RbAp48 levels (unpaired t tests, p = 0.0179; n = 4 mice per group).
(C) Images of the DG from 16-month-old WT mice stained for CBP (green) and BDNF (red), 48 hr after injection of PBS (20 ng) (top) or OCN (bottom) (scale bar, 100 μm).
(D and E) Quantification of CBP (D) and BDNF (E) protein levels after injection of OCN or PBS in the DG of 16-month-old WT mice. OCN increases CBP and BDNF levels (unpaired t tests, p < 0.0001 and p = 0.034, respectively; n = 6 mice per group).
(F) No differences were observed between the genotypes in total exploration time in the familiarization or the testing phase.
(G) Discrimination index for novel object after injections of PBS or OCN in the DG of 16-month-old WT mice (two-way RM ANOVA comparisons, post hoc Bonferroni, n = 16 mice per group treatment, p = 0.0036, Fam versus Nov, p = 0.0008). Aged mice injected with OCN in the DG were able to discriminate between the novel and familiar object.
(H and I) Contextual fear discrimination in 16-month-old mice injected with PBS (H) or OCN (I) in the DG/CA3c. Aged mice injected with OCN in the DG were able to discriminate between the two contexts (three-way RM ANOVA, post hoc Bonferroni for OCN or PBS and CTXA and CTXB, three-way RM ANOVA for treatment and context × treatment, p < 0.0001; n = 16 mice per group).
(J) Contextual fear discrimination at day 24 in 16-month-old mice injected with OCN or PBS. Aged mice injected with OCN in the DG were still able to discriminate between the two contexts (unpaired t test, p = 0.0395; n = 16 mice per group).
(K) Freezing levels of mice injected with PBS or OCN in CA3c 30 s after one 1.5-mA foot-shock.
(L) Freezing levels during 1–3 min of exposure to context D 48 hr after injection of OCN or PBS in CA3c (two-way RM ANOVA, post hoc Bonferroni, p = 0.04 for first minute; n = 16 mice per group).
All data represent means ± SEM. *p < 0.05, **p<0.01, ***p < 0.001.
Consistent with our observation in young animals, injection of OCN into the CA3a region of 16-month-old animals also increased their ability to discriminate between a familiar and a novel object (Figures S7B and S7C). When the same animals were subjected to a contextual fear discrimination task, they increased their ability to discriminate between the two contexts; however, OCN administration in the CA3a region could not enhance the discrimination ability of 16-month-old animals compared to PBS-injected controls (Figures S7D and S7E). Ten days after the end of these tasks, 16-month-old, OCN-injected mice in CA3a displayed similar contextual fear discrimination when compared with age-matched PBS-injected mice (Figure S7F).
It is well established in humans and in rodents that hyperactivity in CA3 is an index of network dysfunction in age-related memory loss (Leal and Yassa, 2015). We therefore thought that OCN injection in the CA3a region should not be effective in a pre-exposure-mediated contextual fear conditioning paradigm as described earlier. We trained middle-aged (16-month-old) mice in the same pre-exposure contextual fear conditioning paradigm (Figure S7G). OCN administration had a small effect in aged animals on the first minute of exposure but not in the overall freezing levels (Figures 7K and 7L). These results further confirm the differential effects of OCN on aging in the DG and CA3 regions and demonstrate that RbAp48 regulation in the DG is essential for discrimination memory in young and aged mice.
DISCUSSION
Molecular Consequences of RbAp48 Function in the Hippocampus
In an effort to delineate the DNA targets of RbAp48, we found that RbAp48 binds to the GPR158 locus, a GPCR that mediates pro-cognitive effects of the bone-derived hormone OCN. Our data demonstrate that RbAp48 protein binds to the GPR158 genetic locus and presumably controls its expression to the level of mRNA. Although injection of OCN in the DG of ST or DT animals failed to produce a significant change in GPR158 mRNA levels after 24 hr, it is possible that GPR158 mRNA becomes transiently upregulated at earlier time points. It should also be considered that different amounts of OCN might be required by the different cell types in the DG to produce changes in the levels of GPR158 mRNA (Sutton et al., 2018). Alternatively, GPR158 could be modulated at a post-transcriptional level to balance OCN function (Li et al., 2018) secondary to regulation of a different locus by RbAp48. Thus, deciphering the molecular regulation of OCN/GPR158/RbAp48 signaling is an avenue for further studies. A recent study found that knockdown of RbAp48 protein in T98G cells causes suppression of GPR158 mRNA (Kitange et al., 2016). Our results provide an example where this molecular regulatory pathway governs normal brain function.
We also found that RbAp48 controls BDNF mRNA levels, another critical component of OCN/GPR158 signaling pathway (Khrimian et al., 2017). In addition, OCN administration affects both protein and mRNA levels of RbAp48 in the DG. These results favor the possibility that OCN/GPR158 signaling creates a loop involving RbAp48 and BDNF proteins. Given the fact that RbAp48 is a member of several chromatin complexes (Henriquez et al., 2013; Muralidharan et al., 2017), involved in transcriptional activation and repression, we believe that the transcriptional regulation of GPR158 locus by these complexes can regulate the availability of OCN and GPR158 in hippocampal sub-regions, mediating its beneficial effects in cognition and aging.
RbAp48 Involvement in Discrimination Memory
Normal aging may produce deficits in memory for NOR (Broadbent et al., 2009; Grady et al., 1995; Resnick et al., 1995) and for pattern separation/completion processes (Clark et al., 2017; McAvoy et al., 2016). To correlate our molecular findings with these mnemonic processes, we used a variety of behavioral tasks with high interference in mice, which presumably mimic behaviorally the pattern separation/completion processes in humans. Our results show that OCN administration to either the DG/CA3c or CA3a enhance NOR memory, whereas the OCN beneficial effects in a contextual fear discrimination paradigm are limited to the DG/CA3c regions of young and middle-aged mice. These effects are completely abolished when the RbAp48 function is inhibited in the DG or when RbAp48 levels deteriorate during natural processes such as aging.
We also observe similar effects when GPR158 is knocked down in the DG/CA3c region, which in turn reduces the RbAp48 protein levels. We demonstrate that GPR158/OCN/RbAp48 function is essential mainly in the granule neurons of the DG and may play a similar role in the immature neurons of adult mice (Oury et al., 2013). Consistent with other studies (Arruda-Carvalho et al., 2014; Sahay et al., 2011), our results show that inhibition of OCN/GPR158/ RbAp48 loop in neuronal progenitor cell types (Nesting and in the granule cell types of the DG (Nestin−) can affect contextual fear discrimination memory. Alternatively, cells outside the neuronal lineage (e.g., parvalbumin) could exert additional control in Nestin cells (Bao et al., 2017).
In addition, we found that OCN has a different role in a behavioral pattern completion task when administered in the CA3a region, where it enhances pre-exposure-mediated fear conditioning only in young animals. This enhancing effect is abolished upon inhibition of RbAp48 and more importantly is enhanced only upon peripheral OCN administration in the CA3a region when the GPR158 protein levels are knocked down in the DG. The above data highlight at the molecular level the importance of circuit integrity between the DG and the CA3 hippocampal sub-regions in pattern separation/completion processes.
This investigation provides more direct evidence for the central role of RbAp48 in age-related memory loss. Our results corroborate the connection between blood circulating factors and normal brain function (Castellano et al., 2017; Villedaet al., 2014) and show that deterioration of humoral factors outside the brain can potentially lead to abnormal neuronal function (Khrimian et al., 2017; Oury et al., 2013). We further show that one of these blood-derived factors, OCN, controls RbAp48 via GPR158 and that in turn exerts regulation in the GPR158/BDNF proteins and probably OCN. Taken together, these results show that RbAp48 function is indispensable to NOR memory and to memory discrimination tasks, including behavioral pattern discrimination and completion. This is interesting because it provides the first example of a nucleosome remodeling factor involved in histone acetylation that is directly associated with these behavioral processes.
On the circuit level, OCN’s regulation of RbAp48 protein may also act as a modulator of the mnemonic processes. Binding of OCN in the DG seems to enhance the ability of the hippocampal formation to enhance discrimination between memories, whereas binding in the CA3a helps to retrieve a memory when partial cues are presented. Both of these processes were attenuated when RbAp48 function is inhibited. This is further supported by the fact that OCN administration in the aged CA3a produces a minor effect in a behavioral pattern completion task highlighting that age-related memory loss might be a combined defect of both decreased discrimination and increased generalization (Besnard and Sahay, 2016). It is therefore of particular interest and importance that this phenomenon is also observed in aged humans (Yassa et al., 2011).
Furthermore, since OCN increases RbAp48 and BDNF mRNA levels and produces an activity-dependent effect on the synapses of the outer molecular layer of the DG (Figure 2D), we believe that upregulation of RbAp48 in the aging DG potentially activates a transcriptional program potentially leading to enhance synapse maintenance. This notion is consistent with our behavioral results and also with the fact that knockdown of RbAp48 protein, as a part of NuRD complex, decreases the maturation of granule neurons and their incorporation into the cerebral circuits (Yamada et al., 2014).
Finally, our results suggest that any therapeutic intervention regarding age-related memory loss needs to be studied extensively on several mnemonic processes to understand the impact of these interventions on the neuronal circuitry.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric R. Kandel (erk5@cumc.columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were maintained under standard conditions approved by the IACUC. All WT mice C57/BL6 were purchased from Jackson Laboratories (RRID:IMSR_JAX:000664). 16-month-old mice (C57/BL6) were purchased from Taconic (RRID:IMSR_TAC:b6) at 12 months of age and used at 16 months old. The tetO-Flag RbAp48-DN mice (backcrossed more than 10 times to C57BL76J background) were bred with CaMKIIa-tTA mice (backcrossed more than 10 times to 129SvEvTac background) as described previously (Pavlopoulos et al., 2013). For regulating tetO-driven gene expression, mice were fed chow supplemented with doxycycline (40 mg/kg) (Mutual Pharmaceutical) as previously described (Pavlopoulos et al., 2011). c-Fos-CreERT2 (Luo) (RRID:IMSR_JAX:021882) was bred with tdTomato (Ai 14) (RRID:IMSR_JAX:007914) and both lines were purchased from Jackson Laboratories. NestinCreERT2/Rosa26/EYFP was a generous gift from Dr. Alex Dranovsky (Dranovsky et al., 2011). Male and female mice were used for all behavioral and experimental testing. All experiments involving animals were approved by the Institutional Animal Care and Use Committee of Columbia University Medical Center.
Neuronal Cultures
Neuronal cultures were prepared from P0 pups according to Beaudoin et al. (Beaudoin et al., 2012).
HEK293FT cell culture
HEK293FT were purchased from Life Sciences and cultured according to manufacturer’s instructions (RRID:CVCL_6911).
Production of OCN and E13Q and E13F
WT and mutant Osteocalcin (OCN) were produced in E. coli (C2566, T7 express competent cells, NEB C2566I). Briefly, the transformed cells were grown on a shaker at 37°C until OD6OO was measured to be 0.8, after which they were induced with 1mm IPTG for 4 hours.
METHOD DETAILS
Stereotaxic Injections
Stereotaxic injections were performed as described previously (Pavlopoulos et al., 2013). Mice were anaesthetized with a Ketamine/Xylazine mixture (100mg/ml/kg) and (20mg/ml/kg) respectively. Injections were targeted bilaterally to the DG (dentate mainly and CA3c) (−1.96 mm anteroposterior (AP), ± 1.28 mm mediolateral (ML), −1.98 mm dorsoventral (DV)), CA3 (−1.96 mm AP, ± 1.8 mm ML, −2 .0 mm DV) (CA3a mainly and CA3b). Injection volumes ranged from 0.5 to 1 μL and contained 5ng of purified OCN. This amount was used for mice in all behavior experiments unless otherwise stated in the figure legends. Injection time was 10min. After the injection the needle remained in place for another 10 min before it was withdrawn.
Tamoxifen (TAM)
NestinCreERT2/Rosa26/EYFP animals aged 8–12 weeks were administered 5 mg of tamoxifen (Sigma), suspended in 100 μL 1:1 honey:water mixture by gavage once a day for 4 days, at least 12 hr apart according to Dranovsky et al. (Dranovsky et al., 2011) and handled for at least 3 min each day. Immediately after the last dose of tamoxifen (1–3 days) all the animals were injected in the DG with pSico/pSicoR GPR158 or shGFP viral particles. Ten days later all groups were subjected to further experimental manipulations as described below. c-Fos-CreERT2/tdTomato received 2 doses of 5 mg Tamoxifen prior to any other experimental manipulations.
Tail Vein Injections
Mice were restrained in a plastic chamber with ample ventilation for less than 30 s in order to receive tail injections. All mice prior to behavioral analyses were habituated to the restrain process for at least five days before the experiments in order to minimize stress to this procedure. Successful tail vein injections were verified by injecting 100 μL of 0.9% Evans blue in NaCI.
ChIP (Chromatin Immuno-Precipitation)
For ChIP assays LowCell ChIP kit (C01010071 Diagenode) was used according to manufacturer instructions. Because RbAp48 protein does not bind directly to DNA we used dual crosslinking with 1.5 mM EGS (Invitrogen). For DNA isolation the iPURE v2 (C03010015 Diagenode) kit was used according to manufacturer instructions. Fold enrichment was calculated using the 2−ΔΔCt method.
Behavior
All mice were under constant environmental enrichment and were handled for 3–5 days for at least 3 min before the beginning of the behavioral tasks. Male and female mice were used for all behavioral testing. After handling at Day 1 mice were injected with Osteocalcin or (PBS) control. After 3 days of recovery mice were tested in an elevated plus maze task. Next all the cohorts were exposed to an empty arena followed by exposure to identical objects (familiar phase) and 24 hours later to a familiar and a novel object (novel phase). 24 hours later mice were exposed to context C and one day later they were first exposed to neutral context B and then to context A (shock) for fourteen consecutive days. Ten days after the end of the task (Day24) groups of mice were re-exposed to both contexts without any shock.
Mice injected with Lenti-viruses were left without any behavioral manipulation ~10–21 days before subjected to the same battery of behavioral tasks. Different cohorts of mice were used for the pre-exposure mediated contextual fear conditioning.
Elevated Plus Maze (EPM)
To exclude any anxiety related effects each cohort of mice was first subjected to an elevated plus maze task (Table S3, detailed Elevated Plus Maze data related to Figures 3, 4, 6, 7, S3, and S7). Mice were tested in an elevated plus maze (EPM). Mice were placed in the testing room one hour prior to testing. Mice were then placed in the center of an elevated plus maze (arm length across, 60×60cm) with a set of open arms and a set of closed arms and allowed to freely explore the maze for 6 minutes before being removed and placed back in their home cage. Videos were recorded using Ethovision software and were additionally scored manually with experimenter blind to the treatments. Measurements were scored based on the location of the mouse (center, open arm, closed arm).
NOR
NOR assay was performed as previously described (Pavlopoulos et al., 2013) with minor modifications. Briefly, mice were left to explore an empty rectangular arena (45 × 45 cm) with visual cues the first day for 6 min. The following day mice were presented with 2 identical objects (2 rectangular black boxes) (familiar phase) and time spent with each object was recorded using Ethovision software. 24 hours after, mice were put back in the same arena with a novel object (white cylinder) and a familiar one for 6 min (novel phase). Videos were additionally scored manually with experimenter blind to the treatments. Discrimination index was calculated measuring the amount of time spend with novel object minus the amount of time spend with familiar versus the total exploration time for both objects. Total exploration time was used as a control measurement in each phase to ensure that any observed difference is not due to anxiety or hyperactivity of the tested subjects. Discrimination index was calculated as: time spent with the first object minus time spent with the second objected, divided by the sum of time spent with both objects.
One Trial Contextual Fear Discrimination learning (Pattern separation)
Pattern separation was performed as previously described with minor modifications (Sahay et al., 2011). Med Associates Fear conditioning chambers were used. The shock associated training Context A had features including an empty chamber with an exposed floor grid, light and fan on, inner and outer chamber doors completely closed, and the mouse placed in the chamber oriented away from the door. The no-shock Context B was the same chamber with a white plastic insert covering the floor grid, a black triangle shaped insert above the floor, a mild lemon scent (0.4% v/v lemon extract in water), inner and outer doors slightly ajar, light and fan off, and the mouse was placed in the chamber oriented toward the door. One day prior to the experiment mice were exposed for 3 min to a Context C which featured a white plastic insert covering the floor grid, a concave white insert covering the left, right, and back walls, light and fan were off, inner and outer chamber doors were completely closed, and the mouse placed in the chamber oriented away from the door. A non-alcoholic antiseptic was used to clean the chamber after each run in all conditions. Mice were brought into the entry room next to the testing room and left there for an hour to acclimate before any testing. Mice were exposed for 3 min to the training context A in which they received a single 2 s foot shock of 0.75mA. 15s after shock delivery mice were taken out of the chamber and returned to their home cage and placed in the entry room. One hour later, mice were placed in the similar context B in which they were left for 3 min and were never shocked. Measurement of freezing levels in both training context (three minutes pre shock) and in the similar context (three minutes) each day allowed assessment of discrimination between the two contexts and was computed as a Discrimination index: (% Freezing context shock − % Freezing no shock)/ (% Freezing context shock + % Freezing no shock). Freezing levels were automatically measured using Med Associates software.
Pre-exposure Contextual Fear Conditioning
Mice were pre-exposed to a Context D in which the chamber had an exposed grid, a concave white insert covering the left, right, and back walls, and a peppermint scent (4% v/v extract in water). A non-alcoholic antiseptic was used to clean the chamber after each run. Mice were placed in the chamber for 10 minutes for 5 consecutive days. On the sixth day they were placed in the chamber for 10 s, given a strong foot shock 1.5mA, and remained in the chamber an additional 30 s before being removed. 24 hours after this the mice undergone surgery or received tail injections. 48 hours after, they were re-exposed to Context D for 3 minutes.
Contextual Fear Conditioning in c-Fos-CreERT2/tdTomato mice
Mice received 2 doses of tamoxifen orally prior to any behavioral manipulation. Two weeks later, mice were exposed for 4 min to the training context A in which they received 3 foot shocks of 0.7 mA for 2 s, 1 min apart (Learning). Twenty-four hours later, mice were injected with OCN or PBS in the DG. Seven days later animals were exposed to the same context without any shock and freezing levels were measured with Med Associates software.
RNA Extraction and Gene expression analysis
DG were dissected as previously described (Hagihara et al., 2009), flash frozen and stored at −80°C. RNA was extracted using the RNA easy mini kit (QIAGEN) according to standard protocols. qPCR was performed on a C1000 Touch Real-Time PCR Detection System (BioRad). Fold enrichment was calculated using the 2−ΔΔCt method. All primer sequences are available upon reasonable requests.
Site Directed mutagenesis and production of OCN and E13Q and E13F
ORF of Osteocalcin was a generous gift from Drs. Frank Ouri and Gerard Karsenty. cDNA was amplified with the appropriate primers and it was sub cloned to a pRSET expression vector (Invitrogen V35120). Mutant OCN was created using primers for site directed mutagenesis with the Lightning kit (Agilent). WT and mutant Osteocalcin (OCN) were produced in E. coli (C2566, T7 express competent cells, NEB C2566I). Briefly, the transformed cells were grown until OD6OO was measured to be 0.8, after which they were induced with 1mm IPTG for 4 hour. Cells were lysed with lysis buffer (50 mm Tris pH 7.65,10% glycerol, 1M NaCI, 0.01% NP40 substitute, 10 mm Imidazole, with 100ug/ml lysozyme, 100ug/ml DNase, 100ug/ml RNase). The lysed solution was then centrifuged and the supernatant was applied to a column containing Ni-NTA resin and allowed to rotate in suspension for one hour to allow for binding of the protein. The protein was eluted with elution buffer (50 mm Tris pH 7.65, 10% glycerol, 1M NaCI, 0.01% NP40 substitute, 300 mm Imidazole). Protein purification was done using the QIAGEN Ni-NTA Fast Start Kit (30600). For Fluorescent tags of mutant and WT OCN the above ORF were cloned on the C-terminal end of the mStrawberry vector (Clontech 632530) in order to produce mStrawberry wt OCN, E13Q and E13F. Proteins were produced and purified using the methods described above. Additionally fluorescently tagged proteins were further purified using the mStrawberry isolation kit (ChromoTek, RFP-Trap_MA kit, rtmak-20) according to manufacturer’s instructions.
GPR158 mouse ORF was purchased from DNASU and subcloned on the pAc-GFP1-N (Clontech 632485) and pAc-GFP1-C vectors (Clontech 630458).
shRNA’s against the GFP protein or GPR158 were cloned in the pSICO (Addgene 11578) and pSICOR (Addgene 11579) vectors according to standard procedures (Ventura et al., 2004). For pSICO mCherry shGPR158, GFP was substituted with the mCherry ORF (pmCherry-N1 632523). Target sequences: for GFP: GCAAGCTGACCCTGAAGTTCAT and for GPR158: GCTCATTATCACGGCTATATT.
HEK293FT cell culture, transfection, and production of Lentiviral particles
HEK293FT were purchased from Life Sciences and cultured according to manufacturer’s instructions (R700–07). For transfections we used FuGENE 6 (Promega) transfection reagent according to manufacturer’s instructions. Viral particles were produced using the HEK293FT cell line according to standard procedures in a BSL-2 safety cabinet. Briefly, T175 flasks were seeded with 2.0 × 107 cells on day 1 with HEK293FT cells. Twenty-four hours later and two hours before transfection, medium was replaced in each flask with 15 mL of pre-warmed Optimem medium (Life Sciences). Transfection DNA was prepared by adding 18 μg of the vector of interest, 9 μg of pMDLg/pRRE (Addgene 12251), 3.6 μg of pRSV-REV (Addgene 12253) and 5.6 μg of pCMV-VSV-G (Addgene 8454) (Dull et al., 1998) in 3.9 mL Optimem. One hundred ul of Lipofectamine 2000 (Life Sciences) was added to 3.9 mL of Optimem and incubated at room temperature for 5 min. Transfection DNA and Lipofectamine mix were combined in a single tube and incubated for at room temperature for 20 min. Transfection mix was added to the plates and incubated for at 37°C. Six hours later transfection mix was replaced by DMEM medium 1% FBS without antibiotics. 2–3 days after transfection supernatants were concentrated via Ultracentrifugation. The viral titers were 109/ml.
Fluorescent binding assays with GPR158
For fluorescent binding between GPR158 and Osteocalcin we have used a FRET based assay according to Emami et al. (Emami-Nemini et al., 2013). Briefly, we used HEK293 FT cells transiently transfected in 96 well black coated plates (CoSTAR) with N- or C-terminal GPR158 GFP constructs. For each well of a coated 96-well plate we have used 30,000 HEK293FT cells per well, 50 μL of medium and 60 ng of plasmid DNA. Cells were transfected in Optiprep solution using FuGENE 6 transfection reagent according to manufacturer’s instructions. 12 hours after, the medium was changed with complete DMEM medium and the cells were left in the incubator for additional 24 hours. The following day the medium was discarded and replaced with assay buffer (20 mM HEPES, 100 mM NaCI, 3 mM MgCI2 and 0.2% (wt/vol) nonfat milk (pH 7.5) at 25°C. Fluorescent recombinant proteins were added at the indicated concentrations and incubated at 25°C for 20 min. To measure the bound fluorescence signal the Clariostar reader was used. An appropriate filter setup for FRET was used based on the GFP and mStrawberry fluorophores. Raw FRET was calculated by dividing the amount of fluorescence transferred to GFP from mStrawberry with the amount of the GFP fluorescence. All described experiments were performed multiple times. Single experiments with all the experimental conditions present are described in the figures (n = 6–8 per condition). For competition assays using unlabeled OCN, cells were pre-incubated with the unlabeled ligand (His-tagged Osteocalcin 0.25 ng/ul). 30 min later fluorescent proteins were added at the described concentrations.
ChlP-seq library preparation and analysis
Library preparations for ChlP-seq were carried out in the Greek Genome Center (GGC) of Biomedical Research Foundation of Academy of Athens (BRFAA) according to lllumina protocol (Ford et al., 2014). Single-end 75 bp reads were generated with NextSeq500 in the GGC. Two independent ChIP and Input experiments were prepared for library construction. Libraries were filtered for low quality reads and trimmed to 50 bp with the use of FASTX-Toolkit. Bowtie2 (Langmead et al., 2009) with -very-sensitive option was used for aligning the data to the mm10 genome version and Samtools (version 0.1.19) (Li et al., 2009) were used for data filtering (rmdup) and file format conversion. Both replicate ChIP and Input were merged in order to increase read depth and MACS14 algorithm (Zhang et al., 2008) was used for peak calling with default settings. Data visualization was performed with IGV browser (Nicol et al., 2009) while gene ontology and pathway analysis were performed with DAVID knowledgebase (Huang et al., 2009). Only pathways and biological processes with p value ≤ 0.05 were considered to be significantly enriched. ChlP-seq data have been deposited in the Short Read Archive (SRA) under the accession codes SRP141688.
Western blot analysis
Mouse DG (2 per animal) were lysed in a mammalian extraction buffer (M-PER) (Thermoscientific) (150 μl) supplemented with and Protease and Phosphatase inhibitor tablets (Roche). 10 to 20 μg of lysates were separated by SDS-polyacrylamide gel electrophoresis in 4%−20% gradient gels (Biorad) and transferred to PVDF Fluorescent membranes (Millipore). Membranes were blocked with Odyssey Blocking buffer. Membranes were cut in strips to detect multiple antigens according to the MW. Membranes were stripped and reprobed for multiple antigen detection also, using stripping buffer from Thermoscientific. Immunoblots were analyzed with the Odyssey Infrared Imaging System (LI-COR Biosciences). The whole hippocampus from 3- and 16-month-old animals was used. 60 to 80 μg of total protein extracts were separated by SDS-polyacrylamide gel electrophoresis in 10%−20% Tricine gels (Biorad). For Osteocalcin detection Supersignal Western Blot Enhancer kit was used (Thermoscientific 46640). Antibodies used: Anti-RbAp48 antibodies (GeneTex Cat# GTX70232, RRID:AB_372869) and (Thermo Fisher Scientific Cat# PA1–869, RRID:AB_2177636), Tubulin-1 (Sigma-Aldrich Cat# T7816, RRID:AB_261770), OCN antibody from Millipore (MABD123), BDNF (Santa Cruz Biotechnology Cat# sc-546, RRID:AB_630940), CBP (Santa Cruz Biotechnology Cat# sc-7300, RRID:AB_626817) and (Santa Cruz Biotechnology Cat# sc-583, RRID:AB_2245237), and GPR158 (GeneTex Cat# GTX87693, RRID:AB_10723755). The image studio (Li-COR) was used for quantification.
Immunohistochemistry
Briefly, coronal sections (50 μm) along the entire rostrocaudal extension of the hippocampus were cut on a vibratome and stored in 0.1 M tris pH 7.4, 30% ethylene glycol, and 30% glycerol at −20°C until further processed. Immunofluorescence was performed on every fifth section. Sections were washed 3 times with 0.1 M Tris pH 7.4, 0.15M NaCI and 0.3% Triton X-100 (TBSTX) at room temperature. Antigen retrieval was performed at 80°C for 30 min in 50mM Sodium Citrate buffer pH 8.8. Sections were additionally washed 3 times with TBSTX buffer and blocked for 1 hour at room temperature in blocking buffer. (TBSTX supplement with mouse IgG (1 μg/ml) and 1 %BSA to reduce non-specific binding of mouse antibodies). Primary antibodies were incubated at 4°C for 24 to 48h. After primary antibody incubation, sections were washed at least 3–5 times with TBSTX buffer and then were incubated with the appropriate Alexa fluorescent secondary antibodies (Invitrogen). Sections were washed 3–5 times with TBSTX buffer and then mounted on Superfrost slides (Fisher Scientific) with FluorSave Reagent (Calbiochem). Antibodies used: RbAp48 antibodies (GeneTex Cat# GTX70232, RRID:AB_372869) and (Thermo Fisher Scientific Cat# PA1–869, RRID:AB_2177636), OCN antibodies (Thermo Fisher Scientific Cat# PA1–85754, RRID:AB_2065062) and Millipore (MABD123), PSD95 (UC Davis/NIH NeuroMab Facility Cat# 75–028, RRID:AB_2292909), anti-MAP2 (Abeam Cat# ab5392, RRID:AB_2138153), GPR158 (GeneTex Cat# GTX87693, RRID:AB_10723755), anti-GFP from Abeam (Cat# ab13970, RRID:AB_300798) and Invitrogen (Molecular Probes Cat# A-11122, RRID:AB_221569), anti-mCherry (Abeam Cat# ab205402, RRID:AB_2722769), BDNF Promega Cat#G1641, CBP/KAT3A Abeam Cat#ab2832, and anti-parvalbumin (Swant Cat# GP72, RRID:AB_2665495). The secondary antibody against streptavidin was the Streptavidin, Alexa Fluor 647 conjugate (Thermo Fisher Scientific Cat# S-21374, RRID:AB_2336066). For CldU staining antigen retrieval was performed for 30 min in 2N HCI (in PBS) at 37°C and then the sections were washed with 0.1 M Boric Acid at room temperature followed by three washes in TBSTX. Blocking was performed for 1 hour in 12.5% Normal Donkey serum with 0.3% Triton X-100. Primary antibodies were diluted in the same solution.
QUANTIFICATION AND STATISTICAL ANALYSIS
Spine counting
Spine Counting was performed in the outer molecular layer (OML) of the DG according to Pavlopoulos et al. (2011) and McAvoy et al. (2016).
DG Section Quantification
Between 5–8 brain sections (50 μm) were analyzed from each experimental animal. For each brain both hemispheres of the hippocampus were analyzed. For RbAp48, the region of interest in each section was manually circled and the mean intensity of the fluorescence of this area was measured using ImageJ. This was then subtracted from the background intensity to standardize the expression across all sections to obtain the corrected intensity. For BDNF and CBP analysis, the DG was manually circled in each brain section and the mean intensity of the fluorescently expressed markers was measured. These values were then averaged across all sections for each animal. The same quantification was performed to measure osteocalcin in the CA3a region. For cFos+ neuron analysis, each individual cell expressing a cfos fluorescent marker was counted in each section of both hemispheres. These numbers were then averaged based on the number of sections analyzed.
ChIP (Chromatin Immuno-Precipitation) and RNA Extraction Gene expression analysis
Fold enrichment was calculated using the 2−ΔΔCt method.
Statistics
Statistics were performed with GraphPad Prism 6 software. Repeated-measures ANOVA (RMANOVA), ANOVA, and t tests were used as indicated. The results are presented as means ± SEM. Statistical significance was set at p < 0.05. Full statistical analysis can be found in Table S4, Detailed statistics for each figure related to all Figures).
DATA AND SOFTWARE AVAILABILITY
The accession number for the ChlP-seq data reported in this paper is SRA: SRP141688.
Supplementary Material
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal anti-RbAp48 | GeneTex | Cat#GTX70232; RRID:AB_372869 |
| Rabbit Polyclonal anti-RbAp48 | Thermo Fisher Scientific | Cat#PA1–869; RRID:AB_2177636 |
| Mouse Monoclonal anti-β-Tubulin I | Sigma-Aldrich | Cat#T7816; RRID:AB_261770 |
| Mouse monoclonal anti-Osteocalcin | Millipore | Cat#MABD123 |
| Rabbit polyclonal anti-BDNF (N-20) | Santa Cruz Biotechnology | Cat#sc-546; RRID:AB_630940 |
| Mouse monoclonal anti-CBP (C-1) | Santa Cruz Biotechnology | Cat#sc-7300; RRID:AB_626817 |
| Rabbit polyclonal anti-CBP (C-20) | Santa Cruz Biotechnology | Cat#sc-583; RRID:AB_2245237 |
| Rabbit Polyclonal anti-GPR158 | GeneTex | Cat#GTX87693; RRID:AB_10723755 |
| Goat polyclonal anti-Osteocalcin | Thermo Fisher Scientific | Cat#PA1–85754; RRID:AB_2065062 |
| Mouse monoclonal anti-PSD-95 | UC Davis/NIH NeuroMab Facility | Cat#75–028; RRID:AB_2292909 |
| Chicken polyclonal anti-MAP2 | Abeam | Cat#ab5392; RRID:AB_2138153 |
| Chicken polyclonal anti-GFP | Abeam | Cat#ab13970; RRID:AB_300798 |
| Rabbit polyclonal anti-GFP | Invitrogen | Cat#A-11122; RRID:AB_221569 |
| Chicken polyclonal anti-mCherry | Abeam | Cat#ab205402; RRID:AB_2722769 |
| Chicken polyclonal anti-BDNF | Promega | Cat#G1641; RRID:AB_430850 |
| Rabbit polyclonal anti-KAT3A/CBP | Abeam | Cat#ab2832; RRID:AB_303342 |
| Guinea pig polyclonal anti-Parvalbumin | Swant | Cat#GP72; RRID:AB_2665495 |
| Streptavidin, Alexa Fluor 647 conjugate antibody | Thermo Fisher Scientific | Cat#S-21374; RRID:AB_2336066 |
| Bacterial and Virus Strains | ||
| T7 Express Competent E. coli (High Efficiency) | New England BioLabs | Cat#C2566l |
| Chemicals, Peptides, and Recombinant Proteins | ||
| His-tagged Osteocalcin | This paper | N/A |
| FuGENE 6 Transfection Reagent | Promega | Cat#E2691 |
| Lipofectamine 2000 | Life Sciences | Cat#11668019 |
| Critical Commercial Assays | ||
| LowCell ChIP Kit | Diagenode | Cat#C01010071 |
| iPURE Kit v2 | Diagenode | Cat#C03010015 |
| RNA Easy Mini Kit | QIAGEN | Cat#74104 |
| QuikChange Lightning | Agilent | Cat#210515 |
| Ni-NTA Fast Start Kit | QIAGEN | Cat#30600 |
| RFP-Trap_MA Kit | Chromotek | Cat#rtmak-20 |
| Supersignal Western Blot Enhancer Kit | ThermoFisher | Cat#46640 |
| Deposited Data | ||
| ChIP-seq data | Short Read Archive | SRA: SRP141688 |
| Experimental Models: Cell Lines | ||
| Human: HEK293FT | ThermoFisher | RRID:CVCL_6911 |
| Mouse: Neuronal cultures from P0 pups | This Paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: c57/BL6 | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: c57/BL6 | Taconic Biosciences | RRID:IMSR_TAC:b6 |
| Mouse: tetO-Flag RbAp48-DN | Pavlopoulos et al., 2013 | N/A |
| Mouse: CaMKIIa-tTA | Pavlopoulos et al., 2013 | N/A |
| Mouse: c-Fos-CreERT2 | The Jackson Laboratory | RRID:IMSR_JAX:021882 |
| Mouse: tdTomato (Ai14) | The Jackson Laboratory | RRID:IMSR_JAX:007914 |
| Mouse: NestinCreERT2/Rosa26/EYFP | Dr. Alex Dranovsky (Dranovsky et al., 2011) | N/A |
| Oligonucleotides | ||
| GFP targeting sequence: GCAAGCTGACCCTGAAGTTCAT | This paper | N/A |
| GPR158 targeting sequence: GCTCATTATCACGGCTATATT | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pRSET expression vector | Invitrogen | Cat#V35120 |
| Plasmid: mStrawberry | Clontech | Cat#632530 |
| Plasmid: GPR158 ORF | DNASU | Clone: MmCD00081142 |
| Plasmid: pAc-GFP1-N | Clontech | Cat#632485 |
| Plasmid: pAc-GFP1-C | Clontech | Cat#630458 |
| Plasmid: pSICO | Ventura et al., 2004 | Addgene Plasmid #11578 |
| Plasmid: pSICOR | Ventura et al., 2004 | Addgene Plasmid #11579 |
| Plasmid: pmCherry-N1 | Clontech | Cat#632523 |
| Plasmid: pMDLg/pRRE | Dull et al., 1998 | Addgene Plasmid #12251 |
| Plasmid: pRSV-REV | Dull et al., 1998 | Addgene Plasmid #12253 |
| Plasmid: pCMV-VSV-G | Dull et al., 1998 | Addgene Plasmid #8454 |
| Software and Algorithms | ||
| Ethovision | Noldus | RRID:SCR_000441 |
| Med Associates Video Freeze Software | Med Associates Inc. | RRID:SCR_014574 |
| FASTX-Toolkit | Cold Spring Harbor Laboratory | RRID:SCR_005534 |
| Bowtie2 | Langmead et al., 2009 | RRID:SCR_016368 |
| Samtools (version 0.1.19) | Li et al., 2009 | RRID:SCR_002105) |
| MACS14 algorithm | Zhang et al., 2008 | RRID:SCR_013291 |
| IGV browser | Nicol et al., 2009 | http://bioviz.org/ |
| DAVID | Huang et al., 2009 | https://david.ncifcrf.gov/gene2gene.jsp |
| ImageJ | ImageJ | RRID:SCR_003070 |
| LI-COR Image Studio Software | LI-COR | RRID:SCR_015795 |
| Graphpad Prism 6 | Graphpad | RRID:SCR_002798 |
Highlights.
RbAp48 is important for discrimination memory
RbAp48 controls beneficial actions of osteocalcin by regulating GPR158 and BDNF
RbAp48 upregulation via osteocalcin signaling ameliorates age-related memory loss
ACKNOWLEDGMENTS
We thank Dr. Gerard Karsenty and Lori Khrimian for providing us with recombinant OCN, Dr. Frank Oury for providing us with the open reading frame of OCN plasmid, Dr. Nefeli Slavi for critical reading of the manuscript and assisting with the figures, and Dr. Miduturu Srinivas for the critical reading of the manuscript. Funding was provided by the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.09.077.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, and Barco A (2004). Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42, 947–959. [DOI] [PubMed] [Google Scholar]
- Arruda-Carvalho M, Restivo L, Guskjolen A, Epp JR, Elgersma Y, Josselyn SA, and Frankland PW (2014). Conditional deletion of α-CaMKII impairs integration of adult-generated granule cells into dentate gyrus circuits and hippocampus-dependent learning. J. Neurosci 34, 11919–11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach ME, Barad M, Son H, Zhuo M, Lu YF, Shih R, Mansuy I, Hawkins RD, and Kandel ER (1999). Age-related defects in spatial memory are correlated with defects in the late phase of hippocampal long-term potentiation in vitro and are attenuated by drugs that enhance the cAMP signaling pathway. Proc. Natl. Acad. Sci. USA 96, 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H, Asrican B, Li W, Gu B, Wen Z, Lim SA, Haniff I, Ramakrishnan C, Deisseroth K, Philpot B, et al. (2017). Long-range GABAergic inputs regulate neural stem cell quiescence and control adult hippocampal neurogenesis. Cell Stem Cell 21, 604–617.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin GM 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, and Arikkath J (2012). Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc 7,1741–1754. [DOI] [PubMed] [Google Scholar]
- Besnard A, and Sahay A (2016). Adult hippocampal neurogenesis, fear generalization, and stress. Neuropsychopharmacology 47, 24–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent NJ, Gaskin S, Squire LR, and Clark RE (2009). Object recognition memory and the rodent hippocampus. Learn. Mem 17, 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Mosher KI, Abbey RJ, McBride AA, James ML, Berdnik D, Shen JC, Zou B, Xie XS, Tingle M, et al. (2017). Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RE, Zola SM, and Squire LR (2000). Impaired recognition memory in rats after damage to the hippocampus. J. Neurosci 20, 8853–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R, Tahan AC, Watson PD, Severson J, Cohen NJ, and Voss M (2017). Aging affects spatial reconstruction more than spatial pattern separation performance even after extended practice. Hippocampus 27, 716–725. [DOI] [PubMed] [Google Scholar]
- Dillon SE, Tsivos D, Knight M, McCann B, Pennington C, Shiel AI, Conway ME, Newson MA, Kauppinen RA, and Coulthard EJ (2017). The impact of ageing reveals distinct roles for human dentate gyrus and CA3 in pattern separation and object recognition memory. Sci. Rep 7,14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranovsky A, Picchini AM, Moadel T, Sisti AC, Yamada A, Kimura S, Leonardo ED, and Hen R (2011). Experience dictates stem cell fate in the adult hippocampus. Neuron 70, 908–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MR, Denny CA, and Hen R (2010). Arrest of adult hippocampal neurogenesis in mice impairs single- but not multiple-trial contextual fear conditioning. Behav. Neurosci 124, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, and Naldini L (1998). A third-generation lentivirus vector with a conditional packaging system. J. Virol 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami-Nemini A, Roux T, Leblay M, Bourrier E, Lamarque L, Trinquet E, and Lohse MJ (2013). Time-resolved fluorescence ligand binding for G protein-coupled receptors. Nat. Protoc 8, 1307–1320. [DOI] [PubMed] [Google Scholar]
- Ford E, Nikopoulou C, Kokkalis A, and Thanos D (2014). A method for generating highly multiplexed ChlP-seq libraries. BMC Res. Notes 7, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady CL, McIntosh AR, Horwitz B, Maisog JM, Ungerleider LG, Mentis MJ, Pietrini P, Schapiro MB, and Haxby JV (1995). Age-related reductions in human recognition memory due to impaired encoding. Science 269,218–221. [DOI] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, and Kandel ER (2002). Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 777, 483–493. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, and Luo L (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H, Toyama K, Yamasaki N, and Miyakawa T (2009). Dissection of hippocampal dentate gyrus from adult mouse. J. Vis. Exp 2009 (33), 1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez B, Bustos FJ, Aguilar R, Becerra A, Simon F, Montecino M, and van Zundert B (2013). Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Mol. Cell. Neurosci 57,130–143. [DOI] [PubMed] [Google Scholar]
- Holden HM, and Gilbert PE (2012). Less efficient pattern separation may contribute to age-related spatial memory deficits. Front. Aging Neurosci 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Jinde S, Zsiros V, Jiang Z, Nakao K, Pickel J, Kohno K, Belforte JE, and Nakazawa K (2012). Hilar mossy cell degeneration causes transient dentate granule cell hyperexcitability and impaired pattern separation. Neuron 76,1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrimian L, Obri A, Ramos-Brossier M, Rousseaud A, Moriceau S, Nicot AS, Mera P, Kosmidis S, Karnavas T, Saudou F, et al. (2017). Gpr158 mediates osteocalcin’s regulation of cognition. J. Exp. Med 214 2859–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitange GJ, Mladek AC, Schroeder MA, Pokomy JC, Carlson BL, Zhang Y, Nair AA, Lee JH, Yan H, Decker PA, et al. (2016). Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep 14, 2587–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal SL, and Yassa MA (2015). Neurocognitive aging and the hippocampus across species. Trends Neurosci 38, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25,2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang Y, Sidlauskas K, Ellis M, Evans I, Frankel P, Lau J, El-Hassan T, Guglielmi L, Broni J, et al. (2018). Inhibition of GPR158 by microRNA-449a suppresses neural lineage of glioma stem/progenitor cells and correlates with higher glioma grades. Oncogene 37, 4313–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu IY, Lyons WE, Mamounas LA, and Thompson RF (2004). Brain-derived neurotrophic factor plays a critical role in contextual fear conditioning. J. Neurosci 24, 7958–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, and Almouzni G (2004). Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677, 3–11. [DOI] [PubMed] [Google Scholar]
- McAvoy KM, Scobie KN, Berger S, Russo C, Guo N, Decharatana-chart P, Vega-Ramirez H, Miake-Lye S, Whalen M, Nelson M, et al. (2016). Modulating neuronal competition dynamics in the dentate gyrus to rejuvenate aging memory circuits. Neuron 91, 1356–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, and Tonegawa S (2007). Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science 317, 94–99. [DOI] [PubMed] [Google Scholar]
- Muralidharan B, Khatri Z, Maheshwari U, Gupta R, Roy B, Pradhan SJ, Karmodiya K, Padmanabhan H, Shetty AS, Balaji C, et al. (2017). LHX2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11. J. Neurosci 37, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Buhl DL, McHugh TJ, and Tonegawa S (2009). Hippocampal CA3 output is crucial for ripple-associated reactivation and consolidation of memory. Neuron 62, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ, et al. (2012). Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149,188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr., Raja A, and Loraine AE (2009). The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oury F, Khrimian L, Denny CA, Gardin A, Chamouni A, Goeden N, Huang YY, Lee H, Srinivas P, Gao XB, et al. (2013). Maternal and offspring pools of osteocalcin influence brain development and functions. Cell 155, 228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, and Kandel ER (2011). Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 147, 1369–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E, Jones S, Kosmidis S, Close M, Kim C, Kovalerchik O, Small SA, and Kandel ER (2013). Molecular mechanism for age-related memory loss: the histone-binding protein RbAp48. Sci. Transl. Med 5, 200ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, and Misteli T (2009). The central role of chromatin maintenance in aging. Aging (Albany N.Y.) 1, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, and Barnes CA (2011). Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging 32, 2198–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Trotman KM, Kawas C, and Zonderman AB (1995). Age-associated changes in specific errors on the Benton Visual Retention Test. J. Gerontol. B Psychol. Sci. Soc. Sci 50, 171–178. [DOI] [PubMed] [Google Scholar]
- Rowe WB, Blalock EM, Chen KC, Kadish I, Wang D, Barrett JE, Thibault O, Porter NM, Rose GM, and Landfield PW (2007). Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J. Neurosci 27, 3098–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O’Carroll CM, Kheirbek MA, Burghardt NS, Fenton AA, Dranovsky A, and Hen R (2011). Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature 472,466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Chawla MK, Buonocore M, Rapp PR, and Barnes CA (2004). Imaging correlates of brain function in monkeys and rats isolates a hippocampal subregion differentially vulnerable to aging. Proc. Natl. Acad. Sci. USA 707, 7181–7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark SM, Yassa MA, Lacy JW, and Stark CE (2013). A task to assess behavioral pattern separation (BPS) in humans: Data from healthy aging and mild cognitive impairment. Neuropsychologia 57, 2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton LP, Orlandi C, Song C, Oh WC, Muntean BS, Xie K, Filippini A, Xie X, Satterfield R, Yaeger JDW, et al. (2018). Orphan receptor GPR158 controls stress-induced depression. Elife 7, e33273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, and Jacks T (2004). Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA 101,10380–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, Smith LK, Bieri G, Lin K, Berdnik D, et al. (2014). Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med 20, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Hemberg M, Yoshida T, Cho HY, Murphy JP, Fioravante D, Regehr WG, Gygi SP, Georgopoulos K, and Bonni A (2014). Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 83, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Mattfeld AT, Stark SM, and Stark CE (2011). Age-related memory deficits linked to circuit-specific disruptions in the hippocampus. Proc. Natl. Acad. Sci. USA 108, 8873–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChlP-seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the ChlP-seq data reported in this paper is SRA: SRP141688.


