Abstract
Backgrounds
Aside from Gleason score few factors accurately identify the subset of prostate cancer (PCa) patients at high risk for metastatic progression. We hypothesized that copy number alterations (CNAs), assessed using CpG methylation probes on Illumina Infinium® Human Methylation450 (HM450K) BeadChip arrays, could identify primary prostate tumors with potential to develop metastatic progression.
Methods
Epigenome-wide DNA methylation profiling was performed in surgically resected primary tumor tissues from two cohorts of PCa patients with clinically localized disease who underwent radical prostatectomy (RP) as primary therapy and were followed prospectively for at least five years: 1) a Fred Hutchinson (FH) Cancer Research Center-based cohort (n= 323 patients); and 2) an Eastern Virginia (EV) Medical School-based cohort (n= 78 patients). CNAs were identified using the R package ChAMP. Metastasis was confirmed by positive bone scan, MRI, CT or biopsy, and death certificates confirmed cause of death.
Results
We detected 15 recurrent CNAs were associated with metastasis in the FH cohort and replicated in the EV cohort (p< 0.05) without adjusting for Gleason score in the model. Eleven of the recurrent CNAs were associated with metastatic progression in the FH cohort and validated in the EV cohort (p<0.05) when adjusting for Gleason score.
Conclusions
This study shows that CNAs can be reliably detected from HM450K-based DNA methylation data.
There are 11 recurrent CNAs showing association with metastatic-lethal events following RP and improving prediction over Gleason score. Genes affected by these CNAs may functionally relate to tumor aggressiveness and metastatic progression.
Introduction
Prostate cancer (PCa) is the second most common cause of cancer-related deaths in American men. It is estimated by the American Cancer Society that in 2020, there will be about 191,930 new cases of PCa, and about 33,330 deaths from the disease [1]. Although localized PCa is highly curable by definitive therapy such as radical prostatectomy (RP) or radiation therapy (RT), a subset of the patients who receive RP or RT as their primary treatment will experience disease relapse. Studies show that among patients who undergo RP, about 15% and 35% of them subsequently develop biochemical recurrence (BCR) within 5 years and 10 years after surgery, respectively; furthermore, about 35% of the patients who experience BCR will eventually develop metastatic disease [2–4]. Due to the biologically and clinically heterogeneous nature of PCa, current practice is to stratify risk guided by Gleason score, grade group, tumor stage, PSA level, PSA density, and number of positive cores [5]. This still leads to some intermediate risk patients being over-treated for indolent disease and some men who are under-treated for aggressive disease [6–8]. In the current study we consider PCa that recurred in a metastatic and/or lethal form as “aggressive.” Ideally, intensive treatment should be given only to patients who will develop aggressive disease and in whom treatment will prevent these adverse outcomes. Thus, prognostic biomarkers that can improve upon existing risk stratification methods and better identify patients at risk of developing aggressive disease are greatly needed [9].
Recent genome-wide studies have resulted in the identification of somatic DNA CNAs in both primary PCa [10,11] and advanced PCa tissues [12,13] that occur at a sufficient frequency to be used as biomarkers. The most frequently detected CNAs in primary tumors include amplifications of oncogenes such as MYC (8q24.21, 10%−40%), deletions of tumor suppressor genes such as NKX3–1 (8q21.2, 40%−70%), PTEN (10q23.31, 10%−40%), CDKN1B (12q13.1, 20%−30%), RB1 (13q14.2, 30%−50%), and TP53 (17p13.1, 20%−30%), while the landscape of genomic alterations for advanced metastatic PCa is usually characterized by high copy amplifications of AR in patients that are resistant to castration-based therapy [14].
Investigation of the association between potential CNA-based biomarkers and clinical outcomes in PCa patients has been the focus of recent investigations. To date, most studies have focused on biochemical (i.e., PSA) recurrence as the clinical outcome as it is the most common outcome event following RP [15–18]. Metastatic and/or lethal progression is a more serious clinical outcome than BCR. However, the relationship between CNAs and metastatic-lethal events has only been reported in a few studies [19–21]. Metastatic-lethal events are difficult to observe in patients with localized tumors treated by RP due to the long progression time. One study of RP patients estimated that the median time to BCR was less than 5 years; the median time to development of clinical metastasis after BCR was 8 years; and, after development of metastatic disease, the median time to death was 5 years [2]. Despite the small sample size and limited number of outcome events, Liu et al. successfully confirmed a significant association of MYC gain or CNAs at PTEN and/or MYC with PCa-specific mortality following RP [20].
While microarray-based technologies such as comparative genomic hybridization (CGH) arrays and SNP arrays are usually the standard approach for CNA profiling, the Illumina Infinium® Methylation BeadChip assays can also detect CNA because they both assess the amount of DNA at a site [22,23]. Kwee et al. studied 27 samples derived from lymphoid tumors, and showed that the Illumina Infinium® Human Methylation27 BeadChip, the predecessor of HM450K, was able to estimate CNAs; and, the results were consistent with those obtained using the Affymetrix 10K SNP array, especially for large genomic segments (size > 10MB) [24]. Feber et al. showed that the HM450K could be used to accurately assess CNAs to the same degree of reliability and sensitivity as standard SNP array platforms, such as the Affymetrix SNP 6.0 or Illumina CytoSNP arrays on bladder cancer and glioblastoma multiforme tumor samples from the TCGA data portal [22]. In a more recent benchmark study, Chao et al. showed that HM450K-based CNA detection achieves a sensitivity similar to results obtained from competing SNP microarray platforms [25]. In this study we applied the method originally developed by Feber and colleagues [22] to detect CNAs using HM450K methylation data.
In this study we used HM450K data to identify CNAs and initially examined their association with metastatic-lethal recurrence in a FH-based cohort of patients (n= 323). Another nested case-control dataset from EV (n= 78) was used for validation. A number of studies have been carried out to develop biomarker panels for predicting metastatic-lethal events using these two datasets, either based on gene expression [26,27] or DNA methylation [28,29]. In our study, we first calibrated the HM450K-based CNA detection method implemented in the R package ChAMP using the TCGA prostate cancer dataset, which has both HM450K and SNP 6.0 array data that are typically used for CNA detection. We then applied the calibrated method to HM450K data in the FH and EV datasets to evaluate the potential association of recurrent CNAs with metastatic-lethal progression.
Materials and methods
Study populations
We used data from two independent study populations diagnosed with clinically localized adenocarcinoma of the prostate and treated with RP. The first cohort includes 479 European-American PCa patients previously enrolled in population-based studies conducted at the Fred Hutchinson Cancer Research Center (FH) [30,31]. Patients in the first study were diagnosed between January 1993 and December 1996, while the patients in the second study were diagnosed between January 2002 and December 2005. Gleason score, PSA at diagnosis, and tumor stage were collected from the Seattle-Puget Sound Surveillance, Epidemiology, and End Results Program cancer registry. Vital status and underlying cause of death were also obtained from the cancer registry. PCa outcomes were determined from prospectively collected information from follow-up surveys that were completed by patients in 2004–2005, 2010–2011 and 2015–2016, review of medical records, and/or physician follow-up as needed. Metastatic progression was confirmed by a positive bone scan, MRI, CT or biopsy. Prostate cancer-specific deaths were confirmed by review of death certificates. Patients who developed metastases or died from PCa were combined into a metastatic-lethal category. These 479 patients had a mean follow-up time of 11.4 years. Among these patients, 295 had no evidence of recurrence, 108 had BCR only, and 28 patients developed metastatic-lethal events. Forty-eight patients who were missing follow-up or were ineligible for this analysis were excluded. The FH Institutional Review Board approved the study and all participants signed informed consent statements.
The second study population used for validation includes 84 European-American patients treated at Eastern Virginia Medical School (EV). The group includes men who experienced disease progression to metastatic or lethal PCa (n= 32) and patients with no evidence of recurrence for five or more years after surgery (n= 46) who were diagnosed during the same time period (1992–2009). Metastatic-lethal events were identified the same way as described in the FH cohort. The patients in the EV dataset had an average follow-up period of 9.0 years.
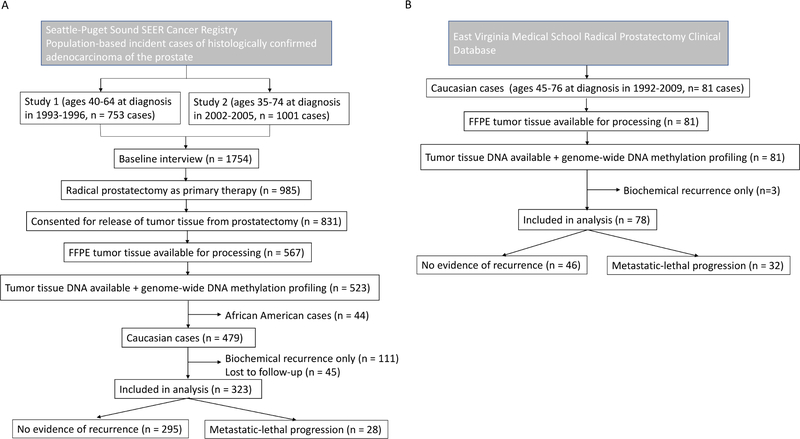
CONSORT diagrams of FH and EV studies are shown in Figure 1.
Figure 1.
CONSORT diagrams. A) FH study, B) EV study
Tumor tissue sample preparation and DNA extraction
Formalin-fixed paraffin-embedded prostate tumor tissue blocks were obtained from RP specimens and used to make hematoxalin and eosin stained slides, which were reviewed by pathologists to confirm the presence and location of adenocarcinoma. For each patient, two 1-mm tumor tissue cores from the dominant lesion that were enriched with ≥75% tumor cells were taken for DNA purification. Tumor adjacent benign samples from 20 patients in FH and 10 patients in EV were also obtained. The RecoverAll Total Nucleic Acid Isolation Kit (Ambion/Applied Biosiences, Austin, TX) was used to extract DNA, which was then quantified with PicoGreen, aliquoted onto 96-well plates and shipped to Illumina (Illumina, Inc., San Diego, CA) for DNA methylation profiling.
DNA methylation profiling
The EZ DNA Methylation Kit (Zymo Research, Irvine, CA) was used to bisulfite convert all the DNA samples. Controls on the array were used to track the bisulfite conversion efficiency. The Infinium® HumanMethylation450 BeadChip (Illumina) was used to measure epigenome-wide methylation using beads with target-specific probes designed to interrogate individual CpG sites (>485,000) [32]. Samples from the FH cohort were assayed as one batch (8 plates) and the EV samples were assayed as a second batch (2 plates). Across the 96-well plates, blind duplicate (FH, n = 16; EV, n = 7) and replicate (FH, n = 2; EV, n = 3) samples placed on all plates were incorporated for each cohort. Correlation in terms of DNA methylation levels for duplicate/replicate samples in the FH and EV cohorts were all great than 0.96. We also evaluated the DNA methylation levels of a few pairs of tumor samples from different tissue cores (FH, n=3; EV, n=2); correlations between each pair of samples were all greater than 0.95. All plates also contained Illumina controls and negative controls. PCa outcome events were randomly distributed across plates, and laboratory personnel were blinded to the location of duplicate and replicate samples. The DNA methylation data were stored in the database of Genotypes and Phenotypes (dbGaP) (study accession: phs001921.v1.p1).
CNA detection
The R package ChAMP [33,34] was used to profile CNAs using raw data (idat files). Default parameter settings were used to run ChAMP, which first applies a number of QC filtering steps on the data: it removes failed samples with more than 5% CpG sites having a detection p-value >0.05; a QC plot including beta distributions from all the samples was generated and used to find samples that deviated significantly from others and which may not be of good quality (e.g., incomplete bisulfite conversion); it removes probes that (a) have a beadcount <3 in at least 5% of samples, (b) are non-CpG probes, (c) overlap with SNPs, (d) align to multiple locations, and (e) are on the X or Y chromosome. ChAMP then adjusts for batch effects on the methylation intensity of the remaining probes using COMBAT, and further performs quantile normalization on the intensity data. For each tumor sample, it computes the log2 ratio of CpG intensity to the mean CpG intensity of normal control samples, which are a group of normal samples or tumor adjacent benign samples. CNA segments for each tumor sample are then generated by using the circular binary segmentation (CBS) algorithm from the R package DNAcopy [35] on the log2 ratio data, with the parameter α (significance level for the test to accept change-points) setting as 0.001.
Recurrent CNA detection
Recurrent CNAs (RCNAs) are regions of the genome that are significantly amplified or deleted across a set of samples. We considered RCNAs as candidate regions of interest (ROI) whose CNAs can be used to predict a clinical outcome (e.g., no recurrence vs. metastatic-lethal progression). We used GISTIC2.0 [36] to detect RCNA regions on tumor samples of the FH cohort. For a tumor sample, GISTIC2.0 quantifies the CNA, or copy number change, in a region by taking the difference of the median copy number within the region and the median copy number of the whole genome. GISTIC2.0 measures both the frequency and the magnitude of the CNA in each tumor sample and identifies RCNA regions that have a statistically higher frequency of CNA over background aberrations on the population level. GISTIC2.0 detects both arm level regions and focal regions, and these regions are classified as either amplified regions or deleted regions. Default parameters were applied to detect RCNA using GISTIC2.0. For an amplified RCNA region, if a tumor sample has a copy-number-change greater than the cutoff (0.1), GISTIC2.0 sets its CNA in this region as 1, and 0 otherwise. For a deleted RCNA region, if a tumor sample has a copy number change less than the GISTIC2.0 cutoff (−0.1), GISTIC2.0 sets its CNA in this region as 1, and 0 otherwise. Thus, CNA of a tumor sample in each studied region is quantified as a binary variable. Focal RCNA regions with a residual q-value less than 0.25 and arm-level regions with a q-value less than 0.05 were considered candidates, according to the default setting of GISTIC2.0.
RCNAs associated with metastatic-lethal outcomes
We computed the association between the detected RCNA regions and patient outcomes (i.e., no recurrence vs. metastatic-lethal progression) using a logistic regression model. We evaluated the association between the metastatic-lethal outcome and a number of clinical variables, including Gleason score, age at diagnosis, diagnostic PSA level, and pathological tumor stage using Chi-square tests. Gleason score, PSA, and tumor stage were found to be significantly associated with the outcome (p-value <0.05). Gleason score is the variable most strongly associated with the outcome of interest, and further inclusion of other clinical variables did not improve upon Gleason score alone (p-value <0.05, likelihood ratio test); therefore we only included Gleason score as a covariate in the model. For each RCNA region discovered in the FH dataset, we conducted two sets of association analyses; one assessed the association directly based on RCNA, and the other quantified the association considering Gleason score as well. The FDR cutoff of 0.2 was applied to control for multiple comparisons, and the remaining RCNA regions were used as the RCNAs detected in the FH dataset. We applied the same association analysis on the EV dataset to validate RCNAs based on a significance level α = 0.05 and the same direction of effect.
For each validated RCNA, we computed area under the curve (AUC) and partial AUC (pAUC, 95% specificity) to evaluate its performance for predicting metastatic-lethal progression. P-values for AUC and pAUC were computed using 2000 stratified bootstrap replicates using the R package pRoC [37]. Likelihood ratio test (LRT) was used to compare a model fit with Gleason score only and a model fit with Gleason score and a validated RCNA.
Results
Patient characteristics
Selected characteristics of the PCa patients are shown in Table 1. The age at diagnosis for patients with metastatic-lethal events was similar to that of patients with no recurrence, while patients with metastatic-lethal progression were more likely to have higher Gleason scores, regional stage disease, and higher diagnostic PSA levels.
Table 1.
Selected characteristics of prostate cancer patients
| FH training dataset (n = 323) | EV validation dataset (n = 78) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metastatic-lethal (n=28) | No recurrence (n=295) | p-valuea | Metastatic-lethal (n=32) | No recurrence (n=46) | p-valuea | |||||||||
| Characteristics | No. | % | Mean (SD) | No. | % | Mean (SD) | No. | % | Mean (SD) | No. | % | Mean (SD) | ||
| Age at diagnosis (year) | 58.0 (7.2) | 58.2 (7.1) | 0. 89 | 60.1 (5.9) | 59.8 (6.6) | 0.86 | ||||||||
| Gleason score | ||||||||||||||
| <7 | 5 | 17.9 | 171 | 58 | 1.10E-07 | 2 | 6.2 | 14 | 30.4 | 8.55E-06 | ||||
| 7(3+4) | 9 | 32.1 | 99 | 34 | 9 | 28.1 | 27 | 58.7 | ||||||
| 7(4+3) | 6 | 21.4 | 12 | 4.1 | 7 | 21.9 | 2 | 4.3 | ||||||
| 8–10 | 8 | 28.6 | 13 | 4.4 | 14 | 43.8 | 3 | 6.5 | ||||||
| Pathological stageb | ||||||||||||||
| Local | 13 | 46.4 | 230 | 71 | 5.30E-04 | 0 | 0 | 26 | 56.5 | 6.90E-07 | ||||
| Regional | 15 | 53.6 | 65 | 29 | 32 | 100 | 20 | 43.5 | ||||||
| PSA (ng/mL) at diagnosis | ||||||||||||||
| <4.0 | 4 | 14.3 | 51 | 17 | 1.50E-05 | 5 | 15.6 | 11 | 23.9 | 0.32 | ||||
| 4.0–9.9 | 8 | 28.6 | 193 | 65 | 19 | 59.4 | 30 | 65.2 | ||||||
| 10.0–19.9 | 6 | 21.4 | 20 | 6.8 | 6 | 18.8 | 3 | 6.5 | ||||||
| ≥20 | 7 | 25 | 12 | 4.1 | 2 | 6.2 | 1 | 2.2 | ||||||
| Missing | 3 | 10.7 | 19 | 6.4 | 0 | 0 | 1 | 2.2 | ||||||
A t-test (age) or chi-square test was used (all categorical variables).
Local = pT2, N0/NX, M0; Regional = pT3-T4 and/or N1, M0.
Benchmarking the HM450K-based CNA detection method
The TCGA PRAD data were used to calibrate the CNA detection method, including both DNA methylation raw intensity data (level 1) and copy number segmentation (level 3) from 494 primary tumor samples based on Affymetix SNP 6.0 array data, and DNA methylation raw intensity data from 50 tumor adjacent benign samples.
The SNP 6.0 array data and the HM450K methylation array data from 494 TCGA primary PCa samples were used to evaluate the performance of the methylation array-based CNA detection method. We used ChAMP to detect CNAs for each tumor sample on DNA methylation raw intensity data. The methylation array-derived CNA profiles were compared to the SNP 6.0 array-derived copy number segments (level 3 data) downloaded from the TCGA data portal in the same tumor samples as the benchmark. We checked how many SNP 6.0 segments could be recovered by HM450K segments, where each SNP 6.0 segment overlapped with a HM450K segment for at least half of its length. A lot of small segments appeared in the HM450K results, which are probably technical artifacts due to unique probe design of the methylation array.
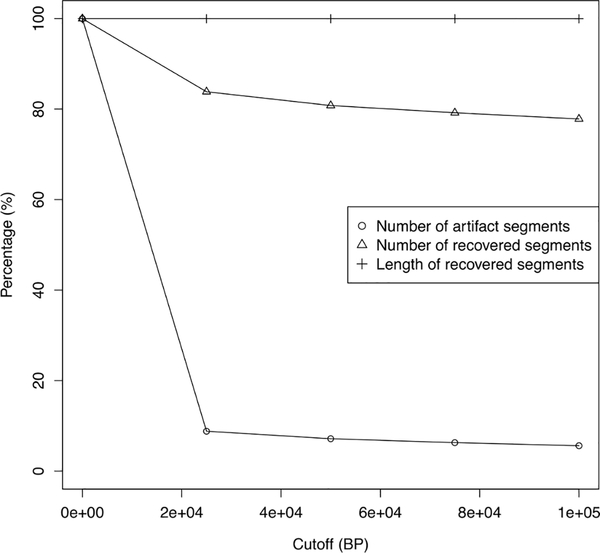
To remove the potential artifact CNAs detected in HM450K data, we conducted a number of analyses by removing short segments with the length less than the thresholds of 0, 25KB, 50KB, 75KB, and 100KB, and evaluated the proportions of recovered segments and artifact segments. The results are shown in Figure 2. By applying the cutoff of 25KB, 92% of artifact segments were removed and at the same time 84% and 99% of recovered segments in terms of segments number and length were retained in the remaining segments. Further increasing the threshold would not remove many more artifact segments and would further reduce the number of recovered segments. We thereafter removed short segments (<25 KB) in our analyses to detect CNAs using HM450K data.
Figure 2.
Tuning of cutoffs for the HM450K-based CNA detection method. The elbow point appears at 25KB.
Detection of RCNAs associated with metastatic-lethal events
We first detected CNA segments on the raw methylation intensity data of 323 tumor samples, including 295 patients with no recurrence and 28 patients with metastatic-lethal outcomes, from the FH dataset using ChAMP. Twenty adjacent benign samples were used as normal controls. We then defined RCNA regions using GISTIC2.0. at the population level on all the tumor samples.
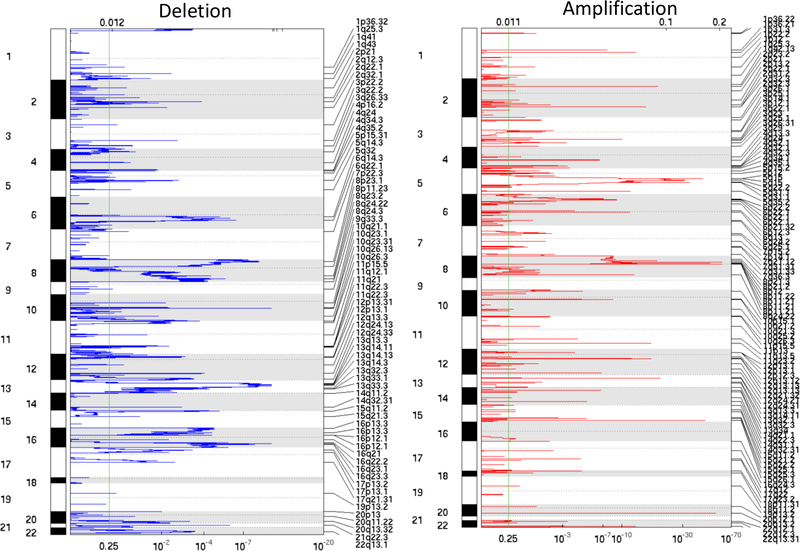
At the focal level (i.e., the length of a RCNA is less than 95% of the chromosome arm it resides in), we detected 106 amplified RCNA regions (Focal1-Focal106, Supplementary Table 1) and 67 deleted RCNA regions (Focal107-Focal173, Supplementary Table 1) among all the tumor samples. The obtained amplification regions cover 445 MB across all autosomal chromosomes (~3000 MB) except chr9 and chr21, with the 25th, 50th, and 75th percentiles as 77 KB, 141 KB, and 347 KB, respectively. The deletion regions cover 1051 MB across all autosomal chromosomes except chr18, with 25th, 50th, and 75th percentiles as 400 KB, 1.8 MB, and 24.2 MB, respectively. More than 10,600 genes reside in these regions, including genes found to be frequently mutated in PCa, such as SPOP, TP53, PTEN, HRAS, ATM, CDKN1B, NKX3–1, PIK3CA, BRCA1, and BRCA2. The amplification and deletion regions are shown in Figure 3, and a description of these focal RCNAs is provided in Supplementary Table 1.
Figure 3.
RCNA regions detected by GISTIC2.0 on the FH dataset.
At chromosomal arm-level, ten arms (3q, 4q, 5p, 5q, 6p, 6q, 7p, 7q, 8p, and 8q) were detected as amplification arms; nine arms (8p, 8q, 13q, 16p, 16q, 18q, 19p, 19q, and 22q) were detected as deletion arms.
We performed two sets of association analyses between RCNAs and patient outcome. The first one is to study which RCNAs are directly associated with the outcome, and the second one is to study which RCNAs are associated with the outcome adjusting for Gleason score. The former analysis aims to discover RCNAs that are associated with the progression outcome regardless of Gleason score. These RCNAs may uncover novel mechanisms of PCa metastatic-lethal progression, and they can be used to predict the outcomes when Gleason score is not available. The latter is to assess the added predictability on top of Gleason score, as Gleason score is the best clinical variable for determining PCa metastatic potential in our study, we are most interested in identifying RCNAs that improve the prognostic discrimination of patients beyond that provided by Gleason score. The numbers of RCNAs found in the RCNA detection and association analyses are summarized in Table 2.
Table 2.
The number of RCNAs found in the RCNA detection and association analyses
| Focal-level | Arm-level | |||
|---|---|---|---|---|
| Deletion | Amplification | Deletion | Amplification | |
| #RCNA detected in FH | 67 | 106 | 10 | 9 |
| #RCNA association discovered in FH | 27, 15 | 34, 15 | 4, 2 | 1, 1 |
| #RCNA association validated in EV | 10, 7 | 3, 2 | 2, 2 | 0, 0 |
The number before a comma represents the association result without adjusting for Gleason score, the number after a comma represents the association result adjusting for Gleason
The first analysis identified 66 RCNAs, including 34 focal amplifications, 27 focal deletions, 1 arm with amplifications (3q) and 4 arms with deletions (13q, 16p, 16q, and 18q) with FDR <0.2 in the FH dataset. Of these regions, 3 focal amplification regions, 11 focal deletion regions, and 2 arm deletions (16p and 16q) were validated in the EV dataset (p-value <0.05). Among these regions, a short one (Focal165) was part of another region (Focal166). Because we are more interested in independent RCNAs, this short region was removed from downstream analysis. As a result, 3 focal amplifications, 10 focal deletions, and 2 arm deletions (16p and 16q) remained, as shown in Table 3. These confirmed 15 regions were further evaluated as the RCNAs associated with metastatic-lethal progression (Table 3). The focal deletion CNA (Focal160) in the p arm of chromosome 16 was found to have a moderate correlation (~0.6) with the arm level deletion 16p based on the two datasets. Similarly, Focal161-Focal164 on the q arm of chromosome 16 were all found to have a moderate correlation (0.38–0.66) with the arm level deletion 16q based on the two datasets. In addition to the metastatic-lethal endpoint, we also studied BCR and PCa-specific death in the FH dataset to verify if CNAs in the validated regions were also associated with these additional clinical outcomes. Eight focal regions (Focal21, Focal129, Focal143, Focal144, Focal161, Focal163, Focal164, and Focal166) were found to have significant association (p-value <0.05) with BCR (Supplementary Table 1). Furthermore, the cases with CNAs tend to have a worse PCa-specific survival than the cases without CNAs (p-value <0.05), as shown in Supplementary Figure 1 None of the identified CNAs overlap with the CpGs selected in the DNA methylation panel [28,29], and the correlations between CNAs and these CpGs were all low (<0.36) in the FH dataset.
Table 3.
Validated RCNAs associated with metastatic-lethal PCa progression (without adjusting for Gleason score).
| CNA | Type | Chromosome | Start (MB) | End (MB) | Width (MB) | #genes | Genetic element of interest | Metastatic-lethal (%) FH, EV | No recurrence (%) FH, EV | AUC* | pAUC* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Focal18 | AMP | chr3 | 62.28 | 119 | 56.58 | 200 | ATP6V1A | 17.9, 34.4 | 4.1, 13 | 0.61 | 0.003 |
| Focal21 | AMP | chr3 | 90.09 | 180 | 89.58 | 508 | GSK3B | 21.4, 46.9 | 3.1, 15.2 | 0.66 | 0.004 |
| Focal61 | AMP | chr8 | 135.7 | 136 | 0.11 | 3 | MIR30D | 21.4, 53.1 | 3.7, 19.6 | 0.67 | 0.003 |
| Focal118 | DEL | chr4 | 1B | 25.8 | 25.75 | 194 | CTBP1, PPP2R2C | 10.7, 25 | 1.7, 6.5 | 0.59 | 0.005 |
| Focal129 | DEL | chr8 | 1.77 | 41.9 | 40.14 | 304 | EPHX2 | 39.3, 56.2 | 10.8, 32.6 | 0.62 | 0.002 |
| Focal143 | DEL | chr12 | 9.8 | 10.1 | 0.31 | 7 | 17.9, 31.2 | 4.4, 8.7 | 0.61 | 0.004 | |
| Focal144 | DEL | chr12 | 12.77 | 12.9 | 0.11 | 2 | CDKN1B | 17.9, 37.5 | 4.1, 13 | 0.62 | 0.004 |
| Focal160 | DEL | chr16 | 27.79 | 28.1 | 0.32 | 1 | 35.7, 46.9 | 16.3, 15.2 | 0.66 | 0.004 | |
| Focal161 | DEL | chr16 | 58.33 | 58.6 | 0.22 | 2 | NDRG4 | 32.1, 62.5 | 9.2, 19.6 | 0.71 | 0.004 |
| Focal162 | DEL | chr16 | 72.05 | 72.2 | 0.1 | 4 | DHODH | 42.9, 62.5 | 22, 19.6 | 0.71 | 0.004 |
| Focal163 | DEL | chr16 | 77.76 | 78.1 | 0.32 | 2 | 39.3, 59.4 | 11.9, 28.3 | 0.66 | 0.003 | |
| Focal164 | DEL | chr16 | 82.13 | 83.8 | 1.71 | 4 | 42.9, 68.8 | 12.2, 43.5 | 0.63 | 0.002 | |
| Focal166 | DEL | chr17 | 1B | 17.7 | 17.72 | 342 | TP53, PELP1 | 21.4, 37.5 | 4.7, 15.2 | 0.61 | 0.003 |
| 16p | DEL | chr16 | 1B | 36.6 | 36.6 | 606 | 28.6, 31.2 | 9.8, 8.7 | 0.61 | 0.004 | |
| 16q | DEL | chr16 | 36.6 | 90.4 | 53.75 | 488 | 28.6, 37.5 | 7.1, 8.7 | 0.64 | 0.005 |
AUC and pAUC are computed based on the EV dataset.
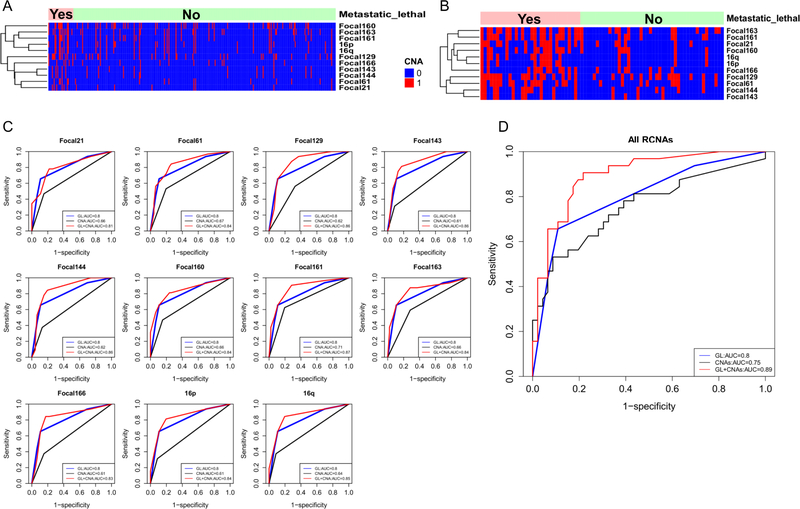
The second analysis detected 33 RCNAs, including 15 focal amplifications, 15 focal deletions, 1 arm with amplification (3q) and 2 arms with deletions (16p and 16q) with FDR <0.2 in the FH dataset. Of these regions, two focal amplification regions, 7 focal deletion regions, and 2 arm deletions (16p and 16q) were validated in the EV dataset. The remaining 11 regions were all listed in Table 4, Figures 4A and 4B, and were included in Table 3 as well. These regions were further evaluated as the RCNAs associated with metastatic-lethal progression after accounting for Gleason score. For each validated RCNA, we built three logistic regression models using the FH dataset; one used only the CNA marker as a predictor of outcome, one used only Gleason score, and the other used a combination of the CNA marker and Gleason score. These trained models were applied to predict metastatic-lethal events in the EV dataset (Table 4 and Figure 4C). AUC and pAUC were used to assess the prediction performance of these models. For the Gleason score alone model, the AUC= 0.8 and pAUC= 0.0084. The inclusion of each RCNA in a model with Gleason score increased the AUCs, which ranged from 0.81–0.87, and Focal161 has the best AUC performance among all 11 regions. Four RCNAs (Focal160, Focal161, 16p and 16q) in chromosome 16 were found to significantly improve the AUC (at the significance level of 0.05) of the Gleason score model, and 3 RCNAs (Focal160, 16p, and 16q) in chromosome 16 were found to significantly improve the pAUC (at the significance level of 0.05). We also used LRT to compare the prediction performance of the two models (i.e., Gleason score alone, and the combined model including RCNA). All regions have significant p-values, which confirms that these regions are complimentary to Gleason score for predicting metastatic-lethal outcomes. We further built another model including Gleason score and all the validated RCNAs to predict patient outcomes. The AUC further increased to 0.89 (p-value=0.017) as shown in Figure 4D. However, the model may have the overfitting issue as EV data has been used to validate the RCNA associations. The development of a CNA biomarker model requires another independent dataset. We also evaluated if these validated RCNAs can add to the National Comprehensive Cancer Network (NCCN) risk group factor to predict patient outcomes. The NCCN risk group classification is mainly determined by Gleason score, PSA level, and tumor stage. We accordingly classified all the cases into the following three risk groups: cases with local stage and PSA level <10 ng/ml, and Gleason score <= 6 were assigned to a low-risk group; cases with regional stage, or Gleason score >7, or PSA > 20 ng/ml were assigned to a high-risk group; and all other cases were assigned to an intermediate-risk group. As shown in Supplementary Figure 2, AUC equals to 0.77 when only the risk group factor is used as the predictor variable; the addition of each individual RCNA into the model increases the AUC in most of the models, in the range of 0.79~0.83. If all RCNAs are included in the model, the AUC increases to 0.87 (p-value= 0.05), which indicates these RCNAs can substantially improve the prediction of patient outcomes beyond the risk group factor.
Table 4.
Validated RCNAs associated with metastatic-lethal PCa progression (adjusting for Gleason score).
| CNA | Type | Chromosome | Start (MB) | End (MB) | AUC* | AUC* p-value | pAUC* | pAUC* p-value | p-value** |
|---|---|---|---|---|---|---|---|---|---|
| Focal21 | AMP | chr3 | 90.09 | 179.67 | 0.81 | 0.768 | 0.019 | 0.179 | 0.007 |
| Focal61 | AMP | chr8 | 135.73 | 135.84 | 0.84 | 0.203 | 0.007 | 0.785 | 0.009 |
| Focal129 | DEL | chr8 | 1.77 | 41.91 | 0.86 | 0.072 | 0.005 | 0.484 | 0.002 |
| Focal143 | DEL | chr12 | 9.8 | 10.11 | 0.86 | 0.075 | 0.009 | 0.973 | 0.023 |
| Focal144 | DEL | chr12 | 12.77 | 12.88 | 0.86 | 0.077 | 0.004 | 0.226 | 0.019 |
| Focal160 | DEL | chr16 | 27.79 | 28.11 | 0.84 | 0.039 | 0.02 | 0.004 | 0.01 |
| Focal161 | DEL | chr16 | 58.33 | 58.55 | 0.87 | 0.009 | 0.015 | 0.165 | 0.009 |
| Focal163 | DEL | chr16 | 77.76 | 78.08 | 0.84 | 0.057 | 0.015 | 0.159 | 0.003 |
| Focal166 | DEL | chr17 | 1B | 17.72 | 0.83 | 0.238 | 0.005 | 0.691 | 0.005 |
| 16p | DEL | chr16 | 1B | 36.6 | 0.84 | 0.027 | 0.014 | 0.037 | 0.012 |
| 16q | DEL | chr16 | 36.6 | 90.35 | 0.85 | 0.009 | 0.015 | 0.03 | 0.009 |
Significant p-values are shown in boldface.
AUC and pAUC are based on combined models (i.e., the CNA and Gleason score), computed based on the EV dataset.
The p-value is based on the LRT comparing the prediction performance of two models (i.e., Gleason score alone, and the combined model).
Figure 4.
The validated 11 RCNAs (adjusting for Gleason score).
A) CNAs detected in the FH dataset
B) CNAs confirmed in the EV dataset
C) ROC curves for predicting metastatic-lethal prostate cancer based on the EV dataset. Curves are shown for each validated region: CNA alone, Gleason score (GL) alone, and Gleason score plus the CNA combined (GL+CNA )
D) ROC curves for predicting metastatic-lethal prostate cancer based on the EV dataset. Three curves are shown for all the CNAs (CNAs), Gleason score (GL) alone, and Gleason score plus all the CNAs (GL+CNAs)
Discussion
In this study we conducted a comprehensive CNA analysis using HM450 DNA methylation data to detect and validate somatic RCNAs associated with metastatic-lethal outcomes in men diagnosed with clinically localized PCa. Our Seattle-based cohort is one of the largest methylome studies for assessing progression in men treated for localized stage disease. Previously, gene expression [26,27] and epigenetic markers [28,29] for the prediction of cancer aggressiveness have been developed in this cohort of PCa patients. The current work aimed to explore the possibility of using DNA methylation data to infer CNAs and, if feasible, investigate the associations between CNAs and metastatic-lethal outcomes. We believe that we are the first group to use methylome data to analyze CNAs in PCa. Comparing the CNAs generated by the DNA methylation-derived method and the existing ones generated on Affymetix SNP 6.0 arrays for TCGA PRAD samples, we showed that it is feasible to call CNAs using DNA methylation data, and CNAs evaluated from the epigenetic data can independently predict adverse patient outcomes. We identified 15 RCNAs that predicted tumor aggressiveness. Four of them were not detected in previous CNA studies, including Focal143 (12p13.31) in chromosome 12, Focal160 (16p12.1), Focal163 (16q23.1), and Focal164 (16q23.3) in chromosome 16. These novel findings may help us to understand the mechanisms of tumor progression in PCa. More interestingly, 11 of the CNAs (Table 4) improved upon Gleason score for the prediction of outcome events. These CNAs could be used to develop an optimal CNA marker panel to predict these outcomes. Although for the majority of the individual validated CNAs the increment of prediction performance in terms of AUC appears to be limited, combining these CNAs in a marker panel substantially improved AUC performance.
High throughput array platforms such as SNP arrays and CGH arrays have been extensively used to detect CNA. CNA can also be inferred using genome-wide DNA methylation data based on methylation arrays including HM450K. Feber et al. demonstrated satisfactory concordance in CNAs detected by HM450K and by SNP arrays: over 94% and 75% agreement were observed, respectively, for large genomic rearrangements (>10 MB) and focal genomic alterations (<1 MB) [22]. Despite the good concordance in detected CNAs between methylation arrays and SNP arrays, there are substantial differences between the designs of the two platforms. For example, the genomic probe distributions are quite different in the two arrays: the probes on the SNP 6.0 array are evenly spaced across the whole human genome while the probes on the HM450K array are gene-centric. The majority (~95%) of CpG probes are within 2 kb for 95% of the known genes and, on average, there are >9 probes per gene [23]. The unique probe design of the HM450K array makes the methylation-derived CNA profiles more susceptible to technical artifacts. We evaluated the methylation-derived CNAs using the TCGA PRAD data and SNP array-derived CNAs, which were used as the benchmark. At the individual CNA level, we found good concordance between the two sets of results: 81% of the benchmark segments were detected by the methylation-derived method. However, many small, probably artifact segments were generated by the methylation-derived method. We therefore decided to remove those small segments (with length <25 KB) before detecting RCNA. Our study found that, by careful calibration, HM450K methylation data could be utilized to detect CNAs, so that both CNAs and DNA methylation levels can be measured in a single assay in a timely and cost-effective manner.
In this study we detected 15 RCNAs associated with the metastatic-lethal events in PCa, and 11 of them were associated with the outcomes adjusting for Gleason score. Several of the detected regions had been discovered in previous studies, and a lot of them contain genes contributing to metastatic-lethal progression of PCa. The amplification region Focal18 (62.3MB-118.9MB, 3p12.1) contains ATP6V1A, which was reported to promote invasion and metastasis in prostate cancer [38]; and the CNA is positively correlated with gene expression in the FH dataset (p-value= 2.64e-4). The amplification region Focal21 (90.09MB-179.67MB, 3p12.1) contains 508 genes, including GSK3B, which was demonstrated as a positive regulator in AR transactivation and prostate cancer growth [39]. The amplification region Focal61 (135.7MB-135.8MB; 8q24.22) contains 3 genes, including MIR30D. This gene was previously found to be overexpressed in PCa and associated with biochemical recurrence [40], and the gene was reported to regulate androgen receptor signaling in PCa [41]. The deletion region Focal118 (1B-25.75MB, 4p16.2) contains CTBP1, a gene previously reported to regulate the expression of tumor suppressors and genes involved in cell death, and was found to be over-expressed in metastatic prostate cancer [42]. This CNA was found to be negatively correlated with CTBP1 gene expression in the FH dataset (p-value= 0.013). The region also includes PPP2R2C; loss of the gene was found to be associated with increased prostate cancer-specific mortality [43]. The deletion region Focal129 was detected in chromosome 8 (1.77MB-41.9MB, 8p11.23). Parts of the deletion region were previously reported in the literature, including chr8: 20.11MB-27.67MB [11], chr8: 15.40MB-15.41MB [13], and chr8: 21.99MB-42.01MB [10]. Among the 304 genes in the region, EPHX2 gene expression was negatively associated with its CNA in the FH dataset (p-value= 8.48e-7); the gene was previously reported to be correlated with AR activity and was identified as a therapeutic target with biomarker potential for PCa [44], which was confirmed in our study. EPHX2 gene expression was negatively correlated with metastatic-lethal outcomes (p-value= 0.014) in the FH dataset. The deletion region Focal144 (12.77–12.9MB, 12p13.1) was included in a wider region (5.7MB-17.6MB) detected by Taylor et al. [10], and the region contains CDKN1B, which was found to be frequently down-regulated and associated with poorer prognosis in prostate cancer [45]. The deletion region Focal161 (58.33MB-58.55MB;16q21) contains NDRG4, a tumor suppressor and prognostic marker of gastric cancer [46]. The deletion region Focal162 (72.05MB-72.15MB; 16q22.2) was identified as one of the most recurrently aberrant deletion regions found in the PCa patient cohort evaluated by Lalonde and colleagues [17]. DHODH is among the five genes residing in the region, and there was a significant positive correlation between the CNA and gene expression in the FH dataset (p-value= 5.23e-6). Its protein product is an enzyme required for de novo pyrimidine synthesis, which is an essential link between the enhanced mitochondrial bioenergetics and aberrant proliferation in transformed prostate epithelial cells [47]. The deletion region Focal166 (1B-17.7MB spans 5 cytobands,17p13.3–17p11.2) includes the tumor suppressor gene TP53. In addition, PELP1 in the region was found to have a significant correlation between the CNA and gene expression level in the FH dataset (p-value= 1.05e-7), and it plays a central role in nonandrogenic activation of the AR in PCa [48]. Part of the region (7.50MB-7.59MB, 17p13.1) was detected in the TCGA and Taylor et al. studies [10,11]. For arm-level RCNA regions, deletion of 16q was reported in a few studies [11,13]; deletion of 16p was also reported in a prior study by Hieronymus and coworkers [21].
Many efforts have been taken to investigate biomarkers to predict metastatic-lethal progression in patients initially diagnosed with clinically localized PCa. Earlier studies have been carried out to find biomarkers of metastatic-lethal progression using the same datasets used here. Twenty three gene transcripts [26] and eight CpG sites [28] were discovered in the FH dataset and validated in the EV dataset. Further validation has been completed for these biomarkers, leading to development of a 5-CpG DNA methylation score [29] and a four gene expression score [27]. In our current analyses, the 11 validated RCNAs in Table 4 had individual AUCs ranging between 0.81–0.87. Compared to the gene expression and methylation biomarkers, some RCNAs encompass a larger number of genes; two focal RCNAs (Focal21 and Focal129) were found to encompass more than 250 genes. In addition, we detected two arm level events (16p and 16q). These RCNAs might be more robust in predicting clinical outcomes.
Several RCNAs predicting clinical outcomes when adjusting for Gleason score (Table 4) are in chromosome 16, including 3 focal deletions (Focal160:16p12.1, Focal161:16q21, and Focal163:16q23.1) and 2 arm-level deletions (16p and 16q). Part of these hot spot deletion areas (in 16q) were identified by Kuth et al., and 16q deletion length was found to be strongly linked to PCa progression [49]. In another study, Osman et al. suggested that the long arm of chromosome 16 (16q) might harbor tumor suppressor genes involved in PCa progression [50]. We confirmed in this study that deletions in chromosome 16 may play an important role in PCa metastatic-lethal progression, and the findings justify future research to identify potential tumor suppressor genes located on chromosome 16.
There are a number of limitations of this study. A major limitation is the paucity of patients who eventually developed metastatic-lethal outcomes. However, metastatic-lethal outcome events are rare in PCa patients diagnosed with localized tumors and treated surgically; therefore we used a cohort with long-term follow-up (mean=11.4 years) to accrue the outcomes. To our knowledge, our FH cohort is one of the best resources for studying primary tumor biomarkers in relation to prognosis of localized PCa patients. Moreover, when inferring copy numbers from DNA methylation data, some potential confounding factors such as tumor cell fraction and genome ploidy were not considered to call CNAs, in part because the CpG probes were not designed for calling allele specific copy numbers. To the best of our knowledge, there is no method available to call CNAs on DNA methylation data considering these factors.
The use of DNA methylation array (HM450K) data may result in artifacts when calling CNAs. For example, DNA methylation is measured using bisulfite sequencing, which relies on the conversion of every single unmethylated cytosine residue to uracil. Incomplete conversion can cause incorrect interpretation of unconverted unmethylated cytosines as methylated cytosines and would generate false-positive results when determining methylation status at CpG dinucleotides. In this study we used ChAMP, which includes a number of QC steps to remove samples of low quality. One of these QC steps is specifically used to remove samples with the incomplete bisulfite conversion problem by plotting beta distributions from all the samples. Furthermore, when calling CNAs, ChAMP actually compares the DNA methylation on the studied tumor samples with a group of normal samples from the same batch of samples processed; this step should further remove the potential effect of incomplete sulfite conversion. Another potential problem is the low genome-wide coverage (~2%) of total CpG sites in HM450K. However, it should not be a major issue in the current analysis since we were not focused on detecting very short CNA regions (e.g., less than 1kB). We compared the CNAs generated from the DNA methylation-based technology and CNAs generated using an existing method based on SNP array data (from TCGA PRAD) in our benchmark study. These two sets of results showed a relatively high concordance (~81%). Other researchers have also shown that the DNA methylation-based method can achieve comparable performance with the existing SNP microarray-based methods [24, 25].
There are several strengths of this study. We searched for RCNAs associated with clinical outcomes based on a well-characterized population cohort, and the findings were validated in an independent dataset. Patients involved in the discovery and validation datasets had long-term follow-up to accrue metastatic-lethal outcomes in men diagnosed with localized tumors. To minimize the platform/batch variability, we used the same platform (HM450K) and the same downstream analysis procedures for the discovery (FH) and validation (EV) datasets. Potential batch effects within each dataset were removed using the COMBAT algorithm included in the R package ChAMP, and results were confirmed by the high correlation between the DNA methylation levels of duplicates/replicates across plates that were incorporated for both cohorts. The potential bias introduced by tumor heterogeneity was well controlled by careful selection of tissue cores from the dominant lesion that was enriched with ≥75% tumor cells. Future studies will be needed to test the best combination of these CNA biomarkers for their association with PCa outcomes using an independent patient cohort.
None of the validated RCNAs overlap with any of the gene expression biomarkers [26] or DNA methylation biomarkers [28] previously found to predict metastatic-lethal events in the FH and EV datasets, and the correlation between the CNAs and the other types of biomarkers were generally low (e.g., the magnitudes of correlation were less than 0.39 based on the FH dataset). Thus, the detected CNAs are independent from other types of biomarkers and may be complimentary for predicting metastatic-lethal progression in patients diagnosed and treated for clinically localized PCa. Future studies may consider integrating these different types of somatic signals to further improve the prediction performance.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Cancer Institute (R01 CA222833, R01 CA056678, R01 CA092579, K05 CA175147 (JLS), and P50 CA097186), with additional support provided by the Fred Hutchinson Cancer Research Center (P30 CA015704). We acknowledge the support of the Eastern Virginia Medical School Biorepository, Norfolk, Virginia.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA : a cancer journal for clinicians 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. Jama 1999;281(17):1591–7. [DOI] [PubMed] [Google Scholar]
- 3.Roehl KA, Han M, Ramos CG, Antenor JAV, Catalona WJ. Cancer progression and survival rates following anatomical radical retropubic prostatectomy in 3,478 consecutive patients: long-term results. The Journal of urology 2004;172(3):910–4. [DOI] [PubMed] [Google Scholar]
- 4.Bruce JY, Lang JM, McNeel DG, Liu G. Current controversies in the management of biochemical failure in prostate cancer. Clin Adv Hematol Oncol 2012;10(11):716–22. [PubMed] [Google Scholar]
- 5.American Urological Association. 2017. AUA Clinical Guidelines. https://www.auanet.org/guidelines/prostate-cancer-clinically-localized-guideline.
- 6.Moyer VA. Screening for prostate cancer: US preventive services task force recommendation statement. Annals of internal medicine 2012;157(2):120–34. [DOI] [PubMed] [Google Scholar]
- 7.Delpierre C, Lamy S, Kelly-Irving M, Molinié F, Velten M, Tretarre B, et al. Life expectancy estimates as a key factor in over-treatment: the case of prostate cancer. Cancer epidemiology 2013;37(4):462–8. [DOI] [PubMed] [Google Scholar]
- 8.Lee YJ, Park JE, Jeon BR, Lee SM, Kim SY, Lee YK. Is prostate-specific antigen effective for population screening of prostate cancer? A systematic review. Annals of laboratory medicine 2013;33(4):233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klotz J The future of active surveillance. Transl Androl Urol 2018;7(2):256–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell 2010;18(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163(4):1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso CS, Wu Y, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castrate resistant prostate cancer. Nature 2012;487(7406):239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson D, Van Allen EM, Wu Y, Schultz M, Lonigro RJ, Mosquera J, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W DNA alterations in the tumor genome and their associations with clinical outcome in prostate cancer. Asian J Androl 2016;18(4):533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Gammal AT, Brüchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Köllermann J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clinical Cancer Research 2010;16(1):56–64. [DOI] [PubMed] [Google Scholar]
- 16.Kluth M, Runte F, Barow P, Omari J, Abdelaziz ZM, Paustian L, et al. Concurrent deletion of 16q23 and PTEN is an independent prognostic feature in prostate cancer. International Journal of Cancer 2015;137(10):2354–63. [DOI] [PubMed] [Google Scholar]
- 17.Lalonde E, Ishkanian A, Sykes J. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. The Lancet Oncology 2014;15(13):1521–32. [DOI] [PubMed] [Google Scholar]
- 18.Choucair K, Ejdelman J, Brimo F, Aprikian A, Chevalier S, Lapointe J. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. Bmc Cancer 2012;12(1):543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clinical cancer 2011;17(20):6563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Xie CC, Thomas CY, Kim ST, Lindberg J, Egevad L, et al. Genetic markers associated with early cancer-specific mortality following prostatectomy. Cancer 2013;119(13):2405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, et al. Copy number alteration burden predicts prostate cancer relapse. Proceedings of the National Academy of Sciences 2014;111(30):11139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feber A, Guilhamon P, Lechner M, Fenton T, Wilson GA, Thirlwell C. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biology 2014;15:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordlund J, Bäcklin CL, Zachariadis V, Cavelier L, Dahlberg J, Öfverholm I, et al. DNA methylation-based subtype prediction for pediatric acute lymphoblastic leukemia. Clin Epigenetics 2015;7(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwee I, Rinaldi A, Rancoita P, Rossi D, Capello D, Forconi F, et al. Integrated DNA copy number and methylation profiling of lymphoid neoplasms using a single array. British Journal of Haematology 2012;156(3):354–7. [DOI] [PubMed] [Google Scholar]
- 25.Chao S, Kim H, Zeiger MA, Umbricht CB, Cope LM. Measuring DNA copy Number variation using high-density methylation microarrays. Journal of Computational Biology 2019;26(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubicz R, Zhao S, Wright JL, Coleman L, Grasso CS, Geybels MS, et al. Gene expression panel predicts metastatic-lethal prostate cancer outcomes in men diagnosed with clinically localized prostate cancer. Mol Oncol 2017;11(2):140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng A, FitzGerald LM, Wright JL, Kolb S, Karnes RJ, Kenkins RB, et al. A four-gene transcript score to predict metastatic-lethal progression in men treated for localized prostate cancer: development and validation studies. The Prostate 2019;79(14):1589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S, Geybels MS, Leonardson A, Rubicz R, Kolb S, Yan Q, et al. Epigenome-wide tumor DNA methylation profiling identifies novel prognostic biomarkers of metastatic-lethal progression in men diagnosed with clinically localized prostate cancer. Clinical Cancer Research 2017;23(1):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S, Leonardson A, Geybels MS, McDaniel AS, Yu M, Kolb S, et al. A five-CpG DNA methylation score to predict metastatic-lethal outcomes in men treated with radical prostatectomy for localized prostate cancer. The Prostate 2018;78(14):1084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiology Biomarkers & Prevention 1999;8(10):881–6. [PubMed] [Google Scholar]
- 31.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. American journal of epidemiology 2008;168(3):250–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98(4):288–95. [DOI] [PubMed] [Google Scholar]
- 33.Morris TJ, Butcher LM, Feber A. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics 2014;30(3):428–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Y, Morris TJ, Webster AP. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017;33(24):3982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshan VE, Olshen A. DNAcopy: DNA copy number data analysis. R package version 1.56.0; 2018. [Google Scholar]
- 36.Merme lGH, Schumacher SE, Hill B. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JZ. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitton B, Okamoto H, Packham G, Crabb SJ. Vacuolar ATPase as a potential therapeutic target and mediator of treatment resistance in cancer. Cancer Med 2018;7(8):3800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Thrasher JB, Terranova P. Glycogen synthase kinase-3: a potential preventive target for prostate cancer management. Urol Oncol 2015;33(11):456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi N, Uemura H, Nagahama K. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget 2012;3(11):1455–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y, Song C, Yang S. Tumor-suppressive function of miR-30d-5p in prostate cancer cell proliferation and migration by targeting NT5E. Cancer Biotherapy and Radiopharmaceuticals 2018;33(5):203–11. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Asangani IA, Chakravarthi BV, Ateeq B, Lonigro RJ, Cao Q, et al. Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia 2012;14(10):905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bluemn EG, Spencer ES, Mecham B, Gordon RR, Coleman I, Lewinshtein D, et al. PPP2R2C loss promotes castration-resistance and is associated with increased prostate cancer-specific mortality. Mol Cancer Res 2013;11(6):568–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vainio P, Gupta S, Ketola K, Mirtti T, Mpindi J, Kohonen P, et al. Arachidonic acid pathway members PLA2G7, HPGD, EPHX2, and CYP4F8 identified as putative novel therapeutic targets in prostate cancer. Am J Pathol 2011;178(2):525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature Reviews Cancer 2008;8:253–67. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, She J, Yang J. NDRG4 in gastric cancer determines tumor cell proliferation and clinical outcome. Molecular Carcinogenesis 2018;57(6):762–71. [DOI] [PubMed] [Google Scholar]
- 47.Hail N, Chen P, R BL. Teriflunomide (leflunomide) promotes cytostatic, antioxidant, and apoptotic effects in transformed prostate epithelial cells: evidence supporting a role for teriflunomide in prostate cancer chemoprevention. Neoplasia 2010;12(6):464–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Ravindranathan P, Ramanan M, Kapur P, Hammes SR, Hsieh J, et al. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol Endocrinol 2012;26(4):550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kluth M, Jung S, Habib O, Eshagzaiy M, Heinl A, Amschler N, et al. Deletion lengthening at chromosomes 6q and 16q targets multiple tumor suppressor genes and is associated with an increasingly poor prognosis in prostate cancer. 2017;8(65):108923–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osman I, Scher H, Daibacni G, Reuter V, Zhang Z, Cordon-Cardo C. Chromosome 16 in primary prostate cancer: A microsatellite analysis. International Journal of Cancer 1997;71:580–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.