Abstract
Human African Trypanosomiasis (HAT) is a potentially fatal parasitic infection caused by the trypanosome sub-species Trypanosoma brucei gambiense and T. b. rhodesiense transmitted by tsetse flies. Currently, global HAT case numbers are reaching less than 1 case per 10,000 people in many disease foci. As such, there is a need for simple screening tools and strategies to replace active screening of the human population which can be maintained post-elimination for Gambian HAT and long-term for Rhodesian HAT. Here, we describe the proof of principle application of a novel high-resolution melt assay for the xenomonitoring of Trypanosoma brucei gambiense and T. b. rhodesiense in tsetse. Both novel and previously described primers which target species-specific single copy genes were used as part of a multiplex qPCR. An additional primer set was included in the multiplex to determine if samples had sufficient genomic material for detecting genes present in low copy number. The assay was evaluated on 96 wild-caught tsetse previously identified to be positive for T. brucei s. l. of which two were known to be positive for T. b. rhodesiense. The assay was found to be highly specific with no cross-reactivity with non-target trypanosome species and the assay limit of detection was 104 tryps/mL. The qPCR successfully identified three T. b. rhodesiense positive flies, in agreement with the reference species-specific PCRs. This assay provides an alternative to running multiple PCRs when screening for pathogenic sub-species of T. brucei s. l. and produces results in less than 2 hours, avoiding gel electrophoresis and subjective analysis. This method could provide a component of a simple and efficient method of screening large numbers of tsetse flies in known HAT foci or in areas at risk of recrudescence or threatened by the changing distribution of both forms of HAT.
Author summary
With global cases of Human African Trypanosomiasis (HAT) reaching pre-elimination numbers, identifying areas of trypanosome transmission becomes increasingly important. With decreasing numbers of cases, the cost of identifying each positive case increases. Screening for trypanosomes in tsetse flies instead of in the human host allows for a faster, more cost-effective and efficient method of detecting areas in which parasite transmission is on-going. Molecular tools such as PCR play a critical role in molecular identification of parasites in vectors. However, at present, multiple PCRs are required to confirm the presence or absence of human infective trypanosomes. Here we present a novel assay which allows for the screening of both species of trypanosomes responsible for HAT in one assay. This method eliminates the need for multiple assays per sample, produces results which are easier to interpret, and reduces the risk of contamination of samples. We found the assay to be as sensitive as the current gold-standard PCR when evaluated on a subset of wild tsetse flies. With further field evaluation, this method could provide an alternative monitoring method for HAT, which can be used leading up to elimination and maintained once elimination has been achieved.
Introduction
Human African Trypanosomiasis (HAT) is a potentially fatal disease caused by subspecies of Trypanosoma brucei s. l. transmitted by the bite of an infected tsetse fly (Glossina spp). HAT consists of two forms of the disease, each with its own distinct parasite, vectors, disease pathology, treatment and geographical distribution. Gambian HAT (gHAT), caused by Trypanosoma brucei gambiense, is a largely anthroponotic disease found across central and west Africa and accounts for the large majority of HAT cases (>97%) [1]. Gambian HAT can remain asymptomatic for months to years with symptoms often presenting once the infection has significantly advanced. Conversely, Rhodesian HAT (rHAT), caused by Trypanosoma brucei rhodesiense, is a zoonosis with occasional human infection, and represents less than 3% of all HAT cases. The World Health Organisation (WHO) has targeted the elimination of HAT as a public health problem by 2020, defined as less than 1 new case per 10, 000 inhabitants in at least 90% of endemic foci and fewer than 2000 cases reported globally [2]. Due to the zoonotic nature of rHAT, this WHO target is applicable to gHAT only. With gHAT on the brink of elimination and rHAT persisting in several foci across East and Southern Africa [3], it is crucial to identify any remaining active cases, foci of transmission and areas of resurgence. The epidemiology of the two forms of HAT differ greatly and therefore monitoring and screening strategies also differ. Monitoring gHAT is largely reliant on the screening of the at-risk human population and treatment of cases [4]. Accurate estimates of disease prevalence require high rates of coverage, which can be difficult to achieve, particularly in areas affected by conflict and political instability [5] and where prevalence approaches <1 case per 10,000. As a result, there has been emphasis on the development of rapid diagnostic tests (RDTs) and field-friendly screening tools [6–10]. In comparison to gHAT, there has been little progress or investment into the development of a screening tool for rHAT with current emphasis on passive case detection and control of the vector population. A reliance on passive detection results in a delay in the identification and treatment of infected individuals, both of which are crucial for the control of disease transmission.
With declining numbers of cases, active screening programmes are no longer cost-effective [11] and there is a need for a monitoring tool which can be maintained sustainably for rHAT and post-elimination for gHAT. Xenomonitoring, the screening of vectors for the presence of parasites, provides a potential alternative to host sampling. This method has already been successfully utilised as a surveillance tool within the Lymphatic Filariasis elimination programme [12–14]. Vectors are often routinely collected as part of vector control programmes and as such, making use of this material may prove to be more economical than active screening of either human or animal populations. Additionally, screening vectors for infection is often less time-consuming and with efficient processing, can provide a view of disease transmission in real-time. Microscopy has been traditionally used for vector screening due to its low cost, high specificity and ease of use in-field. However, the sensitivity of microscopy is variable, it is labour intensive and morphological identification of T. brucei gambiense and T. b. rhodesiense trypanosomes is not possible [15,16].
The development of molecular tools has provided alternatives to traditional screening methods. PCR is widely used for the detection of trypanosome DNA, with highly sensitive assays developed for T. brucei s. l. [17] which includes T. b. gambiense and T. b. rhodesiense along with the animal trypanosome T. b. brucei. Successful amplification indicates the presence of one of the members of T. brucei s. l. but does not identify which member of T. brucei s. l. is present. Identification of T. b. gambiense and T. b. rhodesiense is reliant on the detection of specific single-copy genes (TgsGP [18] and SRA [19,20] respectively) for each subspecies. For detection of these low copy genes, the presence of sufficient genetic material is crucial. A negative result may indicate the absence of the target species or simply that insufficient DNA is present. To differentiate between these two scenarios, primers have been designed to screen for other single-copy genes [21], namely the single-copy phospholipase-C (PLC) gene, present in all members of T. brucei s. l. [22–25]. Samples found to have sufficient DNA can then be screened using primers specific for T. brucei s. l. subspecies.
High-resolution melt analysis (HRM) is a post-qPCR analysis method which can be used to detect heterogeneity within nucleotide sequences and has previously been used for the detection of other parasite species [26–28]. A fluorescent dye is added to the PCR reaction which intercalates into double stranded DNA. Following amplification, the amplicon is heated gradually causing the strands to separate. Separation of the double strands releases the incorporated dye causing a drop in fluorescence. The rate of DNA strand disassociation and the temperature at which it separates (Tm) is dependent on the amplicon’s nucleotide sequence. Different sequences will have different melting temperatures which can then be used as a diagnostic identifier. HRM is a closed tube process resulting in a reduced risk of contamination and produces results in approximately two hours circumventing gel electrophoresis, making it a faster, more specific alternative to traditional PCR. Multiplexing allows for the screening of a number of targets simultaneously, making sample processing more efficient.
Objectives
We aim to develop a qPCR HRM assay designed to distinguish between the two human infective sub-species of T. brucei s. l. To improve the reliability of the assay primers were included to determine if sufficient DNA is present for the sub-species primers to work. To meet this objective we first designed and identified suitable primers that are compatible with the HRM qPCR, followed by assay optimisation and evaluation of sensitivity and specificity. Finally, the assay was compared to the current gold standard PCR using field samples available from other studies (S1 Fig).
Methods
Ethics statement
Ethical approval for this work was obtained from the Commission for Science and Technology (Costech) in Tanzania (permit number 2016-33-NA-2014-233). The samples from Tanzania were collected under the auspices of a BBSRC funded project (“Life on the edge: tackling human African trypanosomiasis on the edge of wilderness areas”, BB/L019035/1; COSTECH research permit number 2014-280-NA-2014-223).
Primers
In order to be compatible with a HRM qPCR assay, primers that produced an amplicon of 150–350 base pairs with distinct melt temperatures were sought. Based on the literature, novel T. b. rhodesiense primers were derived from the sequence of a sub-species-specific serum-resistance-associated (SRA) protein gene (accession number AF097331.1). The primers for T. b. gambiense, previously designed and published by Radwanska et al. [29] were found to be compatible with the HRM qPCR method. These primers target the sub-species-specific glycoprotein (TgsGP: accession number AJ277951). The third primer set was designed to identify the presence of sufficient genetic material. These primers amplify a single-copy phospholipase-C (PLC) gene found in all members of the Trypanozoon group [22–25].
Assay optimisation
To optimise the new HRM assay, the specificity of the multiplexed assay was evaluated using DNA from a range of non-target trypanosome species: Trypanosoma congolense Savannah (Gam2), T. congolense Forest (ANR3), T. congolense Kilifi (WG84), T. simiae (TV008), T. godfreyi (Ken7), T. vivax (Y486) and T. grayi (ANR4). Additionally, cross-reactivity of T. b. gambiense and T. b. rhodesiense primers against non-target T. brucei s. l. sub-species (i. e. T. b. rhodesiense, T. b. brucei and T. b. gambiense) DNA was tested. Each species’ DNA was run in triplicate along with negative water controls and target species positive controls.
The analytical sensitivity of the assay was assessed using a tenfold dilution series of T. b. gambiense and T. b. rhodesiense DNA obtained from culture which ranged from an estimated 106 to 101 tryps/mL, with each concentration run in duplicate. Quantification of DNA in terms of trypanosomes was calculated using concentrations of DNA as read by a Qubit 2.0 fluorometer (Invitrogen) and an assumed approximate DNA quantity of 0.1pg per trypanosome [30]. TgsGP and SRA PCR were run alongside on the same dilution series for a direct comparison of sensitivities [21,29].
Screening of field samples
A subsample of wild-caught tsetse were selected to validate the HRM performance on field samples. The flies had been captured in 2015–2016 in Tanzania as part of the study reported by Lord et al [31]. Full methods are described in Lord et al [31], but in brief, a total of 5,986 tsetse were trapped using odour-baited Nzi traps deployed at sites in Grumeti and Ikorongo wildlife reserve, and Serengeti National Park. Captured flies were stored individually in 100% ethanol at room temperature and brought to LSTM for analysis. DNA extraction was carried out using Genejet DNA purification kit (Thermo K0721) according to the manufacturer’s instructions. Flies were screened for T. brucei s. l. and T. congolense Savannah using TBR [17] and TCS PCR [32] respectively. Both primer sets target satellite repeat sequences specific to each species. Flies positive for T. brucei s. l. were then screened for T. b. rhodesiense using SRA PCR.
PCR
A subsample of 96 flies positive by TBR PCR were used in this study, of which two had been previously found to be positive for T. b. rhodesiense [31]. The species composition of the tsetse was 69.8% Glossina pallidipes (67/96) and 30.2% G. swynnertoni (29/96). Of the 96 T. brucei s. l. positive flies, 11.5% (11/96) were also found to be positive for T. congolense Savannah. Flies were re-screened using TBR [17] and SRA PCR [21] to reconfirm the presence and integrity of parasite DNA. TBR PCR conditions were as follows: initial denaturation at 95°C for 10 minutes, 40 cycles of 95°C for 30 seconds, 63°C for 90 seconds and 72°C for 70 seconds followed by an extension step at 72°C for 10 minutes. SRA PCR conditions were: initial denaturation at 95°C for 10 minutes, 40 cycles of 94°C for 30 seconds, 63°C for 90 seconds and 72°C for 70 seconds followed by an extension step at 72°C for 10 minutes. Additionally, flies were screened for T. b. gambiense using TgsGP PCR [29] using the following conditions: denaturation at 95°C for 10 minutes, 40 cycles of 95°C for 30 seconds, 63°C for 90 seconds and 72°C for 70 seconds followed a 10 minute extension at 72°C.
HRM qPCR
All 96 flies were screened using the novel multiplexed HRM qPCR. HRM reactions were run in a total volume of 12.5μl consisting of 2.5μl DNA template, 6.25 μl HRM Master Mix (Thermo-start ABgene, Rochester, New York, USA), 3.25 μl sterile DNase/RNase free water (Sigma, ST. Louis, USA) and 400nM of all forward and reverse primers were included in a multiplex. Reactions were carried out on a Rotor-Gene 6000 real-time PCR machine (Qiagen RGQ system). The following protocol was followed: denaturation at 95°C for 5 minutes followed by 40 cycles and denaturation for 10 seconds at 95°C per cycle, annealing and extension for 30 seconds at 58°C, and final extension for 30 seconds at 72°C. The melting step ran from 75°C to 90°C with a temperature increase of 0.1°C every 2 seconds. A threshold was set at 10% of the maximum normalized fluorescence (dF/dT) of the highest peak, with all peaks that both occurred at the diagnostic temperature (Tm) and crossed this threshold classified as positive.
Results
Primers
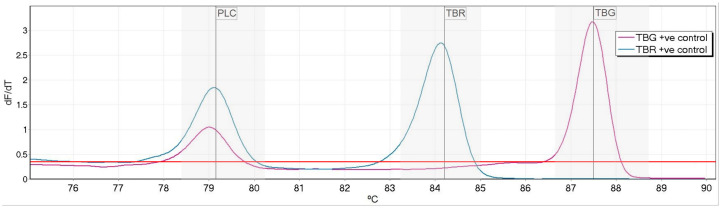
One primer-pair per target was selected based on successful amplification, distinct Tm and production of a clear peak. Product sizes for each amplicon ranged from 134–319 base pairs (Table 1) with peak temperatures ranging from 79.2°C to 87.5°C. To allow for automated calling of peaks, bin widths of 1.5°C (0.75°C either side of diagnostic Tm) were set for each target (Fig 1).
Table 1. HRM and PCR primers used for identification of single-copy gene targets of T. b. rhodesiense, T. b. gambiense and T. brucei s. l.
HRM primers were run in multiplex whilst PCR primers were run in singleplex. *This primer set was run in multiplex in HRM and in singleplex in PCR.
| Primer | Species | Primer sequence 5’-3’ | Product size (bp) | Product Tm (°C) | Reference | Assay |
|---|---|---|---|---|---|---|
| PLC1 | Trypanozoon | CAGTGTTGCGCTTAAATCCA | 319 | 79.1 | This study | HRM |
| PLC2 | CCCGCCAATACTGACATCTT | |||||
| TbRh1 | T. b. rhodesiense | GAAGCGGAAGCAAGAATGAC | 134 | 84.2 | This study | HRM |
| TbRh2 | GGCGCAAGACTTGTAAGAGC | |||||
| TgsGP1 | T. b. gambiense | GCTGCTGTGTTCGGAGAGC | 308 | 87.5 | [29] | HRM and PCR* |
| TGsGP2 | GCCATCGTGCTTGCCGCTC | |||||
| 657 | Trypanozoon | CGCTTTGTTGAGGAGCTGCAA GCA | 324 | - | [21] | PCR |
| 658 | TGCCACCGCAAAGTCGTTATT TCG | |||||
| SRA 02 | T. b. rhodesiense | AGCCAAAACCAGTGGGCA | 669 | - | [21] | PCR |
| SRA 03 | TAGCGCTGTCCTGTAGACGCT |
Fig 1. High resolution melt profile of phospholipase-C (PLC), T. b. rhodesiense (TBR) and T. b. gambiense (TBG) targets.
Normalized fluorescence dF/dt is plotted against degrees °C (deg.) The melt temperatures for each peak were 79.1°C for PLC, 84.2°C for T. b. rhodesiense and 87.5°C for T. b. gambiense. The positivity threshold is shown in red and target specific bins indicated in grey.
Analytical specificity and sensitivity
No non-specific amplification was seen when the assay was challenged with a range of non-target trypanosome species, namely T. congolense (Savannah, Kilifi and Forest subgroups), T. vivax, T. simiae, T. simiae Tsavo, T. godfreyi and T. grayi. The limit of detection was found to be an estimated 104 trypanosomes/mL for T. b. gambiense and T. b. rhodesiense using purified DNA from culture (S4 Fig). When the performance of the HRM was compared to TgsGP and SRA PCR on the same DNA dilution series, the HRM was as sensitive at detecting T. b. gambiense DNA and tenfold more sensitive for T. b. rhodesiense DNA (S2 and S3 Figs).
Screening of field samples
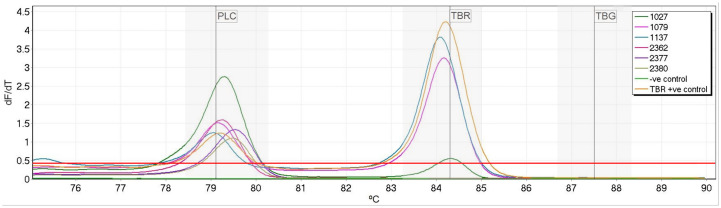
All flies (96/96) were positive by TBR PCR when initially rescreened. Using the multiplexed HRM, PLC positive control peaks were produced by 43 samples (44.8%), indicating sufficient target DNA for identification of either T. b. gambiense or T. b. rhodesiense. PLC PCR identified 19 samples (19.8%) with sufficient DNA quantity (Table 2). All samples which were positive by PLC PCR were also positive by HRM. None of the flies positive for T. congolense Savannah produced any non-specific amplification. Three flies (3.1%) were identified as positive for T. b. rhodesiense DNA both by HRM and PCR (Table 2), all of which also produced PLC peaks and were positive by PLC PCR (Fig 2). No flies were found to be positive for T. b. gambiense through either PCR or HRM. Flies that were positive for PLC but negative for T. b. gambiense or T. b. rhodesiense were considered to be infected with livestock trypanosome T. b. brucei.
Table 2. Breakdown of results of HRM and PCR on 96 field caught tsetse.
| HRM | PCR | ||||||
|---|---|---|---|---|---|---|---|
| Tsetse species | Total screened (N) | PLC positive (%) | SRA positive (%) | TgsGP positive (%) | PLC positive (%) | SRA positive (%) | TgsGP positive |
| G. pallidipes | 67 | 34 (50.7) | 2 (3.0) | 0 | 15 (22.4) | 2 (3.0) | 0 |
| G. swynnertoni | 29 | 9 (31.0) | 1 (3.4) | 0 | 4 (13.8) | 1 (3.4) | 0 |
Fig 2. Melt profile of field samples.
Three samples were identified as T. b. rhodesiense due to the production of both PLC and TBR peaks (samples:1027, 1079 and 1137). Samples which only produced PLC peaks were considered to be T. b. brucei (samples: 2362,2377 and 2380). The TBR positive control was T. b. rhodesiense DNA and the negative control used was nuclease free water. Normalized fluorescence dF/dt is plotted against degrees °C (deg.) with threshold indicated in red and species-specific bins shown in grey.
Discussion
Here we describe a novel high-resolution melt analysis for the detection and differentiation of T. b. rhodesiense and T. b. gambiense. This multiplexed assay screens for both pathogenic trypanosome species simultaneously with the addition of a third primer set to identify the presence of sufficient DNA to detect single-copy genes, acting as a positive control. The assay demonstrated high specificity with no cross-reaction with other non-target trypanosome species also transmitted by tsetse. The limit of detection of the HRM was lower than those reported in the literature for TgsGP [29] and SRA PCR [21]. However, when the three assays were compared directly against a dilution series of DNA, HRM was found to be as sensitive as TgsGP PCR and tenfold more sensitive than SRA PCR. The assay identified three wild caught tsetse flies to be positive for T. b. rhodesiense DNA. The three T. b. rhodesiense positives included two flies that had been previously identified by Lord et al [31]. All three flies including the additional fly that was identified by the HRM were positive by SRA PCR. However, the Ct value of the additional SRA positive was 30.5 in comparison to the two previously recorded positives which had Ct values of 21.1 and 21.8. This may indicate a lower DNA quantity and may represent a sample at the threshold of PCR detection. Of the 96 flies screened, 44.8% were positive for the universal-Trypanozoon PLC gene by HRM, indicating that they contained sufficient DNA for single-copy gene detection and therefore the detection of the SRA or TgsGP targets; if present. The remaining 55.2% of tsetse that were negative for the single-copy PLC gene can be assumed to have insufficient DNA present for the detection of the sub-species targets, we can therefore not rule these out as being negative for either T. b. gambiense or T. b. rhodesiense. Similar results have been reported in previous studies [33] and such a result is unsurprising when comparing the copy numbers, with TBR primers targeting a ~10,000 copy region and the sub-species primers targeting a single-copy gene, and presents an ongoing challenge. The two methods also differed in the number of flies positive for the single-copy PLC gene with the HRM method identifying a higher number of flies, indicating a greater sensitivity. This is likely due to the difficulty in identifying faint bands on a gel compared to the ability to better quantify a positive result using qPCR. When the Ct values of flies positive by HRM and PCR were compared to those that were only positive by HRM, the Cts of those positive by HRM only were slightly higher suggesting a potential failure of the PCR due to lower quantities of DNA in these samples (S5 Fig).
The method has three advantages over traditional PCR methods. First, the HRM time to result of ≤2 hours is faster than PCR followed by gel electrophoresis which can take over three hours for product amplification and visualisation. Second, this is a closed-tube assay which reduces contamination risk. Finally, it does not require interpretation of gel electrophoresis results. Through the use of detection bins, sample processing can also be automated, further speeding up and simplifying data analysis. The simple and fast nature of this method indicates it could be suitable for the high-throughput processing of tsetse. With prevalence of T. b. gambiense in tsetse from HAT foci predicted to be as low as 1 in 105 [34], there is a need for a xenomonitoring tool which can be applied to large numbers of samples. At present, this method is not suitable to a low-resources setting because it requires a qPCR machine, however with further optimisation this technique could be applicable to more field-friendly technologies such as the Magnetic Induction Cycler (Mic) (Bio molecular Systems). This method has the potential to provide the basis of a real-time trypanosome transmission monitoring platform, enabling timely reactive measures by disease control programmes. Furthermore, with the risk of traditionally distinct geographical distributions of both Rhodesian and Gambian HAT changing due to the movement of livestock [35], human migration [36] and climate change [37–40], it may become increasingly important to screen simultaneously for both trypanosome species. The HRM allows for this and removes the risk of presumptive screening based on historic disease distributions.
Study limitations
The authors acknowledge that the reliance of low copy genes for target identification is a limiting factor of the described diagnostic assay. However, at present these single copy genes are the only identifiers of members of the T. brucei s. l. and so pose a challenge to any diagnostic method based on these targets. This study was also challenged by a small number (96) of field caught flies from a rHAT focus and none from a gHAT focus where tsetse infected with T. b. gambiense might be present. Consequently, assay performance on field samples could only be evaluated for T. b. rhodesiense. Due to trypanosome prevalence in HAT foci predicted to be very low [34], obtaining sufficient field samples for any assay validation will be difficult. As control efforts such as active screening and treating the human population, combined with tsetse control, the numbers of infected tsetse will likely decline even further [41–44]. The authors are currently in planning to validate this method in a gHAT focus. Additionally, the field samples had only been screened for the presence of T. brucei s. l. and T. congolense Savannah. Studies of flies caught in this area suggest that these flies would have likely been infected with other trypanosome species [45,46] however this cannot be confirmed. Further validation using field caught flies with known infections with other non-target species would provide further evidence of the specificity of this method. Another limitation of this study was the comparison of only two methods for the identification of T. b. rhodesiense when there are alternative molecular methods available for HAT detection such as Trypanozoon sub-species specific LAMP [47,48] assays, which have demonstrated a high degree of sensitivity. Comparison between a wider range of methods would help assess HRM’s potential as a screening tool for human trypanosomes. A final limitation of this assay, as with all assays targeting DNA, is the inability to differentiate between a tsetse with an active infection or one with a transient one. Similarly, the assay is unable to determine whether a fly has a mature infection and is therefore capable of transmitting the disease. By using RNA as a target it may be possible to identify an active infection and determine what stage of infection it is; either mature or immature. Detailed analyses of RNA transcripts would be required to overcome this challenge.
In order to overcome these limitations follow-up studies will be conducted in order to more fully validate the HRM assay against larger field samples for both T. b. gambiense and T. b. rhodesiense.
In summary, we describe the development of a novel HRM assay for the detection and discrimination of human African trypanosomes in tsetse flies. The assay also incorporates an internal control, identifying samples with sufficient genomic material. The closed tube nature of the assay in addition to the relatively fast time and potential for automated calling lends itself to use in high throughput xenomonitoring surveillance campaigns for HAT.
Supporting information
(TIF)
Marker (M) is 100bp (Invitrogen).
(TIF)
Marker (M) is 100bp (Invitrogen).
(TIF)
Positivity threshold is shown in red and target specific bins indicated in grey.
(TIF)
(TIF)
Acknowledgments
We thank Drs Liam Morrison and Jennifer Lord for providing the Tanzanian tsetse samples and screening data collected under the auspices of the Zoonosis and Emerging and Livestock Systems (ZELS) programme (BB/L019035/1).
Data Availability
Data are fully available and found within the manuscript.
Funding Statement
This work was supported by the Medical Research Council via the LSTM-Lancaster Doctoral Training Partnership [grant number MR/N013514/1], received by GG. Samples were collected as part of the Zoonoses and Emerging Livestock Systems (ZELS) programme supported by the UK’s Biotechnology and Biological Sciences Research Council (BB/L019035/1), received by ST, JL and IS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Trypanosomiasis, human African (sleeping sickness) [Internet]. [cited 2018 Sep 6]. http://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness)
- 2.Holmes P. First WHO Meeting of Stakeholders on Elimination of Gambiense Human African Trypanosomiasis. Büscher P, editor. PLoS Negl Trop Dis [Internet]. 2014. October 23 [cited 2018 Jun 18];8(10):e3244 Available from: http://dx.plos.org/10.1371/journal.pntd.0003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Accelerating work to overcome the global impact of neglected tropical diseases: a roadmap for implementation: executive summary. 2012;
- 4.World Health Organization. Control and surveillance of human African trypanosomiasis. [cited 2018 Jun 18]; http://apps.who.int/iris/bitstream/handle/10665/95732/9789241209847_eng.pdf?sequence=1&isAllowed=y [PubMed]
- 5.Tong J, Valverde O, Mahoudeau C, Yun O, Chappuis F. Challenges of controlling sleeping sickness in areas of violent conflict: experience in the Democratic Republic of Congo. Confl Health [Internet]. 2011. December 26 [cited 2019 Sep 12];5(1):7 Available from: https://conflictandhealth.biomedcentral.com/articles/10.1186/1752-1505-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matovu E, Kitibwa A, Picado A, Biéler S, Bessell PR, Ndung’u JM. Serological tests for gambiense human African trypanosomiasis detect antibodies in cattle. Parasit Vectors [Internet]. 2017. November 3 [cited 2018 Jun 20];10(1):546 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29100526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamonneau V, Camara O, Ilboudo H, Peylhard M, Koffi M, Sakande H, et al. Accuracy of Individual Rapid Tests for Serodiagnosis of Gambiense Sleeping Sickness in West Africa. Büscher P, editor. PLoS Negl Trop Dis [Internet]. 2015. February 2 [cited 2018 Jun 20];9(2):e0003480 Available from: http://dx.plos.org/10.1371/journal.pntd.0003480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisser S, Lumbala C, Nguertoum E, Kande V, Flevaud L, Vatunga G, et al. Sensitivity and Specificity of a Prototype Rapid Diagnostic Test for the Detection of Trypanosoma brucei gambiense Infection: A Multi-centric Prospective Study. PLoS Negl Trop Dis. 2016;10(4). 10.1371/journal.pntd.0004608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büscher P, Mertens P, Leclipteux T, Gilleman Q, Jacquet D, Mumba-Ngoyi D, et al. Sensitivity and specificity of HAT Sero-K-SeT, a rapid diagnostic test for serodiagnosis of sleeping sickness caused by Trypanosoma brucei gambiense: A case-control study. Lancet Glob Heal. 2014. June 1;2(6):e359–63. 10.1016/S2214-109X(14)70203-7 [DOI] [PubMed] [Google Scholar]
- 10.Boelaert M, Mukendi D, Bottieau E, Kalo Lilo JR, Verdonck K, Minikulu L, et al. A Phase III Diagnostic Accuracy Study of a Rapid Diagnostic Test for Diagnosis of Second-Stage Human African Trypanosomiasis in the Democratic Republic of the Congo. EBioMedicine. 2018. January 1;27:11–7. 10.1016/j.ebiom.2017.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keating J, Yukich JO, Sutherland CS, Woods G, Tediosi F. Human African trypanosomiasis prevention, treatment and control costs: A systematic review Vol. 150, Acta Tropica. Elsevier B.V.; 2015. p. 4–13. [DOI] [PubMed] [Google Scholar]
- 12.Farid HA, Morsy ZS, Helmy H, Ramzy RMR, El Setouhy M, Weil GJ. A critical appraisal of molecular xenomonitoring as a tool for assessing progress toward elimination of lymphatic filariasis. Am J Trop Med Hyg. 2007. October;77(4):593–600. [PMC free article] [PubMed] [Google Scholar]
- 13.Schmaedick MA, Koppel AL, Pilotte N, Torres M, Williams SA, Dobson SL, et al. Molecular Xenomonitoring Using Mosquitoes to Map Lymphatic Filariasis after Mass Drug Administration in American Samoa. Bockarie M, editor. PLoS Negl Trop Dis [Internet]. 2014. August 14 [cited 2020 Jan 15];8(8):e3087 Available from: https://dx.plos.org/10.1371/journal.pntd.0003087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau CL, Won KY, Lammie PJ, Graves PM. Lymphatic Filariasis Elimination in American Samoa: Evaluation of Molecular Xenomonitoring as a Surveillance Tool in the Endgame. Rinaldi G, editor. PLoS Negl Trop Dis [Internet]. 2016. November 1 [cited 2020 Jan 15];10(11):e0005108 Available from: https://dx.plos.org/10.1371/journal.pntd.0005108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morlais I, Grebaut P, Bodo JM, Djoha S, Cuny G, Herder S. Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 1998. June 15;70(1):109–17. 10.1016/s0001-706x(98)00014-x [DOI] [PubMed] [Google Scholar]
- 16.Mukadi P, Gillet P, Lukuka A, Atua B, Sheshe N, Kanza A, et al. External quality assessment of Giemsa-stained blood film microscopy for the diagnosis of malaria and sleeping sickness in the Democratic Republic of the Congo. Bull World Health Organ. 2013. June;91(6):441–8. 10.2471/BLT.12.112706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moser DR, Ochs DE, Bailey CP, McKane MR, Donelson JE, Cook GA. Detection of trypanosoma congolense and trypanosoma brucei subspecies by dna amplification using the polymerase chain reaction. Parasitology [Internet]. 1989. August 6 [cited 2020 Feb 12];99(1):57–66. Available from: https://www.cambridge.org/core/product/identifier/S0031182000061023/type/journal_article [DOI] [PubMed] [Google Scholar]
- 18.Berberof M, Pérez-Morga D, Pays E. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol Biochem Parasitol [Internet]. 2001. March 1 [cited 2019 Oct 9];113(1):127–38. Available from: https://www.sciencedirect.com/science/article/pii/S0166685101002080. [DOI] [PubMed] [Google Scholar]
- 19.De Greef C, Imberechts H, Matthyssens G, Van Meirvenne N, Hamers R. A gene expressed only in serum-resistant variants of Trypanosoma brucei rhodesiense. Mol Biochem Parasitol. 1989. September 1;36(2):169–76. 10.1016/0166-6851(89)90189-8 [DOI] [PubMed] [Google Scholar]
- 20.De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol. 1994. December 1;68(2):277–84. 10.1016/0166-6851(94)90172-4 [DOI] [PubMed] [Google Scholar]
- 21.Picozzi K, Carrington M, Welburn SC. A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol. 2008. January;118(1):41–6. 10.1016/j.exppara.2007.05.014 [DOI] [PubMed] [Google Scholar]
- 22.Hereld D, Hart GW, Englund PT. cDNA encoding the glycosyl-phosphatidylinositol-specific phospholipase C of Trypanosoma brucei (glycolipid anchor/variant surface glycoprotein). Vol. 85, Proc. Natl. Acad. Sci. USA. 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrington M, Bülow R, Reinke H, Overath P. Sequence and expression of the glycosyl-phosphatidylinositol-specific phospholipase C of Trypanosoma brucei. Mol Biochem Parasitol. 1989. March 15;33(3):289–96. 10.1016/0166-6851(89)90091-1 [DOI] [PubMed] [Google Scholar]
- 24.Winkle S. Geißeln der Menschheit. Kulturgeschichte der Seuchen. Kult der Seuchen. 1997;Düsseldorf.
- 25.Mensa Wilmot K, Hereld D, Englund PT. Genomic Organization, Chromosomal Localization, and Developmentally Regulated Expression of the Glycosyl-Phosphatidylinositol-Specific Phospholipase C of Trypanosoma brucei [Internet]. MOLECULAR AND CELLULAR BIOLOGY. 1990. [cited 2020 Feb 17]. Available from: http://mcb.asm.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higuera SL, Guhl F, Ramírez JD. Identification of Trypanosoma cruzi Discrete Typing Units (DTUs) through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasites and Vectors [Internet]. 2013. [cited 2020 Jun 24];6(1). Available from: https://pubmed.ncbi.nlm.nih.gov/23602078/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosseini-Safa A, Mohebali M, Hajjaran H, Akhoundi B, Zarei Z, Arzamani K, et al. High resolution melting analysis as an accurate method for identifying Leishmania infantum in canine serum samples. J Vector Borne Dis. 2018. December 1;55(4):315–20. 10.4103/0972-9062.256568 [DOI] [PubMed] [Google Scholar]
- 28.Talmi-Frank D, Nasereddin A, Schnur LF, Schönian G, Töz SÖ, Jaffe CL, et al. Detection and Identification of Old World Leishmania by High Resolution Melt Analysis. Louzir H, editor. PLoS Negl Trop Dis [Internet]. 2010. January 12 [cited 2020 Jun 24];4(1):e581 Available from: https://dx.plos.org/10.1371/journal.pntd.0000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radwanska M, Claes F, Magez S, Magnus E, Pérez-Morga D, Pays E, et al. Novel primer sequences for Polymerase Chain Reaction-based detection of [Internet]. 2014 [cited 2018 Sep 10]. https://www.researchgate.net/publication/11055435. [DOI] [PubMed]
- 30.Borst P, Van der Ploeg M, Van Hoek JFM, Tas J, James J. On the DNA content and ploidy of trypanosomes. Mol Biochem Parasitol. 1982. July 1;6(1):13–23. 10.1016/0166-6851(82)90049-4 [DOI] [PubMed] [Google Scholar]
- 31.JS Lord, RS. Lea, FK. Allan, M Byamungu, DR Hall, J Lingley, F Mramba, E Paxton, GA Vale, JW Hargrove, LJ Morrison, SJ Torr HA. Assessing the effect of insecticide-treated cattle on tsetse abundance and trypanosome transmission at the wildlife-livestock interface in Serengeti, Tanzania. bioRxiv doi. 2020; [DOI] [PMC free article] [PubMed]
- 32.Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. SENSITIVE DETECTION OF TRYPANOSOMES IN TSETSE FLIES BY DNA AMPLIFICATION. Vol. 22. 1992. [DOI] [PubMed]
- 33.Cunningham LJ, Lingley JK, Tirados I, Esterhuizen J, Opiyo M, Mangwiro CTN, et al. Evidence of the absence of human African trypanosomiasis in two northern districts of Uganda: Analyses of cattle, pigs and tsetse flies for the presence of Trypanosoma brucei gambiense. Mireji P, editor. PLoS Negl Trop Dis [Internet]. 2020. April 7 [cited 2020 Jul 1];14(4):e0007737 Available from: https://dx.plos.org/10.1371/journal.pntd.0007737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rock KS, Torr SJ, Lumbala C, Keeling MJ. Quantitative evaluation of the strategy to eliminate human African trypanosomiasis in the Democratic Republic of Congo. Parasites and Vectors. 2015. October 22;8(1):1–13. 10.1186/s13071-015-1131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selby R, Bardosh K, Picozzi K, Waiswa C, Welburn SC. Cattle movements and trypanosomes: restocking efforts and the spread of Trypanosoma brucei rhodesiense sleeping sickness in post-conflict Uganda. Parasit Vectors [Internet]. 2013. September 27 [cited 2019 Sep 12];6(1):281 Available from: http://parasitesandvectors.biomedcentral.com/articles/10.1186/1756-3305-6-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picado A, Ndung’u J. Elimination of sleeping sickness in Uganda could be jeopardised by conflict in South Sudan Vol. 5, The Lancet Global Health. Elsevier Ltd; 2017. p. e28–9. [DOI] [PubMed] [Google Scholar]
- 37.Lord JS, Hargrove JW, Torr SJ, Vale GA. Climate change and African trypanosomiasis vector populations in Zimbabwe’s Zambezi Valley: A mathematical modelling study. Thomson M, editor. PLOS Med [Internet]. 2018. October 22 [cited 2020 Feb 18];15(10):e1002675 Available from: http://dx.plos.org/10.1371/journal.pmed.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore S, Shrestha S, Tomlinson KW, Vuong H. Predicting the effect of climate change on African trypanosomiasis: integrating epidemiology with parasite and vector biology. [cited 2020 Feb 18]; http://rsif.royalsocietypublishing.org. [DOI] [PMC free article] [PubMed]
- 39.Fèvre EM, Coleman PG, Odiit M, Magona JW, Welburn SC, Woolhouse MEJ. The origins of a new Trypanosoma brucei rhodesiense sleeping sickness outbreak in eastern Uganda. Lancet [Internet]. 2001. August 25 [cited 2020 Feb 17];358(9282):625–8. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0140673601057786. [DOI] [PubMed] [Google Scholar]
- 40.Picozzi K, Fèvre EM, Odiit M, Carrington M, Eisler MC, Maudlin I, et al. Sleeping sickness in Uganda: a thin line between two fatal diseases. 2001 [cited 2020 Feb 17]; http://www.bmj.com/. [DOI] [PMC free article] [PubMed]
- 41.Mahamat MH, Peka M, Rayaisse J-B, Rock KS, Toko MA, Darnas J, et al. Adding tsetse control to medical activities contributes to decreasing transmission of sleeping sickness in the Mandoul focus (Chad). Dinglasan RR, editor. PLoS Negl Trop Dis [Internet]. 2017. July 27 [cited 2020 Jul 1];11(7):e0005792 Available from: https://dx.plos.org/10.1371/journal.pntd.0005792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courtin F, Camara M, Rayaisse J-B, Kagbadouno M, Dama E, Camara O, et al. Reducing Human-Tsetse Contact Significantly Enhances the Efficacy of Sleeping Sickness Active Screening Campaigns: A Promising Result in the Context of Elimination. Aksoy S, editor. PLoS Negl Trop Dis [Internet]. 2015. August 12 [cited 2020 Jul 1];9(8):e0003727 Available from: https://dx.plos.org/10.1371/journal.pntd.0003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirados I, Stanton MC, Selby R, Mpembele F, Mwamba Miaka E, Boelaert M, et al. Impact of Tiny Targets on Glossina fuscipes quanzensis, the primary vector of Human African Trypanosomiasis in the Democratic Republic of Congo. bioRxiv [Internet]. 2020. April 7 [cited 2020 Jul 1];1–31. Available from: 10.1101/2020.04.07.029587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tirados I, Esterhuizen J, Kovacic V, Mangwiro TNC, Vale GA, Hastings I, et al. Tsetse Control and Gambian Sleeping Sickness; Implications for Control Strategy. Dumonteil E, editor. PLoS Negl Trop Dis [Internet]. 2015. August 12 [cited 2020 Jul 1];9(8):e0003822 Available from: https://dx.plos.org/10.1371/journal.pntd.0003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams ER, Malele II, Msangi AR, Gibson WC. Trypanosome identification in wild tsetse populations in Tanzania using generic primers to amplify the ribosomal RNA ITS-1 region. Acta Trop [Internet]. 2006. November 1 [cited 2018 Sep 7];100(1–2):103–9. Available from: https://www.sciencedirect.com/science/article/pii/S0001706X06001902. [DOI] [PubMed] [Google Scholar]
- 46.Adams ER, Hamilton PB. New molecular tools for the identification of trypanosome species. Futur Microbiol [Internet]. 2008. [cited 2020 Jun 18];3(2):167–76. Available from: www.futuremedicine.com. [DOI] [PubMed] [Google Scholar]
- 47.Njiru ZK, Mikosza ASJ, Matovu E, Enyaru JCK, Ouma JO, Kibona SN, et al. African trypanosomiasis: Sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol [Internet]. 2008. April [cited 2019 Jul 10];38(5):589–99. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0020751907003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wastling SL, Picozzi K, Kakembo ASL, Welburn SC. LAMP for Human African Trypanosomiasis: A Comparative Study of Detection Formats. Masiga DK, editor. PLoS Negl Trop Dis [Internet]. 2010. November 2 [cited 2020 Jun 24];4(11):e865 Available from: https://dx.plos.org/10.1371/journal.pntd.0000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Marker (M) is 100bp (Invitrogen).
(TIF)
Marker (M) is 100bp (Invitrogen).
(TIF)
Positivity threshold is shown in red and target specific bins indicated in grey.
(TIF)
(TIF)
Data Availability Statement
Data are fully available and found within the manuscript.