Abstract
Trimethoprim is a synthetic antibacterial agent that targets folate biosynthesis by competitively binding to the di-hydrofolate reductase enzyme (DHFR). Trimethoprim is often administered synergistically with sulfonamide, another chemotherapeutic agent targeting the di-hydropteroate synthase (DHPS) enzyme in the same pathway. Clinical resistance to both drugs is widespread and mediated by enzyme variants capable of performing their biological function without binding to these drugs. These mutant enzymes were assumed to have arisen after the discovery of these synthetic drugs, but recent work has shown that genes conferring resistance to sulfonamide were present in the bacterial pangenome millions of years ago. Here, we apply phylogenetics and comparative genomics methods to study the largest family of mobile trimethoprim-resistance genes (dfrA). We show that most of the dfrA genes identified to date map to two large clades that likely arose from independent mobilization events. In contrast to sulfonamide resistance (sul) genes, we find evidence of recurrent mobilization in dfrA genes. Phylogenetic evidence allows us to identify novel dfrA genes in the emerging pathogen Acinetobacter baumannii , and we confirm their resistance phenotype in vitro. We also identify a cluster of dfrA homologues in cryptic plasmid and phage genomes, but we show that these enzymes do not confer resistance to trimethoprim. Our methods also allow us to pinpoint the chromosomal origin of previously reported dfrA genes, and we show that many of these ancient chromosomal genes also confer resistance to trimethoprim. Our work reveals that trimethoprim resistance predated the clinical use of this chemotherapeutic agent, but that novel mutations have likely also arisen and become mobilized following its widespread use within and outside the clinic. Hence, this work confirms that resistance to novel drugs may already be present in the bacterial pangenome, and stresses the importance of rapid mobilization as a fundamental element in the emergence and global spread of resistance determinants.
Keywords: trimethoprim, antibiotics, chemotherapeutic agent, resistance, sulfonamides, phylogenetics, evolution
Data Summary
Nucleotide and protein sequences analysed in this study have been downloaded from publicly available National Center for Biotechnology Information databases.
The scripts used for data collection and analysis can be obtained at the GitHub ErillLab repository (https://github.com/ErillLab/).
The Bayesian phylogenetic tree can be visualized online on iTOL (https://itol.embl.de/tree/855674159451585133078) [1].
Impact Statement.
Antibiotic resistance is a pressing and global phenomenon. It is well established that resistance to conventional antibiotics emerged millions of years ago in either antibiotic-producing bacteria or their competitors. Resistance to synthetic chemotherapeutic agents cannot be explained by this paradigm, since these drugs are not naturally produced. Hence, resistance is assumed to have evolved rapidly following the clinical introduction of these drugs. Recently, we showed that resistance to one such drug, sulfonamide, evolved not recently, but millions of years ago, suggesting that the diversity of bacterial genomes may well contain genes conferring resistance to drugs yet to be developed. Here, we analyse the origin of resistance to trimethoprim, another chemotherapeutic agent developed in the 1960s. Using phylogenetic methods, we identify new variants of the trimethoprim-resistance genes that had not previously been reported, and we trace the chromosomal origins for a number of already known resistance variants. Our results show that resistance to trimethoprim is very diverse, and has originated both from recent mutations and from pre-existing ancient variants. These results stress the importance of gene mobilization mechanisms as the main drivers of the current antibiotic-resistance phenomenon.
Introduction
Bacterial resistance to antibacterial agents remains an increasingly challenging and global problem in modern health care [2, 3]. Bacterial cells display a diverse array of mechanisms to cope with exposure to antibacterial compounds, including modification or overexpression of the antibacterial target, efflux or reduction of antibacterial uptake and the use of alternate pathways [4]. Constant exposure to non-lethal concentrations of antibacterial agents may lead to the selection of partial resistance to antibiotics over relatively short time spans [5], and this evolution may be hastened by simultaneous exposure to multiple antibacterials [6]. However, the rapid proliferation of multidrug-resistant nosocomial pathogens in the last 50 years has not been driven by the independent evolution of resistance traits, but through the extensive dissemination of mobile genetic elements carrying resistance genes [4, 7]. It is widely accepted that most genes conferring resistance to antibiotics present in pathogenic bacteria were obtained by successive lateral gene transfer of homologues that originally evolved in the microbes that produce the antibiotic or in their natural competitors [7, 8]. The high plasticity of bacterial genomes, enabled by a large repertoire of mobile genetic elements, and the availability of a large pool of ancient antibiotic-resistance determinants, hence, set the stage for the rapid proliferation of antibiotic resistance, giving rise to multi-resistant clinical strains just a few years after the commercial introduction of antibiotics [7].
Synthetic chemotherapeutic agents predate antibiotics in the clinical setting, and continue to be used synergistically with antibiotics to treat microbial infections [9]. Following the initial discovery and clinical use of arsphenamine in 1907 [10], interest in chemotherapeutic agents quickly took off after the development of sulfa drugs in the 1930s [11]. The discovery of trimethoprim (a di-aminopyrimidine) was received with interest because, like sulfonamides, trimethoprim targets the bacterial synthesis of tetrahydrofolic acid, which is a necessary cofactor in the synthesis of thymine and purines [12]. Sulfonamides are structural analogues of para-aminobenzoic acid (PABA) and inhibit the synthesis of di-hydropteroate by competing with PABA for binding to the di-hydropteroate synthase (DHPS) enzyme, resulting in sulfonamide-bound di-hydropterin [13]. Trimethoprim is a structural analogue of di-hydrofolic acid, derived from di-hydropteroate. It acts by competitively binding to the di-hydrofolate reductase (DHFR) enzyme; hence, inhibiting the production of tetrahydrofolic acid [13, 14]. The synergistic use of trimethoprim and sulfonamides was expected to have a potent bactericidal effect by producing a serial blockade on the tetrahydrofolic acid pathway [12, 15].
Unlike antibiotics, chemotherapeutic agents are not produced by natural organisms, yet resistance to these novel drugs arose quickly after their mass-production and it is today pervasive among clinical isolates [7]. In the case of sulfonamides and trimethoprim, which are usually administered in tandem, resistance via chromosomal mutations to both chemotherapeutics was reported soon after their clinical introduction [13]. Chromosomal resistance to sulfonamides can occur through mutations yielding increased production of PABA [16] or, more commonly, via mutations to the chromosomal folP gene (encoding DHPS), which decrease the affinity of DHPS for sulfonamide without detriment to PABA binding [13, 17]. Such mutations have been reported in multiple bacterial groups and target different conserved regions of DHPS [13]. Similarly, chromosomal resistance to trimethoprim may arise via mutations that increase transcription of the folA gene (encoding DHFR) [18], or through mutations that decrease the affinity of DHFR for trimethoprim [13]. The vast majority of resistant clinical isolates to both sulfonamides and trimethoprim, however, are not due to chromosomal mutations, but to the acquisition of resistance determinants on mobile genetic elements [13]. Parallel to their systematic combined use in both clinical and agricultural settings, genes conferring resistance to sulfonamides and trimethoprim are frequently found together on mobile elements, such as class 1 integrons [19] or conjugative plasmids [13, 20]. The mobile genes conferring resistance to sulfonamide are homologues of the chromosomally encoded folP gene and are collectively known as sul genes (for sulfonamide resistance). Mobile genes conferring resistance to trimethoprim are either homologues or functional analogues of the chromosomally encoded folA gene and are collectively known as dfr genes (for di-hydrofolate reductase) [17].
In spite of their frequent co-occurrence on mobile genetic elements, there are significant differences between the mobile genes conferring resistance to sulfonamides (sul genes) and trimethoprim (dfr genes). To date, only three sul gene classes have been described in clinical isolates [21], whereas more than 30 different dfr genes have been reported in clinically relevant strains [22]. Trimethoprim-resistance (dfr) genes have been further classified into two families (dfrA and dfrB). These two families encode evolutionarily unrelated proteins of markedly different sizes. Sequence similarity indicates that dfrA genes are homologues of the chromosomally encoded folA genes, whereas dfrB genes are functional analogues of unknown origin [23, 24]. Most dfrA genes follow a standard naming convention consisting of dfrA followed by a numerical value indicating their discovery rank order. However, several dfrA genes first identified in Gram-positive bacteria, and thought at the time to be unrelated to the Gram-negative dfrA genes, were originally named following an alphabetical convention (dfrC–K). The disparity in genetic diversity among sulfonamide and trimethoprim mobile resistance determinants is suggestive of different evolutionary processes leading to the onset and spread of resistance to these two chemotherapeutic agents [13]. It was suggested that resistance to sulfonamide had arisen in a few isolated organisms and rapidly spread upon the introduction of sulfa drugs, whereas trimethoprim resistance had independently evolved, and had been subsequently mobilized multiple times [13].
Recently, we examined the origins of sul genes through comparative genomics, phylogenetic analysis and in vitro assays [25]. We identified a well-defined mutational signature in sul-encoded proteins with respect to chromosomally encoded folP genes, and we used this conserved motif to map the origins of sul genes in bacterial chromosomes. Our work revealed that the three groups of sul genes identified in clinical isolates originated in the Leptospiraceae and were transferred to the Rhodobiaceae more than 500 million years ago. These two ancient resistant determinants were later independently mobilized, and rapidly disseminated following the commercial introduction of sulfa drugs. By tracing the phylogenetic lineage of sul genes and demonstrating that these two bacterial families were resistant to sulfonamides long before their discovery and clinical use, our work indicated that resistance to novel drugs could very well pre-exist, and be ready for mobilization, within the vast bacterial pangenome. Here, we apply similar methods to examine the phylogenetic relationships among dfrA and chromosomally encoded folA genes. Our aim is to shed light on the evolutionary processes giving rise to mobile trimethoprim-resistance genes. Our work illustrates significant similarities and differences in the processes leading to the emergence and spread of trimethoprim- and sulfonamide-resistance determinants, reveals previously unreported clusters of dfrA genes, and suggests that systematic analyses of the bacterial pangenome may be of use in the design of novel antibacterials.
Methods
Sequence data collection
To identify homologues of DfrA proteins, we first compiled a panel of Dfr proteins reported in the literature. Dfr proteins belong to two distinct families of unrelated sequences (DfrA and DfrB; Fig. S1, available with the online version of this article). We mapped these sequences to pfam models of DfrA (PF00186) and DfrB (PF06442) (Table S1) using hmmer version 3.1b2 (hmmscan) [26], and we discarded sequences mapping to the DfrB family, retaining only DfrA proteins for analysis (Table S2). We further excluded redundant DfrA sequences (amino acid sequence identity >90 %) using t-coffee version 11.00.8cbe486 seq_reformat command [27], and used the resulting non-redundant panel to identify DfrA homologues in protein records associated with National Center for Biotechnology Information (NCBI) GenBank/RefSeq sequences corresponding to mobile genetic elements. These were defined as sequences containing the keywords ‘plasmid’, ‘integron’ or ‘transposon’ in their title, belonging to complete genome records [28, 29]. Protein records corresponding to blastp hits matching stringent value (<1e−20) and query coverage (>75 %) thresholds were added to the panel if non-redundant (amino acid sequence identity <90 % with respect to existing panel members), and their classification as mobile elements was validated by assessing that the nucleotide record encoding them contained at least one gene encoding an integrase, transposase or plasmid replication protein, as determined by hmmer (hmmscan, E value <1e−05) with reference pfam models (Table S3) [30–34]. To detect putative chromosomally encoded folA genes associated with mobile dfrA genes, we used the sequences in the extended non-redundant panel of DfrA homologues as queries for tblastn searches against NCBI GenBank complete genomes with stringent E value (<1e−40) and query coverage (>75 %) settings. Hits with nearby genes annotated as resistance determinants, transposases or integrases were considered to encode chromosomally integrated mobile DfrA homologues and not considered for further analysis. For each mobile DfrA homologue in the panel, the first, if any, tblastn hit satisfying these thresholds was considered, for the purposes of this study, to be a proxy for the closest putative chromosomally encoded FolA protein. The choice of representative DfrA sequences did not alter the tblastn results. To complete the panel of protein sequences used to reconstruct the evolutionary history of DfrA/FolA sequences, we used the non-redundant panel of mobile DfrA sequences to identify via blastp (E value <1e−20, coverage >75 %) FolA proteins encoded by the chromosomes of NCBI RefSeq representative species for all bacterial orders, and for each bacterial family in the Proteobacteria . In addition, the closest archaeal homologues of bacterial FolA sequences were determined by searching with blastp the NCBI protein database, restricted to Archaea (taxid:2157), with the Escherichia coli FolA protein. A member of each family from the order ( Halobacteriales ) of the identified best archaeal hit of E. coli FolA was sampled to populate the outgroup.
Phylogenetic analysis
For phylogenetic inference, we performed a t-coffee multiple sequence alignment of protein sequences for the complete panel of DfrA and FolA homologues, combining three clustalw (version 2.1) profile amino acid sequence alignments with different (5, 10, 25) gap opening penalties and leveraging the E. coli FolA crystal structure (P0ABQ5) to adjust gap penalties [35]. The resulting amino acid sequence alignment was processed with Gblocks version 0.91b (allowed gap positions, with half; minimum number of sequences for a conserved position, 86; minimum number of sequences for a flanking position, 95; maximum number of contiguous nonconserved positions, 5; minimum length of a block, 4) [36]. Bayesian phylogenetic inference on the trimmed multiple amino acid sequence alignment was carried out with MrBayes version 3.2.6 [37]. Four Metropolis-coupled Markov chain Monte Carlo simulations with four independent chains were run for 20 000 000 generations, using a mixed four-category gamma distributed rate plus proportion of invariable sites model [invgamma] and a JTT (Jones–Taylor–Thornton) amino acid substitution model [38]. Chains were sampled every 100 iterations and stationarity was analysed with Tracer version 1.7.1 [39] by monitoring the estimated sample size (ESS). To determine burn-in, chain results were summarized with MrBayes imposing the restriction that ESS be above 200 and that the potential scale reduction factor be within 0.005 of 1. Based on summarization results, the burn-in was set at 20 % of iterations. A consensus tree was generated with the half-compat option and visualized using the ggtree version 1.14.6 R library [40]. Clades of reported DfrA proteins were determined as branches with posterior probability values higher than 0.8 containing at least five reported DfrA sequences. Ancestral state reconstruction of a single binary trait (mobile/chromosomal) was performed with BayesTraits version 3.0.2 [41]. The mobile/chromosomal state of each sequence was determined through the data collection pipeline outlined above. Known states at tree tips were labelled, and ancestral states were reconstructed using the multistate and maximum-likelihood settings.
DNA techniques and in vitro trimethoprim-susceptibility assay
With the exception of the Ralstonia solanacearum GMI1000 (Marc Valls, Center for Research in Agricultural Genomics, Barcelona, Spain) and E. coli K-12 (CGSC5073) folA genes, which were amplified from genomic DNA, dfrA and folA homologues were adapted to E. coli codon usage, synthesized (ATG:biosynthetics) and then subcloned into a dephosphorylated pUA1108 vector [42] using an NdeI and BamHI (New England Biolabs) double digest when possible. Genes with internal restriction sites for any of these two enzymes were subcloned into the same vector using the HiFi DNA assembly kit (New England Biolabs), following the manufacturer’s protocol. Oligonucleotides used in this work are listed in Table S4. All constructs were validated by sequencing (Macrogen) prior to their use in transforming E. coli K-12 (CGSC5073). The minimum inhibitory concentration (MIC) for trimethoprim (Sigma-Aldrich) for strains of E. coli K-12 (CGSC5073) carrying different versions of pUA1108 encoding folA or dfrA homologues was determined following the Clinical and Laboratory Standards Institute guidelines using microdilution tests in Mueller–Hinton (MH) broth (Merck) [43]. All MIC assays were performed in triplicate. Colonies were grown on Luria–Bertani agar for 18 h and then suspended in sterile 0.9 % NaCl solution to a McFarland 0.5 turbidity level. Suspensions were then diluted at 10−2 in MH broth, and 50 µl (5×104) cells were inoculated into microtitre plates that contained 50 µl MH broth supplemented with 1024–0.250 mg trimethoprim l−1. To determine growth, optical density at 550 nm was measured after 24 h incubation at 37 °C. The dfrA1 gene was used as a positive control [44] and the E. coli folA gene as a negative control [45].
Sequence analysis
To assess whether the identified chromosomal gene associated with a mobile dfrA gene is the canonical folA gene for the genus, and not the product of a subsequent recombination of the mobile dfrA gene into the chromosome, we computed the pairwise amino acid identity among the products of all chromosomal folA homologues and then compared this distribution with the pairwise amino acid identity of the putative origin versus the chromosomal folA homologues. We used a one-sided Mann–Whitney U test to determine whether the two distributions were significantly different. To analyse the mol% G+C content relationship between sul/dfrA genes and their host chromosomes, we used pre-compiled panels of sequences for non-redundant Sul [25] and DfrA homologues to search protein records associated with NCBI GenBank/RefSeq sequences of mobile genetic elements. The nucleotide sequences of the genes encoding these proteins was then retrieved. The mol% G+C content of the corresponding sul and dfrA genes, as well as the overall mol% G+C content in both the mobile genetic element and the chromosome of the species harbouring it, were computed with custom Python scripts. To analyse whether mobile dfrA genes with mol% G+C content close to their hosts’ genomes are more similar to the hosts’ folA genes than expected if dfrA–host associations were arbitrary, we performed a permutation test comparing the mean pairwise amino acid sequence alignment distance between DfrA proteins and host-encoded FolA proteins. We randomly permuted DfrA-host assignments 1000 times and computed the corresponding P value as the rank of the non-permuted mean pairwise alignment distance. The input files and scripts used for data collection and analysis are available in public repositories: (i) input files (json, txt and fasta) and blast database for Python scripts used in data collection and analysis – https://doi.org/10.6084/m9.figshare.12156891.v1, and (ii) GitHub repository containing the Python scripts used for data collection and analysis – https://doi.org/10.5281/zenodo.3760352.
Results and Discussion
A large fraction of reported dfrA genes share a common evolutionary origin
To explore the phylogenetic relationship of trimethoprim-resistance determinants with their chromosomally encoded folA counterparts, we used a non-redundant panel of protein sequences encoded by reported dfr genes (Table S2) to detect putative DHFR homologues in sequenced mobile elements. We discarded sequences associated with the dfrB gene family, and retained for analysis non-redundant sequences mapping to the clades defined by dfrA genes reported in the Proteobacteria and by dfrDEFGK genes associated with Gram-positive bacteria. For convenience, and in accordance with recent reports on dfr nomenclature [46], we hereinafter designate these two groups (dfrA and dfrDEFGK) with the umbrella term dfrA. These reference mobile DfrA homologues were then combined with FolA homologues sampled from representative genomes of all bacterial orders with complete genome assemblies, and of each bacterial family within the Proteobacteria (Table S5). The resulting multiple amino acid sequence alignment was used to perform Bayesian phylogenetic analysis of FolA/DfrA sequences.
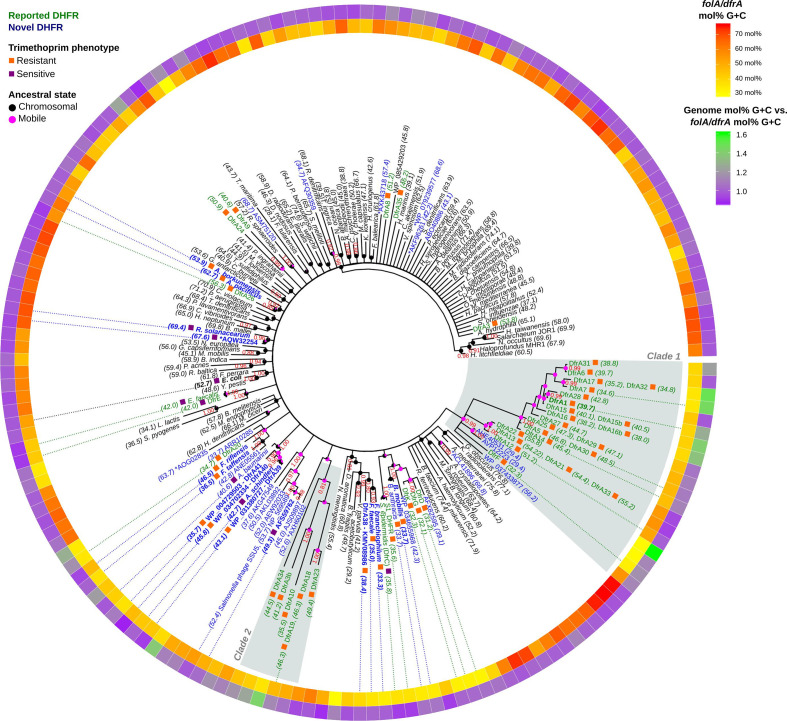
The phylogenetic tree shown in Fig. 1 showcases the genetic diversity of DfrA/FolA proteins, which encompass sequences with pairwise amino acid sequence identities ranging from 99 to 20 % (Table S6). The resulting phylogeny also reveals that the vast majority (70.7%) of known DfrA sequences map to two well-supported (>0.8 posterior probability), distinct clades that likely arose from two different ancestors. The first clade (clade 1), typified by the DfrA1 and DfrA12 proteins [47], includes 22 sequences encoded by previously reported dfrA genes with a mean amino acid identity of 51.19 % ±17.63 sd, divided into two subgroups (containing 17 and 5 known dfrA genes, respectively) and associated with Gammaproteobacteria pathogens. This clade also includes a basal set of taxa composed of the Clostridioides difficile dfrF gene and two newly identified mobile dfrA homologues also from Firmicutes isolates. The second clade (clade 2), exemplified by DfrA18 [48], comprises a group of six highly diverged (34.37%±10.15 sd mean amino acid identity) DfrA sequences from Gammaproteobacteria isolates.
Fig. 1.
Consensus tree of DHFR protein sequences. Branch support values are provided as Bayesian posterior probabilities estimated after four independent runs of 20 000 000 generations. Support values are only shown for branches with posterior probability values higher than 0.8. For chromosomal DHFR, the species name is displayed. Mobile DHFRs are denoted by their established dfrA name or by their NCBI GenBank accession number. Reported dfrA genes deemed redundant (>90 % identity) are listed next to the corresponding non-redundant taxon included in the analysis. Next to each tip label, coloured boxes designate trimethoprim-resistant (orange) and -sensitive (purple) DHFR. Numbers between brackets indicate the mol% G+C content of the sequence for the gene encoding the DHFR. Tip label colouring denotes previously reported (green) and novel (blue) DHFRs. Bold label text indicates that resistance has been experimentally assessed in this work. DHFR variants marked with an asterisk are encoded in megaplasmids (>400 kbp). The internal ring shows the mol% G+C of the gene encoding the DHFR in a yellow−red colour scale, while the external ring displays the ratio between the mol% G+C content of the genome harbouring the DHFR gene and the mol% G+C content of the gene. Dotted lines from the inner ring to tip labels denote genes discussed in the text. Reconstructed mobile/chromosomal states are displayed on ancestral nodes as pink/black pie charts.
Analysis of the dfrA gene sequences in these two clades reveals an unexpected degree of heterogeneity in mol% G+C content. In the first clade, several dfrA homologues, including the C. difficile dfrF gene, show relatively low mol% G+C content (Fig. 1, inner ring), matching the Firmicutes species they were reported on (Table S7, Fig. 1, outer ring). Similarly, dfrA genes in the dfrA12 group show a mol% G+C content (53.28 mol%±1.80 sd) that is well in line with that of the Enterobacteriaceae isolates harbouring them. Conversely, the largest group in this clade, encompassing dfrA1, dfrA7 and dfrA14, shows a mean mol% G+C content of 41.03 mol%±3.99 sd, which is substantially lower than the average mol% G+C content of the Enterobacteriaceae harbouring these mobile elements. The same holds true for the second clade (dfrA18), which also shows lower mol% G+C content (43.88 mol%±4.91 sd) than expected for the Enterobacteriaceae . To ascertain whether this pattern of mol% G+C heterogeneity extended to other previously reported and putative dfrA genes, we examined the mol% G+C content of dfrA (935 genes) and sul (408 genes) homologues identified in this analysis with respect to the genome mol% G+C content of the host species harbouring these mobile resistance genes.
The results shown in Fig. 2(a) and Table S8 reveal that dfrA genes tend to align with host genome mol% G+C content (Pearson ⍴=0.56), whereas sul genes display a two-tiered distribution of mol% G+C content that is essentially independent of host genome mol% G+C (Pearson ⍴=0.14). Available dfrA and sul sequences are dominated by variants of a known dfrA and sul genes that have been isolated predominantly in a select group of bacterial hosts (Fig. 2b). To correct for this skew, we filtered dfrA sequences based on the amino acid identity (<90 %) of their encoded proteins. This filtering resulted in a significantly smaller number of non-redundant representative dfrA (63 genes) and sul (4 genes) sequences (Table S9). The four representative sul genes correspond to one exemplar of the sul1 and sul2 families, and two exemplars of the sul3 family. Among representative dfrA genes, 14 map to the first clade (clade 1) of Fig. 1 and 4 to the second clade (clade 2). The correlation of dfrA genes with host genome mol% G+C increases significantly (Pearson ⍴=0.78) when considering only non-redundant representative dfrA sequences. The fact that the mol% G+C content of representative dfrA sequences aligns well with their host genome mol% G+C could suggest that mol% G+C content in dfrA genes has been ameliorated to match the host’s. Alternatively, it could indicate that the mobile dfrA gene originated via mobilization of a chromosomal folA gene from a bacterium in the same clade as the current host. The later scenario posits that, besides mol% G+C content similarity, representative dfrA genes should also encode proteins with significant sequence similarity to their hosts’ FolA protein. We performed a permutation test to analyse whether representative dfrA gene products show significant similarity with their hosts’ FolA protein (Table S10). Our results indicate that this is the case (P<0.001), suggesting that most mobile dfrA genes are still associated with species from the same clade they originated in.
Fig. 2.
Correlation between the mol% G+C content of mobile dfrA (red circles) and sul (green squares) genes and that of their host genome. Large open circles/squares denote representatives of clusters of redundant sequences (identity >90 %), and dfrA genes from clade 1 and clade 2 in Fig. 1 are marked with an additional corona. A 0.75 % jitter to both x- and y-axis values has been applied for visualization purposes. The red line shows the linear regression for representative dfrA gene values. The Pearson R2 coefficient is superimposed. Vertical background bars in (a) designate DfrA sequences harboured by mobile genetic elements (MGEs) identified in E. coli and K. pneumoniae isolates, which are heavily overrepresented in the dataset. Sequences from clusters with more than 100 sequences (represented by dfrA12, dfrA5 and dfrA1) are shown with specific markers, and highlighted by horizontal background bars. The number of MGEs identified as harbouring dfrA genes, before and after filtering DfrA sequence identity (>90 %), is shown in (b).
The filtering of dfrA sequences based on amino acid identity brings forward three large clusters (represented by dfrA12, dfrA5 and dfrA1, and belonging to clade 1 from Fig. 1) containing more than 100 genes with amino acid identity larger than 90 %. The dfrA genes in these clusters show a distribution of mol %G+C content that is essentially independent of the host genome mol% G+C, as in the case of sul genes (Fig. 2a), and their products show no significant sequence similarity with the hosts’ FolA (permutation test P>0.1). This indicates that the dfrA genes in these large clusters have spread across distantly related bacterial clades, primarily through their association with sul-containing integron-based transposable elements that are widely disseminated among clinically relevant bacteria [49, 50]. This analysis also brings to the fore the presence of multiple dfrA cluster representatives, with widely divergent mol% G+C, on narrow bands of host genome mol% G+C content. These bands correspond to E. coli (50.7 mol% G+C) and Klebsiella pneumoniae (57.4 mol% G+C) isolates, which are heavily oversampled in the dataset (Fig. 2b). The marked divergence in mol% G+C content ( E. coli , 9.55 mol %±6.15 sd; K. pneumoniae , 6.93 mol %±4.63 sd) and amino acid sequence identity ( E. coli , 38.57 % ±15.24 sd; K. pneumoniae , 38.39 % ±14.02 sd; Table S11, Fig. S2) among these representative dfrA genes suggests that they originated via mobilization from a diverse set of chromosomal backgrounds.
The fact that the mol% G+C of dfrA genes aligns with their host genome’s mol% G+C, and that DfrA proteins display higher amino acid sequence identity when aligned to their host genome FolA proteins than to other FolA proteins, strongly supports the notion that dfrA genes have been mobilized multiple times within different bacterial clades [13]. In a few instances, typified by the large dfrA clusters illustrated in Fig. 2, dfrA genes have been captured by highly efficient mobile elements and dispersed widely across unrelated groups of bacteria [49]. These mobile elements often harbour sul genes, which also display a host-independent mol% G+C distribution. Many of the dfrA genes identified here are associated with clinical isolates. The divergent mol% G+C content and amino acid identity of these dfrA genes indicates that pathogenic bacteria have obtained dfrA genes on multiple occasions and from different sources, highlighting the ability of mobilized resistance determinants to reach clinically relevant pathogens [17, 51].
Novel trimethoprim-resistance determinants of Acinetobacter clinical isolates identified through phylogenetic methods
The phylogenetic tree in Fig. 1 includes reported DfrA proteins and their putative homologues, as well as FolA proteins identified via tblastn as putative DfrA homologues or sampled uniformly across bacterial clades. The inferred phylogeny also reveals several groups of previously unreported mobile DHFR homologues that form well-supported clades in association with chromosomal FolA proteins. Hence, these FolA proteins could constitute the chromosomal origins of the associated mobile DHFR homologues, and provide insights into the emergence and dissemination of trimethoprim-resistance genes. To determine whether these mobile DHFR homologues did confer resistance to trimethoprim, we cloned a subset of dfrA/folA genes and performed broth microdilution assays to determine the MIC of trimethoprim. Considering that the clinical breakpoint for trimethoprim in E. coli is 4 mg l−1 [52], the results, shown in Table 1, reveal that most of the mobile DHFR homologues identified here do confer significant resistance to trimethoprim. The sole exception is the protein AQW32254. Close inspection revealed that this DHFR homologue is encoded by a megaplasmid (1.2 Mb) from a Ralstonia isolate, and that this is the only DHFR homologue present in its complete genome. Hence, we determined that this DHFR homologue was a bona fide FolA protein and not a mobile DHFR homologue, and we excluded from further analysis all other DHFR homologues identified in megaplasmids (>400 kbp).
Table 1.
MICs of trimethoprim for wild-type E. coli K-12 (CGSC5073) and derivatives carrying different versions of dfr/folA or the control empty vector
Values are representative of four independent replicates.
|
Strain |
Mobile / chromosomal |
Nucleotide accession no. |
Cloned protein ID |
Trimethoprim (mg l−1) |
|---|---|---|---|---|
|
E. coli CGSC5073 |
– |
– |
– |
0.25 |
|
E. coli pUA1108 |
– |
– |
– |
0.25 |
|
E. coli pUA1108::folA E. coli |
C |
4 |
||
|
E. coli pUA1108::dfrA1 |
M |
>512 |
||
|
E. coli pUA1108::folA Flavobacterium branchiophilum |
C |
256 |
||
|
E. coli pUA1108::folA Flavobacterium faecale |
C |
>512 |
||
|
E. coli pUA1108::dfrA38 Acinetobacter baumannii |
M |
256 |
||
|
E. coli pUA1108::folA Acinetobacter schindleri |
C |
0.25 |
||
|
E. coli pUA1108::dfrA39 Acinetobacter baumannii |
M |
512 |
||
|
E. coli pUA1108::dfrA40 Acinetobacter baumannii |
M |
128 |
||
|
E. coli pUA1108::dfrA41 Acinetobacter defluvii |
M |
>512 |
||
|
E. coli pUA1108::folA Fluviicola taffensis |
C |
>512 |
||
|
E. coli pUA1108::folA 'Candidatus Fluviicola riflensis' |
C |
>512 |
||
|
E. coli pUA1108::folA Alcanivorax pacificus |
C |
32 |
||
|
E. coli pUA1108::folA Alcanivorax borkumensis |
C |
16 |
||
|
E. coli pUA1108::folA Bacillus mobilis |
C |
>512 |
||
|
E. coli pUA1108::folA Ralstonia solanacearum |
C |
0.5 |
||
|
E. coli pUA1108::folA blood disease bacterium A2-HR MARDI |
M |
1 |
||
|
E. coli pUA1108::folA E. coli O104:H4 |
M |
2 |
Two remaining clades of novel mobile DHFR homologues from clinically relevant bacteria associated with chromosomal FolA proteins were shown to confer resistance to trimethoprim on E. coli (Table 1). To investigate whether the sequence determinants conferring resistance had originated in the associated chromosomal background, we cloned the most closely related chromosomal folA gene as well as a gene encoding an additional DHFR homologue from the same genus, and performed broth microdilution assays to determine the MIC of trimethoprim. We also performed ancestral state reconstruction of the molecule encoding the DHFR homologues (chromosomal/mobile trait), as determined during the data collection process (Tables S5 and S12).
The combined results of Table 1 and Fig. 1 reveal different patterns of trimethoprim-resistance acquisition. The protein KMV08986 is a DHFR homologue harboured by a conjugative plasmid from an Acinetobacter baumannii clinical isolate. Its most closely related chromosomally encoded DHFR homologue is the FolA protein of Flavobacterium branchiophilum , which confers resistance to trimethoprim (Table 1). To ascertain whether this chromosomally encoded DHFR homologue was encoded by a bona fide folA gene, instead of a mobile dfrA gene that integrated into the chromosome, we compared the genus-wide distribution of pairwise amino acid sequence alignment distances between FolA proteins to the pairwise distance of the identified homologue versus all other FolA proteins in the genus. The Flavobacterium branchiophilum FolA sequence is significantly different from other Flavobacterium FolA sequences (Mann–Whitney U P<0.05; Table S13), raising the possibility that this chromosomal gene could be in fact a recombined mobile dfrA gene. However, phylogenetic analysis with a broader representation of Flavobacterium sequences (Fig. S3) confirms the well-supported branching of Flavobacterium branchiophilum FolA with other Flavobacterium species FolA proteins, and comparative genomics analysis reveals that the genetic neighbourhood of the chromosomal folA gene is conserved in the genus Flavobacterium (Fig. S4). Furthermore, the FolA protein of a prototypical genus member, Flavobacterium faecale , also confers resistance to trimethoprim on E. coli (Table 1). These results indicate that the FolA protein was likely resistant to trimethoprim in the ancestor of extant Flavobacterium species, which diverged more than 50 million years ago [53]. The branching of the Acinetobacter baumannii protein KMV08986 in the reconstructed phylogeny and the associated ancestral state reconstruction indicates that this mobile DHFR homologue likely originated via mobilization of a chromosomal folA gene within the phylum Bacteroidetes . The encoded FolA protein was likely resistant to trimethoprim, but the exact donor species remains to be elucidated.
In contrast to Flavobacterium proteins, the Acinetobacter schindleri FolA protein does not confer resistance to trimethoprim, in agreement with previous reports of Acinetobacter schindleri susceptibility to trimethoprim [54], and with the well-established susceptibility of Acinetobacter baumannii FolA to trimethoprim [55, 56]. The Acinetobacter schindleri FolA protein is closely related to three mobile DHFR homologues conferring resistance to trimethoprim and harboured by Acinetobacter baumannii (protein ID: WP_031380727, WP_034702334) and Acinetobacter defluvii (protein ID: WP_004729503) clinical and environmental isolates. These mobile DHFR homologues branch within a well-supported clade of chromosomal Acinetobacter FolA proteins, as supported by ancestral state reconstruction (Fig. 1, Table S12). The trimethoprim susceptibility of Acinetobacter chromosomal folA genes and the phylogenetic placement of these DHFR homologues, hence, indicates that the observed resistance to trimethoprim was acquired immediately prior to or after mobilization from an Acinetobacter chromosomal background. This is supported by the observation that these mobile DHFR homologues confer different levels of resistance to trimethoprim (Table 1), and that the largest MIC correlates with the location of the DHFR homologue on a plasmid harbouring multiple antibiotic-resistance determinants (Fig. S5). The gene encoding this DHFR homologue is preceded by an insertion sequence transposase (Fig. S5), in an arrangement that has been reported to drive up expression of the DHFR homologue through promoter enhancement [57]. However, the MIC determined here corresponds to that of the isolated DHFR ORF, indicating that it confers heightened resistance irrespective of the promoter driving its expression. This suggests that these DHFR homologues have acquired mutations conferring heightened resistance to trimethoprim in parallel to their broader dissemination on multi-resistant mobile elements. Based on their validated trimethoprim-resistance phenotype and their level of amino acid sequence identity versus previously reported DfrA proteins (<95 %; Table S14) [13], we propose to designate these Acinetobacter DHFR homologues as DfrA38 (protein ID: KMV08986), DfrA39 (protein ID: WP_031380727), DfrA40 (protein ID: WP_034702334) and DfrA41 (protein ID: WP_004729503).
Here, we report the identification of trimethoprim-susceptible chromosomal folA genes that are closely related to mobile dfrA genes, as well as the discovery of chromosomally encoded folA genes conferring resistance to trimethoprim. This indicates that, in contrast to sulfonamides [25], trimethoprim-resistance mutations with small or negligible fitness cost must occur frequently enough in natural environments. These folA variants can then be selected for and mobilized upon exposure to trimethoprim. It is well-documented that resistance to trimethoprim, mediated by mutations in the chromosomal folA gene, develops very rapidly and in a fairly structured way [58–60], whereas resistance to sulfonamides takes much longer to evolve in a laboratory setting. Moreover, sulfonamide-resistant mutants typically show significantly reduced affinity to PABA. This results in a net fitness cost in the absence of sulfonamide that is only palliated by the emergence of subsequent compensatory mutations [61, 62]. Beyond structural constraints on the respective binding pockets, a crucial difference between both chemotherapeutic agents lies in their respective targets. While trimethoprim directly inhibits DHFR, sulfonamides compete with PABA for access to DHPS, yielding a non-productive sulfonamide-bound di-hydropterin. For sulfonamides, therefore, it is the PABA-to-sulfonamide ratio that limits the production of di-hydropteroate from a limited pool of pteridine di-phosphate, and this cannot be altered via overexpression of DHPS [63]. Conversely, DHFR overexpression can provide partial resistance to trimethoprim, and mutations enhancing DHFR expression have been reported to be the first to appear in directed evolution experiments [60]. The ability to obtain partial resistance through overexpression may provide a stepping stone for the gradual accumulation and refinement of mutations conferring substantial resistance with little fitness cost and, hence, facilitate the development of trimethoprim resistance [59, 60].
Trimethoprim resistance in chromosomally encoded folA genes
Besides uncovering novel dfrA genes, the phylogenetic analysis in Fig. 1 also identifies several chromosomal folA genes associated with previously reported dfrA genes. Two of these chromosomal folA genes have already been reported in the literature as putative origins of dfrA genes, and their identification here provides some degree of validation for the phylogenetic approach implemented in this work. The putative chromosomal origin for Staphylococcus aureus Tn4003 S1-DHFR has been identified as the chromosomally encoded dfrC gene ( Staphylococcus epidermidis ) and is reported to be susceptible to trimethoprim [64]. The Enterococcus faecalis dfrE gene, identical to the chromosomally encoded folA gene of Enterococcus faecalis , was reported to confer moderate resistance to trimethoprim in E. coli , but only when cloned in a multicopy plasmid, which could easily result in overexpression-mediated resistance [63, 65].
To ascertain whether the chromosomal folA genes found here to be associated with other known dfrA genes (dfrA20, dfrA26 and the dfrDGK cluster) confer resistance to trimethoprim, we performed broth microdilution assays to determine the MIC of trimethoprim on these chromosomally encoded FolA proteins and on another FolA protein from the same genus. In all cases, both related FolA proteins confer resistance to trimethoprim (Table 1). The most closely associated chromosomal folA genes are not significantly different from other folA genes in their respective genera (Mann–Whitney U P>0.05; Table S13), as reflected also by substantial conservation of the folA genomic neighbourhood (Fig. S4). Together, these data indicate that resistance to trimethoprim was present in the ancestor of these genera. The dfrA26 gene was identified in a K. pneumoniae clinical isolate and its most closely associated chromosomal folA gene is a member of the genus Alcanivorax . The branching pattern of dfrA26 within this clade and ancestral state reconstruction results (Fig. 1, Table S12) suggest that it arose via mobilization of a chromosomal folA gene from the genus Alcalinivorax. The dfrDGK genes have been reported in Enterococcus faecalis , Enterococcus faecium and Staphylococcus aureus , and ancestral state reconstruction results indicate that these mobile dfrA genes originated through mobilization of a member of closely related genus Bacillus , members of which have been reported to be naturally resistant to trimethoprim [66]. In both cases, therefore, the phylogenetic evidence and the similarity in mol% G+C content among chromosomal and mobile genes (Fig. 1, Table S15) point towards a mobilization event that has to date remained circumscribed to related genera. Conversely, the dfrA20 gene was identified in a Pasteurella multocida isolate, yet the chromosomal folA gene most closely associated to it is encoded by Fluviicola taffensis , a Bacteroidetes ; hence, suggesting a much more distant mobilization event (Fig. 1, Table S15). In all three cases, however, we find evidence that pre-existing resistant folA genes can be readily mobilized from both close (e.g. dfrDGK) or distant (e.g. dfrA20) species.
The resistance to trimethoprim reported here for the chromosomal folA genes of two different genera of Bacteroidetes , two distinct Alcanivorax species and a Bacillus strain underscores the deep ancestry of chromosomal mutations yielding resistance to trimethoprim. The folA genes of Flavobacterium and Fluviicola were shown here to confer resistance to trimethoprim. These two genera are thought to have diverged more than 500 million years ago and define major lineages within the Flavobacteriales , suggesting that resistance to trimethoprim emerged in an ancestor of this bacterial order. It is worth noting that several of the chromosomal folA genes shown here to be associated with mobile DHFR homologues ( Alcanivorax , Flavobacterium and Fluviicola ) appear to be resistant at the genus level and correspond to genera of aquatic bacteria. This parallels our recent identification of soil and subterranean water bacteria as the likely originators of clinical sulfonamide-resistance genes [25], and suggests that the intensive use of trimethoprim/sulfamethoxazole in agriculture, aquaculture and animal husbandry in the last 50 years may have acted as a trigger for the selection and mobilization of pre-existing folA and folP genes conferring resistance to trimethoprim and sulfonamides. Conversely, trimethoprim-susceptible chromosomal folA genes found here to be associated with dfrA genes belong to clinically relevant genera ( Staphylococcus and Acinetobacter ) that may have been under more direct trimethoprim pressure. This suggests that among relatively isolated bacterial populations, frequent exposure to high levels of trimethoprim may trigger the mobilization of spontaneous folA mutants, whereas longer term exposure to sub-lethal doses of trimethoprim in ecological rich habitats might instead rely predominantly on the mobilization of naturally resistant folA genes (Fig. 3).
Fig. 3.
Schematic representation of the two proposed evolutionary processes (based on the results presented in Figs 1 and 2, and Table 1) leading to the dissemination of trimethoprim-resistance determinants. Left panel: upon the introduction of trimethoprim, mobilization events involving pre-existing resistant chromosomal folA genes can be favourably selected. Right panel: following the introduction of trimethoprim, mobilization events involving folA genes with novel mutations that confer resistance to this chemotherapeutic agent may be selected for and disseminated among closely related bacteria.
Phage-encoded folA genes do not confer resistance to trimethoprim
Our phylogenetic analysis also identifies a well-defined clade of Enterobacteriaceae cryptic plasmids derived from Salmonella phage SSU5 and encoding DHFR homologues [67–70]. Genes encoding DHFR homologues occur frequently in many bacteriophage families, often in tandem with thymidylate synthase genes [71], but their functional role has not been fully elucidated. We performed broth microdilution assays to determine the MIC of trimethoprim of E. coli O104:H4 DHFR (protein ID: AFS59762). This phage-encoded DHFR does not confer resistance to trimethoprim (Table 1). The high amino acid sequence identity and neighbourhood conservation among the DHFR enzymes encoded by these Enterobacteriaceae cryptic plasmids and phages (Table S16, Fig. S4) would presumably suggest that these DHFR enzymes are susceptible to trimethoprim.
Bacteriophages can transfer substantial amounts of genetic material via generalized transduction, and their potential as reservoirs of antibiotic-resistance determinants has gained increased attention with the advent of metagenomics [72, 73]. However, recent studies have shown that many potential resistant determinants encoded by phages do not confer resistance against their putative targets. Furthermore, only a small proportion of complete phage genomes contain putative antibiotic-resistance genes [74]. Enzymes participating in the folate biosynthesis pathway, however, are relatively frequent in phage genomes. These include homologues of the folP gene encoding DHPS, of the thyX gene encoding flavin-dependent thymidylate synthase [75–77] and, predominantly, homologues of the folA gene encoding DHFR, often found in tandem with the thyA gene encoding type 1 thymidylate synthase [71].
Early work on Enterobacteria phage T4 showed that the phage-encoded thyA and folA gene products are functional and also participate in the phage baseplate structure [78], and thyX has been shown to be functional in a number of phages [75–77]. It has been proposed that these genes help bacteriophages overcome shortages in the deoxynucleotide pool during replication, but their potential in conferring resistance to sulfonamides or trimethoprim remains largely unexplored. The detection here of DHFR homologues in Enterobacteriaceae cryptic plasmids and phages, and the subsequent assessment of their trimethoprim susceptibility, reinforces the notion that these genes have been functionally co-opted by phages principally for deoxynucleotide synthesis. Nonetheless, these genes may still confer partial trimethoprim resistance as a by-product of folA overexpression, as recently reported for Stenotrophomonas maltophilia phage DLP4 [79].
Conclusions
Recent work has shown that resistance to sulfonamide, a synthetic chemotherapeutic agent, can be present in the bacterial pangenome well before the discovery of the agent. Here, we have used a combination of in silico and in vitro techniques to identify novel trimethoprim-resistance genes, and to identify chromosomal folA genes that are strongly associated with novel and previously reported dfrA genes. We find that most of the chromosomal folA genes associated with mobile dfrA genes confer resistance to trimethoprim, but we detect cases of novel mutations being rapidly mobilized. Hence, our work shows that the observations from sulfonamide resistance extend to trimethoprim, with generalized chromosomal resistance determinants predating the origin of several genera and several clusters of resistance genes disseminated broadly among clinical isolates. Moreover, this work also reveals that, unlike sulfonamides, resistance to trimethoprim is relatively easy to generate and frequently associated with species from the same clade it originated in. The identification of ancient resistance determinants for two synthetic chemotherapeutic agents strongly suggests that resistance to any novel drugs is likely to be already present in the bacterial pangenome. Systematic screening of existing natural variants could provide, therefore, the means to pre-emptively identify derivatives presenting widely distributed natural resistance determinants and, conversely, to engineer derivatives that circumvent most, if not all, natural resistant variants.
Supplementary Data
Funding information
This work was supported by grant BIO2016-77011-R from the Spanish Ministerio de Economia y Competitividad to J.B. M.S.-O. was the recipient of a predoctoral fellowship from the Ministerio de Educación, Cultura y Deporte de España.
Acknowledgements
The authors wish to thank Joan Ruiz and Susana Escribano for their technical support during some of the experimental procedures, as well as Ángela Martínez-Mateos for her continued support. The authors also express their gratitude to Dr Marc Valls for kindly providing the R. solanacearum GMI1000 strain.
Author contributions
Conceptualization, J. B., I. E.; data curation, M.S.-O., I. E.; formal analysis, M.S.-O., I. E.; funding acquisition, M. L., J. B.; investigation, M.S.-O., P. C., I. E.; methodology, M. L., J. B., I. E.; project administration, P. C., M. L., J. B., I. E.; resources, P. C., M. L., J. B.; software, M.S.-O., I. E.; supervision, P. C., J. B., I. E.; visualization, M.S.-O., I. E.; manuscript preparation – original draft, M.S.-O., I. E.; manuscript preparation – review and editing, M.S.-O., P. C., M. L., J. B., I. E.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: MIC, minimum inhibitory concentration; NCBI, National Center for Biotechnology Information; PABA, para-aminobenzoic acid.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Sixteen supplementary tables and five supplementary figures are available with the online version of this article.
References
- 1.Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlet J, Rambaud C, Pulcini C. Save antibiotics: a call for action of the World Alliance Against Antibiotic Resistance (WAAAR) BMC Infect Dis. 2014;14:436. doi: 10.1186/1471-2334-14-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossolini GM, Arena F, Pecile P, Pollini S. Update on the antibiotic resistance crisis. Curr Opin Pharmacol. 2014;18:56–60. doi: 10.1016/j.coph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baym M, Lieberman TD, Kelsic ED, Chait R, Gross R, et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc Natl Acad Sci USA. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aminov RI, Mackie RI. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol Lett. 2007;271:147–161. doi: 10.1111/j.1574-6968.2007.00757.x. [DOI] [PubMed] [Google Scholar]
- 8.Sengupta S, Chattopadhyay MK, Grossart H-P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol. 2013;4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kmeid JG, Youssef MM, Kanafani ZA, Kanj SS. Combination therapy for Gram-negative bacteria: what is the evidence? Expert Rev Anti Infect Ther. 2013;11:1355–1362. doi: 10.1586/14787210.2013.846215. [DOI] [PubMed] [Google Scholar]
- 10.Williams KJ. The introduction of ‘chemotherapy’ using arsphenamine – the first magic bullet. J R Soc Med. 2009;102:343–348. doi: 10.1258/jrsm.2009.09k036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masters PA, O'Bryan TA, Zurlo J, Miller DQ, Joshi N. Trimethoprim-sulfamethoxazole revisited. Arch Intern Med. 2003;163:402–410. doi: 10.1001/archinte.163.4.402. [DOI] [PubMed] [Google Scholar]
- 13.Sköld O. Resistance to trimethoprim and sulfonamides. Vet Res. 2001;32:261–273. doi: 10.1051/vetres:2001123. [DOI] [PubMed] [Google Scholar]
- 14.Quinlivan EP, McPartlin J, Weir DG, Scott J. Mechanism of the antimicrobial drug trimethoprim revisited. FASEB J. 2000;14:2519–2524. doi: 10.1096/fj.99-1037com. [DOI] [PubMed] [Google Scholar]
- 15.Hitchings GH. Mechanism of action of trimethoprim-sulfamethoxazole-I. J Infect Dis. 1973;128:S433–S436. doi: 10.1093/infdis/128.Supplement_3.S433. [DOI] [PubMed] [Google Scholar]
- 16.Landy M, Larkum NW, Oswald EJ, Streightoff F. Increased synthesis of p-aminobenzoic acid associated with the development of sulfonamide resistance in Staphylococcus aureus . Science. 1943;97:265–267. doi: 10.1126/science.97.2516.265. [DOI] [PubMed] [Google Scholar]
- 17.Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39:279–289. doi: 10.1128/AAC.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flensburg J, Sköld O. Massive overproduction of dihydrofolate reductase in bacteria as a response to the use of trimethoprim. Eur J Biochem. 1987;162:473–476. doi: 10.1111/j.1432-1033.1987.tb10664.x. [DOI] [PubMed] [Google Scholar]
- 19.Shin HW, Lim J, Kim S, Kim J, Kwon GC, et al. Characterization of trimethoprim-sulfamethoxazole resistance genes and their relatedness to class 1 integron and insertion sequence common region in Gram-negative bacilli. J Microbiol Biotechnol. 2015;25:137–142. doi: 10.4014/jmb.1409.09041. [DOI] [PubMed] [Google Scholar]
- 20.Rådström P, Swedberg G, Sköld O. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob Agents Chemother. 1991;35:1840–1848. doi: 10.1128/AAC.35.9.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perreten V, Boerlin P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother. 2003;47:1169–1172. doi: 10.1128/AAC.47.3.1169-1172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagg KA, Watkins LF, Moore MD, Bennett C, Joung YJ, et al. Novel trimethoprim resistance gene dfrA34 identified in Salmonella Heidelberg in the USA. J Antimicrob Chemother. 2019;74:38–41. doi: 10.1093/jac/dky373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White PA, Rawlinson WD. Current status of the aadA and dfr gene cassette families. J Antimicrob Chemother. 2001;47:495–496. doi: 10.1093/jac/47.4.495. [DOI] [PubMed] [Google Scholar]
- 24.Toulouse JL, Edens TJ, Alejaldre L, Manges AR, Pelletier JN. Integron-associated DfrB4, a previously uncharacterized member of the trimethoprim-resistant dihydrofolate reductase B family, is a clinically identified emergent source of antibiotic resistance. Antimicrob Agents Chemother. 2017;61:e02665-16. doi: 10.1128/AAC.02665-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez-Osuna M, Cortés P, Barbé J, Erill I. Origin of the mobile di-hydro-pteroate synthase gene determining sulfonamide resistance in clinical isolates. Front Microbiol. 2018;9:3332. doi: 10.3389/fmicb.2018.03332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taly JF, Magis C, Bussotti G, Chang JM, Di Tommaso P, et al. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat Protoc. 2011;6:1669–1682. doi: 10.1038/nprot.2011.393. [DOI] [PubMed] [Google Scholar]
- 28.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2017;45:D37–D42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics. 1998;14:423–429. doi: 10.1093/bioinformatics/14.5.423. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaushik S, Mutt E, Chellappan A, Sankaran S, Srinivasan N, et al. Improved detection of remote homologues using cascade PSI-BLAST: influence of neighbouring protein families on sequence coverage. PLoS One. 8:e56449. doi: 10.1371/journal.pone.0056449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, et al. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 34.Chen Q, Zobel J, Verspoor K. Benchmarks for measurement of duplicate detection methods in nucleotide databases. Database. 2017;2017:baw164. doi: 10.1093/database/baw164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 37.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 38.Erill I. Dispersal and regulation of an adaptive mutagenesis cassette in the bacteria domain. Nucleic Acids Res. 2006;34:66–77. doi: 10.1093/nar/gkj412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G, Smith DK, Zhu H, Guan Y, Lam TT‐Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 41.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 42.Mayola A, Irazoki O, Martínez IA, Petrov D, Menolascina F, et al. RecA protein plays a role in the chemotactic response and chemoreceptor clustering of Salmonella enterica . PLoS One. 2014;9:e105578. doi: 10.1371/journal.pone.0105578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard. 6th edn. Wayne, PA: Clinical and Laboratory Standards Institute; 2003. [Google Scholar]
- 44.Fling ME, Richards C. The nucleotide sequence of the trimethoprim-resistant dihydrofolate reductase gene harbored by Tn7. Nucleic Acids Res. 1983;11:5147–5158. doi: 10.1093/nar/11.15.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith DR, Calvo JM. Nucleotide sequence of the E. coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980;8:2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faltyn M, Alcock B, McArthur A. Evolution and nomenclature of the trimethoprim resistant dihydrofolate (dfr) reductases. Preprints. 2019:2019050137 [Google Scholar]
- 47.van Hoek AHAM, Mevius D, Guerra B, Mullany P, Roberts AP, et al. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villa L, Visca P, Tosini F, Pezzella C, Carattoli A. Composite integron array generated by insertion of an ORF341-type integron within a Tn21-like element. Microb Drug Resist. 2002;8:1–8. doi: 10.1089/10766290252913692. [DOI] [PubMed] [Google Scholar]
- 49.Grape M, Farra A, Kronvall G, Sundström L. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. Clin Microbiol Infect. 2005;11:185–192. doi: 10.1111/j.1469-0691.2004.01059.x. [DOI] [PubMed] [Google Scholar]
- 50.Ho PL, Wong RC, Chow KH, Que TL. Distribution of integron-associated trimethoprim-sulfamethoxazole resistance determinants among Escherichia coli from humans and food-producing animals. Lett Appl Microbiol. 2009;49:627–634. doi: 10.1111/j.1472-765X.2009.02717.x. [DOI] [PubMed] [Google Scholar]
- 51.Volz C, Ramoni J, Beisken S, Galata V, Keller A, et al. Clinical resistome screening of 1,110 Escherichia coli isolates efficiently recovers diagnostically relevant antibiotic resistance biomarkers and potential novel resistance mechanisms. Front Microbiol. 2019;10:1671. doi: 10.3389/fmicb.2019.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacGowan AP, Wise R. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J Antimicrob Chemother. 2001;48:17–28. doi: 10.1093/jac/48.suppl_1.17. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 54.Sigala J-C, Suárez BP, Lara AR, Borgne SL, Bustos P, et al. Genomic and physiological characterization of a laboratory-isolated Acinetobacter schindleri ACE strain that quickly and efficiently catabolizes acetate. Microbiology. 2017;163:1052–1064. doi: 10.1099/mic.0.000488. [DOI] [PubMed] [Google Scholar]
- 55.Falagas ME, Vardakas KZ, Roussos NS. Trimethoprim/sulfamethoxazole for Acinetobacter spp.: a review of current microbiological and clinical evidence. Int J Antimicrob Agents. 2015;46:231–241. doi: 10.1016/j.ijantimicag.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Pérez-Varela M, Corral J, Aranda J, Barbé J. Roles of efflux pumps from different superfamilies in the surface-associated motility and virulence of Acinetobacter baumannii ATCC 17978. Antimicrob Agents Chemother. 2019;63:e02190-18. doi: 10.1128/AAC.02190-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leelaporn A, Firth N, Byrne ME, Roper E, Skurray RA. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother. 1994;38:2238–2244. doi: 10.1128/AAC.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vickers AA, Potter NJ, Fishwick CWG, Chopra I, O'Neill AJ. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. J Antimicrob Chemother. 2009;63:1112–1117. doi: 10.1093/jac/dkp090. [DOI] [PubMed] [Google Scholar]
- 59.Watson M, Liu J-W, Ollis D. Directed evolution of trimethoprim resistance in Escherichia coli . FEBS J. 2007;274:2661–2671. doi: 10.1111/j.1742-4658.2007.05801.x. [DOI] [PubMed] [Google Scholar]
- 60.Toprak E, Veres A, Michel J-B, Chait R, Hartl DL, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swedberg G, Fermér C, Sköld O. Point mutations in the dihydropteroate synthase gene causing sulfonamide resistance. In: Ayling JE, Nair MG, Baugh CM, editors. Chemistry and Biology of Pteridines and Folates. Boston, MA: Springer; 1993. pp. 555–558. [DOI] [PubMed] [Google Scholar]
- 62.Griffith EC, Wallace MJ, Wu Y, Kumar G, Gajewski S, et al. The structural and functional basis for recurring sulfa drug resistance mutations in Staphylococcus aureus dihydropteroate synthase. Front Microbiol. 2018;9:1369. doi: 10.3389/fmicb.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmer AC, Kishony R. Opposing effects of target overexpression reveal drug mechanisms. Nat Commun. 2014;5:4296. doi: 10.1038/ncomms5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dale GE, Broger C, Hartman PG, Langen H, Page MG, et al. Characterization of the gene for the chromosomal dihydrofolate reductase (DHFR) of Staphylococcus epidermidis ATCC 14990: the origin of the trimethoprim-resistant S1 DHFR from Staphylococcus aureus? J Bacteriol. 1995;177:2965–2970. doi: 10.1128/JB.177.11.2965-2970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coque TM, Singh KV, Weinstock GM, Murray BE. Characterization of dihydrofolate reductase genes from trimethoprim-susceptible and trimethoprim-resistant strains of Enterococcus faecalis . Antimicrob Agents Chemother. 1999;43:141–147. doi: 10.1128/AAC.43.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrow EW, Bourne PC, Barrow WW. Functional cloning of Bacillus anthracis dihydrofolate reductase and confirmation of natural resistance to trimethoprim. Antimicrob Agents Chemother. 2004;48:4643–4649. doi: 10.1128/AAC.48.12.4643-4649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 68.Ahmed SA, Awosika J, Baldwin C, Bishop-Lilly KA, Biswas B, et al. Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS One. 2012;7:e48228. doi: 10.1371/journal.pone.0048228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim M, Kim S, Ryu S. Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar typhimurium rough strains. J Virol. 2012;86:10894. doi: 10.1128/JVI.01796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Octavia S, Sara J, Lan R. Characterization of a large novel phage-like plasmid in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2015;362:fnv044. doi: 10.1093/femsle/fnv044. [DOI] [PubMed] [Google Scholar]
- 71.Asare PT, Jeong T-Y, Ryu S, Klumpp J, Loessner MJ, et al. Putative type 1 thymidylate synthase and dihydrofolate reductase as signature genes of a novel bastille-like group of phages in the subfamily Spounavirinae . BMC Genomics. 2015;16:582. doi: 10.1186/s12864-015-1757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muniesa M, Colomer-Lluch M, Jofre J. Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human-body associated bacterial populations? Mob Genet Elements. 2013;3:e25847. doi: 10.4161/mge.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balcazar JL. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 10:e1004219. doi: 10.1371/journal.ppat.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11:237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhattacharya B, Giri N, Mitra M, Gupta SKD. Cloning, characterization and expression analysis of nucleotide metabolism-related genes of mycobacteriophage L5. FEMS Microbiol Lett. 2008;280:64–72. doi: 10.1111/j.1574-6968.2007.01047.x. [DOI] [PubMed] [Google Scholar]
- 76.Wittmann J, Gartemann K-H, Eichenlaub R, Dreiseikelmann B. Genomic and molecular analysis of phage CMP1 from Clavibacter michiganensis subspecies michiganensis . Bacteriophage. 2011;1:6–14. doi: 10.4161/bact.1.1.13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang S, Zhang S, Jiao N, Chen F. Comparative genomic and phylogenomic analyses reveal a conserved core genome shared by estuarine and oceanic Cyanopodoviruses. PLoS One. 2015;10:e0142962. doi: 10.1371/journal.pone.0142962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kozloff LM, Lute M, Crosby LK. Bacteriophage T4 virion baseplate thymidylate synthetase and dihydrofolate reductase. J Virol. 1977;23:637–644. doi: 10.1128/JVI.23.3.637-644.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peters DL, McCutcheon JG, Stothard P, Dennis JJ. Novel Stenotrophomonas maltophilia temperate phage DLP4 is capable of lysogenic conversion. BMC Genomics. 2019;20:300. doi: 10.1186/s12864-019-5674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.