Summary
Selective and targeted removal of individual species or strains of bacteria from complex communities can be desirable over traditional, broadly acting antibacterials in several contexts. However, generalizable strategies that accomplish this with high specificity have been slow to emerge. Here we develop programmed inhibitor cells (PICs), that direct the potent antibacterial activity of the type VI secretion system (T6SS) against specified target cells. The PICs express surface-displayed nanobodies that mediate antigen-specific cell-cell adhesion to effectively overcome the barrier to T6SS activity in fluid conditions. We demonstrate the capacity of PICs to efficiently deplete low abundance target bacteria without significant collateral damage to complex microbial communities. The only known requirements for PIC targeting are a Gram-negative cell envelope and a unique cell surface antigen; therefore, this approach should be generalizable to a wide array of bacteria and find application in medical, research and environmental settings.
Graphical Abstract
eTOC Blurb
Few tools exist for the selective depletion of specific bacteria from complex mixtures, such as the gut microbiome. Ting et al. demonstrate that nanobody-based cell-cell adhesion enables efficient depletion of bacteria of interest from a complex community via the targeted delivery of toxic proteins by a programmable inhibitor cell.
Introduction
The application of traditional antibiotics can have many undesirable consequences. Most notably, their activity can promote the emergence of resistance and disrupt health-beneficial microbial communities (Bush et al., 2011; Looft and Allen, 2012; Sommer and Dantas, 2011). Since these drawbacks are a direct result of off-target effects, the scientific community has sought to develop alternative antimicrobial strategies, with a focus on enhanced and tunable selectivity. Recent examples of such efforts include bacteriophage harboring programmable Cas9 and bacteria that deliver Cas9 or regulated toxins by conjugation (Hamilton et al., 2019; Lopez-Igual et al., 2019; Ram et al., 2018). While these and other strategies hold promise, there are many hurdles to overcome prior to their widespread implementation. For instance, phage-based approaches suffer from the rapid evolution of resistance in target populations, the particles can be rendered inactive in transit to the target site (e.g. in the gastrointestinal (GI) tract), and the often narrow host range of phage can lead to challenges in identifying clones active against a particular bacterium (Chan et al., 2013; Loc-Carrillo and Abedon, 2011). Selective antibacterial methodologies relying on conjugation circumvent some of these limitations. In particular, a donor strain derived from species evolved to transit the GI tract can be utilized (Lopez-Igual et al., 2019). However, a disadvantage of these approaches is the relatively low efficiency of plasmid transfer (Ronda et al., 2019).
We sought to develop a generalizable platform for selectively depleting target bacteria within mixed communities. Recent findings by our laboratory and others have revealed potent mechanisms of contact-dependent antagonism between bacteria (Aoki et al., 2005; Cao et al., 2016; Hood et al., 2010; Souza et al., 2015; Whitney et al., 2017). Among these, the type VI secretion system (T6SS) is a widespread pathway that catalyzes the delivery of antibacterial toxins between neighboring Gram-negative cells (Cherrak et al., 2019). A hallmark of this system is its capacity to target cells indiscriminately, a behavior attributable to its promiscuous delivery mechanism and toxins that disrupt broadly conserved cellular processes (Coulthurst, 2019). We reasoned that if the antibacterial activity of the T6SS could be specifically directed toward target cell populations, these properties of the system could facilitate its development into a flexible, alternative antibacterial platform.
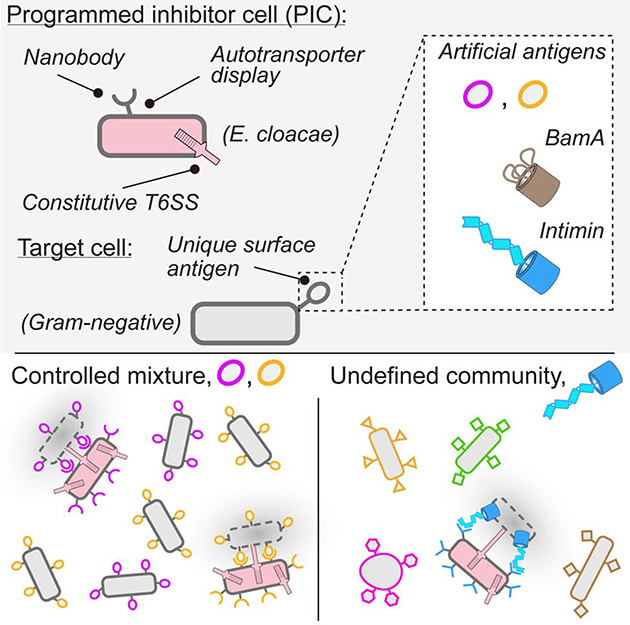
Beyond the requirement for a Gram-negative target cell, the only known barriers to T6S-based intoxication are close (< 200 nm) and long-term (> minute) cell-cell association (Leroux et al., 2012). The stringency of these requirements is illustrated by the observation that cells susceptible to the T6SS of a strain under conditions of dense co-cultivation on solid media, where intimate cell-cell contacts are enforced, are fully protected from the system in liquid media, where cell-cell contacts are transient (Hood et al., 2010). Therefore, we posited that selective T6S-based depletion of bacteria could be achieved by promoting the specific adhesion of a T6S+ inhibitor cell to the targeted population. The antigens expressed on the surface of a bacterial cell are variable and can resolve the identity of these organisms at multiple taxonomic levels. In this study, we demonstrate that this principle can be exploited to achieve selective and programmable T6S-based killing by generating strains expressing surface-displayed antibodies directed at unique cell surface epitopes of target cell populations.
Results
Nanobody-antigen pairs enable programmable T6S-mediated killing in liquid medium
To test the hypothesis that specific adhesion of a T6S+ cell to a defined target population can promote selective killing under fluid conditions, we took advantage of the observation that bacteria are able to present functional camelid-derived single domain antibodies (nanobodies) on their cell surface (Fleetwood et al., 2013) (Figure 1A). For proof-of-concept studies, we utilized characterized nanobody (Nb)-antigen (Ag) pairs in conjunction with a previously developed autotransporter display system (Pinero-Lambea et al., 2015). Elegant work by Glass et al. demonstrated that cognate Nb-Ag pairs displayed by this system facilitate specific cell-cell adhesion, allowing bacterial cell patterning (Glass and Riedel-Kruse, 2018).
Figure 1: Cell-cell adhesion mediated by surface-expressed nanobody-antigen pairs promotes targeted killing via the T6SS in liquid medium.
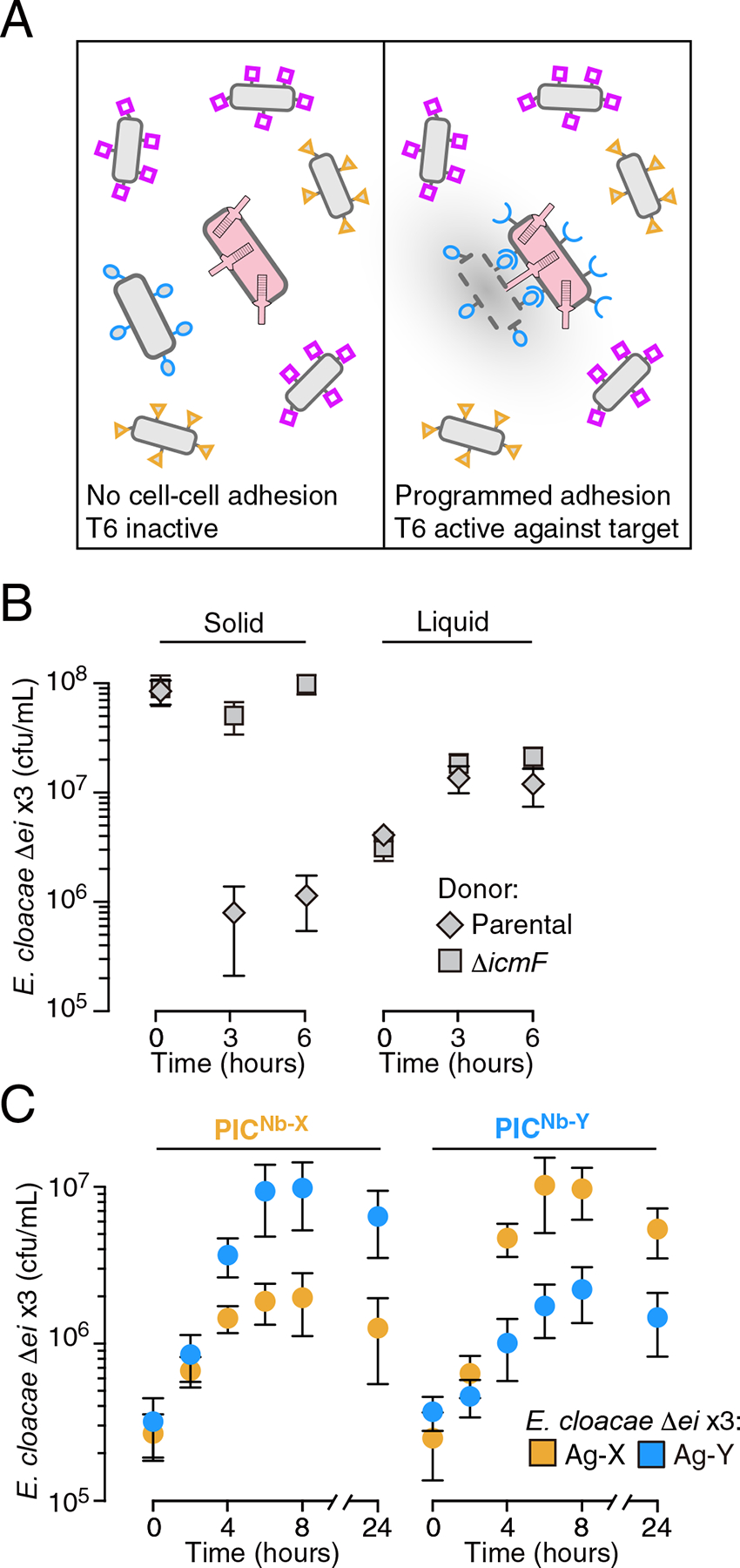
(A) Schematic of the strategy to achieve programable and selective cell killing. Bacteria with unique surface antigens (shapes) and a programmed T6S+ inhibitor cell (PIC, pink) are represented. (B) Growth of E. cloacae ΔrhsA rhsIA rhsB rhsIB tae4 tai4 (Δei x3), under contact-promoting (solid) or well-mixed (liquid) growth conditions in co-culture with the indicated donor E. cloacae strain (n=3 technical replicates ± S.D.; * p<0.05, t-test). E. cloacae ΔicmF is deficient in T6SS activity due to lack of a required structural component. (C) Impact of E. cloacae PICs expressing the indicated cell surface-displayed nanobody on growth of a T6S-susceptible strain (E. cloacae Δei x3, containing deletions of the T6SS effector genes rhsA, rhsB and tae4 and their corresponding immunity genes) expressing cognate or orthogonal antigens during co-culture in liquid medium (n=2 biological replicates with 3 technical replicates each, ± S.D.). See also Figure S1.
We began by examining the feasibility of this approach to promote intraspecific killing by the γ-proteobacterium Enterobacter cloacae. This bacterium is genetically tractable, its T6SS is active under standard laboratory conditions, and three toxic substrates of its T6SS, RhsA, RhsB, and Tae4 have been described (Russell et al., 2013; Whitney et al., 2014). Although it is a member of the normal human gut microbiota, E. cloacae is also considered an opportunistic pathogen and an emerging antibiotic resistant threat (Band et al., 2016; Mezzatesta et al., 2012).
In wild-type bacteria, self-intoxication by the T6SS is prohibited by immunity proteins encoded by genes adjacent to those encoding the effectors they inactivate (Russell et al., 2011). Therefore, we generated a susceptible target strain for these studies by deleting rhsA, rhsB, tae4 and their adjacent cognate immunity genes, rhsIA, rhsIB, and tai4 (Δei x3). As expected, this strain was susceptible to T6S-based intoxication by the parent strain when cell-cell contacts are enforced by growth on solid media, but not in liquid media, where contacts are transient (Figure 1A and Figure S1A).
Next, we introduced nanobody and antigen expression systems into the parent and T6S-susceptible strains, respectively. To facilitate mismatched control experiments, we utilized two characterized Nb-Ag pairs: Nb-X–Ag-X and Nb-Y–Ag-Y (Glass and Riedel-Kruse, 2018). We found that in liquid cultures, the growth of antigen-expressing T6S-susceptible strains was specifically inhibited by parental strains expressing cognate, but not non-cognate nanobodies (Figure 1C and Figure S1B). This inhibition was not observed when the T6SS of the nanobody-expressing strain was inactivated (Figure S1C). Immunoblotting analysis and cell surface accessibility assays ruled out the possibility that T6S inactivation interfered with nanobody expression or localization (Supplementary Figure S1D,E). Together, these data suggest that cell adhesion via Nb–Ag interactions can direct the antibacterial activity of the T6SS at target cells. We henceforth refer to E. cloacae strains expressing nanobodies that promote adhesion to target cells as PICs (programmed inhibitor cells).
To determine whether our approach could be utilized more broadly, we tested the capacity of PICs to deplete E. coli. E. coli does not possess immunity factors against E. cloacae T6S effectors; therefore, the strain is likely to be inherently susceptible to RhsA, RhsB, Tae4, as well as other, yet undescribed effectors that E. cloacae may deliver. Using co-culture on solid media, we confirmed E. coli susceptibility to T6S-mediated intoxication by E. cloacae (Figure S2A). Congruent with the intraspecies competition assays described above, we found in this interspecies mixture that PICs inhibited target cell growth in liquid media only when they possessed both an active T6SS and expressed a matched nanobody (Figure 2A, and Figure S2A–C). The magnitude of the growth inhibitory effect of PICs targeting E. coli (PICNb-X, 570-fold; PICNb-Y, 220-fold) was significantly higher than that observed for a self-derived strain sensitized only to RhsA, RhsB, and Tae4 (PICNb-X, 6.4-fold; PICNb-Y, 3.3-fold), suggesting E. cloacae indeed utilizes T6S effectors beyond those that we have identified. PICs could be inhibiting the proliferation of E. coli by inducing bacteriostasis, killing, or by a combination of these mechanisms. We found that within 30 minutes of addition, PICs reduced the colony forming units (c.f.u.) of stationary phase non-growing target cells by 16.1-fold (Figure 2D and Figure S2D). This experiment indicates PICs kill E. coli, though it does not rule-out a mixed mechanism that also involves bacteriostasis. These data demonstrate the capacity of programmable cell-cell adhesion to promote efficient interspecies killing by the T6SS.
Figure 2: PICs achieve potent and specific killing of a cognate antigen expressing species in liquid culture and under aggregation-promoting conditions.
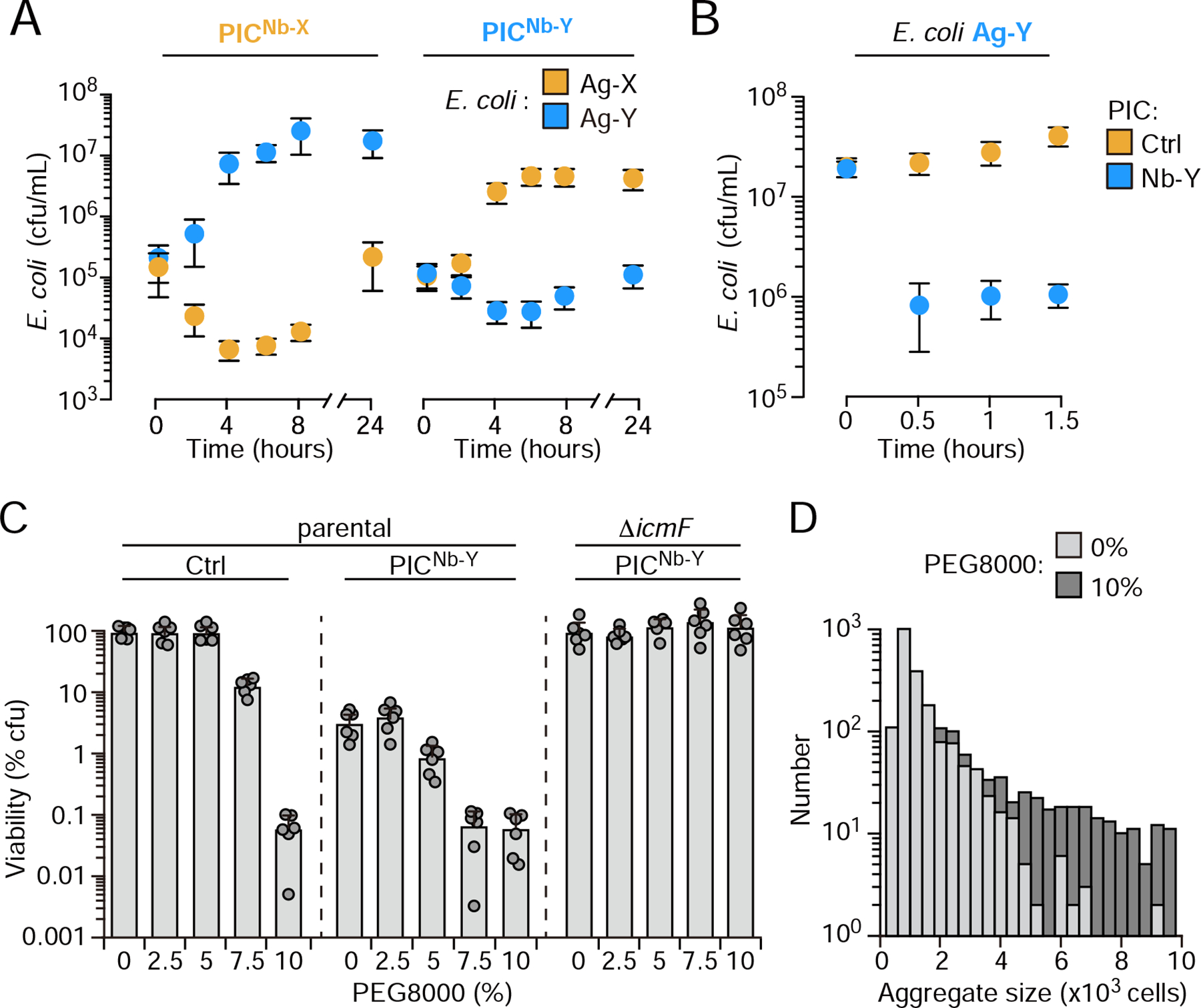
(A) Growth of E. coli MG1655 expressing the indicated antigens during co-culture in liquid medium with PICs expressing cognate or orthogonal nanobodies. (n=2 biological replicates with 3 technical replicates each, ± S.D.). (B) Survival of Ag-Y expressing E. coli grown to stationary phase in pure culture, then mixed with the indicated PIC. The control PIC expressed the empty surface-display construct used for nanobody presentation (n=2 biological replicates with 3 technical replicates each, ± S.D). (C) Viable CFUs (percentage of control PIC treated cultures incubated without PEG800) of Ag-Y expressing E. coli after 6 hrs growth with the PICs indicated in liquid medium amended with the indicated concentration of PEG 8000 (n=3 technical replicates, representative of 2 biological replicates, ± S.D.). (D) Quantification of aggregate sizes (number of aggregates counted containing the indicated number of cells) observed by phase contrast microscopy of cultures grown with indicated concentration of PEG 8000 as described in (D). See also Figure S2.
Polymer-mediated aggregation can enhance PIC activity
The specificity of PICs is dependent on fluidity; cell-cell contacts that occur independently of Nb–Ag interactions lead to indiscriminate killing (Figure S2a). However, in many natural environments, high molecular weight linear polymers are present which can affect the degree to which cells aggregate by both chemical and physical mechanisms (Ding et al., 2015; Preska Steinberg et al., 2019; Secor et al., 2018). To quantitatively describe the sensitivity of the system to polymer-mediated aggregation, we used a high molecular weight polymer (PEG 8K) to systematically vary the degree of cell aggregation through a process termed depletion aggregation (Schwarz-Linek et al., 2010; Secor et al., 2018). With this method, we identified a concentration of the polymer that enhances the efficiency of PICs without observably impacting their specificity (5.0%) (Figure 2C). As expected, at high concentrations of the polymer (>10% w/v), cellular aggregation is enhanced independently of Nb–Ag interaction and under these conditions PIC targeting is indiscriminate (Figure 2C–D and Figure S2E). A previous report similarly demonstrated that the high molecular weight linear polymer polyvinylpyrrolidone can also induce T6S-mediated targeting by Vibrio fischeri in liquid media; however, the authors of this study attributed the effect they observed to an increase in viscosity (Speare et al., 2020). Our findings suggest that PIC efficacy could benefit from natural polymer-mediated aggregation. However, a threshold effect exists, wherein conditions that strongly promote aggregation could generate off-target effects.
PICs function efficaciously in multi-strain and multi-species mixtures
A critical feature of our approach is its potential to distinguish target from non-target cells. As a first step toward addressing the specificity of the system, we exposed PICs to fluorescently-labeled E. coli cells expressing matched antigens diluted to varying degrees within differentially fluorescently-labeled cells expressing control antigens. In this scenario, we found no evidence of off-target activity, including when control cells outnumbered target cells by 1000-fold (Figure 3A,B and Figure S3A). Moreover, the efficiency of PICs was unaffected by the degree of target cell dilution. The detection limit of our assay prohibited us from testing beyond this dilution of target cells; however, these data clearly demonstrate that PICs can exhibit high specificity for target cells found within mixed populations.
Figure 3: PICs selectively deplete target cells from defined bacterial mixtures.
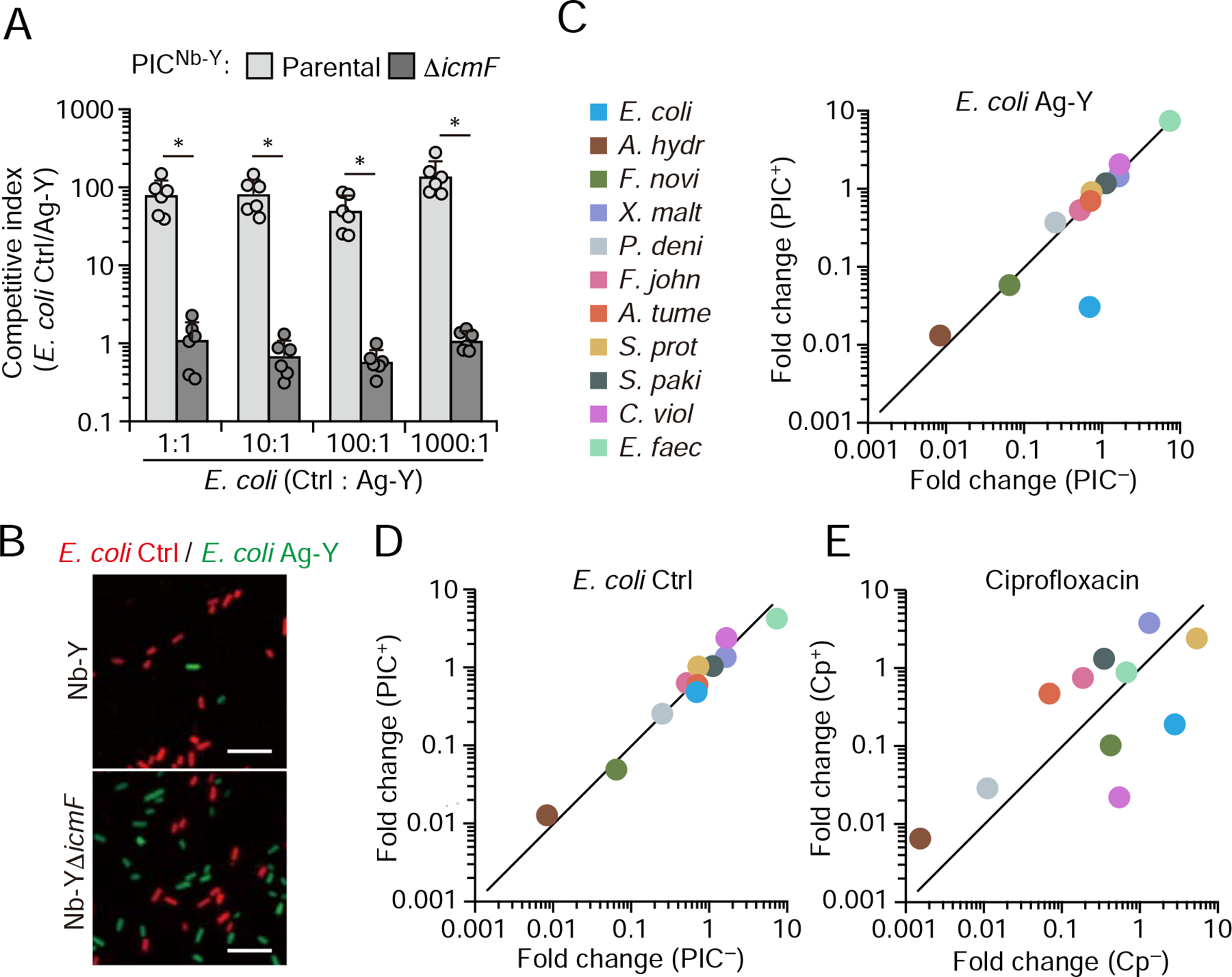
(A), Relative fitness of a control E. coli strain (control strain as described in Figure 1) compared to a strain displaying Ag-Y when grown in liquid co-culture with PICNb-Y with (parental) or without (ΔicmF) an active T6SS (n=3 technical replicates, representative of 2 biological replicates, ± S.D.; * p<0.05, t-test). The initial ratio (by OD600 nm) of the E. coli control to Ag-Y expressing strain is indicated below. (B) Representative fluorescence micrographs indicating relative abundance of E. coli control (red) and Ag-Y expressing (green) cells after growth with parental or T6S-inactive strain (ΔicmF) PICNb-Y. The initial ratio of E. coli control to Ag-Y was 1:1. PIC cells are not fluorescently labeled. Scale bar, 5 μm. Full micrographs in Figure S3. (C, D), Comparison of the change in relative abundance of community members after 8 hr growth with or without PIC addition. The E. coli strain included is indicated at top. Initial abundance (by OD600 nm): PICs, 20%; target E. coli, 6%. (E), Change in abundance of community members with and without 8 hr treatment with ciprofloxacin at 40 ng/mL (2.5x MIC of E. coli). Initial and final read counts for each species in C-E presented in Table S1. See also Figure S3.
Next, we challenged our PIC system by substantially increasing the diversity of bacteria present with the target cell. Specifically, we introduced PICs and the target cells, initially at 20% and 6% abundance (by OD600 nm), respectively, to a synthetic community consisting of 12 species derived from four phyla, including both Gram-positive and Gram-negative representatives. Quantification of community constituents by 16S rRNA gene sequencing before and after cultivation revealed no detectable PIC off-target activity, despite the susceptibility of several members to the E. cloacae T6SS (Figure 3C, Figure S3B, and Table S1). In contrast, the level of target cells within the community was substantially reduced (96%) by PICs. Reduction of the initial abundance of target and PIC cells to 3% and 9%, respectively, did not decrease PIC efficacy (98%) (Figure S3C and Table S1). As a reference, we also exposed our synthetic community to ciprofloxacin, an antibiotic used to treat serious E. coli infections, among many other indications. In our experimental regime, ciprofloxacin displayed similar potency toward E. coli as PICs; however, the antibiotic caused substantial collateral effects within the community (Figure 3E). These findings demonstrate the feasibility of using PICs to deplete target cells from multispecies environments. Moreover, they highlight the improved specificity achievable by PICs in this context relative to conventional antibiotics.
PICs selectively target via natural surface antigens
The antigens we employed to this point are not naturally found on the cell surface of bacteria. However, the utility of PICs hinges on their ability to kill target cells via the recognition of native antigens. BamA is a widely conserved, essential and cell surface-accessible outer-membrane protein that is required for the biogenesis of transmembrane β-barrel proteins (Konovalova et al., 2017). Naturally occurring molecules targeting BamA have validated the protein as an antibacterial target and, most pertinent to our study, nanobodies have been reported that recognize the E. coli protein in its membrane-integrated state (Imai et al., 2019; Kaur et al., 2019). We thus assessed whether display of these BamA-targeting nanobodies (Nb-BamA) could facilitate PIC-mediated killing of E. coli. Indeed, we found that PICNb-BamA effectively suppressed the proliferation of the E. coli laboratory strains DH5α and DH10B in a Nb-BamA- and T6SS-dependent manner (Figure 4A). Interestingly, the parent of these strains, MG1655, which possesses an identical bamA sequence, was not inhibited by the PICs despite its susceptibility to the T6SS of E. cloacae (Figure 2A). BamA is an integral membrane protein lacking a significant ectodomain; therefore, we hypothesized that structures protruding from the cell surface of MG1655 could reduce PIC efficacy by blocking access to their target antigen. We tested this hypothesis by evaluating PIC susceptibility of an MG1655 derivative bearing a truncated LPS core (ΔrfaD) (Coleman, 1983). This mutation rendered MG1655 vulnerable to PICNb-BamA-mediated inhibition, suggesting that the PIC platform will be most efficacious when targeting antigens that protrude from the cell surface.
Figure 4: PICs targeting native a natural surface antigen achieves selective killing in a complex community and resistance to killing is slow to emerge.
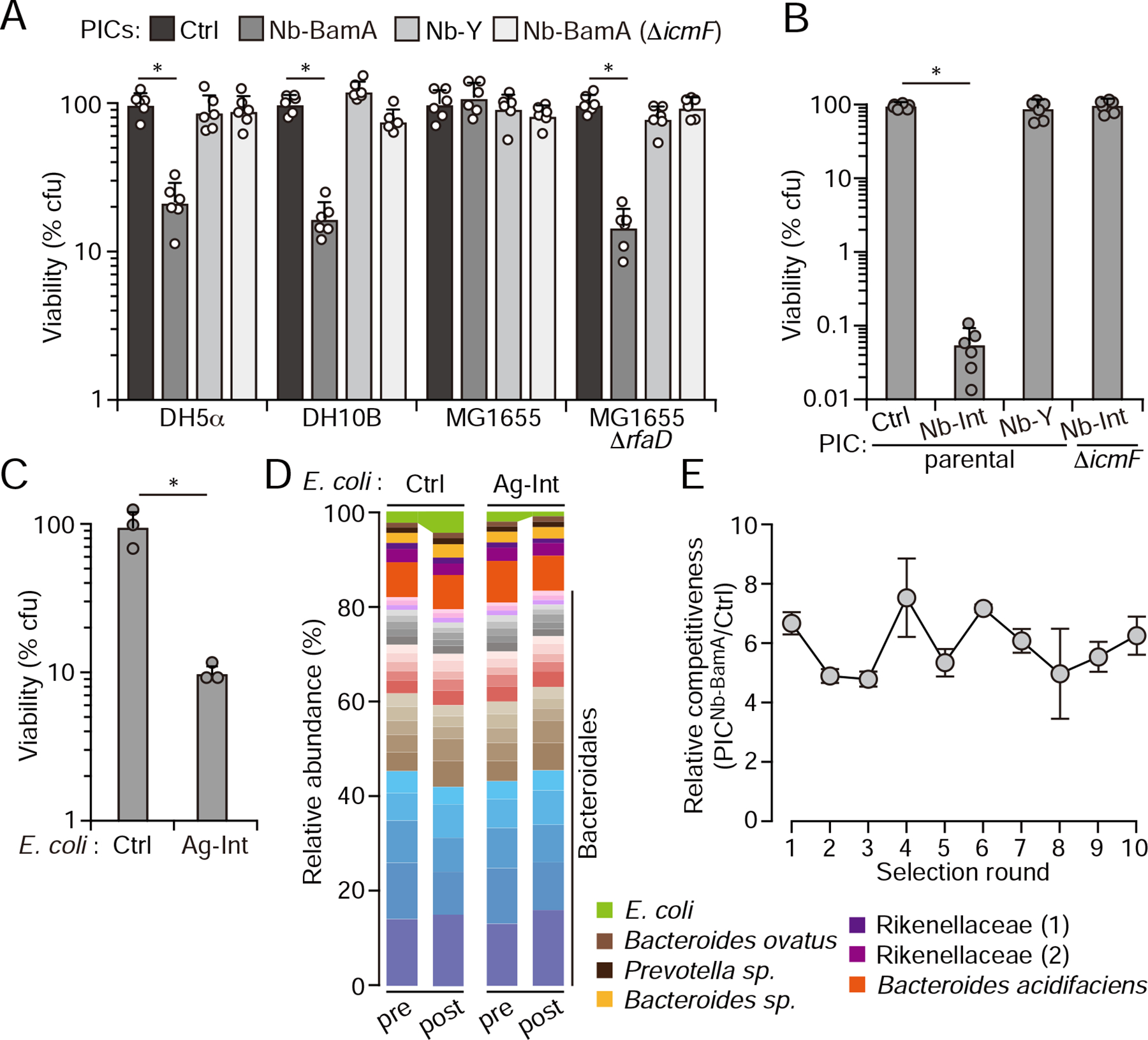
(A) Viable CFUs (percentage of control PIC treated cultures) remaining after growing the indicated E. coli strains for 6 hrs with the PICs noted at top in liquid medium amended with 5% PEG8000 to mimic a natural polymer-rich environment. Data represent 2 biological replicates each containing 3 technical replicates, ± S.D, * p<0.05, t-test. (B) Viable CFUs (percentage of control PIC-treated cultures) remaining after growing E. coli MG1655 expressing full-length intimin from E. coli O157:H7 for 6hrs with the indicated PICs in liquid medium amended with 5% PEG8000 (Data represent 2 biological replicates, each containing 3 technical replicates ± S.D.; * p<0.05, t-test). Nb-Int targets the carboxyl-terminal 280 amino acids of intimin, which are not found in the surface display construct. (C) E. coli MG1655 expressing full-length intimin recovered (% of control E. coli in parallel mixes) after incubating for 1 hour with PICNb-Int (50-fold E. coli by OD600nm) together with a complex bacterial community (200-fold E. coli by OD600nm) isolated directly from fresh mouse fecal samples. (D) Stacked bar plot showing the relative abundance of mouse fecal community members before and after incubation with PICNb-Int, as determined by sequencing the V3–V4 region of the 16S rRNA gene. OTUs were clustered at 97% identity, and the 30 most abundant OTUs (by sequence count, E. cloacae excluded) are shown. The E. coli strains added to the communities indicated at top. Normalized OUT counts are presented in Table S2. (E) Competitiveness of PICNb-BamA toward E. coli DH5a relative to the competitiveness of control PICs after the indicated number of rounds of incubating E. coli in the presence of PICNb-BamA for 6 hr. Each round of selection was initiated with a fresh population of the parent PIC. n= 3 technical replicates ± SD. See also Figure S4.
Our finding that bacterial surface structures can interfere with PIC targeting motivated us to examine the suitability of another native E. coli target protein, intimin. Owing to the critical role intimin plays in adherence to epithelia by certain pathogenic strains of E. coli, the protein has been the subject of considerable study (Celli et al., 2000; McWilliams and Torres, 2014). Unlike BamA, intimin adopts an extended structure and protrudes significantly (14 nm) from the cell surface (Batchelor et al., 2000; Kelly et al., 1999; Luo et al., 2000). Indeed, the intimin-derived display system we utilize in this study - despite containing a significantly truncated form of the protein - effectively presents synthetic antigens for PIC targeting (Figure 1) (Pinero-Lambea et al., 2015). To target intimin, we took advantage of a nanobody that recognizes the C-terminus of the protein (Nb-Int), which localizes distal to the cell surface and is not present in our display system (Ruano-Gallego et al., 2019). We found that PICNb-Int efficiently and specifically depletes E. coli producing the cell surface adhesin (Figure 4B). It is worth noting that the degree of E. coli targeting via interaction with intimin exceeds that of Ag-X, Ag-Y or BamA. A multitude of factors could contribute to the magnitude of targeting achievable by different Nb-Ag interactions, but taken together with the variability in BamA-targeting between E. coli strains, these results are consistent with distance of target antigens from the cell surface, and thus their accessibility for nanobody binding, being an important variable influencing PIC activity.
PICs selectively deplete target cells from gut-derived communities
Intimin expression is a characteristic trait of enteropathogenic E. coli strains (Celli et al., 2000; McWilliams and Torres, 2014). Thus, we sought to determine whether robust and selective intimin-mediated PIC targeting could be achieved in the context of a complex and undefined community derived from the mammalian GI tract. Toward this end, we introduced intimin-producing E. coli into freshly isolated total fecal bacteria from conventionally reared mice, and measured the capacity of PICNb-Int to deplete these strains from the mixture. To mimic conditions in vivo, we conducted these experiments under a regime that limits proliferation (e.g. high cell density and short timescale). One hour after the addition of PICNb-Int, E. coli c.f.u. levels dropped by approximately 90%, whereas in separate experiments using E. coli expressing only the truncated version of intimin not recognized by Nb-Int, E. coli levels were unaltered (Figure 4C). This degree of targeting was maintained across a wide range of target cell abundance (1–0.05%) and in fecal microbiota deriving from two independently reared mouse colonies (Figure S4A). We also examined the effects of PICs on the fecal community using 16S rRNA gene sequencing. Among OTUs with >10 counts (our cutoff for analysis), those corresponding to E. coli underwent the greatest extent of PIC-mediated depletion in both fecal microbiome samples (Figure 4D, Figure S4B, Table S2). The magnitude of this depletion was less than that determined by c.f.u. enumeration, which we attribute to the persistence of DNA following cell lysis. The overall effect of PICs on the community composition was minor, and mostly limited to low abundance OTUs. We cannot ascertain from our current data whether the small changes that were observed derived from indirect consequences of E. coli depletion or bona fide off-target killing. The community profiling also revealed that the two murine microbiomes we tested diverge greatly in the phylogenetic distribution of their constituents, suggesting that PIC-mediated targeting in this environment may be largely insensitive to the specific bacteria present.
Resistance to PICs that target BamA is slow to emerge
A concern with any antimicrobial approach is the emergence of resistance. In our PIC approach, we considered two potential broad routes by which target cells could acquire resistance: target cell mutations diminishing susceptibility to PIC toxins or mutations that influence nanobody recognition of target cell surface antigens. In general, the T6SS delivers a payload of biochemically diverse toxins that act simultaneously on a range of essential cellular structures (Alcoforado Diniz et al., 2015; LaCourse et al., 2018), and the PICs we employ are not an exception. Based on this, we have speculated that target modification represents an unlikely physiological route to high level T6SS resistance (LaCourse et al., 2018). Rather, toxin neutralization via the horizontal acquisition of genes encoding toxin-specific resistance determinants appears to be widespread in microbial communities (Ross et al., 2019). Since this resistance mechanism cannot be accurately modeled in an in vitro setting, we designed our resistance study to capture mutations that could impact cell surface antigen recognition. Specifically, we employed PICs that recognize BamA, as this protein has been shown to accumulate antibiotic-resistance conferring substitutions within the β-barrel region of the protein that is recognized by Nb-BamA (Imai et al., 2019; Kaur et al., 2019). To evaluate the potential for PIC resistance to emerge, we passaged E. coli serially in the presence of PICNb-BamA. After each passage, a population derived from all surviving colonies was used as inoculum for the subsequent passage and the sensitivity of the population to PICNb-BamA-based targeting was quantitatively assessed. Remarkably, after 10 passages, we found no change in the sensitivity of E. coli to PICNb-BamA, suggesting that resistance to PICs - at least those targeting BamA - can be slow to emerge (Figure 4E). Based on the structure of the BamA-Nb-BamA complex (Kaur et al., 2019), which shows that Nb-BamA makes extensive contacts with the inner surface of the β-barrel, we posit that the substitutions necessary to break this interaction are incompatible with BamA function.
Discussion
Our results represent a first step toward harnessing the potent antibacterial activity of the T6SS to eradicate specific bacteria from polymicrobial assemblages. We believe this system - once further optimized and tailored for individual targets - could find applications in basic research, biotechnology, and the clinic. Optimization will be explored in future studies; however, we envision this process might include i) tailoring the effector repertoires of PICs to achieve maximal killing efficiency and specificity toward the target, ii) defining the features of cell surface antigens that best promote PIC adherence, iii) increasing the expression of the T6SS within PICs, iv) identifying or engineering alternative PIC strains (e.g. inherently avirulent or engineered attenuation, adapted to particular environments of interest),and v) engineering PICs to express immunity determinants that provide protection from antibacterial toxins produced by target cells encoding their own T6SS or another antagonistic pathway.
We were unable to detect resistance of target cells that are sensitized to PIC-mediated growth suppression via nanobody-BamA interactions. It is difficult to know how well these in vitro experiments will predict resistance in a natural system. Clearly, the function and essentiality of the antigen targeted, and the nature of the interaction of the antigen with the nanobody, will impact the rate by which resistance mutations arise. Proteins and other non-proteinaceous antigens present within or immediate to the bacterial outer-membrane likely have a higher probability of being essential than those that extend from the surface, yet our data suggest that those protruding from the surface best support PIC targeting. An effective means of circumventing this tradeoff could be to develop PICs that recognize multiple antigens on target cells. In addition to stemming the emergence of resistance, the initial binding of a PIC cell to an antigen distal to the cell surface may facilitate interactions with more proximal antigens. The identity of the target cell will also exert significant influence on the rate of resistance to PICs. Recent work suggests that cell surface features such as exopolysaccharides can interfere with T6S-based intoxication (Toska et al., 2018). If target bacteria produce such structures constitutively or if readily acquired mutations can activate their expression, the efficacy of PICs could be impaired through both reduced efficacy of T6S-mediated targeting and occlusion of surface antigens.
It is likely that no single alternative antimicrobial technology will prove most useful in all, or even a majority of scenarios. Nevertheless, PICs have several potential advantages that are worth noting. First and foremost among these is their generality. Owing to the promiscuity of the T6SS, the single specialized reagent required to target a new Gram-negative bacterium by the approach is a nanobody that recognizes a unique epitope on the cell surface of that bacterium. PICs targeting Gram-positive bacteria could also be developed. In this case, target cell killing could be achieved using the Esx pathway. Though divergent in sequence and mechanism from the T6SS, the Esx pathway - present widely in Gram-positive bacteria - can also catalyze indiscriminate cell contact-dependent toxin delivery into neighboring cells (Cao et al., 2016; Whitney et al., 2017).
Other notable advantages of PICs are the limited biological information needed for targeting and the short timescale theoretically required for PIC preparation and deployment. Surface-exposed candidate antigens on target bacteria can be defined with high confidence from most bacteria using commonplace bioinformatic tools that require only genomic (or metagenomic) sequence data (Almagro Armenteros et al., 2019; Yu et al., 2010). A set of candidates could then be applied to currently available pipelines for the in vitro selection of specific nanobodies from highly diverse libraries (~1013), allowing functional screening of these to begin within a period of three weeks (McMahon et al., 2018; Moutel et al., 2016; Zimmermann et al., 2018). Altogether, these advantages bode well for the development of PICs into viable alternatives to traditional antimicrobials in myriad applications.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph Mougous (mougous@uw.edu).
Materials Availability
Plasmids and bacterial strains generated in this study are available upon request from the lead contact.
Data and Code Availability
Sequence data associated with this study has been deposited to the NCBI Sequence Read Archive under Bioproject PRJNA625997. Code generated in this study is available from Github at https://github.com/kevinjohncutler/CellAggregateAnalysis.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in the KEY RESOURCES TABLE. PICs were derived from Enterobacter cloacae ATCC-13047. Escherichia coli strains used in this study included MG1655, DH5α, and DH10B for targeting by PICs, DH5α and EC100 λ pir for plasmid maintenance, and S17–1 λ pir for conjugal transfer of plasmids into E. cloacae. Other bacterial strains used in the synthetic community competition include Serratia proteamaculans 568, Agrobacterium tumefaciens FACH, Paracoccus denitrificans ATCC-177441, Flavobacterium johnsoniae UW101, Sphingobacterium pakistanense, Chromobacterium violaceum, Mycobacterium smegmatis MC2 155, Enterococcus faecalis OG1RF, Listeria monocytogenes 10403S, Francisella novicida U112, Xanthomonas maltophilia ATCC-13637, and Aeromonas hydrophila An65A68. Bacteria were cultured routinely at 37°C in Luria Bertani (LB) medium supplemented with 0.5% (w/v) glucose, unless noted otherwise. Antibiotics and chemicals were used at the following concentrations: 50 μg ml−1 streptomycin; 50 μg ml−1 spectinomycin; 150 μg ml−1 carbenicillin; 50 μg ml−1 kanamycin; 25 μg ml−1 chloramphenicol; 40 ng ml−1 ciprofloxacin; 400 ng ml−1 anhydrotetracycline.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-myc | Developmental Studies Hybridoma Bank | Cat# 9E10 |
| Mouse monoclonal anti-RpoB | Biolegend | Cat# 663903 |
| Sheep anti-Mouse HRP conjugated | Millipore | Cat# AC111P |
| Goat anti-Mouse Alexa fluor 488-conjugated | Thermo Fisher Scientific | Cat# A-11001 |
| Bacterial and Virus Strains | ||
| Escherichia coli MG1655 galK::cat-J23101-dTomato | Gift from Dr. Erik Gullberg | N/A |
| Escherichia coli MG1655 galK::cat-J23101-mTagBFP2 | Gift from Dr. Erik Gullberg | N/A |
| Escherichia coli MG1655 ΔrfaD::CMR | This paper | N/A |
| Escherichia coli EHEC O157:H7 strain EDL933, Δstx1Δstx2 | Gift from Dr. John Leong | N/A |
| Escherichia coli DH5α | Thermo Fisher Scientific | Cat# 18258012 |
| Escherichia coli DH10B | Thermo Fisher Scientific | Cat# 18297010 |
| Escherichia coli S17–1 | Biomedal Lifescience | Cat# BS-3234 |
| Escherichia coli EC100D | Lucigen | Cat# ECP09500 |
| Enterobacter cloacae ATCC-13047 | ATCC | ATCC 13047 |
| Enterobacter cloacae ATCC-13047 ΔicmF | Whitney et al., 2014 | N/A |
| Enterobacter cloacae ATCC-13047 RifR, ΔrhsBΔrhsIB::spec | Gift from Dr. Christopher Hayes | N/A |
| Enterobacter cloacae ATCC-13047 RifR, ΔrhsBΔrhsIB::spec, ΔrhsAΔrhsIA | This study | N/A |
| Enterobacter cloacae ATCC-13047 RifR, ΔrhsBΔrhsIB::spec, ΔrhsAΔrhsIA, Δtae4Δtai4 | This study | N/A |
| Serratia proteamaculans 568 Tn7::sfGFP (KanR) | Ting et al., 2018 | N/A |
| Agrobacterium tumefaciens FACH | Gift from Dr. Eugene Nester | Cat#18258012 |
| Paracoccus denitrificans ATCC-17741 | ATCC | ATCC 17741 |
| Flavobacterium johnsoniae UW101 | ATCC | ATCC 17061 |
| Sphingobacterium pakistanense | Lab collection | N/A |
| Chromobacterium violaceum | Lab collection | N/A |
| Mycobacterium smegmatis MC2155 | ATCC | ATCC 700084 |
| Enterococcus faecalis OG1RF | ATCC | ATCC 47077 |
| Listeria monocytogenes 10403S | Gift from Dr. Joshua Woodward | N/A |
| Francisella novicida U112 Tn7::sfGFP (KanR) | Ledvina et al., 2018 | N/A |
| Xanthomonas maltophilia ATCC-13637 | ATCC | ATCC 13637 |
| Aeromonas hydrophila An65A68 | Lab collection | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Anhydrotetracycline | Cayman Chemical Company | Cat# 10009542 |
| Kanamycin | Gold Biotechnology | Cat# K-120-50 |
| Chloramphenicol | Fisher Bioreagents | Cat# BP904-100 |
| Streptomycin | Research Products International | Cat# S62000–50 |
| Ciprofloxacin | Sigma-Aldrich | Cat# 17850 |
| Spectinomycin | Gold Biotechnology | Cat# S-140-25 |
| Carbenicillin | Gold Biotechnology | Cat# C-103-25 |
| NdeI | New England Biolabs | Cat# R0111 |
| PstI | New England Biolabs | Cat# R0140 |
| Gibson Assembly Master Mix | New England Biolabs | Cat# E2611 |
| T4 DNA ligase | New England Biolabs | Cat# M0202 |
| p-chlorophenylalanine | Alfa Aesar | Cat# A13323 |
| L-arabinose | Sigma-Aldrich | Cat# A3256-500 |
| DNase I | Sigma-Aldrich | Cat# 04716728001 |
| Nycodenz | Progen | Cat# 1002424 |
| Polyethylene glycol 8000 (PEG8000) | Sigma-Aldrich | Cat# 89510 |
| Phosphate buffered saline (PBS) | VWR | Cat# E404-100 |
| Agarose | VWR | Cat# 0710-500 |
| Radian HRP substrate | Azure biosystems | Cat# AC2101 |
| Critical Commercial Assays | ||
| InstaGene Matrix | Bio-Rad | Cat# 7326030 |
| DNeasy PowerSoil Kit | Qiagen | Cat# 12888-100 |
| Deposited Data | ||
| Raw 16S rRNA gene sequencing data | This study | NCBI BioProject ID: PRJNA625997 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | The Jackson Laboratory | Stock# 000664 RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primers listed in Table S3 | Integrated DNA Technology | N/A |
| Recombinant DNA | ||
| pDSG_null_control (pDSG323) | Glass and Riedel-Kruse et al., 2018 | N/A |
| pDSG_Nb-X (pDSG372) | Glass and Riedel-Kruse et al., 2018 | N/A |
| pDSG_Nb-X_myc | This paper | N/A |
| pDSG_Nb-Y (pDSG398) | Glass and Riedel-Kruse et al., 2018 | N/A |
| pDSG_Nb-Y_myc | This paper | N/A |
| pDSG_Nb-Int | This paper | N/A |
| pDSG_Nb-BamA | This paper | N/A |
| pDSG_Ag-X (pDSG358) | Glass and Riedel-Kruse et al., 2018 | N/A |
| pDSG_Ag-Y (pDSG360) | Glass and Riedel-Kruse et al., 2018 | N/A |
| pDSG_intimin_full_length | This paper | N/A |
| pRE118-pheS | Gift from Dr. Christopher Hayes | N/A |
| pRE118-pheS-ΔrhsAΔrhsIA | This paper | N/A |
| pRE118-pheS-Δtae4Δtai4 | This paper | N/A |
| pKD3 | Datsenko and Wanner et al., 2000 | RRID:Addgene_45604 |
| pKD46 | Datsenko and Wanner et al., 2000 | N/A |
| pBAD18 | Guzman et al., 1995 | N/A |
| pBAD33 | Guzman et al., 1995 | N/A |
| Software and Algorithms | ||
| GraphPad Prism version 7.0 for Mac | GraphPad, Software, La Jolla, California, USA | www.graphpad.com; RRID:SCR_022798 |
| ImageJ | NIH | Imagej.nih.gov; RRID:SCR_003070 |
| Adobe Illustrator CC 2015 | Adobe Systems Incorporated, San Jose, California, USA | www.adobe.com/products/illustrator; RRID:SCR_010279 |
| SuperSegger | Wiggins Lab, Software, Seattle, Washington, USA | mtshasta.phys.washington.edu/website/SuperSegger.php |
| Geneious 10.0.9 | Geneious, Software, Newark, New Jersey, USA | www.geneious.com RRID:SCR_010519 |
| Cell Aggregate Analysis algorithm | This study | github.com/kevinjohncutler/CellAggregateAnalysis |
| FlowJo | FlowJo LLC, Software, Ashland, Oregon, USA | www.flowjo.com RRID: SCR_008520 |
| MATLAB | MathWorks, Software, Natick, Massachusetts, USA | www.mathworks.com RRID:SCR_001622 |
| VSEARCH | Rognes et al., 2016 | github.com/torognes/vsearch |
| MICCA 1.7.2 | Albanese et al., 2015 | micca.org |
Mice
Murine fecal samples employed in this study were obtained from two separately-reared colonies of C57BL/6J mice maintained in specific pathogen free (SPF) conditions. Mice used to establish the colonies were originally purchased from Jackson Laboratories. The rearing and maintenance of these mouse colonies was approved by the University of Washington Institutional Animal Care and Use Committee. Daily care of the colonies was provided and SPF conditions were ensured through the rodent health monitoring program overseen by the Department of Comparative Medicine at the University of Washington. Fecal samples were collected at random without regard to sex or mouse age.
METHOD DETAILS
Plasmid construction
All primers used in plasmid construction and generation of mutant strains are listed in Table S3. Tet expression plasmids used for nanobody and antigen presentation in E. cloacae and E. coli in this study were generated from pDSG323-derived expression plasmids previously described (Glass and Riedel-Kruse, 2018). The expression plasmids for synthetic antigens and cognate nanobodies described by Glass et al. were modified to introduce a 3’ myc tag to the expressed proteins by cloning of the amplified tag into the NdeI and PstI restriction sites. Nb1/Ag1 and Nb3/Ag3 were renamed Nb-X/Ag-X and Nb-Y/Ag-Y, respectively for the purposes of our study. The Nb-Int and Nb-BamA expression plasmids display the previously characterized nanobodies IB10 and Nb_B12, respectively (Ruano-Gallego et al., 2019)(Kaur et al., 2019). Each was constructed by synthesizing the nanobody-encoding sequence as a Gblock (IDT) and subsequently inserting it (by Gibson assembly) downstream of the gene encoding the truncated version of intimin in pDSG323. To generate the intimin expression plasmid, the full-length intimin gene (eae, EDL933_4947) was PCR amplified from genomic DNA of EHEC O157:H7 strain EDL933 and substituted for the truncated version of intimin used for nanobody or antigen display in pDSG323 (Glass and Riedel-Kruse, 2018). For the production of E. cloacae in-frame deletion constructs, 750 bp regions flanking the deletion were amplified, joined using splicing by overlap extension (SOE) PCR, and subsequently cloned into the E. cloacae suicide vector pRE118-pheS using the SacI and XbaI restriction sites (pRE118-pheS was a gift from Christopher Hayes of UC Santa Barbara).
Generation of mutant bacterial strains
To generate mutations in E. cloacae, deletion constructions in pRE118-pheS were transformed into E. coli S17–1 λ pir. E. coli S17–1 λ pir donor cells carrying the deletion constructs and E. cloacae recipient strains to be mutated were grown overnight on LB plates containing antibiotics as appropriate, then scraped together to create a 2:1 mixture of each donor-recipient pair that was spread on an LB agar plate and incubated at 37°C for 6 hours to facilitate plasmid transfer via conjugation. Cell mixtures were then scraped into PBS and plated on LB medium agar plates supplemented with kanamycin and streptomycin to select for E. cloacae containing the deletion construct inserted into the chromosome. E. cloacae merodiploid strains were then grown overnight in non-selective LB medium at 37°C, followed by counter selection on M9 minimal medium agar plates with 0.4% glucose and 0.1% (w/v) p-chlorophenylalanine. Kanamycin sensitive colonies were screened for allelic replacement by colony PCR and mutations were confirmed by Sanger sequencing of PCR products.
E. coli bearing a deletion of rfaD was generated by the lambda red recombinase system (Datsenko and Wanner, 2000). In brief, PCR products containing a chloramphenicol resistance cassette flanked by 50 bp of homology to the 5’ and 3’ termini of the rfaD gene were electroporated into E. coli MG1655 carrying pKD46 induced to expressed the recombinase with 0.2 % (w/v) L-arabinose for 5 hours at 30°C. E. coli was then incubated in LB for 1 hour, plated on chloramphenicol containing LB agar and incubated overnight at 37°C.
Bacterial competition assays
Competitions between two bacterial strains
Bacterial competitions on solid media were performed as previously described (LeRoux et al., 2015). Briefly, overnight cultures were spun for 1 min at 17,900 x g to pellet cells and culture supernatant was removed. Cell pellets were washed once with LB medium, spun again, and finally resuspended in LB medium. Cell suspensions were then normalized to OD600 = 2.0. Mixtures of E. cloacae and target cells were then established at 20:1 (E. cloacae vs. E. cloacae Δei x3) or 3:1 (E. cloacae vs. E. coli) v/v ratios. Starting ratios were established by performing 10-fold serial dilutions and plating on appropriate selective media. Competitions were initiated by spotting 3 × 5 ul of each mixture on nitrocellulose filters placed on 3% (w/v) agar LB plates and incubated at 37°C for 3 or 6 hours. Cells were then harvested by scraping individual spots from excised sections of the nitrocellulose filter into LB medium. Suspensions were serially diluted and plated on selective media for quantification of CFUs. For intraspecies competitions, E. cloacae Δei x3 was marked by chromosomal insertion of a spectinomycin resistance gene, while donor E. cloacae strains were unmarked; E. cloacae Δei x3 populations were quantified by enumerating CFU obtained on LB with spectinomycin, and donor populations quantified by subtracting this number from the total CFU enumerated on non-selective LB plates. For interspecies competitions, E. coli was marked by chromosomal insertion of a chloramphenicol resistance gene; S. proteamaculans, F. novicida, and S. pakistanense were marked by a kanamycin resistance gene; E. cloacae strains were unmarked as in the intraspecies competition experiments; E. cloacae was distinguished by plating on LB containing streptomycin (intrinsic resistance). Competitive indices for each experiment were determined by dividing the final donor to recipient ratio by the initial donor to recipient ratio.
For bacterial competitions in liquid media, overnight cultures were pelleted, washed and resuspended in LB as described above. For both intra- and interspecies competitions, E. cloacae donor and target cells were diluted to OD600 = 0.03 and 0.0003, respectively. Competitions were performed in LB with 0.5% glucose and 400 ng/mL of anhydrotetracycline to induce nanobody or antigen expression. PEG 8000 was added to the medium when indicated in the figure legends. Starting ratios of donor and recipient strains were established as described above. Competitions were incubated at 37°C with shaking at 200 rpm for 6 hours. Cells were collected at indicated time points (2, 4, 6, 8, or 24 hours), serially diluted, and plated on selective media for quantification of CFUs.
Competitions between three bacterial strains
E. cloacae expressing Nb-Y was cocultured with two E. coli strains, those displaying Ag-Y or a null control expressing only the intimin display construct (pDSG323) (Glass and Riedel-Kruse, 2018). Suspensions of E. cloacae and each E. coli strain were prepared as described above for interspecies liquid media competitions. Competitions were then initiated in which the starting concentration of E. cloacae and the null control strain were held constant at OD600 = 0.03 and 0.01, respectively, while the starting concentration of E. coli expressing Ag-Y was varied by 10 fold dilutions from 0.01 to 0.00001, to establish starting ratios for the two E. coli strains of 1:1, 10:1, 100:1 and 1000:1. The two E. coli populations were distinguished by chromosomal insertion at galK of genes encoding chloramphenicol resistance and dTomato (control strain) or mTagBFP (Ag-Y expressing strain) under the control of the constitutive promoter pLlacO (strains were a gift from Erik Gullberg of Uppsala University). Anhydrotetracycline and glucose were supplemented as describe above. Competitions were incubated at 37°C with shaking at 200 rpm for 6 hours. Initial and after competition samples were collected, plated on LB containing chloramphenicol, and visualized using an Azure Biosystems c600.
Bacterial survival experiments
To assess survival of bacteria targeted by nanobody-producing E. cloacae, strains were grown overnight then diluted to OD600 = 0.1 and grown for 4 hours (until stationary phase) in LB with anhydrotetracyline to induce nanobody or antigen production. Cells were then pelleted, washed and resupended as described above, then normalized to OD600 = 5.0. Mixtures of E. cloacae and E. coli were then established at 100:1 v/v ratios in LB with glucose and anhydrotetracycline. Starting ratios of cells were established as described above. Competitions were incubated at 37°C with shaking at 200 rpm, and samples were collected at 30, 60, and 90 minutes. Cells were harvested at each time point and then serially diluted and plated on selective media for quantification of CFUs as described above.
Synthetic community competition experiment
Synthetic community competition experiments contained the following 12 species in addition to E. cloacae and E. coli: Serratia proteamaculans, Agrobacterium tumefaciens, Paracoccus denitrificans, Flavobacterium johnsoniae, Sphingobacterium pakistanense, Chromobacterium violaceum, Mycobacterium smegmatis, Enterococcus faecalis, Listeria monocytogenes, Francisella novicida, Xanthomonas maltophilia, and Aeromonas hydrophila. Overnight cultures of each of the species were pooled together at equal concentration (normalized to OD600 = 0.01) and mixed with E. cloacae expressing Nb-Y (OD600 = 0.03 or 0.01) and E. coli expressing Ag-Y or the null control described above (OD600 = 0.01 or 0.003). The mixtures were grown in LB medium with glucose at 30°C (to permit growth of organisms included in the mixture that are unable to grow at 37°C) with shaking at 200 rpm for 8 hours. After the incubation period, competitions were harvested and washed with fresh LB medium. Cells were then incubated with 2mg mL−1 DNase for 30 minutes at 37°C to remove extracellular DNA and washed a final time with LB containing 10 μM EDTA to inactivate DNase. Total DNA was extracted using the InstaGene Matrix (Bio-Rad). To calculate the abundance ratio of bacterial strains in the mixed population, the V3 and V4 regions of the 16S rRNA gene were amplified using proprietary primers (Genewiz) and sequenced with an Illumina MiSeq. 40,000 to 200,000 paired end reads were generated from each sample; paired end reads were merged using VSEARCH(Rognes et al., 2016). Reads were then trimmed and filtered with MICCA 1.7.2, using a minimum length of 250 and an expected error rate of 0.5. MICCA also was used to de novo cluster the sequences, as well as for taxonomic classification with the Greengenes core set database, downloaded from the MICCA FTP site 2019–06-28, http://micca.org (Albanese et al., 2015). Species for which <10 read counts were obtained across the majority of samples post cultivation (M. smegmatis, L. monocytogenes) were excluded from downstream analyses. The fraction change in relative read abundance for each community member from before to after competition was calculated by dividing the final read proportion deriving from a given community member by the initial proportion.
Complex, undefined community competition experiment
A complex, undefined microbial community was derived from fresh mouse fecal samples by homogenization followed by Nycodenz density gradient centrifugation as described previously, with the modifications indicated below (Hevia et al., 2015). Fecal samples for the two replicates of the experiment were each collected from a distinct, separately reared colony of C57BL/6J mice maintained in specific pathogen free (SPF) conditions. Mice used to establish the colonies were originally purchased from Jackson Laboratories. Daily care of the colonies was provided and SPF conditions were ensured through the rodent health monitoring program overseen by the Department of Comparative Medicine at the University of Washington. Upon collection, mouse fecal samples were resuspended in Phosphate-buffered saline (PBS) buffer and homogenized by Tissue-Tearor homogenizer (BioSpec). Samples were then gently added to the top of 80 % (w/v) Nycodenz, followed by ultra-centrifugation at 10,000 g for 40 minutes. The top layer of PBS buffer was carefully removed, and the high density fecal bacterial community from the middle layer was collected and normalized to OD600 = 20 (Figure 4C, D, Figure S4A, E. coli at 0.25% or 0.05%, Figure S4B) or 5 (Figure S4A, E. coli at 1%) in PBS buffer. The resulting fecal bacterial community was mixed with equal volumes of E. cloacae expressing Nb-Int and normalized to OD600 = 5 (Figure 4C,D, Figure S4A, E. coli at 1% or 0.25%, Figure S4B) or 1 (Figure S4, E. coli at 0.05%), and E. coli marked with a chromosomally encoded chloramphenicol resistance gene and expressing intimin or the null control (OD600 = 0.05, Figure 4C,D, Figure S4A, E. coli at 1% or 0.25%, Figure S4B or 0.01, Figure S4A, E. coli at 0.05%) in LB amended with glucose and anhydrotetracyline. The mixtures were grown at 37°C with shaking at 200 rpm for 1 hour. Initial and post-incubation samples of the mixtures were plated on LB with chloramphenicol and incubated aerobically to selectively quantify E. coli populations. After the incubation period, cells were pelleted and washed with fresh LB medium. Total DNA was extracted using the DNeasy PowerSoil Kit (Qiagen). The V3–V4 region of the 16S rRNA gene was amplified from the samples using proprietary primers (Genewiz) and sequenced with an Illumina MiSeq as described above. OTU counts were determined using the Genewiz 16S-EZ analysis pipeline.
Assessing conventional antibiotic specificity
The specificity and potency of conventional antibiotics was compared to that of PICs using the 12-member synthetic microbial community described above. Overnight cultures of each of the species were pooled together at equal concentration (normalized to OD600 = 0.01) and E. coli (OD600 = 0.01). The mixtures were grown in LB with glucose and ciprofloxacin at 30°C with shaking at 200 rpm for 8 hours. After the incubation period, cells were harvested and washed with fresh LB medium. Extracellular DNA was removed as described above. Total DNA was extracted using the InstaGene Matrix (Bio-Rad) and V3–V4 region of the 16S rRNA genes were amplified and sequenced as described above. Sequence data was analyzed as described for the synthetic community competition experiments.
Assessing potential for the emergence of resistance to PICNb-BamA
E. cloacae expressing Nb-BamA or the null control and E. coli DH5α containing pBAD33 and pBAD18 (providing resistance to chloramphenicol and carbenicillin for separating E. coli from E. cloacae) were grown overnight in medium amended with appropriate antibiotics. The cultures were then washed, pelleted, and resuspended as described above. Three replicate competitions were then initiated in LB medium amended with glucose, anhydrotetracycline and PEG8000, in which the starting concentration of E. cloacae and E. coli DH5α at OD600 were 0.03 and 0.0003, respectively. Competitions were incubated at 37°C with shaking at 200 rpm for 6 hours. Post-competition samples were pooled and plated densely on 150 mm petri plates containing LB with chloramphenicol and carbenicillin to remove E. cloacae. All E. coli DH5α colonies obtained (~10,000/sample) were collected, diluted, and used to initiate the next round of competition along with fresh cultures of E. cloacae expressing Nb-BamA or the null control, prepared as above. This regime was repeated for ten passages. The populations of E. coli and E. cloacae were quantified before and after each round of competition by plating on selective media. The relative competitiveness of PICNb-BamA at each round was calculated by dividing the competitive index (final E. cloacae/ final E. coli divided by initial E. cloacae/ initial E. coli) for this strain by that of the null control.
Phase contrast and fluorescence microscopy
Imaging was performed on a Nikon Eclipse Ti-E wide-field epi-fluorescence microscope, equipped with a sCMOS camera (NEO, Andor Technology) and X-cite LED for fluorescence imaging. We image through 60X 1.4 NA oil-immersion objective, and maintain a constant focal plane using a Nikon Perfect Focus system. The microscope is controlled by NIS-Elements.
Samples from bacterial competition experiments were imaged by phase contrast and fluorescence microscopy as previously described (Ting et al., 2018). Briefly, mixtures of E. cloacae and E. coli were spotted on a PBS with 2% agarose pad placed on a microscope slide. Still images of the cells were acquired before and after competition, including a phase-contrast image (to visualize cell morphology) and fluorescence image (to distinguish E. coli, which expressed dTomato or mTagBFP, from unlabeled E. cloacae) for each field of view. To visualize bacterial aggregates from cultures amended with varying concentrations of PEG8000, liquid cultures of E. cloacae normalized to OD600 0.03 and E. coli normalized to OD600 0.01 were mixed, grown in LB with glucose and PEG8000 at the concentrations indicated for 6 hrs, diluted, spotted onto an agarose pad, and imaged via phase microscopy. Aggregate sizes were quantified by a masking algorithm in MATLAB, the main stages of which are local nominalization, gradient thresholding, and hole-filling.
Protein expression level analyses
To analyze the expression of nanobodies, E. cloacae strains expressing the proteins from pDSG323-derivatives were grown in LB medium supplemented with or without anhydrotetracycline at 37°C for 6 hours and harvested at an OD600 of 1.0. For each quantification assay, cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1% (v/v) Triton X100, 1 mM EDTA, and 1 mM DTT) and with 2X SDS-PAGE sample loading buffer. Samples were boiled at 95°C for 10 minutes and loaded at equal volumes to resolve using SDS-PAGE, then transferred to nitrocellulose membranes. Membranes were blocked in TBST (10 mM Tris-HCl pH 7.5, 150 mM NaCl2, and 0.1% w/v Tween-20) with 5% (w/v) non-fat milk for 30 min at room temperature, followed by incubation with primary antibodies (anti-myc or anti-RpoB) diluted 1:1000 in TBST for 1 hour at room temperature. Blots were then washed by TBST, followed by incubation with secondary antibody (Goat anti-mouse HRP conjugated) diluted 1:5000 in TBST for 30 minutes at room temperature. Finally, blots were washed by TBST again and were developed using the Radiance HRP substrate (Azure Biosystems) and visualized using the Azure Biosystems c600.
Cell surface accessibility assays (Flow cytometry and immunofluorescence assay)
Overnight grown cells were diluted to an OD600 of 0.03 in LB and incubated either with or without anhydrotetracycline for 6 hours at 37°C. Cells were then harvested at an OD600 of 1.0 and washed with filtered phosphate buffered saline (PBS; 8 mM of Na2HPO4, 1.5 mM of KH2PO4, 3 mM of KCl and 137 mM of NaCl, pH7). An aliquot of 200 μl of cells was incubated for 1 hour on ice with anti-myc antibodies diluted 1:1000 in PBS and 10% (v/v) goat serum in a final volume of 500 μl. Cells were followed by washed with filtered PBS and resuspended in 500 μl of filtered PBS containing 10% (v/v) goat serum. Then, bacteria were incubated for 30 minutes at 4°C in the dark with goat Alexa fluor 488-conjugated anti-mouse IgG (1:250; ThermoFisher Scientific). Finally, cells were washed once and resuspended in a final volume of 1 ml of filtered PBS and fluorescence analyzed in a MACSQuantTM VYB flow cytometer (Miltenyi Biotec; Bergisch Gladbach, Germany). Fluorescence was excited at 488 nm and recorded with a 525/50 nm band-pass filter. The results were processed using FlowJo (FlowJo LLC; Ashland OR, USA). Two biological replicates were performed and 100,000 events acquired for each experiment.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance in bacterial competition experiments was assessed by unpaired t-tests between relevant samples. Details of statistical significance is provided in the figure legends.
Supplementary Material
Highlights.
Programmed inhibitor cells (PICs) recognize target bacteria via surface nanobodies.
PICs selectively and potently deplete low abundance targets via toxin delivery.
PICs deplete target cells from complex communities with minimal off-target effects.
Resistance to PICs recognizing an essential surface structure is slow to emerge.
Acknowledgements
We thank Ingmar Riedel-Kruse, Christopher Hayes, John Leong, Joshua Woodward, Dan Stetson, Jason Smith, Eugene Nester, Marlen Adler, and Erik Gullberg for providing reagents. Support for this research was provided by the Defense Threat Reduction Agency (HDTRA1-13-1-0014 to J.D.M), the NIH (R01-AI080609 to J.D.M), the Public Health Service Grants (AI126277, AI114625, AI145325 to M.R.), the Chiba University-UCSD Center for Mucosal Immunology, Allergy, and Vaccines (to M.R.), the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (to M.R.), the HELIOS (BIO2015-66960-C3-2R) and SETH (RTI2018-095584-B-C42) Projects of the Spanish Ministry of Science (to V.d.L.), the MADONNA (H2020-FET-OPEN-RIA-2017-1-766975), BioRoboost (H2020-NMBP-BIO-CSA-2018), and SYNBIO4FLAV (H2020-NMBP/0500) Contracts of the European Union (to V.d.L.) and the S2017/BMD-3691 InGEMICS-CM funded by the Comunidad de Madrid (European Structural and Investment Funds) (to V.d.L.). J.D.M. is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The University of Washington has filed a provisional patent application regarding this manuscript.
References
- Albanese D, Fontana P, De Filippo C, Cavalieri D, and Donati C (2015). MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 5, 9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoforado Diniz J, Liu YC, and Coulthurst SJ (2015). Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cellular microbiology 17, 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, and Nielsen H (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37, 420–423. [DOI] [PubMed] [Google Scholar]
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, and Low DA (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248. [DOI] [PubMed] [Google Scholar]
- Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, et al. (2016). Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1, 16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor M, Prasannan S, Daniell S, Reece S, Connerton I, Bloomberg G, Dougan G, Frankel G, and Matthews S (2000). Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J 19, 2452–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, et al. (2011). Tackling antibiotic resistance. Nat Rev Microbiol 9, 894–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Casabona MG, Kneuper H, Chalmers JD, and Palmer T (2016). The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Deng W, and Finlay BB (2000). Enteropathogenic Escherichia coli (EPEC) attachment to epithelial cells: exploiting the host cell cytoskeleton from the outside. Cellular microbiology 2, 1–9. [DOI] [PubMed] [Google Scholar]
- Chan BK, Abedon ST, and Loc-Carrillo C (2013). Phage cocktails and the future of phage therapy. Future microbiology 8, 769–783. [DOI] [PubMed] [Google Scholar]
- Cherrak Y, Flaugnatti N, Durand E, Journet L, and Cascales E (2019). Structure and Activity of the Type VI Secretion System. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman WG Jr. (1983). The rfaD gene codes for ADP-L-glycero-D-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J Biol Chem 258, 1985–1990. [PubMed] [Google Scholar]
- Coulthurst S (2019). The Type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Bourven I, Guibaud G, van Hullebusch ED, Panico A, Pirozzi F, and Esposito G (2015). Role of extracellular polymeric substances (EPS) production in bioaggregation: application to wastewater treatment. Applied microbiology and biotechnology 99, 9883–9905. [DOI] [PubMed] [Google Scholar]
- Fleetwood F, Devoogdt N, Pellis M, Wernery U, Muyldermans S, Stahl S, and Lofblom J (2013). Surface display of a single-domain antibody library on Gram-positive bacteria. Cell Mol Life Sci 70, 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DS, and Riedel-Kruse IH (2018). A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns. Cell 174, 649–658 e616. [DOI] [PubMed] [Google Scholar]
- Hamilton TA, Pellegrino GM, Therrien JA, Ham DT, Bartlett PC, Karas BJ, Gloor GB, and Edgell DR (2019). Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun 10, 4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia A, Delgado S, Margolles A, and Sanchez B (2015). Application of density gradient for the isolation of the fecal microbial stool component and the potential use thereof. Sci Rep 5, 16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe 7, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Meyer KJ, Iinishi A, Favre-Godal Q, Green R, Manuse S, Caboni M, Mori M, Niles S, Ghiglieri M, et al. (2019). A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Hartmann JB, Jakob RP, Zahn M, Zimmermann I, Maier T, Seeger MA, and Hiller S (2019). Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J Biomol NMR 73, 375–384. [DOI] [PubMed] [Google Scholar]
- Kelly G, Prasannan S, Daniell S, Fleming K, Frankel G, Dougan G, Connerton I, and Matthews S (1999). Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat Struct Biol 6, 313–318. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Kahne DE, and Silhavy TJ (2017). Outer Membrane Biogenesis. Annu Rev Microbiol 71, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCourse KD, Peterson SB, Kulasekara HD, Radey MC, Kim J, and Mougous JD (2018). Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat Microbiol 3, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, and Mougous JD (2012). Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proceedings of the National Academy of Sciences of the United States of America 109, 19804–19809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, et al. (2015). Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc-Carrillo C, and Abedon ST (2011). Pros and cons of phage therapy. Bacteriophage 1, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T, and Allen HK (2012). Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Igual R, Bernal-Bayard J, Rodriguez-Paton A, Ghigo JM, and Mazel D (2019). Engineered toxin-intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat Biotechnol 37, 755–760. [DOI] [PubMed] [Google Scholar]
- Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, Finlay BB, and Strynadka NC (2000). Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405, 1073–1077. [DOI] [PubMed] [Google Scholar]
- McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, Erlandson SC, Hilger D, Rasmussen SGF, Ring AM, et al. (2018). Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nature structural & molecular biology 25, 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams BD, and Torres AG (2014). Enterohemorrhagic Escherichia coli Adhesins. Microbiol Spectr 2. [DOI] [PubMed] [Google Scholar]
- Mezzatesta ML, Gona F, and Stefani S (2012). Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future microbiology 7, 887–902. [DOI] [PubMed] [Google Scholar]
- Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, Ligat L, Rain JC, Favre G, Olichon A, et al. (2016). NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero-Lambea C, Bodelon G, Fernandez-Perianez R, Cuesta AM, Alvarez-Vallina L, and Fernandez LA (2015). Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth Biol 4, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preska Steinberg A, Datta SS, Naragon T, Rolando JC, Bogatyrev SR, and Ismagilov RF (2019). High-molecular-weight polymers from dietary fiber drive aggregation of particulates in the murine small intestine. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram G, Ross HF, Novick RP, Rodriguez-Pagan I, and Jiang D (2018). Conversion of staphylococcal pathogenicity islands to CRISPR-carrying antibacterial agents that cure infections in mice. Nat Biotechnol 36, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, and Mahe F (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C, Chen SP, Cabral V, Yaung SJ, and Wang HH (2019). Metagenomic engineering of the mammalian gut microbiome in situ. Nature methods 16, 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Verster AJ, Radey MC, Schmidtke DT, Pope CE, Hoffman LR, Hajjar AM, Peterson SB, Borenstein E, and Mougous JD (2019). Human gut bacteria contain acquired interbacterial defence systems. Nature 575, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano-Gallego D, Fraile S, Gutierrez C, and Fernandez LA (2019). Screening and purification of nanobodies from E. coli culture supernatants using the hemolysin secretion system. Microb Cell Fact 18, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, and Mougous JD (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, and Mougous JD (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Linek J, Dorken G, Winkler A, Wilson LG, Pham NT, French CE, Schilling T, and Poon WCK (2010). Polymer-induced phase separation in suspensions of bacteria. Europhysics Letters 89. [Google Scholar]
- Secor PR, Michaels LA, Ratjen A, Jennings LK, and Singh PK (2018). Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 115, 10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer MO, and Dantas G (2011). Antibiotics and the resistant microbiome. Current opinion in microbiology 14, 556–563. [DOI] [PubMed] [Google Scholar]
- Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LR, Salinas RK, et al. (2015). Bacterial killing via a type IV secretion system. Nat Commun 6, 6453. [DOI] [PubMed] [Google Scholar]
- Speare L, Smith S, Salvato F, Kldiner M, and Septer AN (2020). Environmental viscosity modulates interbacterial killing during habitat transition. MBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SY, Bosch DE, Mangiameli SM, Radey MC, Huang S, Park YJ, Kelly KA, Filip SK, Goo YA, Eng JK, et al. (2018). Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska J, Ho BT, and Mekalanos JJ (2018). Exopolysaccharide protects Vibrio cholerae from exogenous attacks by the type 6 secretion system. Proc Natl Acad Sci U S A 115, 7997–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Molecular Microbiology 92, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, et al. (2017). A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 6, e26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics (Oxford, England) 26, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I, Egloff P, Hutter CA, Arnold FM, Stohler P, Bocquet N, Hug MN, Huber S, Siegrist M, Hetemann L, et al. (2018). Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data associated with this study has been deposited to the NCBI Sequence Read Archive under Bioproject PRJNA625997. Code generated in this study is available from Github at https://github.com/kevinjohncutler/CellAggregateAnalysis.


