Abstract
Efficient delivery of hydrophilic drugs, nucleic acids, proteins, and any combination thereof is essential for various biomedical applications. Herein, we report a straightforward, yet versatile approach to efficiently encapsulate and deliver various hydrophilic payloads using a pH-responsive silica–metal–organic framework hybrid nanoparticle (SMOF NP) consisting of both silica and zeolitic imidazole framework (ZIF). This unique SMOF NP offers a high loading content and efficiency, excellent stability, and robust intracellular delivery of a variety of payloads, including hydrophilic small molecule drugs (e.g., doxorubicin hydrochloride), nucleic acids (e.g., DNA and mRNA), and genome-editing machineries (e.g., Cas9-sgRNA ribonucleoprotein (RNP), and RNP together with donor DNA (e.g., RNP+ssODN)). The superior drug delivery/gene transfection/genome-editing efficiencies of the SMOF NP are attributed to its pH-controlled release and endosomal escape capabilities due to the proton sponge effect enabled by the imidazole moieties in the SMOF NPs. Moreover, the surface of the SMOF NP can be easily customized (e.g., PEGylation and ligand conjugation) via various functional groups incorporated into the silica component. RNP-loaded SMOF NPs induced efficient genome editing in vivo in murine retinal pigment epithelium (RPE) tissue via subretinal injection, providing a highly promising nanoplatform for the delivery of a wide range of hydrophilic payloads.
Keywords: Silica, metal-organic framework, hybrid nanoparticle, gene delivery, genome editing machinery delivery
Graphical Abstract
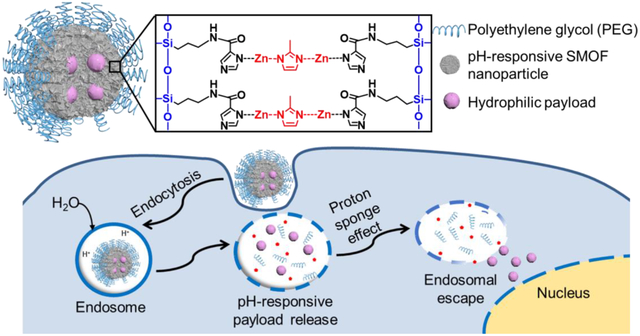
1. Introduction
Intracellular delivery of hydrophilic drugs, nucleic acids, and proteins plays an important role in medicine, including targeted drug delivery, gene therapy, and genome editing[1–4]. Nevertheless, efficient encapsulation and delivery of hydrophilic payloads while retaining their active conformations remains a significant challenge. A desirable delivery system would be able to encapsulate bioactive molecules with a high loading efficiency without compromising their biological activity.
The use of inorganic nanoparticles (NPs) (e.g., gold NPs, calcium phosphate NPs, silica-based NPs) has emerged as a promising strategy for efficient drug/gene delivery[5–7]. Silica NPs have shown remarkable potential in nanomedicine due to their unique properties, such as high stability, multifunctionality, and biocompatibility[8–10]. However, stimuli-responsive silica NPs are either difficult to design (i.e., pH-responsive), or require unstable/hard-to-synthesize silica monomers (i.e., GSH-responsive)[11, 12], thus hindering their delivery efficiency.
Metal–organic frameworks (MOFs), a class of materials formed by the covalent assembly of polydentate bridging ligands and metal-connecting points, have been studied for drug, gene, and genome-editing machinery delivery[13–15]. Zeolitic imidazolate framework (ZIF) is a subclass of metal–organic frameworks formed by coordination between Zn2+ ions and 2-methylimidazole (2-MIM) with good biocompatibility[16, 17]. Moreover, the excellent pH-buffering capacity of the imidazole bridging ligand confers pH-responsive capabilities to the ZIF, as well as an enhanced ability to escape the endocytic pathway[18]. ZIF-based nanosystems have been used for drug, gene, and protein delivery[18–20].
Herein, we report the fabrication of a pH-responsive silica–metal–organic framework hybrid NP (SMOF NP) consisting of both silica and ZIF via a facile water-in-oil emulsion approach, allowing for the delivery of a variety of hydrophilic payloads including small molecule drugs, nucleic acids, and genome-editing machineries. Hydrophilic payloads can be encapsulated in the NPs with a high loading content (> 9 wt%) and a high loading efficiency (> 90%). The proton sponge effect of imidazole in the ZIF moiety of the SMOF NPs contributes to the pH-controlled release and endosomal escape capabilities, while the surface of the SMOF NP is customizable via functional groups in the silica component.
We studied the genome editing efficiency in murine retinal pigmented epithelium (RPE), as genetic disorders in the RPE can cause a variety of eye diseases (e.g., retinal degeneration and blindness) [21]. SMOF NPs were decorated with a targeting ligand, all-trans retinoic acid (ATRA) (i.e., SMOF-ATRA). ATRA binds to the inter-photoreceptor retinoid-binding protein, a major protein in the inter-photoreceptor matrix that selectively transports all-trans-retinol to the RPE and 11-cis-retinal to photoreceptor[22, 23]. SMOF-ATRA generated robust gene editing in murine retinal pigment epithelium (RPE) after local administration. This stable, biocompatible, and multifunctional SMOF NP is a promising nanoplatform capable of efficiently delivering a variety of hydrophilic payloads.
2. Experimental Section
Materials
1H-imidazole-4-carboxylic acid, thionyl chloride (SOCl2), tetraethyl orthosilicate (TEOS), tetrahydrofuran (THF), Triton X-100, acetone, ethanol, ammonia (30% in water) 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), and N-hydroxysuccinimide (NHS) were purchased from Fisher Scientific, USA. Hexanol, cyclohexane, 2-methyl-1H-imidazole (2-MIM), (3-aminopropyl)triethoxysilane (APTES), and doxorubicin hydrochloride (DOX·HCl) were bought from Tokyo Chemical Industry Co., Ltd., USA. Methoxypolyethylene glycol-N-succinimidyl ester (mPEG-NHS, Mn = 5000) and hydroxyl-polyethylene glycol-N-succinimidyl ester (HO-PEG-NHS, Mn = 5000) were obtained from Jenkem Technology, USA. Anhydrous zinc nitrate (ZnNO3) and all-trans retinoic acid (ATRA) were purchased from Sigma–Aldrich, USA. Nuclear localization signal (NLS)-tagged streptococcus pyogenes Cas9 nuclease (sNLS–SpCas9–sNLS) was provided by Aldevron, USA.
Synthesis of N-(3-(Triethoxysilyl)propyl)-1H-Imidazole-4-Carboxamide (TESPIC)
A 1H-imidazole-4-carboxylic acid (500 mg, 3.85 mmol) solution in SOCl2 (8 ml) was heated while stirring to reflux overnight. The reaction mixture was cooled to room temperature and added into toluene. The precipitate was then collected by filtration and dried in vacuo at room temperature to yield the acid chloride intermediate, 1H-imidazole-4-carbonyl chloride. The freshly synthesized 1H-imidazole-4-carbonyl chloride was suspended in anhydrous THF (5 ml), followed by addition of Triethylamine (855 mg, 8.47 mmol) and APTES (851 mg, 3.85 mmol). The mixture was stirred at room temperature overnight under a nitrogen atmosphere. The mixture was subsequently filtered, and the solvent was then removed by rotary evaporation to yield the final product TESPIC. Since the silica reactants have the tendency to undergo hydrolysis/polymerization during column purification, TESPIC was synthesized and used without purification for SMOF NP formation[11, 24]. 1H NMR (400 MHz, DMSO-D6): δ 0.60 (dd, 2.4 H, J = 14.6, 6.2 Hz), δ 1.12 (t, 0 H, J = 7.0 Hz), δ 1.57 (dt, 2 H, J = 15.9, 8.0 Hz), δ 2.83–2.61 (m, 2 H), δ 3.70 (q, 6 H, J = 6.0 Hz), δ 7.03 (s, 1 H), δ 7.40 (s, 1 H). 13C NMR (100 MHz, DMSO-D6): δ 165.76, 135.50, 132.89, 128.21, 58.01, 42.55, 22.88, 18.64, and 7.55.
Preparation of Silica–Metal–Organic Framework Hybrid Nanoparticles (SMOF NPs)
SMOF NPs were synthesized by a water-in-oil emulsion method. Triton X-100 (1.75 ml) and hexanol (1.75 ml) were dissolved in cyclohexane (7.5 ml) to form the organic phase. An aqueous ZnNO3 (0.5 M) solution (20 μl) containing the desirable payload (e.g., DOX·HCl, DNA, mRNA, RNP, and RNP+ssODN; 5 mg/ml) was mixed with 400 μl of the organic phase. This mixture was vortexed for 15 s and then sonicated in an ultrasonic water bath for 15 s to form the water-in-oil emulsion, which was then magnetically stirred at 1500 rpm. To this emulsion, TEOS, TESPIC, APTES, and 2-MIM with different feed weight ratios were dissolved in 100 μl organic phase and added to the above emulsion. For example, to achieve a feed weight ratio of silica reactants (i.e., TEOS, TESPIC, APTES) to MOF reactant (i.e., 2-MIM) of 60:40, the total weight of TEOS+TESPIC+APTES added to the emulsion would be 1.2 mg, while the weight of 2-MIM would be 0.8 mg. Upon the addition of 3 μL of 30% ammonia aqueous solution, the mixture was stirred for 4 h at room temperature. Thereafter, mPEG-NHS (100 μg in 100 μl hexanol) was added to the above emulsion and was stirred for another 2 h. To prepare ATRA-modified SMOF NPs (i.e., SMOF-ATRA), HO-PEG-NHS was used instead of mPEG-NHS. The final payload-encapsulated SMOF NPs were precipitated by 600 μl acetone, and then washed by ethanol and water three times each.
ATRA was conjugated to the SMOF NP surface via EDC/NHS catalyzed esterification. Payload-encapsulated SMOF NPs (0.5 mg) were re-dispersed in 0.5 ml DI water. EDC (60 μg), NHS (60 μg) and a DMSO solution of ATRA (6 μg in 3 μl DMSO) were added to the above solution, and the pH was adjusted to 8. The solution was stirred at room temperature for 6 h, and then the SMOF-ATRA was washed by water three times.
Characterization
The chemical structure of TESPIC was analyzed by nuclear magnetic resonance (NMR) spectroscopy (Avance 400, Bruker Corporation, USA). The hydrodynamic diameter and zeta potential of the SMOF NPs were characterized by a dynamic light scattering (DLS) spectrometer (Malvern Zetasizer Nano ZS) at a 90° detection angle with a concentration of 0.1 mg/ml. The morphologies of SMOF NPs were characterized by transmission electron microscopy (TEM, Tecnai 12, Thermo Fisher, USA) and scanning electron microscopy (SEM, Zeiss/LEO 1530, Carl Zeiss Microscopy, USA). X-ray powder diffraction of SMOF NPs was performed by Bruker D8 Discovery (Bruker Corporation, USA).
Loading Content/Loading Efficiency Study
To calculate the loading content and loading efficiency of the payloads in the SMOF NPs, 1 mg∙mL−1 of SMOF NP stock solution with different payloads were prepared. Thereafter, 10 μL of SMOF NP was incubated with 40 μL of acetate buffer (0.1 M, pH 5.5) for 30 min to allow for complete dissociation of SMOF NPs. The DOX·HCl loading content/efficiency was studied by UV-Vis spectroscopy. The RNP and RNP-ssODN loading contents/efficiencies were measured via a bicinchoninic acid assay (BCA assay, Thermo Fisher, USA). DNA and mRNA loading contents/efficiencies were evaluated using a NanoDrop One (Thermo Fisher, USA).
Cell Culture
Cells were cultured in a cell culture incubator (Thermo Fisher, USA) at 37 °C with 5% carbon dioxide at 100% humidity. HEK 293 cells (a human embryonic kidney cell line), NHDF (a normal human dermal fibroblast cell line), and RAW 264.7 cells (a mouse macrophage cell line) were purchased from ATCC (USA) and cultured with DMEM medium (Gibco, USA) with 10% (v/v) fetal bovine serum (FBS, Gibco, USA) and 1% (v/v) penicillin–streptomycin (Gibco, USA). HCT 116 cells (a human colon cancer cell line) were cultured with 89% McCoy’s 5A medium, 10% FBS, and 1% penicillin–streptomycin. All the culture media used for hydrophilic drug delivery/nucleic acid transfection/genome editing efficiency studies were complete culture media containing 10% FBS.
Hydrophilic Drug Delivery Study
The cellular uptake behavior of DOX·HCl-loaded SMOF NPs was analyzed using flow cytometry. HEK 293 cells were seeded onto 96-well plates with 15,000 cells per well 24 h before treatment. The cells were incubated with free DOX·HCl and DOX·HCl-loaded SMOF NPs for 4 h with a DOX·HCl concentration of 5 μg/ml. Thereafter, cells were harvested with 0.25% trypsin-EDTA (Thermo Fisher, USA), spun down, and resuspended with 200 μl PBS (Thermo Fisher, USA). DOX·HCl uptake was detected with an Attune NxT flow cytometer system (Thermo Fisher, USA) and analyzed with FlowJo 7.6.
DNA and mRNA Transfection Efficiency Study
HEK 293, HCT116, NHDF, and RAW 264.7 cells were seeded onto 96-well plates at a density of 15,000 cells per well 24 h prior to treatment. Cells were transfected with either a green fluorescence protein (GFP) plasmid DNA (Addgene #40259, USA)-loaded SMOF NPs, or a GFP-mRNA (OZ Biosciences INC, San Diego, CA)-loaded SMOF NPs, The dosage of DNA or mRNA dosages is 200 ng/well, the SMOF NP concentrations for DNA- and mRNA-loaded SMOF NPs were 21 μg/ml and 22 μg/ml, respectively. DNA or mRNA was also transfected using a commercially available transfection agent, Lipofectamine 2000 (i.e., Lipo 2000, Thermo Fisher, USA), as a positive control group. The Lipofectamine-DNA complex was prepared as suggested by the manufacturer. The amount of Lipo 2000 and DNA (or mRNA) used per well was 0.5 μl and 200 ng, respectively. An untreated group was used as the negative control group. After 48 h, HEK 293, HCT116, and NHDF cells were harvested with 0.25% trypsin-EDTA. RAW264.7 cells were harvested by repeated pipetting. The cells were then spun down and resuspended with 200 μl of PBS. GFP expression efficiencies were obtained with a flow cytometer and analyzed with FlowJo 7.6. Gating strategies are shown in Figure S7.
RNP Genome-Editing Efficiency Study
GFP-expressing HEK 293 cells (GenTarget Inc.) were used as an RNP-transfection cell model. Cells were seeded at a density of 5,000 cells per well onto 96-well plates, 24 h before treatment. RNP was prepared as previously reported [25] by mixing sNLS–SpCas9–sNLS and in vitro transcribed sgRNA (Integrated DNA Technologies, Inc., GFP protospacer: 5’-GCACGGGCAGCTTGCCGG-3’) at a 1:1 molar ratio. Cells were treated with Lipo 2000 (0.5 μl/well) complexed with RNP or RNP-loaded SMOF NPs. The RNP dosage was kept at 150 ng/well (i.e., an equivalent Cas9 protein dosage of 125 ng/well). An untreated group was used as the control group. A quantity of 100 μl of fresh culture medium was added into each well 48 h after treatment and thereafter; half of the culture medium was refreshed every 48 h. Six days after treatment, cells were harvested with 0.25% trypsin-EDTA, spun down, and resuspended with 200 μl of PBS. The RNP genome-editing efficiencies were quantified via flow cytometry. Data were analyzed with FlowJo 7.6.
RNP+ssODN Co-Delivery for Precise Gene Correction
The RNP+ssODN mixture was prepared by simply mixing the as-prepared RNP and single-stranded oligonucleotide DNA (ssODN) donor template at 4 °C for 5 min at a 1:1 molar ratio. Blue fluorescence protein (BFP)-expressing HEK 293 cells generated through lentiviral transduction of a BFP dest clone (Addgene, Cambridge, MA) was employed as a model cell line[25]. When cells are transfected with RNP+ssODN-targeting BFP (Integrated DNA Technologies, Inc., BFP protospacer: 5’-GCTGAAGCACTGCACGCCAT-3’), if precise editing occurs, three nucleotides within the BFP gene are edited and converted to a green fluorescent protein (GFP) gene as described previously (BFP to GFP ssODN sequence: 5’-TCATGTGGTCGGGGTAGCGGCTGAAGCACTGCACGCCATGGGTCAGGGTGGTCACGAGGGTGGGCCAGGGCACCGGCAGCTTGCCGGTGGTGCAGATGAA-3’, changing BFP to GFP via alternation of histidine to tyrosine) [25, 26]. BFP-expressing HEK 293 cells were seeded at a density of 15,000 cells per well onto a 96-well plate 24 h before treatment. Cells were treated with Lipo 2000 (0.5 μl/well) carrying RNP and ssODN or with RNP+ssODN-loaded SMOF NPs. For each treatment, the RNP+ssODN dosage was kept at 175 ng/well (i.e., an equivalent Cas9 protein dosage of 125 ng/well). The precise gene-editing efficiencies were quantified six days after treatment using flow cytometry by counting the percentage of green fluorescence positive cells. Data were analyzed with FlowJo 7.6.
Intracellular Trafficking of RNP SMOF NPs
Intracellular trafficking of RNP SMOF NPs was investigated by confocal laser scanning microscopy (CLSM, Nikon, Japan). In this case, ATTO550-labeled sgRNA (Integrated DNA Technologies, Inc.) was used to form the Cas9/ATTO-sgRNA RNP loaded into the SMOF NPs. HEK 293 cells were seeded onto a Nunc™ Lab-Tek™ II CC2- chamber slide (Thermo Fisher, USA) at 50,000 cells per well 24 h prior to treatment. At each time point (i.e., 0.5, 2, and 4 h) after SMOF NP treatment, the cells were washed by PBS and then stained with endosome/lysosome marker LysoTracker Green DND-26 (100 nM) and nucleus marker Hoechst 33342 (10 μg/mL) for 30 min at 37 °C.
Cell Viability Assay
The cytotoxicity of the DOX·HCl-loaded SMOF NPs was studied using an MTT assay. HEK 293 cells were seeded onto 96-well plates with 20,000 cells per well 24 h prior to treatment. The cells were then treated with free DOX·HCl, DOX·HCl-loaded SMOF NPs, and empty SMOF NPs (DOX·HCl concentrations of 6 and 12 μg/ml). Cells without treatment were used as a control group. After 48 h, the cell viability was measured using a standard MTT assay (Thermo Fisher, USA). Briefly, cells were treated with media containing 500 μg/ml MTT and incubated for 4 h. Then, the MTT-containing media was aspirated. Next, the purple precipitate was dissolved in 150 μl of DMSO. The absorbance at 560 nm was obtained with a microplate reader (GloMax® Multi Detection System, Promega, USA).
The cytotoxicity of SMOF NPs was also studied by MTT assay. Cells were treated with complete medium, Lipo 2000 (0.5 μl/well), and empty SMOF NPs, whose concentrations ranged from 10 to 200 μg/ml. Cell viability was measured using a standard MTT assay 48 h after treatment (Thermo Fisher, USA), as mentioned above.
Subretinal Injection
All animal research was approved by UW-Madison animal care and use committee. Ai14 reporter mice (obtained from The Jackson Laboratory) were used to assess the genome editing efficiency induced by RNP-loaded SMOF NPs. C57BI/6 mice were used as the controls for biocompatibility studies. RNPs were prepared using either a sgRNA targeting the stop cassette composed of 3 SV40 polyA blocks (target sequence: 5’-AAGTAAAACCTCTACAAATG-3’) in Ai14 mice or a mouse negative control sgRNA (Integrated DNA Technologies, guide sequence: CGTTAATCGCGTATAATACG). Subretinal injection and subsequent RPE tissue collection were performed as reported previously [27]. Mice were maintained under tightly controlled temperature (23 ± 5 °C), humidity (40–50%), and light/dark (12/12 h) cycle conditions under a 200 lux light environment. The mice were anesthetized by intraperitoneal injection of ketamine (80 mg/kg), xylazine (16 mg/kg) and acepromazine (5 mg/kg) cocktail. Before the subretinal injection, the cornea was anesthetized with a drop of 0.5% proparacaine HCl, and the pupil was dilated with 1.0% tropicamide ophthalmic solution. Mice were placed on a temperature-regulated heating pad during the injection and for recovery purposes. All surgical manipulations were carried out under a surgical microscope (AmScope, Irvine, CA). SMOF-ATRA encapsulating RNP with a sgRNA targeting the Ai14 stop cassette (i.e., Ai14 RNP SMOF), SMOF-ATRA encapsulating RNP with a negative control sgRNA (i.e., negative control SMOF), and PBS were injected into the eyes subretinally. Two microliters of SMOF-ATRA solutions containing 4 μg RNP or PBS was injected into the subretinal space using a UMP3 ultramicro pump fitted with a NanoFil syringe, and the RPE-KIT (all from World Precision Instruments, Sarasota, FL) equipped with a 34-gauge beveled needle. Successful administration was confirmed by visualization of bleb formation. The tip of the needle remained in the bleb for 10 s after bleb formation, when it was gently withdrawn.
To assess tdTomato expression generated by successful genome editing, the mice were sacrificed, and eyes were collected 13 to 14 days after injection and rinsed twice with PBS. A puncture was made at ora serrata with an 18-gauge needle, and the eye was opened along the corneal incisions. The lens was then removed. The eyecup was incised radially to the center and flattened to give a final floret shape. The RPE layer was then separated and flat-mounted on a cover-glass slide. RPE tissues were imaged with a NIS-Elements using a Nikon C2 confocal microscope (Nikon Instruments Inc.). ImageJ (NIH) was used for image analysis.
Electroretinography of the Retina
To study the biocompatibility of SMOF NP after subretinal injection, we tested visual function before sacrificing the mice via electroretinography (ERG). PBS and Ai14 RNP-loaded SMOF-ATRA were injected to the eyes of C57BI/6 mice subretinally. Fourteen days post-injection, animals were anesthetized as described above. All experiments were performed in a dark room. An Espion Ganzfeld full-field system (Diagnosys LLC, Lowell, MA) was used for ERG recordings. A drop of 2% hypromellose (GONIOVISC, HUB Pharmaceuticals, LLC, Rancho Cucamonga, CA) solution was placed on the cornea to keep it moistened and to provide an electrical contact with the ERG electrode. The mice were placed under the Ganzfeld dome to assure uniform illumination of the eyes. The eyes were exposed to a sequential increment of flash intensities (0.01 to 30 cd.s/m^2) for 400 ms with a 2 s interval between each flash for a and b- wave and eyes were exposed 25 cd.s/m^2 for 4 secs to measure c-wave. The data were analyzed using Diagnosys software version 6.0.54 (Diagnosys LLC, Lowell, MA).
Statistical Analysis
Results are presented as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) with Tukey’s multiple comparisons was used to determine the difference between independent groups. Statistical analyses were conducted using GraphPad Prism software version 6.
3. Results and Discussion
Silica–metal–organic framework hybrid nanoparticles (SMOF NPs) were synthesized via a facile water-in-oil emulsion method (Figure 1A and B). An aqueous solution containing zinc ions at a constant concentration (0.5 M) and the desirable payload was emulsified in the continuous oil phase, followed by additions of the silica reactants and the imidazole reactant (i.e., 2-methylimidazole (2-MIM)), which coordinates with the zinc ions and forms the pH-responsive zeolitic imidazole framework (ZIF)) (Figure 1C). The silica-reactive components included tetraethyl orthosilicate (TEOS), a basic building block that constructs the silica network; imidazole-containing N-(3-(triethoxysilyl)propyl)-1H-imidazole-2-carboxamide (TESPIC) that bridges the silica component with the ZIF component; and amine-containing (3-aminopropyl)triethoxysilane (APTES) that enables surface modification. Polyethylene glycol (PEG) was subsequently incorporated onto the SMOF NP surface after the formation of the SMOF NP, which allowed for further surface functionalization (e.g., conjugation of targeting ligands and imaging agents). The as-prepared SMOF NPs were then collected by precipitation in acetone, centrifuged, and washed by ethanol and deionized (DI) water three times each to remove all impurities.
Figure 1. Design and synthesis of SMOF NPs.
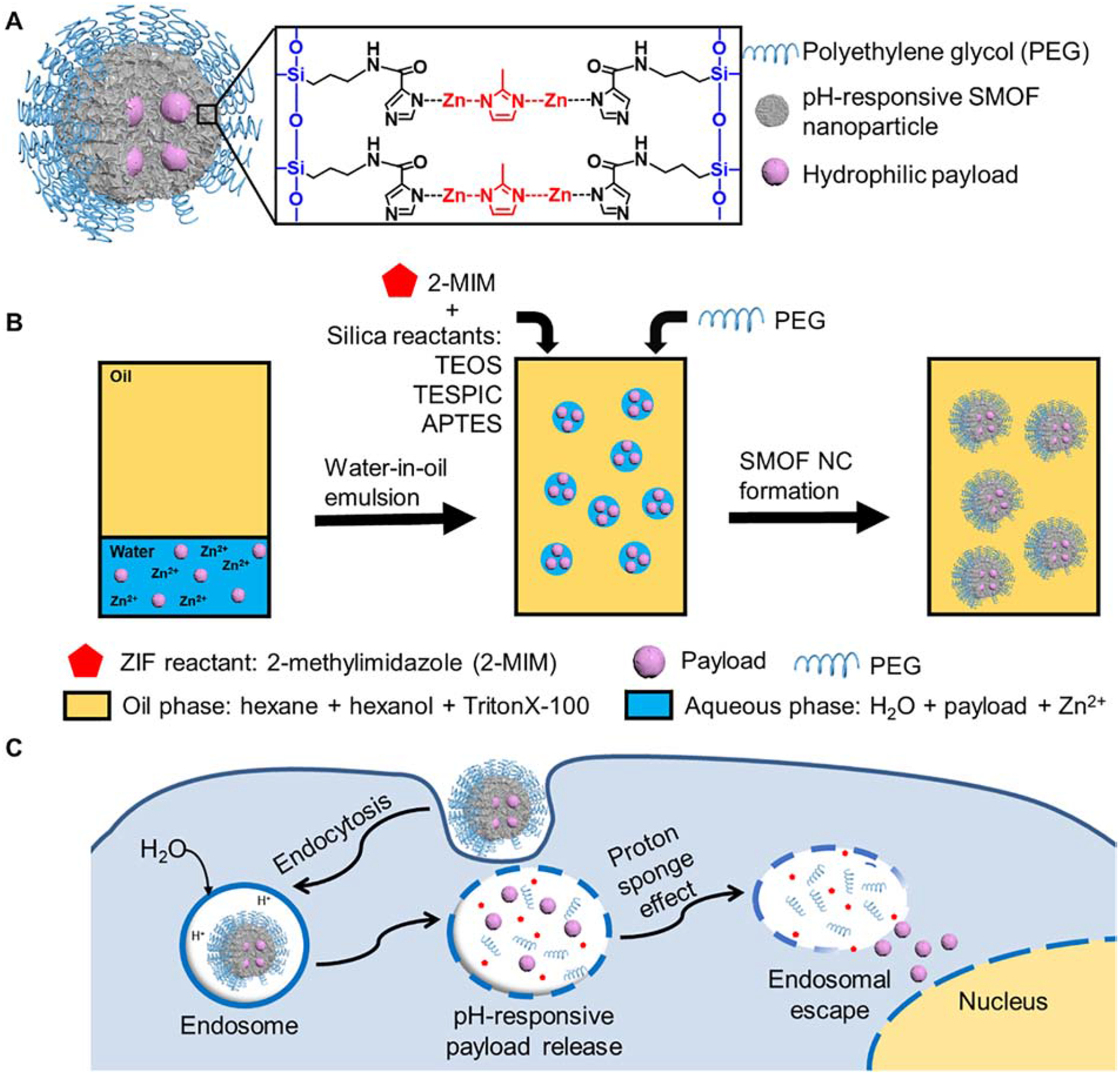
(A) Illustration of the SMOF NP for the delivery of various hydrophilic payloads such as hydrophilic small molecular drugs, DNA, mRNA, proteins, and a combination thereof (e.g., Cas9/sgRNA (RNP) and RNP+ssODN). (B) Synthesis of SMOF NPs via a water-in-oil emulsion method. (C) Schematic illustration of the intracellular trafficking pathways of SMOF NPs. SMOF NP: silica–metal–organic framework hybrid nanoparticle; 2-MIM: 2-methylimidazole; PEG: polyethylene glycol; TEOS: tetraethyl orthosilicate; TESPIC: N-(3-(triethoxysilyl)propyl)-1H-imidazole-2-carboxamide; APTES: (3-aminopropyl)triethoxysilane.
The SMOF NP formulation was first optimized in a human embryonic kidney cell line (HEK 293) using plasmid DNA (Figure 2) and mRNA (Figure S2) as the payloads. The zinc ion concentration in the aqueous phase was fixed at 0.5 M, while various factors were optimized including the feed ratio of the payload to the SMOF NP reactants, the feed ratio of the silica reactants (TEOS+APTES+TESPIC) to the MOF reactant (2-MIM), and the emulsification process (e.g., bath sonication versus probe sonication).
Figure 2. Optimization of the DNA-loaded SMOF NP formulation using HEK 293 cells.
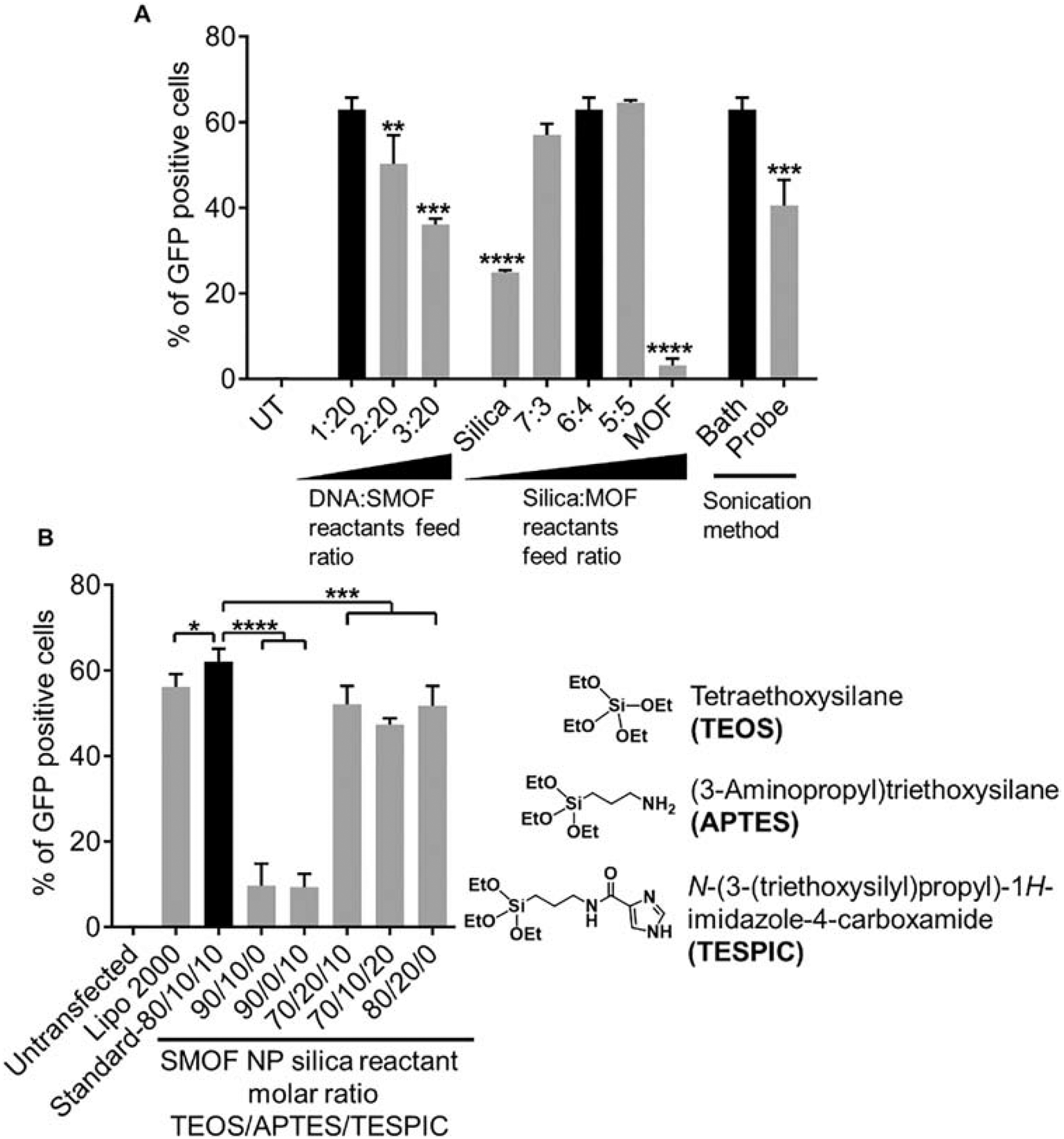
(A) Optimization of the feed weight ratio of the payload over the SMOF reactants, feed weight ratio of the silica reactants to the MOF reactant, and the emulsification method. (B) Optimization of the molar ratio of the three silica reactants TEOS/APTES/TESPIC. The optimal SMOF NP formulation is highlighted by a black bar. NS: not significant; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001; n = 3.
The feed weight ratio between the payload and the SMOF NP reactants is important, as insufficient SMOF NP forming materials may lead to a limited encapsulation volume and subsequently, a low loading efficiency and premature release and degradation of the payloads. The optimal formulation showing the highest DNA transfection efficiency was obtained at a DNA:SMOF reactant feed ratio of 1:20, indicating successful encapsulation of the payload within the SMOF NPs and an efficient intracellular release thereafter (Figure 2A).
The feed weight ratio between the silica reactants (i.e., TEOS+APTES+TESPIC) and the MOF reactant (i.e., 2-MIM) is another critical factor for efficient payload delivery. Without the silica component, surface functionalization of the resulting MOF/ZIF NPs is very challenging because various functional groups can be conveniently introduced into the SMOF NPs through judicious selection of the silica reactants. Without the MOF component, silica NPs alone can neither escape endosomes and lysosomes efficiently nor release the payload rapidly in response to pH, thereby greatly minimizing the delivery efficiency. As shown in Figure 2A, both pure silica NPs and pure MOF NPs formed via water-in-oil emulsions exhibited limited DNA transfection efficiencies. Moreover, pure MOF NPs showed larger particle sizes (>350 nm and polydispersed) after purification, indicating inadequate PEGylation and thus NP aggregation. The molar ratio of the three silica reactants—namely, TEOS, APTES, and TESPIC—was further optimized (Figure 2B). The optimal formulation showing the highest DNA transfection efficiency was obtained when the molar ratio of TEOS:APTES:TESPIC was 80:10:10.
To evaluate the necessity of using a TEOS:APTES:TESPIC ternary composition to form the silica component in the SMOF NP instead of unary or binary counterparts, the SMOF NP formulation without TESPIC (i.e., TEOS:APTES:TESPIC, molar ratio of 90:10:0) was first tested. The resulting SMOF NPs exhibited significantly lower DNA transfection efficiencies, indicating that TESPIC was essential for bridging the silica component to the MOF component within the hybrid SMOF NPs. Moreover, the formulation without APTES (i.e., TEOS:APTES:TESPIC, molar ratio of 90:0:10) also showed large and polydispersed particle sizes and limited the transfection efficacy due to the lack of surface PEGylation and NP aggregation. However, higher APTES or TESPIC ratios did not provide any advantage in achieving higher transfection efficiencies (Figure 2B).
The sonication method also plays an important role in SMOF NP synthesis as it facilitates emulsification and controls the water droplet size in the emulsion. However, sonication that is too strong may affect the integrity of the biomacromolecular payload and thus reduce delivery efficiency. Using probe sonication for as short as 15 s can reduce the DNA transfection efficiency by 50% in comparison with the DNA transfection efficiency achieved via a gentler sonication method (i.e., vortex (15 s) + bath sonication (15 s)) (Figure 2A). This is consistent with previous reports in that probe sonication can damage DNA [28, 29]
The morphology of the DNA-loaded SMOF NP was characterized by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Spherical NPs with uniform sizes around 50–70 nm were observed, as shown in Figure 3A and B. The hydrodynamic diameter of DNA-loaded SMOF NPs, as measured by dynamic light scattering (DLS), was 110 nm (Figure 3C), similar to empty SMOF NPs, (119 nm, Figure S3). Zeta-potential measurements indicated that the DNA-loaded SMOF NPs had a slight positive surface charge (5.6 ± 1 mV), similar to empty SMOF NPs (4.8 mV). Powder X-ray diffraction (XRD) spectra showed that SMOF NPs had similar crystal structures to ZIF (Figure 3D). The ratio of the silica components and ZIF component in the SMOF NPs is controlled by the feed weight ratio of the silica reactants (i.e., TEOS, TESPIC, and APTES) and the ZIF reactant (i.e., 2-MIM). The ratio of the silica component and the ZIF component in the SMOF NPs was studied by energy-dispersive X-ray spectroscopy (EDS). As shown in Figure S1, with the optimal feed weight ratio of the silica reactants to the ZIF reactant at 6:4, the elemental weight ratio between silicon (Si) and zinc (Zn) was 63:37 in the final SMOF NP, which is approximately equivalent to a 1:1 weight ratio of silica to ZIF.
Figure 3. Characterization of SMOF NPs.
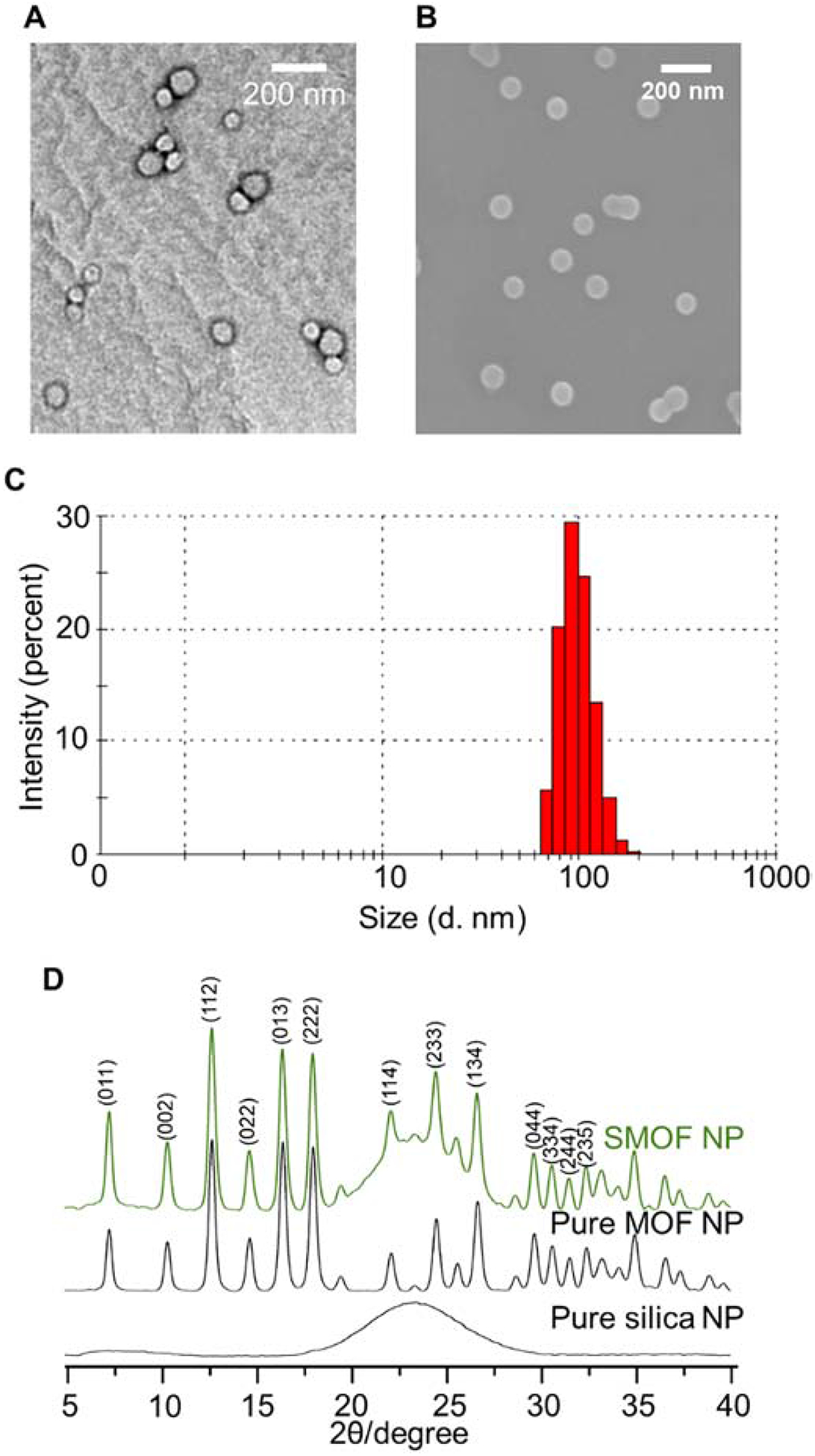
(A) TEM and (B) SEM micrographs of SMOF NPs. (C) Size distribution of SMOF NPs measured by DLS. (D) Powder XRD spectra of SMOF NP, pure MOF NP, and pure silica NP synthesized via the water-in-oil emulsion method.
The SMOF NP contains a pH-responsive ZIF component that degrades in acidic environments, leading to a rapid release of the payload [18]. Meanwhile, the ZIF component can also facilitate the endosomal escape of the payload because the imidazole groups (pKa ~ 6.0) can be protonated in the acidic endocytic compartments (i.e., endosomes), leading to endosomal-membrane disruption by the proton sponge effect. To study the intracellular trafficking of SMOF NPs, confocal laser scanning microscopy (CLSM) was used to image the subcellular distribution of Cas9/ATTO550-labeled sgRNA ribonucleoprotein (RNP) delivered by SMOF NPs (Figure 4). Cells without SMOF NP treatment were used as the control (Figure S4). No significant alternation in cell morphology and integrity was observed in the SMOF NP treated groups in comparison with the control group, indicating that SMOF NP treatment does not affect cell morphology. RNP was observed to co-localize with endo/lysosomes 0.5 h post-treatment, indicating that the uptake of RNP-loaded SMOF NPs occurred via endocytosis. The extent of co-localization of RNP and endo/lysosomes decreased 2 h post-treatment, indicating the efficient endo/lysosomal escape capabilities of SMOF NPs. Assisted by a nuclear localization signal (NLS) fused on both terminuses of the Cas9 nuclease, the RNP signal showed considerable overlap with the nucleus and minimal co-localization with endo/lysosomes as early as 4 h post-treatment, thus indicating the efficient escape from endo/lysosomes and the successful nuclear transportation of RNP.
Figure 4. Intracellular trafficking of RNP-loaded SMOF NPs by CLSM.
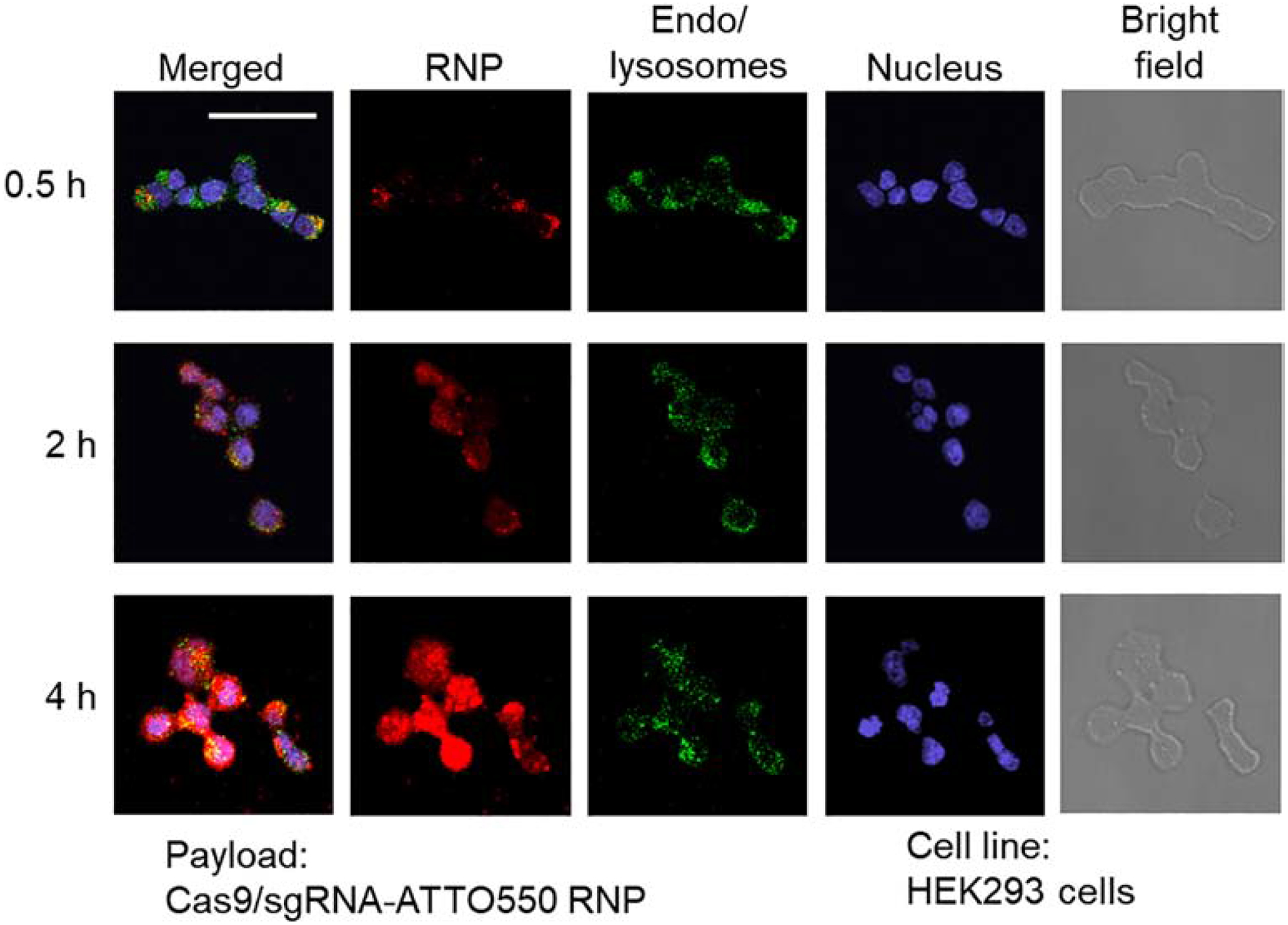
Colocalization of RNP and endo/lysosomes was studied at 0.5 h, 2 h, and 4 h post-treatment. Scale bar: 50 μm.
To investigate the versatility of SMOF NPs for the delivery of different hydrophilic payloads, including small molecule drugs (i.e., doxorubicin hydrochloride (DOX·HCl)), nucleic acids (i.e., DNA and mRNA), and CRISPR-Cas9 genome-editing machineries (i.e., RNP and RNP+ssODN (i.e., a combination of an RNP with a single-stranded oligonucleotide DNA (ssODN) donor template)), the loading content and loading efficiency of different payloads are quantified and summarized in Table 1. For small molecule DOX·HCl, the loading content was 17 wt%, with a loading efficiency of 92%. For hydrophilic biomacromolecules, the loading contents varied between 9.2–9.8 wt%, while the loading efficiencies ranged from 91–97%. The high loading contents and efficiencies can be attributed to the water-in-oil emulsion method that confined the payloads within the water droplet, followed by the formation of the SMOF NP network.
Table 1.
Summary of loading content and loading efficiency of different payloads by SMOF NPs.
| Payload | Loading Content (wt%) | Loading Efficiency (%) |
|---|---|---|
| Doxorubicin hydrochloride | 17 | 92 |
| DNA | 9.5 | 94 |
| mRNA | 9.2 | 91 |
| RNP | 9.8 | 97 |
| RNP+ssODN | 9.5 | 94 |
Efficient delivery of DOX·HCl via SMOF NPs was first studied by flow cytometry in HEK293 cells by taking advantage of the fluorescence of DOX·HCl. Cells without DOX·HCl treatment were used as a control. As shown in Figure 5A, DOX·HCl -loaded SMOF NP-treated cells exhibited a 3.2-fold higher level of DOX·HCl uptake than free DOX·HCl -treated cells 2 h post-treatment, indicating the efficient uptake of SMOF NPs by HEK293 cells. The cellular uptake of DOX·HCl -loaded SMOF NPs was also confirmed by fluorescence microscopy (Figure S3) 4 h post-treatment. The therapeutic effect of DOX·HCl -loaded SMOF NPs was evaluated by an MTT assay (Figure 5B). At both 6 μg/ml and 12 μg/ml DOX·HCl concentrations, the DOX·HCl -loaded SMOF NPs exhibited identical cytotoxicity to free DOX·HCl, while empty SMOF NPs showed no significant cytotoxicity. These results demonstrated the efficient delivery and release of DOX·HCl by SMOF NPs.
Figure 5. Delivery Efficiency of a hydrophilic drug by SMOF NPs.
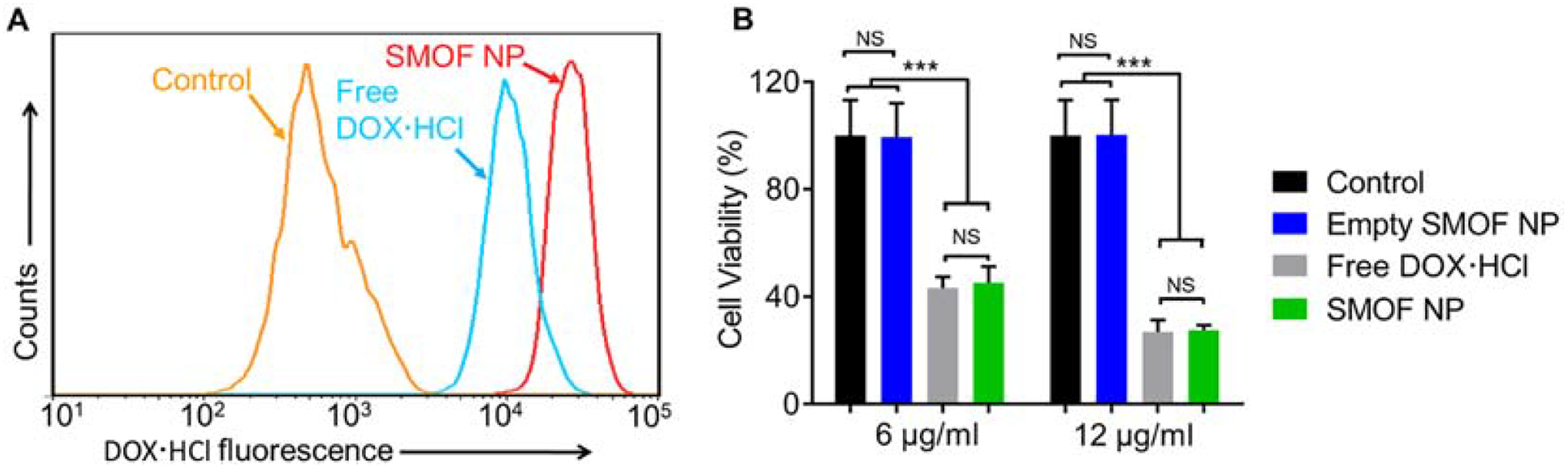
(A) Flow cytometry analysis on HEK293 cells treated with free DOX·HCl (5 μg/ml) and DOX·HCl-loaded SMOF NPs (DOX·HCl concentration, 5 μg/ml) or medium alone (control) for 2 h. (B) Cytotoxicity of empty SMOF NPs, free DOX·HCl and DOX·HCl-loaded SMOF NPs at different DOX·HCl concentrations (i.e., 6 and 12 μg/ml) after co-incubation with HEK293 cells for 48 h.
The transfection efficiency of DNA-loaded SMOF NPs was studied in four different cell types, including a HEK293 cell line, a human colon tumor (HCT116) cell line, a human normal dermal fibroblast (NHDF) cell line, and a rat macrophage (RAW264.7) cell line. In HCT116 cells, DNA-loaded SMOF NPs exhibited a similar transfection efficiency to the commercially available transfection agent Lipofectamine 2000 (Lipo 2000), while in the other three cell lines, SMOF NP showed statistically higher transfection efficiencies than Lipo 2000 (1.4-fold in HEK293 cells, 1.2-fold in NHDF cells and RAW264.7 cells, as shown in Figure 6A),. For mRNA-loaded SMOF NPs, they showed similar transfection efficiencies to Lipo 2000 in HEK293 and NHDF cells, but statistically higher transfection efficiencies in HCT116 and RAW264.7 cells (1.9- and 1.3-fold, respectively, as shown in Figure 6B), indicating efficient delivery of nucleic acids by SMOF NPs.
Figure 6. Delivery efficiency of nucleic acids and CRISPR-Cas9 genome-editing machineries by SMOF NPs.
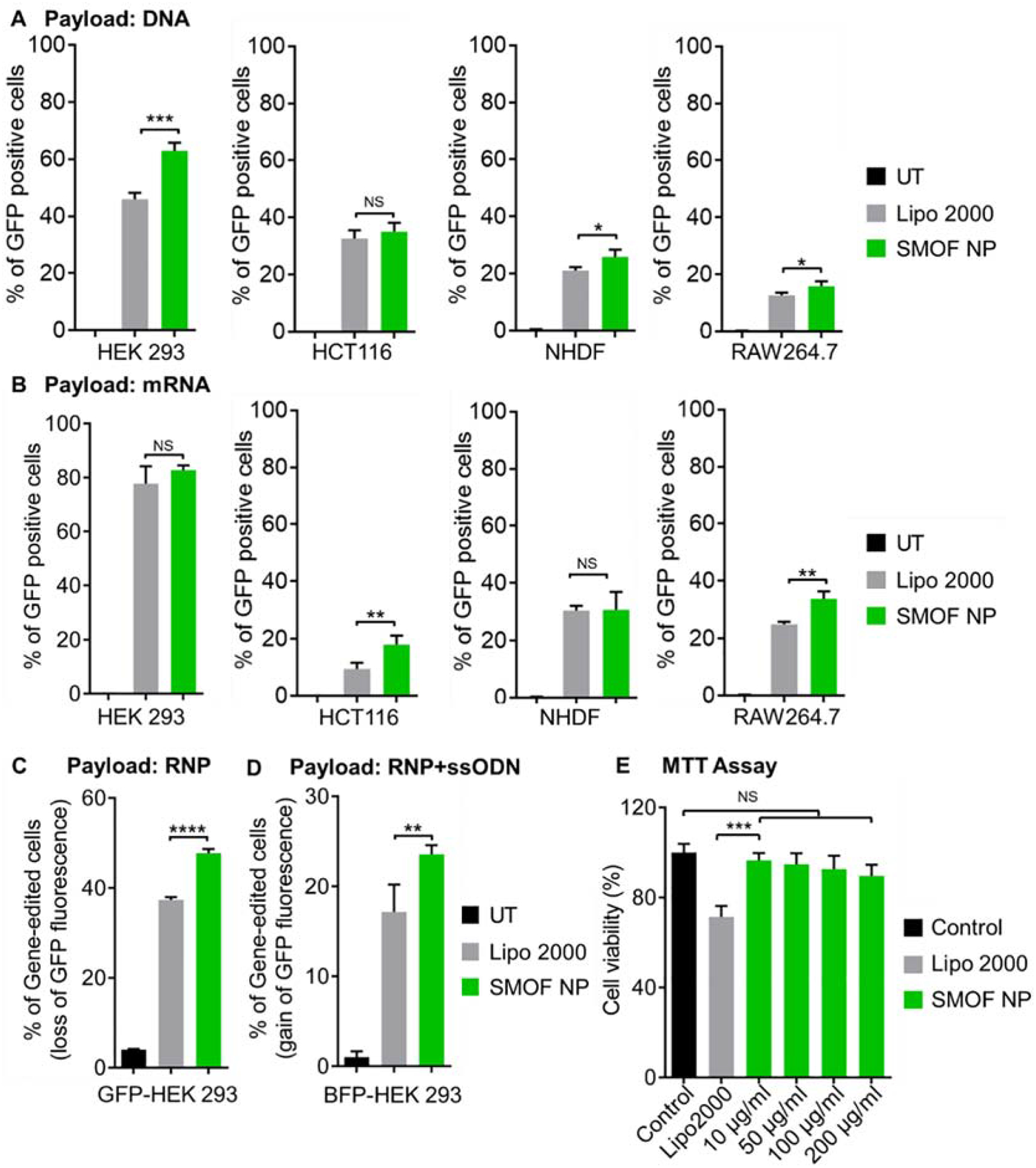
Transfection efficiency of the (A) DNA- and (B) mRNA-loaded SMOF NPs in HEK293, HCT116, NHDF, and RAW264.7 cells. (C) Genome-editing efficiency of RNP-loaded SMOF NPs in GFP-expressing HEK 293 cells. (D) Precise gene-correction efficiency of RNP+ssODN co-loaded SMOF NPs in BFP-expressing HEK 293 cells. The precise gene-correction efficiency of RNP+ssODN repair template converting the BFP to the GFP was assayed by flow cytometry for gain of GFP fluorescence. NS: not significant; *: p < 0.05; **: p < 0.01; ***: p < 0.005; n = 3. (E) Viability of HEK293 cells treated with Lipo 2000 and SMOF NPs with different concentrations. NS: not significant; ***: p < 0.001; n = 5.
Cas9 can cleave double-stranded DNA from a specific genomic locus under the guidance of sgRNA. After the double-stranded DNA break is generated, gene deletion can be achieved by the nonhomologous end-joining (NHEJ) DNA repair pathway[30, 31]. To investigate the genome-editing efficiency of RNP-loaded SMOF NPs, a sgRNA targeting the GFP gene in a transgenic GFP-expressing HEK 293 cell line was used. To enhance nuclear transportation, a Cas9 protein fused with two NLS peptides (sNLS–Cas9–sNLS) was used to form the RNP complexes. As shown in Figure 6C, SMOF NPs exhibited a statistically higher (1.3-fold) geneknockout efficiency than Lipo 2000.
Furthermore, to achieve precise genome editing by co-delivery of RNP and a donor DNA template, gene correction or insertion can be achieved through the homology-directed repair (HDR) pathway[25]. RNP and a donor single-stranded oligonucleotide DNA (ssODN) were loaded into SMOF NPs, and the precise genome-editing efficiency was studied using BFP-expressing HEK293 cells. Precise gene editing will lead to the replacement of three nucleotides in the genome, thereby converting BFP to GFP. The precise genome-editing efficiency was evaluated by the percentage of GFP-positive cells. As shown in Figure 6D, SMOF NPs showed a statistically higher (1.4-fold) gene-correction efficiency than Lipo 2000. These studies indicate that SMOF NPs are suitable for the delivery of CRISPR genome-editing machineries.
To study the biocompatibility of SMOF NPs, the cells were treated with SMOF NPs, and the cell viability was investigated by an MTT assay (Figure 6E and Figure S6). SMOF NPs did not induce significant cytotoxicity in HEK cells with concentrations up to 200 μg/ml, which was at least 9.2-fold of the concentration used for our studies. Similar to previous reports, Lipo 2000 exhibited significant cytotoxicity (with 30% cell death) at the dosage indicated in the user’s manual (i.e., 0.5 μl/well in a 96-well plate, corresponding to 5 μg/ml Lipo 2000) [32].
The genome editing efficiency of RNP-loaded SMOF NPs was further evaluated in vivo via subretinal injection in transgenic Ai14 mice (Figure. 7). All the cells of Ai14 mouse contain a CAGGS promoter and a loxP-flanked stop cassette (three repeats of the SV40 polyA sequence) that prevents expression of the tdTomato fluorescent protein, at the Rosa26 locus. The gain-of-function fluorescent signal in modified cells provides a robust and quantitative readout of genome editing at the stop cassette [27, 33, 34]. RNP targeting the excision of the SV40 polyA blocks can induce tdTomato expression (Figure. 7A). The genome editing efficiency of the RNP-loaded SMOF NPs can be easily monitored through fluorescence imaging. We studied the genome editing efficiency within the targeted retinal pigmented epithelium (RPE) of Ai14 mice, as RPE abnormality can cause a variety of eye diseases (e.g., retinal degeneration, blindness) [21]. SMOF NPs were decorated with ATRA (i.e., SMOF-ATRA). Mice were subretinally injected with SMOF-ATRA NPs loaded with RNPs targeting the Ai14 stop cassette (i.e., Ai14 RNP SMOF) and SMOF-ATRA NPs loaded with negative control RNPs (i.e., negative control SMOF) (Figure. 7B). Thirteen or fourteen days post-injection, the entire RPE tissue was separated from the enucleated eye and flat-mounted to assess genome editing via CLSM (Figure. 7C and Figure S8). As shown in Figure 7C and D, strong tdTomato signals were visualized in the eyes injected with the RNP-loaded SMOF-ATRA targeting the Ai14 stop cassette, while little tdTomato signal was found in eyes treated with negative control SMOF-ATRA (i.e., SMOF-ATRA encapsulating RNP with negative control sgRNA), indicating successful delivery of RNP and robust in vivo genome editing induced by SMOF-ATRA.
Figure 7. SMOF NPs induced efficient genome editing in vivo in Ai14 mice via local administration.
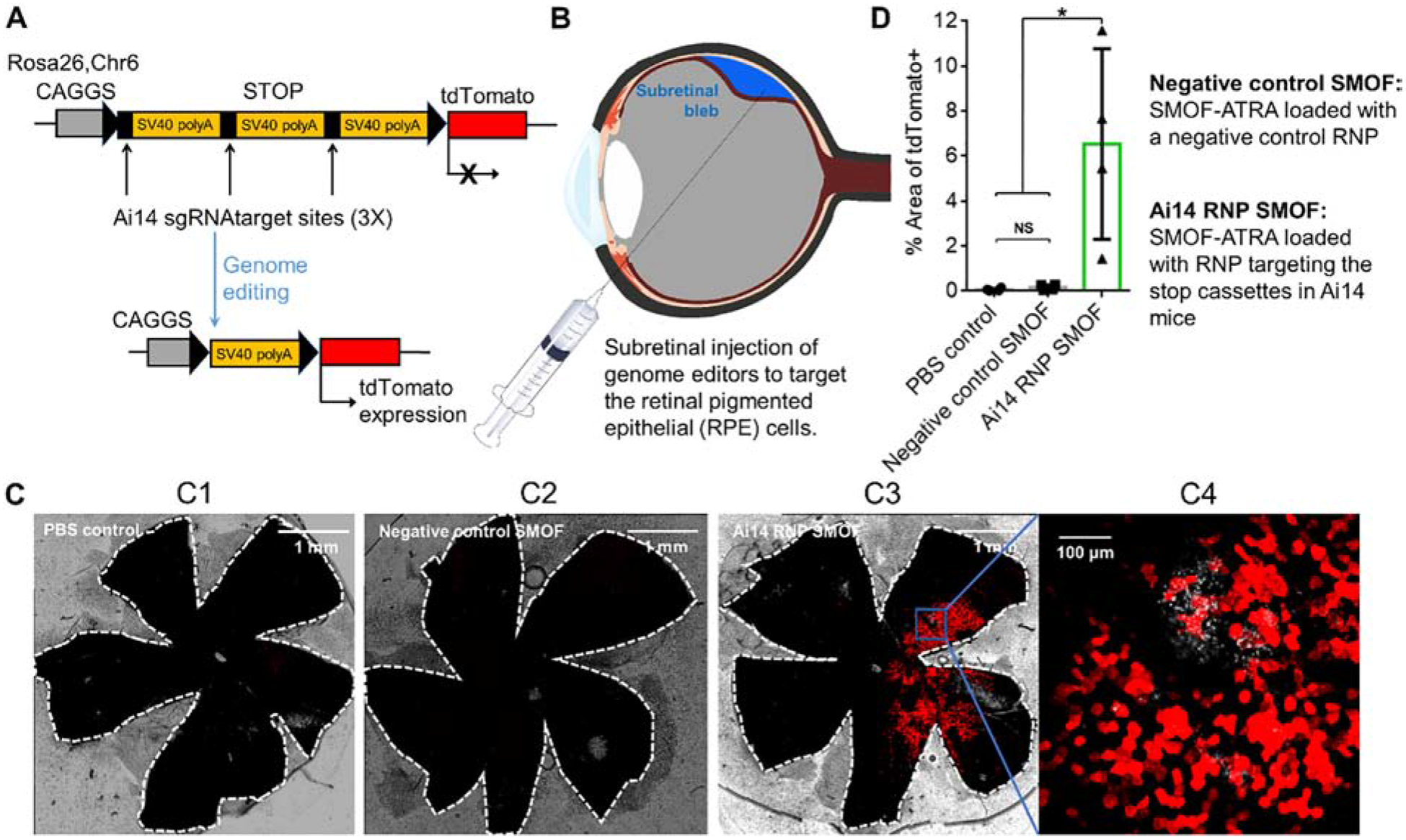
(A) The tdTomato locus in the Ai14 reporter mouse. A stop cassette containing 3 Ai14 sgRNA target sites prevents downstream tdTomato expression. RNP guided excision of the stop cassette results in tdTomato expression. (B) Illustration of SMOF NP subretinal injection targeting the RPE tissue. (C) Representative images of tdTomato+ signal (red) 13 or 14 days after subretinal SMOF-ATRA injection. The whole RPE layer was outlined with a white dotted line. C1: RPE floret of Ai14 mouse subretinally injected with PBS. C2: RPE floret of Ai14 mouse subretinally injected with negative control SMOF-ATRA (SMOF-ATRA encapsulating RNP with negative control sgRNA). C3: RPE floret of Ai14 mouse eye subretinally injected with SMOF-ATRA encapsulating RNP targeting the Ai14 stop cassette (i.e., Ai14 RNP SMOF), and C4: zoom-in image (20X magnification) of genome-edited RPE tissue induced by RNP-loaded SMOF-ATRA. (D) Genome editing efficiency as quantified by percent of the area of whole RPE tissue with tdTomato+ signals. NS: not significant, *: p<0.05.
The biocompatibility of SMOF after subretinal injection was evaluated by electroretinography (ERG) of the retina. ERG evaluates the electronic response of the eye, and any damage to the RPE or retina result in alternations in ERG recording [35–37]. PBS and Ai14 RNP-loaded SMOF-ATRA were injected to the eyes of C57BI/6 mice. As shown in Figure S9, the a-wave and b-wave amplitude of SMOF injected eyes (131.04 ± 37.16 μV, P= 0.713) were similar to the PBS injected eyes (128.1 ± 22.6 μV, P= 0.643) 14 days post-injection. The c-wave amplitude were also not statistically significant when SMOF (329.4 ± 95.2 μV, P = 0.664) was compared with the PBS control (274.2 ± 58.44 μV, P = 0.855). As there were no significant difference in ERG responses between the SMOF injected and PBS injected eyes, these findings suggest that SMOF is either nontoxic or the toxicity is negligible to the retinal/RPE cells.
4. Conclusion
In conclusion, we have demonstrated a novel and versatile silica–metal–organic framework hybrid NP for the efficient delivery of hydrophilic payloads. The protonation of the imidazole component in the SMOF NP network promoted the release and endosomal escape of the payload. SMOF NPs can encapsulate hydrophilic payloads with both a high loading content (17 wt% for small molecules and about 9.5 wt% for nucleic acids/RNP) and high loading efficiency (higher than 90%). The biocompatible and stable SMOF NPs can efficiently deliver a diverse range of hydrophilic payloads, including hydrophilic drug DOX·HCl, nucleic acids, and CRISPR-Cas9 genome-editing machineries. Furthermore, ATRA-conjugated SMOF NP also induced efficient genome editing in mouse retinal pigmented epithelium via subretinal injection, indicating that it is a highly promising nanoplatform for many biomedical applications.
Supplementary Material
Acknowledgements
We acknowledge the generous financial support from the National Institute for Health (1-UG3-NS-111688-01, R01HL129785, R01HL143469, and R01EY024995).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blanco E, Shen H, Ferrari M, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat. Biotechnol, 33 (2015) 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manzoor AA, Lindner LH, Landon CD, Park J-Y, Simnick AJ, Dreher MR, Das S, Hanna G, Park W, Chilkoti A, Overcoming limitations in nanoparticle drug delivery: triggered, intravascular release to improve drug penetration into tumors, Cancer Res, 72 (2012) 5566–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy, Nat. Rev. Genet, 15 (2014) 541–555. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Zhang L, Liu H, Cheng K, Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications, J. Controlled Release, 266 (2017) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scaletti F, Hardie J, Lee Y-W, Luther DC, Ray M, Rotello VM, Protein delivery into cells using inorganic nanoparticle–protein supramolecular assemblies, Chemical Society Reviews, 47 (2018) 3421–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piao Y, Bei HP, Tam A, Yang Y, Zhang Q, Yang M, Zhao X, Calcium Phosphate Nanoparticle-Based Systems for Therapeutic Delivery, in: Theranostic Bionanomaterials, Elsevier, 2019, pp. 147–164. [Google Scholar]
- 7.Slowing II, Vivero-Escoto JL, Wu C-W, Lin VSY, Mesoporous silica nanoparticles as controlled release drug delivery and gene transfection carriers, Adv. Drug Delivery Rev, 60 (2008) 1278–1288. [DOI] [PubMed] [Google Scholar]
- 8.Lee JE, Lee N, Kim T, Kim J, Hyeon T, Multifunctional mesoporous silica nanocomposite nanoparticles for theranostic applications, Acc. Chem. Res, 44 (2011) 893–902. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y-S, Hurley KR, Haynes CL, Critical considerations in the biomedical use of mesoporous silica nanoparticles, The journal of physical chemistry letters, 3 (2012) 364–374. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Xu Z, Chen Z, Liu X, Hou C, Zhang X, Zhang H, Fabrication of single-hole glutathione-responsive degradable hollow silica nanoparticles for drug delivery, ACS Appl. Mater. Interfaces, 6 (2014) 12600–12608. [DOI] [PubMed] [Google Scholar]
- 11.Yuan P, Zhang H, Qian L, Mao X, Du S, Yu C, Peng B, Yao SQ, Intracellular Delivery of Functional Native Antibodies under Hypoxic Conditions by Using a Biodegradable Silica Nanoquencher, Angew. Chem, 129 (2017) 12655–12659. [DOI] [PubMed] [Google Scholar]
- 12.Prasetyanto EA, Bertucci A, Septiadi D, Corradini R, Castro-Hartmann P, De Cola L, Breakable hybrid organosilica nanocapsules for protein delivery, Angew. Chem. Int. Ed, 55 (2016) 3323–3327. [DOI] [PubMed] [Google Scholar]
- 13.Horcajada P, Chalati T, Serre C, Gillet B, Sebrie C, Baati T, Eubank JF, Heurtaux D, Clayette P, Kreuz C, Porous metal–organic-framework nanoscale carriers as a potential platform for drug delivery and imaging, Nature materials, 9 (2010) 172. [DOI] [PubMed] [Google Scholar]
- 14.Della Rocca J, Liu D, Lin W, Nanoscale metal–organic frameworks for biomedical imaging and drug delivery, Acc. Chem. Res, 44 (2011) 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu MX, Yang YW, Metal–organic framework (MOF)-based drug/cargo delivery and cancer therapy, Adv. Mater, 29 (2017) 1606134. [DOI] [PubMed] [Google Scholar]
- 16.Doonan C, Ricco R, Liang K, Bradshaw D, Falcaro P, Metal–organic frameworks at the biointerface: synthetic strategies and applications, Acc. Chem. Res, 50 (2017) 1423–1432. [DOI] [PubMed] [Google Scholar]
- 17.Horcajada P, Gref R, Baati T, Allan PK, Maurin G, Couvreur P, Ferey G, Morris RE, Serre C, Metal–organic frameworks in biomedicine, Chem. Rev, 112 (2011) 1232–1268. [DOI] [PubMed] [Google Scholar]
- 18.Alsaiari SK, Patil S, Alyami M, Alamoudi K, f. Aleisa, J. Merzaban, M. Li, N.M. Khashab, Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework, J. Am. Chem. Soc, (2017). [DOI] [PubMed] [Google Scholar]
- 19.Chen T-T, Yi J-T, Zhao Y-Y, Chu X, Biomineralized metal–organic framework nanoparticles enable intracellular delivery and endo-lysosomal release of native active proteins, J. Am. Chem. Soc, 140 (2018) 9912–9920. [DOI] [PubMed] [Google Scholar]
- 20.Poddar A, Conesa JJ, Liang K, Dhakal S, Reineck P, Bryant G, Pereiro E, Ricco R, Amenitsch H, Doonan C, Encapsulation, Visualization and Expression of Genes with Biomimetically Mineralized Zeolitic Imidazolate Framework-8 (ZIF-8), Small, (2019) 1902268. [DOI] [PubMed] [Google Scholar]
- 21.Berger W, Kloeckener-Gruissem B, Neidhardt J, The molecular basis of human retinal and vitreoretinal diseases, Prog Retin Eye Res, 29 (2010) 335375. [DOI] [PubMed] [Google Scholar]
- 22.Sun D, Sahu B, Gao S, Schur RM, Vaidya AM, Maeda A, Palczewski K, Lu Z-R, Targeted multifunctional lipid ECO plasmid DNA nanoparticles as efficient non-viral gene therapy for leber’s congenital amaurosis, Molecular Therapy-Nucleic Acids, 7 (2017) 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson A, Bok D, Promotion of the release of 11-cis-retinal from cultured retinal pigment epithelium by interphotoreceptor retinoid-binding protein, Biochemistry-us, 31 (1992) 9056–9062. [DOI] [PubMed] [Google Scholar]
- 24.Yuan P, Mao X, Chong KC, Fu J, Pan S, Wu S, Yu C, Yao SQ, Simultaneous Imaging of Endogenous Survivin mRNA and On-Demand Drug Release in Live Cells by Using a Mesoporous Silica Nanoquencher, small, 13 (2017) 1700569. [DOI] [PubMed] [Google Scholar]
- 25.Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K, Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing, Nat. Commun, 8 (2017) 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE, Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA, Nat. Biotechnol, 34 (2016) 339. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Abdeen AA, Wang Y, Shahi PK, Robertson S, Xie R, Suzuki M, Pattnaik BR, Saha K, Gong S, A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing, Nature nanotechnology, 14 (2019) 974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larguinho M, Santos HM, Doria G, Scholz H, Baptista PV, Capelo JL, Development of a fast and efficient ultrasonic-based strategy for DNA fragmentation, Talanta, 81 (2010) 881–886. [DOI] [PubMed] [Google Scholar]
- 29.Pchelintsev NA, Adams PD, Nelson DM, Critical parameters for efficient sonication and improved chromatin immunoprecipitation of high molecular weight proteins, PloS one, 11 (2016) e0148023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doudna JA, Charpentier E, The new frontier of genome engineering with CRISPR-Cas9, Science, 346 (2014) 1258096. [DOI] [PubMed] [Google Scholar]
- 31.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini L, Multiplex genome engineering using CRISPR/Cas systems, Science, (2013) 1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Ma B, Abdeen AA, Chen G, Xie R, Saha K, Gong S, Versatile Redox-Responsive Polyplexes for the Delivery of Plasmid DNA, Messenger RNA, and CRISPR-Cas9 Genome-Editing Machinery, ACS Appl. Mater. Interfaces, 10 (2018) 31915–31927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, Urnes C, Munares GA, Ghosh A, Doudna JA, Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes, Nat. Biotechnol, 35 (2017) 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, A robust and high-throughput Cre reporting and characterization system for the whole mouse brain, Nat. Neurosci, 13 (2010) 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai TYY, Chan W-M, Lam DSC, Transient reduction in retinal function revealed by multifocal electroretinogram after photodynamic therapy, Am J Ophthalmol, 137 (2004) 826–833. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson SG, Acland GM, Aguirre GD, Aleman TS, Schwartz SB, Cideciyan AV, Zeiss CJ, Komaromy AM, Kaushal S, Roman AJ, Safety of recombinant adeno-associated virus type 2–RPE65 vector delivered by ocular subretinal injection, Mol. Ther, 13 (2006) 1074–1084. [DOI] [PubMed] [Google Scholar]
- 37.Becker S, Wang H, Stoddard GJ, Hartnett ME, Effect of subretinal injection on retinal structure and function in a rat oxygen-induced retinopathy model, Mol Vis, 23 (2017) 832. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


