Informational polymers are deciphered by aerolysin pores.
Abstract
Digital data storage is a growing need for our society and finding alternative solutions than those based on silicon or magnetic tapes is a challenge in the era of “big data.” The recent development of polymers that can store information at the molecular level has opened up new opportunities for ultrahigh density data storage, long-term archival, anticounterfeiting systems, and molecular cryptography. However, synthetic informational polymers are so far only deciphered by tandem mass spectrometry. In comparison, nanopore technology can be faster, cheaper, nondestructive and provide detection at the single-molecule level; moreover, it can be massively parallelized and miniaturized in portable devices. Here, we demonstrate the ability of engineered aerolysin nanopores to accurately read, with single-bit resolution, the digital information encoded in tailored informational polymers alone and in mixed samples, without compromising information density. These findings open promising possibilities to develop writing-reading technologies to process digital data using a biological-inspired platform.
INTRODUCTION
DNA has evolved to store genetic information in living systems; therefore, it was naturally proposed to be similarly used as a support for data storage (1–3), given its high-information density and long-term storage (4) with respect to existing technologies based on silicon and magnetic tapes. Alternatively, synthetic informational polymers have also been described (5–9) as a promising approach allowing digital storage. In these polymers, information is stored in a controlled monomer sequence, a strategy that is also used by nature in genetic material. In both cases, single-molecule data writing is achieved mainly by stepwise chemical synthesis (3, 10, 11), although enzymatic approaches have also been reported (12). While most of the progress in this area has been made with DNA, which was an obvious starting choice, the molecular structure of DNA is set by biological function, and therefore, there is little space for optimization and innovation. Alternatively, precise synthesis of sequence-defined abiotic polymers has recently opened up the possibility to tune important parameters such as storage capacity, storage density, erasability, readability, and writing speed (5, 7, 8).
Decoding/reading these data can be achieved for DNA using a variety of sequencing techniques (13), whereas synthetic polymers are, so far, only deciphered by tandem mass spectrometry, a method that is efficient but requires large instruments (14, 15). Nanopores, the next generation of sequencing tool (13, 16), have been exploited for efficiently reading DNA based on the principle that translocation of different nucleobases through a nanopore generates a specific ionic current signature. More recently, nanopores have been explored for sensing digitally encoded DNA nanostructures (17, 18). Inspired by these recent advances, here, we encoded binary information in synthetic polymers tailor-made to be decoded using an engineered pore-forming toxin (PFT), aerolysin (19). By a rational and synergetic development of the pore readout (using specific aerolysin mutants) and the tailored digital analyte (using both DNA nucleotides and non-biological monomers), the translocation speed of the polymer was optimized to have a uniquely identifiable level-by-level signal, which delivered digital reading with single-bit resolution without compromising information density. Current DNA sequencing technologies based on biological nanopores (e.g., using Mycobacterium smegmatis porin A) are still limited by low base-calling accuracy as the maximal resolution is determined by the nearest four nucleotides translocating within the nanopore-sensing constriction (quadromers) (20, 21). Here, we show that aerolysin pores have the potential to achieve the molecular equivalent of single-base resolution for tailored digital analytes, which, in turn, allows for single-bit reading accuracy. Using deep learning, we were able to decode digital sequences encoding up to 4-bit information with a high accuracy while blindly detect the identity and relative concentration of polymer mixtures.
RESULTS AND DISCUSSION
Reading single-bit information encoded in polymers
Single-channel recording experiments were performed to analyze polymer translocation using the PFT aerolysin from Aeromonas hydrophila (Fig. 1A). Aerolysin is one of the best characterized among PFTs, it oligomerizes into a heptameric pore that features a novel and unique fold, constituted by two concentric β barrels held together by hydrophobic interactions (22–25). Aerolysin has been proposed to be a promising nanopore sensor, exhibiting high sensitivity for biomolecular detection and providing excellent current separation and a dwell time range suitable for accurate signal processing (26, 27). Using wild-type aerolysin, four types of nucleobase (28) and 13 natural amino acid have been identified directly (29), establishing this biological pore as a natural candidate for DNA and protein sequencing. Recently, aerolysin mutants have been rationally designed to further enhance the sensing properties of the wild-type pore (19). Notably, the K238A mutant has shown a significantly enhanced resolution for biomolecular recognition and turned out to be, likely because of a partial opening of one of the two main sensitive constrictions (Fig. 1A), the most suitable sensing system to detect the tailor-made molecular structure of the digitally encoded polymers ad hoc developed in this work.
Fig. 1. Aerolysin reading of polymers encoding single-bit information.
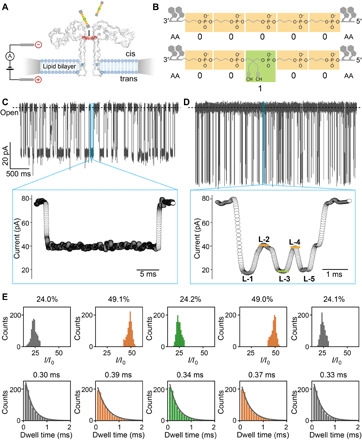
(A) Illustration of single-channel recording setup using an aerolysin pore; the cis and trans chambers are filled with 1.0 M KCl electrolyte buffer, and voltage is applied across the pore using two Ag/AgCl electrodes. Amino acid at 220 and 238 position are highlighted by red and blue, respectively. (B) Schematic structure of two representative polymers: AA00000AA and AA00100AA. (C) Raw current trace of AA00000AA during single-channel recording measurement (top). Magnification of one single event (bottom). (D) Raw current trace of AA00100AA measurement. Magnification of one single event (bottom) showing a multilevel signal: L-1 (gray), L-2 (orange), L-3 (green), L-4 (orange), and L-5 (gray). (E) I/I0 histogram (top) and dwell time distribution (bottom) for L-1, L-2, L-3, L-4, and L-5, respectively. Relative fitted values are reported in each figure. All data were obtained using 1.0 M KCl, 10 mM tris, and 1.0 mM EDTA at pH 7.4, applying a bias potential of 100 mV.
These macromolecules are sequence-defined poly(phosphodiester)s prepared by automated phosphoramidite chemistry, as previously reported (30, 31) (Fig. 1B and fig. S1). The negatively charged backbone of these polymers ensures an efficient aerolysin capture and the efficient translocation from the cis to trans compartment under an applied voltage (32). The polymers were digitally encoded using two monomers of different molecular structure. In the formed chains, the synthons n-propyl-phosphate and (2,2-dipropargyl)-propyl-phosphate represent bit-0 and bit-1, respectively. It shall be noted that this binary alphabet differs from the one that is usually used for mass spectrometry sequencing (33). Here, two monomers of markedly different bulkiness were selected to induce different pore current responses. Furthermore, automated phosphoramidite chemistry allows the use of both biological (i.e. natural DNA nucleotides) (10) and nonbiological monomers (30). Thus, in the present work, biohybrid macromolecules, composed of the aforementioned nonnatural binary alphabet and natural nucleotides, were examined for optimal pore translocation. All the digitally encoded polymers examined in this study are listed in table S1, and their complete characterization is reported in figs. S40 to S85.
Digital decoding in aerolysin pores was first attempted with copolymers containing only bit-0 and bit-1 monomers. The negatively charged backbone of these polymers not only ensures efficient translocation but also speeds up the crossing time that would be too fast to allow decoding (32). As no signals were observed during the single-channel recording after addition of “00000” polymer in the cis side of the chamber, and the signal-to-noise-ratio was too low for “11111” (fig. S2), we introduced a di-deoxyadenosine at the polymer terminals (“AA00000AA,” Fig. 1B). This addition created highly detectable current blockade signals (39.0 ± 3.0 pA as mean residual current, with an open pore current of 76.0 ± 3.0 pA, and 18.9 ± 1.3 ms for dwell time), easily identifying the translocation of “AA00000AA” polymers (Fig. 1C). When a bit-1 monomer was inserted to create an “A00100AA” polymer, the additional moiety induced an obvious lowering of the current levels (Fig. 1D). Compared to AA00000AA, a clear decrease in the mean dwell time (2.8 ± 0.1 ms) was observed, and a fraction of the events (~28%) clearly showed a five-level signal (labeled L-1 to L-5 in Fig. 1D and fig. S3). To understand the relationship between the polymer chemical nature and current levels, we collected more than 10,000 blockade current events for statistical analysis (see Materials and Method and fig. S4). The relative current (I/I0), fitted as Gaussian distribution, showed five distinct levels of values: 24.0 ± 3.2%, 49.1 ± 2.4%, 24.2 ± 2.3%, 49.0 ± 2.5%, and 24.1 ± 2.4%, respectively (Fig. 1E; I0 is the value of the open pore current, and I is the residual current value). The dwell times of each level, fitted with an exponential function, have values of 0.30 ± 0.01, 0.39 ± 0.01, 0.34 ± 0.01, 0.37 ± 0.01, and 0.33 ± 0.01 ms, respectively. The I/I0 values of L-2 and L-4 are nearly identical, while L-1, L-3, and L-5 are also quite similar, and all the current states share a similar characteristic dwell time (Fig. 1E). According to our previous studies, there is a strong correlation between the physical size of the translocated molecules and I/I0 values (28); therefore, L-1 and L-5 can be likely interpreted as the blockade caused by the added volume of the di-deoxyadenosine moieties at the two terminals, while similarly L-3 to the bulky (2,2-dipropargyl)-propyl-phosphate (bit-1). The two lighter n-propyl-phosphates flanked between these bulkier groups (Fig. 1B) contribute instead to higher L-2 and L-4 current states. Therefore, the strategy of including nucleotides in our informational polymer not only prolonged the dwell time but also enhanced the potential resolution of the system to single-bit precision.
Effects of terminal nucleobases on aerolysin capability for polymer reading
To optimize the polymer design and further understand the influence of the terminal nucleotides for decoding, a series of polymers that replaced the terminal di-deoxyadenosine groups with other types of dinucleotides were tested. According to our previous studies, due to backbone stereochemistry, single-stranded DNA prefers to enter the aerolysin pore from the 3′-end (34) (thus all polymers are oriented starting from the 3′-end; Fig. 1B). Translocation events of these polymers showed a qualitatively similar five-level signal (Fig. 2A). While L-1 values for AA00100AA and AA00100CC are nearly identical, in CC00100AA, the relative current is higher, demonstrating eventually that the first current blockade is associated to the 3′ nucleobase and its chemical nature. Among all polymers, L-1 and L-5 of CC00100CC are the highest peaks, which further supports the hypothesis that the first and last current levels are induced by nucleobases at the terminals. Therefore, aerolysin nanopores are able to read not only tailor-made informational polymers but also different types of DNA bases and their order at the terminals. To systemically evaluate the enhancement of different dinucleotides for separation between bit-1 and bit-0 monomers, we compared the mean dwell time (i.e., longer dwell time allows a more accurate determination of blockade current levels) and mean current variation (i.e., higher variation promotes a higher read accuracy for each bit, see Materials and Methods) of all polymers with different terminals (Fig. 2B). As di-deoxyadenosine at both terminals showed the longest dwell time and highest current variation among all polymers, it was chosen as the basic terminal block for the following design.
Fig. 2. Effects of terminal nucleobases on polymer reading.
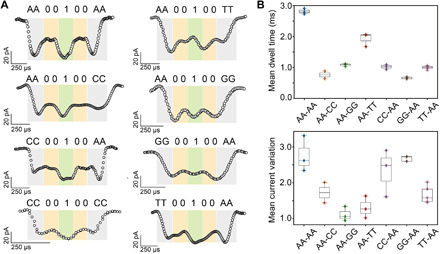
(A) Representative translocation events of the digital macromolecular analyte with different types of nucleotides at the chain termini. Colors scheme as in Fig. 1. (B) Mean dwell time and mean current variation for all polymers with different types of terminal dinucleotides. All data were obtained using 1.0 M KCl, 10 mM tris, and 1.0 mM EDTA buffer at pH 7.4, applying a bias potential of 100 mV. Each value is an average obtained from at least three separate pore measurements.
Decoding polymer sequences by engineered aerolysin pores using deep learning
We then tested the sensitivity of the pore for detecting bit-1 monomers when spanning the five available positions along the n-propyl-phosphate backbone (i.e., AA10000AA, AA01000AA, AA00100AA, AA00010AA, and AA00001AA). For this task, we developed a deep learning approach to process the current signal, which was able to automatically classify a much larger fraction of events (~40%) with high accuracy (~84%; fig. S5). We used long short-term memory (LSTM) (35) recurrent neural network to read the events local extrema followed by a multilayer perceptron (MLP) to classify the polymers (Fig. 3A). These additional results showed that the detection of bit-1 monomers is difficult when flanking directly the terminal nucleobases (fig. S6). When only the innermost positions are considered (i.e., as in AA01000AA, AA00100AA, and AA00010AA), the resulting accuracy of the neural network is as high as 97.6% (Fig. 3B); therefore, these positions were only used for single-bit reading, hereafter indicated in bold (e.g., AA00000AA). We tested between AA0- and -0AA flanking terminals the polymers encoding for 1 to 4 bits of information generating a library of 30 different polymer sequences (i.e., 21 + 22 + 23 + 24; Fig. 3C and figs. S7 to S34). While each of these 30 polymers were measured by a single nanopore independently, we report a reading accuracy of 99.0% for 1-bit polymers, 99.0% for 2-bit, 98.0% for 3-bit, and 86.5% for 4-bit; the accuracy is slightly decreasing for longer polymers that host a larger number of combinations and with higher event selection thresholds (Fig. 3C and figs. S35 to S38).
Fig. 3. Decoding polymer sequences using aerolysin pores and deep learning.
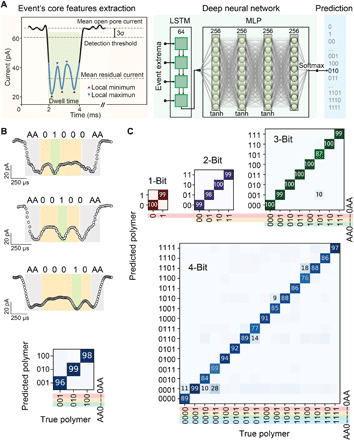
(A) Details for nanopore signal processing and relative deep learning workflow. (B) Characteristic translocation events of polymers containing bit-1 at different positions, i.e., A01000AA, AA00100AA, and AA00010AA, and the corresponding confusion matrix results obtained by deep learning. (C) Confusion matrix of 1-, 2-, 3-, and 4-bit polymer sequences classification shown for a selection percentage of 10% (see figs. S35 to S38 for dependence on selection threshold). Columns represent true polymers from the test set, while rows are the polymers that deep learning assigned them to. All data were obtained using 1.0 M KCl, 10 mM tris, and 1.0 mM EDTA buffer at pH 7.4, applying a bias potential of 100 mV.
To better evaluate the reading capability of aerolysin nanopores, we performed a statistical analysis across N = 546 separate nanopore measurements of all 30 polymers (~6.6 M events in total; Fig. 4A). Notably, few 5-bit and 6-bit polymers were also included to expand the polymer landscape. In general, the averaged dwell time decreased as polymer length increased, tending nonetheless to a value of 2.5 ± 0.8 ms. In particular, if the total coding length is shorter than 3 bits, the dwell time is longer, likely due to the presence of a less abundant negatively charge density per bit, which is steered slower by the applied voltage; while dwell times tend to converge to higher value when the polymer length is longer than 4 bits, and charge density is less affected by the terminal groups (Fig. 4A). On the other hand, although 4-bit polymer dwell times decreased, their current variation increased (Fig. 4B), indicating that more information is encoded in the longer polymers.
Fig. 4. Statistical analysis and assignment of specific polymer’s identity and relative concentration in a mixture.
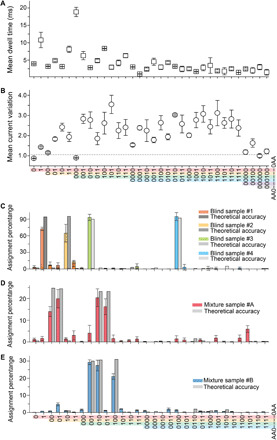
Statistical analysis of (A) mean dwell time and (B) current variation of all polymers. (C) Assignment percentage of blind polymer samples #1 (orange), #2 (yellow), #3 (green), and #4 (blue), respectively. Assignment percentage of mixture sample #A (D) and mixture sample #B (E). The theoretical accuracy is shown by the gray columns. All data were obtained using 1.0 M KCl, 10 mM tris, and 1.0 mM EDTA buffer at pH 7.4, applying a bias potential of 100 mV.
On the basis of the model generated by deep learning, the expectation is that any item in the library of polymer sequences can be identified directly with high confidence. To test this hypothesis, we performed blind tests to identify the given polymers and their relative concentration when mixed. Following this blind procedure, we were able to correctly detect polymer “AA010AA” among all 30 polymers, with a percentage of 72.0 ± 3.0% (Fig. 4C, orange), which is close to the predicted accuracy of the deep learning model (94.0%, fig. S39). Similarly, we correctly assigned polymer “AA0100AA” with an accuracy of 64.0 ± 5.0% (yellow), “AA00010AA” with an accuracy of 93.2 ± 5.2% (green), and “AA001000AA” with an accuracy of 94.5 ± 4.2% (blue). The high-assignment percentages are consistent with the prediction accuracy predicted by deep learning (i.e., 95.0, 90.0, and 92.0%, respectively). We then measured a mixture sample (#A) in which polymers AA0000AA, AA0010AA, AA00100AA, and AA00110AA were blindly mixed at equimolar ratio, recapitulating the composition with an accuracy of 14.0 ± 2.0%, 20.0 ± 4.0%, 20.0 ± 4%, and 16.0 ± 4%, respectively (Fig. 4D). This is similar to the accuracy for a 30-polymer classification given an equimolar concentration ratio of the four polymers (i.e., 24.7, 24.5, 23.2, and 23.2%). Furthermore, a second mixture (#B) containing AA00010AA, AA00100AA, and AA01000AA in an equimolar ratio was tested, and an approximately equal assignment was observed (i.e., 29.4 ± 1.4%, 27.4 ± 3.1%, and 21.0 ± 1.6%), similar to the predicted accuracy obtained by deep learning (30.0, 30.6, and 31.0%; Fig. 4E).
Conclusion
In conclusion, we have demonstrated that tailor-made informational polymers can be efficiently decoded by using a specific variant of the aerolysin pore (K238A). In particular, the design of an optimal bio-inspired writing-reading framework allowed for single-bit resolution, which is unprecedented in analytical chemistry. The aerolysin pore structure can, in principle, be further tuned by engineering its main two sensitive constrictions to optimize the translocation for efficient reading. On the other hand, the vast chemical space accessible to informational polymers can be further explored to enhance optimal decoding by biological nanopore. Informational polymers hybridized with DNA nucleobases keep some of the advantages of synthetic DNA used as support for data storage (36). For instance, different terminal nucleobases, which allow for more efficient capture by the nanopore, can be readily discriminated (Fig. 2), opening the possibility to use canonical DNA bases to define data structure in a format that can enable random access (37–39).
Writing-reading digital data using this biological-inspired nanopore-based platform can offer numerous advantages. First, single-bit resolution on the proposed informational polymer theoretically provides the opportunity to increase the information density of existing DNA-based solutions. For instance, the best resolution in solid-state nanopore platform for digital reading of dsDNA is 8-bps difference for an average mass of ~5 kDa (17), whereas the mass difference between different bit units in our informational polymers is ~70-fold lower (76 Da). Similarly, the minimum distance between two bits in solid-state nanopore solutions is 76 bps, while in the aerolysin framework, we can achieve the molecular equivalent of single-base resolution (140 Da, Fig. 2). Second, nanopore sensing does not require additional labeling, and there is no theoretical upper limit for the reading length (40), further reducing the overall cost and workflow time. Nanopore sensing, which relies on an electrical readout, naturally enables large-scale parallelization based on already established technologies (41), thus allowing the construction of more affordable and portable devices for data management.
Toward practical polymer-based data storage, the next important challenge will entail sequencing longer polymers for which the translocation mechanism has to be optimized to preserve as much as possible single-bit resolution and information density. Possible solutions include (i) reducing negative charge density to obtain longer dwell times for precise reading, (ii) optimizing interbit separation to eliminate the influence from neighboring subunit, and (iii) increasing the rigidity of the polymer backbone to reduce the signal noise. One benefit of informational polymers is that they can be better tailored on the properties of the reader; therefore, new definitions of fundamental bit units and blocks, as well as their sequential assembly or controlled fragmentation, can be designed ad hoc to achieve efficient data storage and retrieval (33, 42, 43). In summary, this bio-inspired platform based on hybrid DNA-polymer analytes that encode digital information read by a biological nanopore sensor opens up new promising perspectives for polymer-based memories, with important advantages for ultrahigh density, long-term storage, and device portability.
MATERIALS AND METHODS
Synthesis of the macromolecular analytes
The polymers used in the nanopore experiments were synthesized by automated phosphoramidite chemistry on an Expedite DNA synthesizer (Perseptive Biosystem 8900), as previously described (30, 31). All polymers were characterized by electrospray ionization–high resolution mass spectrometry (HRMS) (table S1 and figs. S40 to S82), and their purity was controlled by anion-exchange high-performance liquid chromatography (figs. S83 to S85), on an Agilent Apparatus equipped with a column Dionex BioLC DNAPac-PA100 and ultraviolet detectors (260 and 280 nm).
Aerolysin productions
The aerolysin full-length sequence was cloned in the pET22b vector with a C-terminal hexahistidine tag to aid purification as described in our previous work (19). The QuikChange II XL Kit from Agilent Technologies was used for performing site-directed mutagenesis on the aerolysin gene, following manufacturer’s instructions. The recombinant protein K238A was expressed and purified from BL21 DE3 pLys Escherichia coli cells. Cells were grown to an optical density of 0.6 to 0.7 in Luria-Bertani media. Protein expression was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and subsequent growth over night at 20°C. Cell pellets were resuspended in lysis buffer [20 mM sodium phosphate (pH 7.4) and 500 mM NaCl], mixed with cOmplete Protease Inhibitor Cocktail (Roche), and then lysed by sonication. The resulting suspensions were centrifuged (12,000 rpm for 35 min at 4°C), and the supernatants were applied to an HisTrap HP column (GE Healthcare) previously equilibrated with lysis buffer. The protein was eluted with a gradient more than 40 column volumes of elution buffer [20 mM sodium phosphate (pH 7.4), 500 mM NaCl, and 500 mM imidazole], and buffer was exchanged into final buffer (20 mM tris and 500 mM NaCl at pH 7.4) using a HiPrep Desalting column (GE Healthcare). The purified protein was flash-frozen in liquid nitrogen and stored at −20°C.
Single-channel recording experiments
Phospholipid of 1,2-diphytanoyl-sn-glycero-3-phosphocholine powder (Avanti Polar Lipids Inc., Alabaster, AL, USA) was dissolved in octane (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) for a final concentration of 1.0 mg per 100 μl. Purified K238A aerolysin mutant was diluted to the concentration of 0.2 μg/ml and then incubated with Trypsin-agarose (Sigma-Aldrich Chemie GmbH, Buchs, SG Switzerland) for 2 hours under 4°C temperature to activate the toxin for oligomerization. The solution was finally centrifuged to remove trypsin.
Nanopore single-channel recording experiments were performed by Orbit Mini equipment (Nanion, Munich, Germany). Phospholipid membranes were formed across a MECA 4 recording chip that contains a 2 × 2 array of circular microcavities in a highly inert polymer. Each cavity contains an individual integrated Ag/AgCl microelectrode and is able to record four artificial lipid bilayers in parallel. The measurement chamber temperature was set to 25°C for all experiments.
Polymers in powder form were prediluted in water, to a stock concentration of 2.0 mg/ml, and added to the cis side of the chamber in 1.0 M KCl solution buffered with 10 mM tris and 1.0 mM EDTA (pH 7.4) to the final concentration of 20 μmol. All experiments shown here were repeated in 3 to 15 different pores.
Current signal processing
The raw signals are segmented on the basis of voltage discontinuities and large time-scale discontinuities to separate the signals segments where the pore is blocked or where a second pore is inserted into the membrane. For each segment, the open pore current distribution is measured by fitting a Gaussian function on the peak distribution of current with the highest mean current. The signal segments with an open pore current distribution of mean between 67 to 98 pA and SD between 1.5 to 4.2 pA are kept.
The events are extracted using a current threshold at 3σ from the open-pore current distribution (Fig. 3A). The relative current is computed from the mean open pore current (I0). The cores of the events are extracted by removing the current drop at the beginning and end of the events using an adaptive current threshold. The dwell time, average relative current, current variation (σ0 is the value of the open pore current SD, and σ is the residual current SD), and local extrema are computed. The events are selected on the basis of the dwell time (0.4 to 30.0 ms) and the average relative current (15 to 60%) discarding the events that are too short or too long and removing the outliers. In average, this initial filtering procedure discards ~10% of the events (fig. S4).
To detect and label different level in the signal, the local relative current extrema are used to generate a Gaussian mixture model (GMM) with three components: low, high, and transition level. The low and high Gaussian models correspond to the two main modes of the relative current extrema distribution. The transition level describes possible change of state between high and low level. Each event is segmented into low, high, and transition levels of based on the level type with the highest probability predicted by the GMM. Last, the transition levels, which are not transition between high and low such as high-transition-high and low-transition-low, are merged into a single high and low level, respectively.
Last, to classify the current events, a machine learning approach was devised including two steps. The first one is the classification of every events, and the second is the assessment of the quality of the prediction of the classifier (Fig. 3A). The neural network architecture for both the classification and the assessment is an LSTM (35) neural network followed by an MLP using the position in time and relative current of the local extrema for each event as input features. The features are rescaled by a fixed factor to decrease the training time. The classifier is composed of a LSTM with state size of 64 without any activation function, followed by a four fully connected hidden layers of size 256 with hyperbolic tangent as activation functions, and, last, an output layer of size 30 with softmax activation function. The neural networks for the classification and assessment are trained together using a three-part loss functions. The first part is the full-classification cross-entropy loss of the predictions from the classifier and the polymers label. The second part is the assessment cross-entropy loss between the predicted and actual prediction validity from the classifier. The third part is the reinforcement classification loss, which is the full classification cross-entropy loss scaled by the assessment prediction.
Supplementary Material
Acknowledgments
Funding: This research was supported by the Swiss National Science Foundation and EPFL (to M.D.P.) and the European Union’s Horizon 2020 Research and innovation program under the Marie Skłodowska-Curie grant agreement no. 665667 (to C.C.). A.R. acknowledges the support from Swiss National Science Foundation (grant number BSCGI0_157802). J.-F.L. thanks the H2020 program of the European Union (project Euro-Sequences, H2020-MSCA-ITN-2014, grant agreement n°642083) and the CNRS for financial support. The PhD position of N.F.K. was supported by the ITN Euro-Sequences. The authors also thank L. Charles and C. Chendo (Aix Marseille Université) for the electrospray ionization–HRMS analysis of the polymers and M. J. Marcaida for support with aerolysin production. Material can be provided by M.D.P. (pending scientific review and a completed material transfer agreement). Requests for material should be submitted to matteo.dalperaro@epfl.ch. Author contributions: C.C., A.R., J.-F.L., and M.D.P. conceived the idea and designed the study. J-F.L., A.A.O. and N.F.K. conceptualized the digitally-encoded polymers. C.C. and M.D.P. rationally designed the aerolysin readout. C.C. performed the single-channel recording experiments, collected, and analyzed the data. L.F.K. performed signal processing using deep learning. A.A.O. and N.F.K. synthesized and characterized the polymers. N.C. conducted and analyzed molecular modeling and simulations. C.C., L.F.K., A.A.O, A.R., J-F.L., and M.D.P. interpreted the data. C.C., A.R., J-F.L., and M.D.P. wrote the manuscript with input from all authors. Competing interests: M.D.P., C.C., A.R., L.K., J.-F.L., A.A.O., and N.F.K. are inventors on a patent application related to this work filed by Ecole Polytechnique Federale De Lausanne (EPFL) and Centre National de la Recherche Scientifique (no. PCT/EP2020/077229, filed 29 September 2020). The authors declare that they have no other competing interests related to this work. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eabc2661/DC1
REFERENCES AND NOTES
- 1.Church G. M., Gao Y., Kosuri S., Next-generation digital information storage in DNA. Science 337, 1628 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Davis J., Microvenus. Art J. 55, 70–74 (1996). [Google Scholar]
- 3.Lutz J.-F., Ouchi M., Liu D. R., Sawamoto M., Sequence-controlled polymers. Science 341, 1238149 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Grass R. N., Heckel R., Puddu M., Paunescu D., Stark W. J., Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. 54, 2552–2555 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Rutten M. G. T. A., Vaandrager F. W., Elemans J. A. A. W., Nolte R. J. M., Encoding information into polymers. Nat. Rev. Chem. 2, 365–381 (2018). [Google Scholar]
- 6.Trinh T. T., Oswald L., Chan-Seng D., Lutz J.-F., Synthesis of molecularly encoded oligomers using a chemoselective “AB + CD” iterative approach. Macromol. Rapid Commun. 35, 141–145 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Roy R. K., Meszynska A., Laure C., Charles L., Verchin C., Lutz J.-F., Design and synthesis of digitally encoded polymers that can be decoded and erased. Nat. Commun. 6, 7237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martens S., Landuyt A., Espeel P., Devreese B., Dawyndt P., Du Prez F., Multifunctional sequence-defined macromolecules for chemical data storage. Nat. Commun. 9, 4451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan J., Zhang F., Karcher D., Bock R., Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 10, 439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caruthers M., Gene synthesis machines: DNA chemistry and its uses. Science 230, 281–285 (1985). [DOI] [PubMed] [Google Scholar]
- 11.Lutz J.-F., Lehn J.-M., Meijer E. W., Matyjaszewski K., From precision polymers to complex materials and systems. Nat. Rev. Mater. 1, 16024 (2016). [Google Scholar]
- 12.Lee H. H., Kalhor R., Goela N., Bolot J., Church G. M., Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat. Commun. 10, 2383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shendure J., Balasubramanian S., Church G. M., Gilbert W., Rogers J., Schloss J. A., Waterston R. H., DNA sequencing at 40: Past, present and future. Nature 550, 345–353 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Mutlu H., Lutz J.-F., Reading polymers: Sequencing of natural and synthetic macromolecules. Angew. Chem. Int. Ed. 53, 13010–13019 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Colquhoun H., Lutz J.-F., Information-containing macromolecules. Nat. Chem. 6, 455–456 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Jain M., Koren S., Miga K. H., Quick J., Rand A. C., Sasani T. A., Tyson J. R., Beggs A. D., Dilthey A. T., Fiddes I. T., Malla S., Marriott H., Nieto T., O’Grady J., Olsen H. E., Pedersen B. S., Rhie A., Richardson H., Quinlan A. R., Snutch T. P., Tee L., Paten B., Phillippy A. M., Simpson J. T., Loman N. J., Loose M., Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K., Kong J., Zhu J., Ermann N., Predki P., Keyser U. F., Digital data storage using DNA nanostructures and solid-state nanopores. Nano Lett. 19, 1210–1215 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Bell N. A. W., Keyser U. F., Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 11, 645–651 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Cao C., Cirauqui N., Marcaida M. J., Buglakova E., Duperrex A., Radenovic A., Dal Peraro M., Single-molecule sensing of peptides and nucleic acids by engineered aerolysin nanopores. Nat. Commun. 10, 4918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laszlo A. H., Derrington I. M., Ross B. C., Brinkerhoff H., Adey A., Nova I. C., Craig J. M., Langford K. W., Samson J. M., Daza R., Doering K., Shendure J., Gundlach J. H., Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 32, 829–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu B., Fleming S., Szalay T., Golovchenko J., Thermal motion of DNA in an mspa pore. Biophys. J. 109, 1439–1445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degiacomi M. T., Iacovache I., Pernot L., Chami M., Kudryashev M., Stahlberg H., van der Goot F. G., Dal Peraro M., Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nat. Chem. Biol. 9, 623–629 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Iacovache I., De Carlo S., Cirauqui N., Dal Peraro M., van der Goot F. G., Zuber B., Cryo-em structure of aerolysin variants reveals a novel protein fold and the pore-formation process. Nat. Commun. 7, 12062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dal Peraro M., van der Goot F. G., Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Cirauqui N., Abriata L. A., van der Goot F. G., Dal Peraro M., Structural, physicochemical and dynamic features conserved within the aerolysin pore-forming toxin family. Sci. Rep. 7, 13932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao C., Ying Y.-L., Hu Z.-L., Liao D.-F., Tian H., Long Y.-T., Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 11, 713–718 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Piguet F., Ouldali H., Pastoriza-Gallego M., Manivet P., Pelta J., Oukhaled A., Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun. 9, 966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao C., Yu J., Li M.-Y., Wang Y.-Q., Tian H., Long Y.-T., Direct readout of single nucleobase variations in an oligonucleotide. Small 13, 1702011 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Ouldali H., Sarthak K., Ensslen T., Piguet F., Manivet P., Pelta J., Behrends J. C., Aksimentiev A., Oukhaled A., Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 38, 176–181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al Ouahabi A., Charles L., Lutz J.-F., Synthesis of non-natural sequence-encoded polymers using phosphoramidite chemistry. J. Am. Chem. Soc. 137, 5629–5635 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Al Ouahabi A., Kotera M., Charles L., Lutz J.-F., Synthesis of monodisperse sequence-coded polymers with chain lengths above DP100. ACS Macro Lett. 4, 1077–1080 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Boukhet M., König N. F., Al Ouahabi A., Baaken G., Lutz J.-F., Behrends J. C., Translocation of precision polymers through biological nanopores. Macromol. Rapid Commun. 38, 1700680 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Al Ouahabi A., Amalian J.-A., Charles L., Lutz J.-F., Mass spectrometry sequencing of long digital polymers facilitated by programmed inter-byte fragmentation. Nat. Commun. 8, 967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao C., Li M.-Y., Cirauqui N., Wang Y.-Q., Dal Peraro M., Tian H., Long Y.-T., Mapping the sensing spots of aerolysin for single oligonucleotides analysis. Nat. Commun. 9, 2823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochreiter S., Schmidhuber J., Long short-term memory. Neural Comput. 9, 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Goldman N., Bertone P., Chen S., Dessimoz C., LeProust E. M., Sipos B., Birney E., Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yazdi S. M. H. T., Yuan Y., Ma J., Zhao H., Milenkovic O., A rewritable, random-access DNA-based storage system. Sci. Rep. 5, 14138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yazdi S. M. H. T., Gabrys R., Milenkovic O., Portable and error-free DNA-based data storage. Sci. Rep. 7, 5011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Organick L., Ang S. D., Chen Y.-J., Lopez R., Yekhanin S., Makarychev K., Racz M. Z., Kamath G., Gopalan P., Nguyen B., Takahashi C. N., Newman S., Parker H.-Y., Rashtchian C., Stewart K., Gupta G., Carlson R., Mulligan J., Carmean D., Seelig G., Ceze L., Strauss K., Random access in large-scale DNA data storage. Nat. Biotechnol. 36, 242–248 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Zrehen A., Huttner D., Meller A., On-chip stretching, sorting, and electro-optical nanopore sensing of ultralong human genomic DNA. ACS Nano 13, 14388–14398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garalde D. R., Snell E. A., Jachimowicz D., Sipos B., Lloyd J. H., Bruce M., Pantic N., Admassu T., James P., Warland A., Jordan M., Ciccone J., Serra S., Keenan J., Martin S., McNeill L., Wallace E. J., Jayasinghe L., Wright C., Blasco J., Young S., Brocklebank D., Juul S., Clarke J., Heron A. J., Turner D. J., Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Cavallo G., Clément J.-L., Gigmes D., Charles L., Lutz J.-F., Selective bond cleavage in informational poly(alkoxyamine phosphodiester)s. Macromol. Rapid Commun. 41, 2000215 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Cavallo G., Poyer S., Amalian J.-A., Dufour F., Burel A., Carapito C., Charles L., Lutz J.-F., Cleavable binary dyads: Simplifying data extraction and increasing storage density in digital polymers. Angew. Chem. Int. Ed. 57, 6266–6269 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eabc2661/DC1


