Increased activity of the Hippo pathway coactivator Yorkie alters selector gene expression and promotes cell fate plasticity.
Abstract
During development, tissue-specific patterns of gene expression are established by transcription factors and then stably maintained via epigenetic mechanisms. Cancer cells often express genes that are inappropriate for that tissue or developmental stage. Here, we show that high activity levels of Yki, the Hippo pathway coactivator that causes overgrowth in Drosophila imaginal discs, can also disrupt cell fates by altering expression of selector genes like engrailed (en) and Ultrabithorax (Ubx). Posterior clones expressing activated Yki can down-regulate en and express an anterior selector gene, cubitus interruptus (ci). The microRNA bantam and the chromatin regulator Taranis both function downstream of Yki in promoting ci expression. The boundary between Yki-expressing posterior clones and surrounding wild-type cells acquires properties reminiscent of the anteroposterior compartment boundary; Hedgehog signaling pathway activation results in production of Dpp. Thus, at least in principle, heterotypic interactions between Yki-expressing cells and their neighbors could activate boundary-specific signaling mechanisms.
INTRODUCTION
As an organism develops from a fertilized egg, different portions of the embryo begin to exhibit specific patterns of gene expression. Those patterns of gene expression could be a consequence of the position of those cells in the embryo (e.g., Hox genes) or the specification of cells to the primordia of specific tissues (e.g., Pax6 for eye precursors). These patterns of gene expression are initially established by the expression of a specific combination of transcription factors and then stabilized by the attainment of specific chromatin states that make some genes more accessible to the transcription machinery, the so-called epigenetic landscape (1, 2).
For many years, cancer cells have been known to display morphological characteristics that are inappropriate to their tissues of origin. One possible explanation is that cancer cells express tissue-inappropriate genes (3, 4). Recent efforts at characterizing genomes of cancers have shown that a subset of human cancers have mutations in chromatin regulators and splicing factors, which would be predicted to cause changes in gene expression or alterations in the expression of certain splice isoforms (5). However, it is also possible that the activation of a variety of oncogenes or inactivation of tumor suppressor genes could themselves cause tissue-inappropriate gene expression. Currently, we have a poor understanding of how oncogenesis can perturb mechanisms that preserve tissue-specific patterns of gene expression and the biological consequences of these perturbations.
The mechanisms that function during development to establish region-specific and tissue-specific patterns of gene expression have been studied extensively in Drosophila. A hierarchy of transcription factors acts during early embryogenesis to set up patterns of selector gene expression that specify individual regions of the embryo. Examples include the Hox gene Ultrabithorax (Ubx), which is expressed in specific embryonic segments, and the genes engrailed (en) and cubitus interruptus (ci), which are expressed in the posterior and anterior compartments of each segment, respectively. Once their patterns of expression are established, they are maintained by epigenetic mechanisms that include the involvement of the Polycomb and Trithorax groups of genes (6).
The pathways that promote tissue growth are conserved between humans and Drosophila. In Drosophila imaginal discs, the larval primordia of adult structures such as wings and eyes, increased activity of growth-promoting pathways can result in tissue overgrowth. This overgrowth can be elicited by expressing Myc (7), an activated form of Ras (RasV12) (8, 9), or Yorkie (Yki) (10), the coactivator downstream of the Hippo pathway [reviewed in (11)] in clones of cells. This allows us to investigate the effects of each of these genes on the patterns of selector gene expression that have been established in the imaginal discs.
Here, we show that among the genes tested, an activated form of Yki (ykiCA) is especially potent in destabilizing established patterns of selector gene expression. Clones of ykiCA cells in the posterior compartment of the wing disc express the anterior selector gene ci instead of the posterior gene en and sometimes inappropriately express Ubx. We present an investigation of these changes and also show that heterotypic interactions at the boundary between the overgrown clone and surrounding wild-type cells can acquire properties reminiscent of a compartment boundary, a phenomenon that could potentially occur at tumor margins in general.
RESULTS
To test whether oncogenes can also affect the stability of selector gene expression, we created clones of cells expressing either Myc (7), an activated form of Ras (RasV12) (8), or an activated form of yki (ykiCA) with a mutation in its critical serine-168 phosphorylation site, which negatively regulates nuclear localization (10, 12, 13) in the wing imaginal disc. In third instar wing discs, Ci is expressed only in cells anterior to the compartment boundary (Fig. 1A), and Ubx is expressed in the squamous cells of the peripodial epithelium (Fig. 1B) but not in the disc proper. In discs containing multiple ykiCA-expressing clones, we found clones in the posterior (P) compartment expressing Ci (74 of 76 discs) and clones in the disc proper expressing Ubx (23 of 28 discs) (Fig. 1, C and D). Ectopic expression of two other Hox genes Antennapedia and Abdominal-B was not observed in these clones. Many of these clones appear to be extruding from the epithelium, as has been described when patterning gene expression has been altered within clones of cells (14–16). To rule out the possibility that the posterior Ci-expressing clones had originated in the anterior (A) compartment, we generated ykiCA clones using mitotic recombination that marked both the ykiCA-expressing clone and its wild-type sister clone. We observed Ci-expressing ykiCA clones in the posterior compartment adjacent to wild-type twin spots, thus confirming that they originated in the posterior compartment (Fig. 1, E to E″). We did not observe a similar misexpression of these genes in discs expressing either Myc or RasV12 (fig. S1, A, A′, B, B′, and C). Thus, of the oncogenes tested, yki appears especially capable of altering expression patterns of selector genes. We also observed Ci expression in ykiCA clones posterior to the morphogenetic furrow in the eye disc (Fig. 1F), in the posterior compartment of the leg disc, and in the central nervous system (Fig. 1G), indicating that ykiCA can promote Ci expression in diverse tissues. In addition to ectopic expression of Ci, we sometimes observed elevated Ci expression in tissues where Ci is normally expressed, such as in clones in the anterior compartment of the wing disc (Fig. 1, C and H).
Fig. 1. Constitutively active Yorkie (YkiCA) disrupts stable selector gene expression.
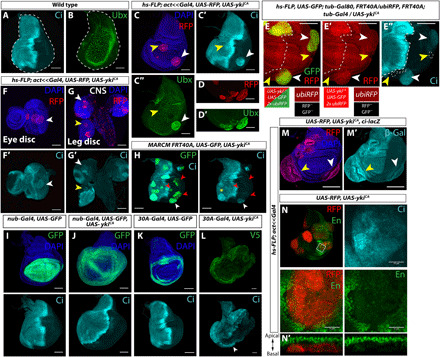
(A and B) Wild-type (WT) wing imaginal discs; Ci is expressed in anterior compartment, and Ubx is expressed in squamous peripodial cells. (C to C″) Clones expressing activated Yki (UAS-ykiCA). Anterior clone (yellow arrowhead) expresses more Ci; posterior clone (white arrowhead) expresses ectopic Ci and Ubx (C″). Discs with Ci-positive clones: n = 74 of 76. Discs with Ubx-positive clones: n = 23 of 28. (D and D′) Ubx-expressing ykiCA clones in the disc proper. (E to E″) Mitotic recombination: ykiCA-expressing clones express red fluorescent protein (RFP) and green fluorescent protein (GFP) (white arrowheads), and neighboring twin spots (yellow arrowheads) express neither. (F and G) ykiCA clones in the eye disc (F and F′), leg disc [(G) and (G′), yellow arrow], and larval brain [(G) and (G′), white arrow]. CNS, central nervous system. (H) Posterior ykiCA marked with GFP expresses Ci in the hinge (white arrowhead) but inconsistently in the pouch (red arrowheads). Anterior clones express more Ci (yellow asterisk). (I to L) nubbin-Gal4, UAS-ykiCA does not cause ectopic Ci [(J), marked by GFP, control disc in (I)], but 30A-Gal4 UAS-ykiCA does [(L), V5 tag on YkiCA, control disc in (K)], especially in the ventral hinge (arrowhead). (M and M′) ci-lacZ is expressed in posterior clones (white arrowheads), and expression is increased in anterior clones (yellow arrowheads). (N) A posterior ykiCA clone with down-regulated En and ectopic Ci expression. (N′) XZ section of a posterior ykiCA that has been extruded basally. Anterior is left in all images. Scale bars, 100 μm. DAPI, 4′,6-diamidino-2-phenylindole; β-Gal, β-galactosidase.
The induction of ectopic Ci expression was not observed in clones overexpressing wild-type yki or clones mutant for the Hippo pathway components hippo (hpo) or warts (wts), (fig. S1, D to F). Thus, ectopic Ci expression depends on especially high levels of Yki activity, caused by overexpression of a form of Yki that cannot be inhibited by Hpo and Wts and hence localizes efficiently to the nucleus. Furthermore, observing clones at different stages of development indicated that ectopic Ci expression required prolonged expression of ykiCA (fig. S2). Even within a given tissue, we observed regional differences in the level of ectopic Ci expression. In the wing disc, clones in the hinge typically expressed higher levels of Ci than clones in the pouch (Fig. 1H). When ykiCA was expressed using nub-Gal4, which is mostly expressed in the pouch, relatively little ectopic Ci was observed (Fig. 1, I and J). In contrast, when the hinge driver 30A-Gal4 was used, ectopic Ci was consistently observed, especially in the ventral hinge posterior to the compartment boundary (Fig. 1, K and L). The hinge is a region of the disc that displays increased plasticity following damage and can be a tumor “hotspot” (17, 18), suggesting that the underlying cause of these phenomena may be related.
ci is normally transcribed exclusively in anterior cells of the wing disc and is repressed in posterior cells by en (19–21). Clones of posterior cells that are mutant for both en and its adjacent paralog invected (inv) express Ci (19). Posterior ykiCA clones up-regulate a transcriptional ci reporter, ci-lacZ, indicating that ci transcription is derepressed (Fig. 1M). Large Ci-positive posterior clones have reduced levels of staining with antibody 4D9, which recognizes both En and Inv (22), suggesting that the derepression of ci transcription could result from a reduction in En and Inv levels (Fig. 1N). En has also been shown to negatively regulate Ubx expression in the wing disc, although that mechanism is less well defined (23, 24); consistent with this, we only observed ectopic Ubx in posterior ykiCA clones. Since Ci is expressed at elevated levels in anterior clones, where En is not normally expressed, ykiCA must also be capable of regulating ci expression by mechanisms that are independent of changes in En levels. Consistent with this notion, ci was identified as a Yki target, and ci RNA was elevated in tissue mutant for wts, which has higher Yki activity than wild-type tissue (25).
How does Yki regulate ci expression? Yki acts together with its binding partner Scalloped (Sd) to activate gene expression, with Sd binding directly to regulatory regions of target genes (26). If Sd is the relevant binding partner, then alterations in gene expression would not occur in Yki-expressing clones that are mutant for sd (27). To determine whether Sd is required for expression of ci, we reduced sd expression in ykiCA clones (Fig. 2, A to I). Anterior ykiCA clones did not show increased Ci expression (Fig. 2, D to F, G to G″, and H to H″), while posterior ykiCA clones no longer expressed ectopic Ci (Fig. 2, D to F and I to I″), indicating that Sd is required for ectopic Ci up-regulation. Yki promotes growth, in large part, by activating expression of the microRNA bantam (ban) (28–30). To address a possible role for ban in mediating ectopic ci expression, we reduced ban levels by generating ykiCA clones in flies heterozygous for a null allele of ban, banΔ1 (Fig. 2, J and K; control in fig. S3A) (31). In banΔ1/+ discs, discs appeared smaller overall, although clone size was unaffected (Fig. 2Q), yet the frequency of Ci-expressing ykiCA clones was reduced both overall and especially in the hinge (Fig. 2R). Thus, the ability of constitutively active Yki (YkiCA) to activate Ci expression in posterior clones is more sensitive to ban levels than its ability to promote overgrowth. We also used a “bantam sponge,” which encodes an RNA that has 10 optimal ban-binding sites upstream of the DsRed coding region; low DsRed expression likely correlates with clones with higher ban levels (32). We found that clones expressing high levels of DsRed had low Ci levels and vice versa, suggesting a correlation between ban levels and induction of Ci (Fig. 2, L and M).
Fig. 2. ykiCA clones require sd, ban, and tara to disrupt patterning gene expression.
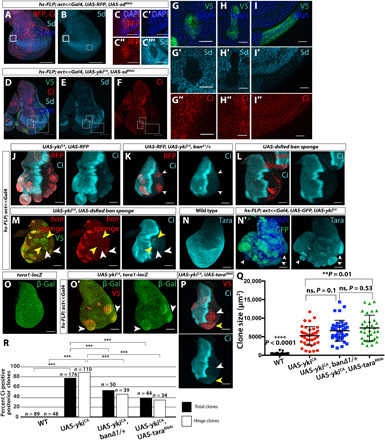
(A to I) RNA interference (RNAi) of sd [validated in (A) to (C‴) using anti-Sd] in anterior ykiCA clones prevents increased Ci expression (D to F, G to G″, and H to H″) and Ci expression in posterior clones (D to F and I to I″). (J and K) ykiCA clones in banΔ1/+ discs (K) are overgrown but express less ectopic Ci [compared to (J)]. (L and M) ban sponge reduces ban microRNA levels. dsRed levels inversely correlate with ban levels. Clones expressing ban sponge express uniformly high dsRed and no ectopic Ci (L). Posterior clones expressing YkiCA and ban sponge show variation in both dsRed and Ci expression (M). High dsRed-expressing posterior clones are smaller and do not express Ci (white arrowheads); low- or no-dsRed clones are overgrown and express Ci (yellow arrowheads). (N and O) YkiCA clones have increased expression of Tara protein (N and N′) and tara1-lacz (O and O′), especially in the hinge (arrowheads). (P) tara RNAi allows overgrowth but reduces Ci expression. (Q) Size of posterior hinge clones of indicated genotypes. ykiCA clones in banΔ1/+ discs are not significantly smaller than those in WT discs; ykiCA + taraRNAi clones are significantly larger than ykiCA clones. (R) Ectopic Ci is observed less often in ykiCA clones when ban or tara levels are reduced. White boxes, hinge clones; black boxes, hinge and pouch clones. Statistics: See Materials and Methods. ns, not significant.
We also investigated taranis (tara), originally classified as a TrxG gene, which is known to modulate homeotic gene expression by hitherto undefined mechanisms (33). A genome-wide chromatin immunoprecipitation screen for Yki targets found an enrichment of Yki binding in the tara promoter, as well as increased tara transcription in wtsP2 discs (25). There is a putative binding site for Sd, Yki’s primary binding partner, approximately 400 base pairs upstream of the tara transcriptional start site. In addition, tara negatively regulates en expression in embryos and in regenerating imaginal discs (34, 35). We found that Taranis protein levels were increased in ykiCA-expressing clones, as was the expression of a tara-lacZ transcriptional reporter (Fig. 2, N and O). These findings are consistent with the possibility that an increase in Taranis in ykiCA clones reduces En levels and thus allows Ci expression. To examine this possibility, we reduced tara expression in ykiCA clones (Fig. 2P; control in fig. S3B). These clones were slightly larger than ykiCA clones (Fig. 2Q), but the expression of ectopic Ci was reduced (Fig. 2R). This result implicates tara in the pathway by which YkiCA induces Ci expression and also shows that ectopic Ci expression does not simply correlate with the extent of overgrowth.
Since our results implicate both ban and tara in the pathway by which YkiCA activates ectopic Ci expression, we tested whether increasing ban and tara levels, either alone or in combination, could induce Ci expression in the posterior compartment. Posterior clones overexpressing either ban or tara alone in wild-type discs expressed very low levels of Ci at most (fig. S3, D and E). However, when expressed in combination with a wild-type version of yki, which normally does not induce Ci expression (fig. S3C), either ban or tara could induce Ci expression, especially in the posterior ventral hinge (Fig. 3, A and B). tara overexpression also enhanced Ci expression in YkiCA clones (Fig. 3C).
Fig. 3. ban and tara in combination induce ectopic patterning gene expression.
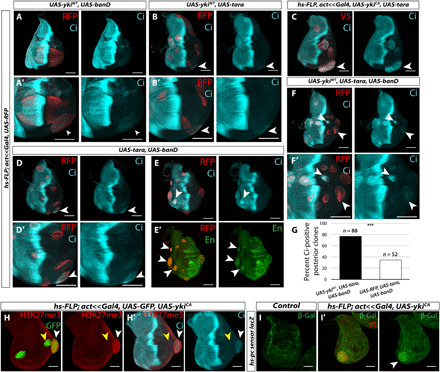
(A and B) Hinge clones expressing either banD (A and A′) or tara (B and B′) together with ykiWT express Ci especially in the ventral hinge. (C) Overexpression of tara in ykiCA clones further increases Ci expression. (D and E) Combined expression of ban and tara causes some overgrowth and also Ci expression in posterior clones (D and D′) in increased Ci expression and ectopic En expression in anterior clones (E and E′). (F and F′) Clones expressing ykiWT, ban, and tara are more overgrown than ykiWT alone or with ban or tara individually and consistently express ectopic Ci in the posterior compartment. (G) A total of 77.3% of posterior clones expressing ykiWT, tara, and ban express ectopic Ci, while 34.6% of posterior clones expressing RFP, tara, and ban are Ci positive. (H and H′) ykiCA clones (GFP positive) show increased H3K27 trimethylation (H3K27me3; red) (H). This increase is only seen in clones in the hinge, where H3K27me3 is already higher than the rest of the disc (white arrowhead) and not in the pouch (yellow arrowhead). (H′) Increased H3K27me3 coincides with ectopic Ci expression. (I and I′) ykiCA clones show decreased Pc-mediated repression of a Polycomb-responsive element from the bxd locus linked to a lacZ reporter (green). YkiCA was tagged with V5. Statistics: See Materials and Methods.
Furthermore, clones expressing ban and tara together could induce ectopic Ci at a low frequency and cause overgrowth reminiscent of ykiCA overexpression (Fig. 3D); anterior clones expressed a higher level of Ci than surrounding wild-type tissue (Fig. 3E). Clones expressing both tara and ban did not have an obvious decrease in En expression (Fig. 3E′), although a reduction in tara has previously been linked to result in increased En expression (34, 35). Thus, tara likely regulates ci expression independently of en. We also found that anterior clones expressing ban and tara expressed ectopic En (Fig. 3E′), thereby generating clones that coexpress Ci and En, which indicates that, at least in this situation, the mere presence of En is insufficient to repress Ci expression and that tara in combination with ban can destabilize selector gene expression in multiple ways. When using the hinge 30A-Gal4 driver, combined expression of both ban and tara, but not either alone, induced a low but reproducible level of posterior Ci and Ptc expression in wild-type discs (fig. S3, H and I; control in fig. S3, F and G). The lower levels of ectopic Ci expression when compared to ykiCA clones and the absence of En down-regulation elicited by ban and tara coexpression indicate that YkiCA must also act via additional unidentified targets. Consistent with this idea, overexpression of ban and tara together in ykiWT clones recapitulated the ykiCA clone phenotype to a greater extent, with consistent although often low-level ectopic Ci expression (Fig. 3F). A total of 77.2% of clones expressing YkiWT in combination with ban and tara expressed ectopic Ci versus 34.6% of clones expressing just ban and tara (Fig. 3G). In addition, while tara overexpression induced ban expression as assessed by a ban-GFP reporter (fig. S3, J and K) (28), ban overexpression did not induce tara (fig. S3, L and M), indicating that the increase in Tara levels does not result from the known ability of Yki to activate ban expression.
To look for evidence of alterations in chromatin in ykiCA clones, we used a panel of antibodies that recognize specific posttranslational modifications of histones. We found that ykiCA clones, especially in the hinge, show elevated levels of H3K27 trimethylation (H3K27me3), which typically correlates with increased Polycomb group (PcG)–mediated gene repression (Fig. 3H) (36, 37). We observed less obvious increases in the level of H3K4me1 and H3K4me3, but not H3K9me3 or H4Ac (fig. S4, A to D). The increase in H3K27me3 could potentially explain the decrease in En expression in ykiCA clones. However, the expression of en in the posterior compartment of imaginal discs is regulated by a particularly large region (approximately 79 kB) (38) composed of multiple regulatory elements that sometimes function antagonistically, which makes it difficult to evaluate the relevance of local alterations in H3K27me3. The apparent global increase in H3K27me3 levels did not always correlate with increased PcG-mediated repression; a reporter that is silenced by a Polycomb (Pc)-responsive element from the bxd locus (39) is derepressed in ykiCA clones (Fig. 3I). Thus, the effects of YkiCA on chromatin state appear to be complex, and hence, effects on individual genes are not easy to predict. This is consistent with the notion that ykiCA activates Ci expression by multiple mechanisms, as demonstrated by effects on Ci in both anterior and posterior compartments.
To test whether the increases in tara and H3K27me3 expression are specific to YkiCA activation or whether these phenomena occur in other overgrown tissue, we stained discs with clones expressing either RasV12 or Myc with antibodies that recognize Tara and H3K27me3. Neither RasV12- nor Myc-expressing clones showed changes in Tara or H3K27me3 relative to surrounding wild-type tissue (fig. S4, E and F), supporting the conclusion that these changes in chromatin regulation are specific to YkiCA activation.
What are the consequences of creating ykiCA clones that have down-regulated en and express ci in the posterior compartment? The juxtaposition of cells that express en and those that do not normally occurs at the anteroposterior compartment boundary. ci is expressed in anterior cells but repressed by En in posterior cells, which instead secrete the short-range morphogen Hedgehog (Hh) (40). Activation of the Hh signaling pathway in cells anterior to the compartment boundary stabilizes the activator form of Ci and results in the transcription of multiple target genes including patched (ptc) and dpp (41, 42). Dpp is a long-range morphogen that diffuses widely from its source and regulates tissue growth and gene expression in both compartments (43, 44). We found that the presence of Ci-expressing ykiCA clones in the posterior compartment generates ectopic sites of Hh pathway signaling near the clone boundary, possibly due to increased levels of the full-length activator form of Ci. The Hh target genes dpp and ptc were both expressed in these clones (Fig. 4, A and B). Evidence of Dpp signaling, as assessed with the presence of the phosphorylated form of the signaling protein Mad (pMAD), was also observed (Fig. 4C). This is noteworthy given previous results showing that Mad and Yki cooperate to activate ban (45), which we demonstrated is necessary for ectopic Ci activation (Fig. 2, K to M and R). Thus, the clone boundary has indeed adopted anteroposterior compartment-boundary–like properties, at least with respect to Hh signaling. However, we did not see consistent evidence of clones establishing ectopic dorsoventral compartment boundaries. We did, however, observe some abnormalities in the pattern of dorsoventral gene expression (fig. S5, A to C). Some dorsal clones had reduced apterous expression (fig. S5C′, arrow), and some ventral clones expressed cut (fig. S5B′), which is normally expressed at the dorsoventral compartment boundary. However, in these ventral clones, cut was expressed throughout the clone and not at the boundary.
Fig. 4. ykiCA clones activate a developmental signaling cascade.
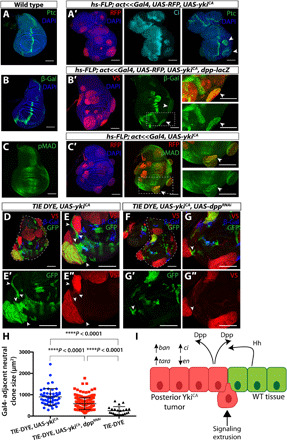
(A) ykiCA clones that express Ci also express Ptc (A′) and dpp-lacZ (B′) at clone margin and have increased pMAD near clones (C′). Control discs without ykiCA clones are shown in (A) to (C). (D and E) ykiCA clones cause nonautonomous overgrowth in neighboring WT tissue. The TIE-DYE system has three independent FLP-out transgenes that express Gal4, GFP, and lacZ. Clones might express none of these or any combination of these depending on the number of FLP-out events in founder cells. Gal4-expressing clones express UAS-ykiCA and are visualized with anti-V5. Neutral GFP-expressing clones are shown in green, and lacZ-expressing clones are shown in blue. Arrowheads indicate two unusually large GFP-positive WT clones that are immediately adjacent to overgrown YkiCA-expressing clones (red). (F and G) Knockdown of dpp in ykiCA clones. In these discs, neutral clones adjacent to ykiCA clones are not as overgrown (arrowheads). (H) Quantification of size of neutral clones directly adjacent to Gal4-expressing clones [expressing UAS-ykiCA (neutral clone, n = 52), UAS-ykiCA and UAS-dppRNAi (neutral clone, n = 184), or UAS-RFP (neutral clone, n = 22)]. (I) Model: YkiCA causes changes in selector gene expression in the posterior compartment via up-regulation of ban and tara. As a result of heterotypic interactions at clone boundaries, ectopic organizing centers are created resulting in the production of morphogens (e.g., Dpp), as well as extrusion of ykiCA tissue. Statistics: See Materials and Methods.
What are the consequences of activating Hh signaling in ykiCA clones? When we inactivated smo in ykiCA clones, we found no impairment in growth, nor did we find growth impairment when we reduced Ci itself in ykiCA clones (fig. S5, D to F and I). We also observed no significant difference in anterior versus posterior clone size for ykiCA clones, smo3, ykiCA clones, or ykiCA, ciRNAi clones (fig. S5, G, H, and J). Thus, their overgrowth is not dependent on Hh signaling. This is consistent with the obvious overgrowth observed in ykiCA clones far from the compartment boundary in the anterior compartment, which are also far from an Hh source. Since the clones were overgrown and in the process of extrusion, it was difficult to ascertain whether clones close to the compartment boundary had crossed it (fig. S5F). Clone overgrowth and changes in selector gene expression were also not dependent on c-Jun N-terminal kinase pathway activation, which has been previously linked to Hippo pathway signaling and thought to facilitate propagation of Yki activity to cells neighboring tumor cells (fig. S5, K to N) (46–49). To investigate potential nonautonomous effects of ykiCA clones, we used the TIE-DYE system (50), which has three independent FLP-out genes, two of which, when activated, express reporters driven by constitutive promoters (ubi-GFP and act-lacZ). The third is a FLP-out Gal4; these clones express UAS-ykiCA. While ykiCA-expressing clones are obviously overgrown, we also observed occasional large neutral clones that appeared to be composed of cells that were close to the perimeter of the ykiCA-expressing clones, including anterior clones that have elevated full-length Ci (Figs. 1C′ and 4, D and E). These clones were not as evident when a UAS-dppRNAi transgene was coexpressed with ykiCA. Neutral clones adjacent to ykiCA-expressing clones were significantly larger than those adjacent to ykiCA, dppRNAi–expressing clones (Fig. 4, F to H, and fig. S5O). This suggests that Dpp secreted by ykiCA clones may be able to promote the growth of adjacent cells, especially in lateral regions of the disc (Fig. 4I).
DISCUSSION
Human cancers are characterized by multiple genetic lesions, a subset of which are driver mutations that are thought to be responsible for their tumorous characteristics. It is estimated that most cancers have two to eight driver mutations (5). This makes it difficult to evaluate the contribution of each mutation to any particular characteristic of the tumor. We have taken advantage of the ability of single-gene manipulations to cause overgrowth in Drosophila imaginal discs to assess the ability of three different oncogenes to destabilize established patterns of selector gene expression and find that yki, the Drosophila ortholog of Yap and Taz, is especially potent in doing so. The patterns of expression of En, Ci, and Ubx are established relatively early in embryogenesis and maintained stably in imaginal discs during the larval stages of development. These patterns of expression can be disrupted in clones expressing an activated form of Yki. Expression of a wild-type form of Yki is capable of disrupting these expression patterns in combination with other genetic manipulations such as overexpression of ban or tara. This latter scenario is more likely to apply to human cancers; increased Yap or Taz activity has been described in multiple human cancers (51), which often also have other genetic lesions.
Our studies show that sd, ban, and tara make important contributions to the pathway by which YkiCA destabilizes gene expression; reducing the expression of any of these in clones expressing YkiCA greatly reduces ectopic Ci expression, and increasing expression of both genes can cause ectopic Ci expression. It is likely that other mechanisms function in parallel to destabilize selector gene expression since combined overexpression of ban and tara increased ectopic Ci expression but did not reduce En expression.
We also demonstrate that changing selector gene expression within an overgrowing clone can create interactions at the clone margin that are reminiscent of compartment boundaries and result in the production of morphogens. A recent study showed that forced expression of En in lgl clones can elicit similar phenomena in anterior clones (52). In addition, ykiCA clones are often extruded, consistent with previous observations that heterotypic interactions caused by overexpressing patterning genes also promotes extrusion (14–16). Previous work found that ci RNA levels were increased in wts mutant tissue (25), yet we did not see ectopic Ci protein expression in wts mutant clones or wild-type Yki-overexpressing clones. Our work shows therefore that sustained expression of very high Yki levels is necessary to destabilize expression of selector genes. However, even under these conditions, the effect on ectopic Ci expression is Sd dependent. Moreover, we have shown that even wild-type Yki can, in combination with increased expression of ban or tara, induce ectopic Ci expression. While these changes in gene expression are most obvious with above-physiological levels of Yki, they nevertheless reflect a previously unknown ability of this pathway to alter patterning gene expression and furthermore to change the growth characteristics of neighboring wild-type cells. Differences in selector gene expression between human cancers or precancerous lesions and their wild-type neighbors have received relatively little attention, and our results call attention to tumor margins as sites where heterotypic interactions could create signaling centers that affect the behavior of tumor cells.
MATERIALS AND METHODS
Experimental design
We set out to characterize the ectopic expression of selector genes seen in some overgrown tissues and to investigate the role of this ectopic gene expression in the biology of overgrowth. To generate overgrown tissue, we used a system with heat shock–induced FLPase and actin-driven FLP-out Gal4 with UAS-ykiS168A. This system generated random overgrown clones throughout the larva, with clone frequency controlled by length of heat shock. All components of this system were contained within one stable stock that could be crossed to other stocks to test the role of candidate genes and use lacZ reporters.
Drosophila stocks
Stocks used in this study include the following: hsFLP; act<stop<Gal4, UAS-ykiS168A.v5, hsFLP;act<stop<Gal4, UAS-RFPNLS, hsFLP, UAS-GFPNLS; tub-Gal80, FRT40A (MARCM FRT 40A), ubi>RFP, FRT40A, banΔ1, banGFP, en-Gal4, dpp-lacZ, UAS-RFP, AP-1-RFP (53), and TIE-DYE (50). Stocks were obtained from the Bloomington Drosophila Stock Center (BDSC): UAS-ykiS168A.v5 (#28818), UAS-yki.v5 (#28819), UAS-yki::GFP (#28815), wtsX1 (#44251), smo3 (#3277), UAS-ciRNAi (#64928), tara1-lacZ (#6403), UAS-taraRNAi (#31634), nub-Gal4 (#42699), 30A-Gal4 (#37534), UAS-dMyc (#9674), UAS-rasv12 (#4847), and UAS-jnkDN (#9311). Ci-lacZ was a gift from D. Kalderon (Columbia University, USA). UAS-bantamsponge and UAS-bantam.D were gifts from S. Cohen (University of Copenhagen, Denmark). hs-Pc-sensor was a gift from V. Pirrotta (Rutgers University, USA). UAS-myc::tara was a gift from R. Smith-Bolton [University of Illinois, Urbana-Champaign, USA (originally from M. Cleary, University of California, Merced, USA)]. MARCM FRT19A and sd47M, FRT19A were gifts from D. Pan.
Temperature shift and clone induction experiments
Experiments using heat shock–controlled FLPase and act<<Gal4 were maintained at 25°C, heat shocked for 7 min at 37°C at 72 hours after egg lay (hAEL) ± 12 hours, and then dissected and analyzed at 144 hAEL ± 12 hours. TIE-DYE experiments were conducted under the same conditions. Because ascertaining clone boundaries in this system can be challenging, we attempted to score only clones where boundaries were reasonably clear. hsFLP-induced mosaic analysis with a repressible cell marker (MARCM) experiments were conducted under the same conditions but with 10- to 15-min heat shocks (except for MARCM 19A experiments in Fig. 2, which were heat shocked for 1 hour). Discs in fig. S5D were dissected at 120 hAEL because of the lack of developmental delay in these larvae, while larvae expressing ykiCA are delayed by approximately 1 day and thus were dissected at 144 hAEL. Crosses using nub-Gal4, 30A-Gal4, and en-Gal4 were incubated at 25°C, and larvae were dissected at wandering third instar (approximately 120 to 144 hAEL).
Immunohistochemistry
Imaginal discs were dissected and fixed in 4% paraformaldehyde for 20 min, washed and permeabilized in phosphate-buffered saline with 0.1% Triton X-100, and blocked in 10% normal goat serum. Primary antibodies used were α-Ci [1:25; Developmental Studies Hybridoma Bank (DSHB)], α-Ubx (1:10; DSHB), α-En (1:25; DSHB), α-β-galactosidase (1:500; Promega), α-V5 (1:500; Sigma-Aldrich), α-Wg (1:100; DSHB), α-Cut (1:100), α-Ptc (1:50; DSHB), α-pMAD (1:500; Abcam), α-Smo (1:10; DSHB), α-MMP1 (1:100; a combination of 14A3D2, 3A6B4, and 5H7B11; DSHB), α-Tara (1:700; K. Koh), α-H3K27me3 (1:500; Active Motif), α-H3K9me3 (1:500; Active Motif), α-H3K4me1 (1:500; Active Motif), α-H3K4me3 (1:500; Active Motif), α-H4ac (1:500; Active Motif), and α-Sd (1:500; K. Guss). Secondary antibodies were from Cell Signaling Technology and used at 1:400. Nuclei were stained with 4′,6-diamidino-2-phenylindole (1:1000; Cell Signaling Technology). Samples were imaged on a Zeiss LSM 700 confocal microscope.
Statistical analysis
Images were processed using Fiji (54), and statistical analysis was completed with GraphPad Prism and VassarStats 2x2 Contingency Table. All scale bars are 100 μm unless otherwise noted. For clone size comparisons in Figs. 2K and 4H, P values were generated through a one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test for wild-type clones, ykiCA clones in a wild-type background, ykiCA clones in a banΔ1/+ background, and ykiCA clones with taraRNAi (2K) or TIE-DYE ykiCA clones, TIE-DYE ykiCA, dppRNAi clones, or TIE-DYE wild-type clones (4H). Error bars are SDs. All comparisons between wild-type clones and other conditions had a P value of <0.0001. For comparisons of the rate of Ci-positive posterior clones in Figs. 2L and 3D, Yates P values were calculated with a χ2 test. All are <0.0001 except ykiCA compared to ykiCA, banΔ1/+ total clones, which is 0.0009. ***P < 0.001, very significant; **P = 0.001 to 0.01, very significant; *P = 0.01 to 0.05, significant; P > 0.05, not significant. Anterior versus posterior clone size comparisons in fig. S5 (G, H, and J) P values were calculated with an unpaired Student’s t test in GraphPad.
Supplementary Material
Acknowledgments
We thank members of the Hariharan Lab for helpful discussion and suggestions; L. Setiawan and M. Worley for feedback on the manuscript; H. Mazdeyasna for experimental assistance; D. Kalderon, S. Cohen, V. Pirrotta, R. Smith-Bolton, D. Pan, and the BDSC for fly stocks; K. Koh for the anti-Taranis antibody; and K. Guss for the anti-Sd antibody. Funding: This work was funded by a grant to I.K.H. from the NIH (R35 GM122490) and a Research Professor Award from the American Cancer Society (RP-16-238-06-COUN). Author contributions: J.C.D.B., M.E.-B., and I.K.H. designed and interpreted experiments. J.C.D.B. and M.E.-B. conducted experiments. J.C.D.B. and M.E.-B. prepared the figures. J.C.D.B. and I.K.H. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Fly stocks generated for this paper may be obtained upon request. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eabe8159/DC1
REFERENCES AND NOTES
- 1.Britten R. J., Davidson E. H., Gene regulation for higher cells: A theory. Science 165, 349–357 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Axelsen J. B., Lotem J., Sachs L., Domany E., Genes overexpressed in different human solid cancers exhibit different tissue-specific expression profiles. Proc. Natl. Acad. Sci. U.S.A. 104, 13122–13127 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flavahan W. A., Gaskell E., Bernstein B. E., Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A. Jr., Kinzler K. W., Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassis J. A., Kennison J. A., Tamkun J. W., Polycomb and Trithorax group genes in Drosophila. Genetics 206, 1699–1725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P., Drosophila myc regulates cellular growth during development. Cell 98, 779–790 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prober D. A., Edgar B. A., Ras1 promotes cellular growth in the Drosophila wing. Cell 100, 435–446 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Karim F. D., Rubin G. M., Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Huang J., Wu S., Barrera J., Matthews K., Pan D., The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Pfleger C. M., The Hippo pathway: A master regulatory network important in development and dysregulated in disease. Curr. Top. Dev. Biol. 123, 181–228 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D., Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh H., Irvine K. D., In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson M. C., Perrimon N., Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 307, 1785–1789 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Shen J., Dahmann C., Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science 307, 1789–1790 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Bielmeier C., Alt S., Weichselberger V., La Fortezza M., Harz H., Jülicher F., Salbreux G., Classen A.-K., Interface contractility between differently fated cells drives cell elimination and cyst formation. Curr. Biol. 26, 563–574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamori Y., Suzuki E., Deng W.-M., Epithelial tumors originate in tumor hotspots, a tissue-intrinsic microenvironment. PLOS Biol. 14, e1002537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verghese S., Su T. T., Drosophila Wnt and STAT define apoptosis-resistant epithelial cells for tissue regeneration after irradiation. PLOS Biol. 14, e1002536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eaton S., Kornberg T. B., Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 4, 1068–1077 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Schwartz C., Locke J., Nishida C., Kornberg T. B., Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development 121, 1625–1635 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Chanas G., Lavrov S., Iral F., Cavalli G., Maschat F., Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev. Biol. 272, 522–535 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Patel N. H., Martin-Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S., Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, 955–968 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Mann R. S., Engrailed-mediated repression of Ultrabithorax is necessary for the parasegment 6 identity in Drosophila. Development 120, 3205–3212 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Emerald B. S., Shashidhara L. S., Negative regulation of Ultrabithorax expression by engrailed is required for proper specification of wing development in Drosophila melanogaster. J. Genet. 79, 61–70 (2000). [Google Scholar]
- 25.Oh H., Slattery M., Ma L., Crofts A., White K. P., Mann R. S., Irvine K. D., Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 3, 309–318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J., Zider A., SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435–441 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Yu J., Pan D., Validating upstream regulators of Yorkie activity in Hippo signaling through scalloped-based genetic epistasis. Development 145, dev157545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M., bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Thompson B. J., Cohen S. M., The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767–774 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Nolo R., Morrison C. M., Tao C., Zhang X., Halder G., The bantam MicroRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895–1904 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Hipfner D. R., Weigmann K., Cohen S. M., The bantam gene regulates Drosophila growth. Genetics 161, 1527–1537 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herranz H., Hong X., Cohen S. M., Mutual repression by bantam miRNA and capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 22, 651–657 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Calgaro S., Boube M., Cribbs D. L., Bourbon H.-M., The Drosophila gene taranis encodes a novel trithorax group member potentially linked to the cell cycle regulatory apparatus. Genetics 160, 547–560 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster K. J., Smith-Bolton R. K., Taranis protects regenerating tissue from fate changes induced by the wound response in Drosophila. Dev. Cell 34, 119–128 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Dutta P., Li W. X., The SERTAD protein Taranis plays a role in Polycomb-mediated gene repression. PLOS ONE 12, e0180026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O’Connor M. B., Kingston R. E., Simon J. A., Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.-M., Bollen M., Esteller M., Croce L. D., de Launoit Y., Fuks F., The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Cheng Y., Brunner A. L., Kremer S., DeVido S. K., Stefaniuk C. M., Kassis J. A., Co-regulation of invected and engrailed by a complex array of regulatory sequences in Drosophila. Dev. Biol. 395, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dellino G. I., Schwartz Y. B., Farkas G., McCabe D., Elgin S. C. R., Pirrotta V., Polycomb silencing blocks transcription initiation. Mol. Cell 13, 887–893 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Tabata T., Eaton S., Kornberg T. B., The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 6, 2635–2645 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Von Ohlen T., Lessing D., Nusse R., Hooper J. E., Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 2404–2409 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aza-Blanc P., Ramírez-Weber F.-A., Laget M.-P., Schwartz C., Kornberg T. B., Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Nellen D., Burke R., Struhl G., Basler K., Direct and long-range action of a Dpp morphogen gradient. Cell 85, 357–368 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Lecuit T., Brook W. J., Ng M., Calleja M., Sun H., Cohen S. M., Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 381, 387–393 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Oh H., Irvine K. D., Cooperative regulation of growth by Yorkie and mad through bantam. Dev. Cell 20, 109–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma X., Wang H., Ji J., Xu W., Sun Y., Li W., Zhang X., Chen J., Xue L., Hippo signaling promotes JNK-dependent cell migration. Proc. Natl. Acad. Sci. U.S.A. 114, 1934–1939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Codelia V. A., Sun G., Irvine K. D., Regulation of YAP by mechanical strain through Jnk and Hippo signaling. Curr. Biol. 24, 2012–2017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enomoto M., Igaki T., Src controls tumorigenesis via JNK-dependent regulation of the Hippo pathway in Drosophila. EMBO Rep. 14, 65–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enomoto M., Kizawa D., Ohsawa S., Igaki T., JNK signaling is converted from anti- to pro-tumor pathway by Ras-mediated switch of Warts activity. Dev. Biol. 403, 162–171 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Worley M. I., Setiawan L., Hariharan I. K., TIE-DYE: A combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development 140, 3275–3284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bajpai A., Sinha P., Hh signaling from de novo organizers drive lgl neoplasia in Drosophila epithelium. Dev. Biol. 457, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee N., Bohmann D., A versatile φC31 based reporter system for measuring AP-1 and NRF2 signaling in Drosophila and in tissue culture. PLOS ONE 7, e34063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eabe8159/DC1


