Microplastic that was exposed to the environment is much more likely internalized by cells than pure unmodified microplastic.
Abstract
Microplastic particles ubiquitously found in the environment are ingested by a huge variety of organisms. Subsequently, microplastic particles can translocate from the gastrointestinal tract into the tissues likely by cellular internalization. The reason for cellular internalization is unknown, since this has only been shown for specifically surface-functionalized particles. We show that environmentally exposed microplastic particles were internalized significantly more often than pristine microplastic particles into macrophages. We identified biomolecules forming an eco-corona on the surface of microplastic particles, suggesting that environmental exposure promotes the cellular internalization of microplastics. Our findings further indicate that cellular internalization is a key route by which microplastic particles translocate into tissues, where they may cause toxicological effects that have implications for the environment and human health.
INTRODUCTION
Microplastic particles are ubiquitous in marine and freshwater ecosystems (1, 2). Once microplastic particles are introduced into these environments, microorganisms and biomolecules attach to their surfaces, forming an eco-corona that can enhance their ingestion by organisms (3). The ingestion of microplastic particles has been shown in a huge variety of organisms (4) ranging from zooplankton (5, 6), to bivalves (7, 8), up to vertebrates (9). From the gastrointestinal tract, which represents the main entry point for ingested foreign particulate matter, microplastic particles can translocate into the circulatory system (7) and the tissues, where their effects include inflammatory responses (8) and necrosis (9). The translocation of microplastic particles into tissues has been suggested to occur via cellular internalization (7, 8). The gastrointestinal system comprises a plethora of different cell types (10), including macrophages. This cell type occurs in all body compartments including the digestive system. Macrophages additionally occupy a central position within the innate immune response to foreign particulate matter and are specialized on the internalization of foreign material (11). Hence, they may play a decisive role for cellular internalization of microplastic particles.
An established approach to investigating the cellular internalization of particles by macrophages is the use of specifically surface-functionalized particles, like carboxylated and, therefore, negatively charged particles (12, 13) or opsonized particles coated with antibodies such as immunoglobulin G (IgG) (14) to induce receptor-mediated phagocytosis (12, 15). Smaller-sized particles are more likely internalized by cells than larger microparticles (16, 17). This may be because of the fact that smaller particles, especially in the nano size range (18), can passively cross cellular membranes, whereas for larger particles, mechanisms of active endocytosis play a crucial role (19). The abovementioned functionalized and pristine particles, predominantly used in toxicological studies, do not resemble microplastic particles found in nature, since plastic in the environment is of different shape and size and is additionally coated with an eco-corona (3). However, in research on the effects of larger microplastic particles, a recent comprehensive review (19) highlighted that “factors other than size have so far hardly been considered when studying tissue translocation.” Recently, Nasser et al. (20) proposed in their comprehensive review that the inclusion of biological matter, like eco-corona coatings, should become a prerequisite for ecotoxicity testing (20). Already seconds after exposure, plastic surfaces are covered by biomolecules (21) forming an initial corona. Over time, biomolecules with higher binding affinities substitute biomolecules with lower binding affinities forming the so called “hard corona,” and biomolecules can adhere to already strongly attached biomolecules forming the “soft corona” (22). This process leads to the formation of a substantial coating that mainly consists of biomolecules, like carbohydrates, lipids, and proteins (3, 23), which raises the intriguing possibility that this eco-corona could promote microplastic particle internalization by host cells. Given that specific protein coatings other than, e.g., IgG can also increase particle internalization into cells (24, 25), we hypothesized that microplastic particles exposed to aquatic environments adsorb biomolecules, forming an eco-corona that enhances the particles’ attachment to and internalization into cells compared with pristine microplastic particles.
RESULTS
To test our hypothesis, we investigated the internalization of fresh water– and salt water–exposed spherical microplastic particles with a diameter of 3 μm, a size class reported to occur in the environment (26) and of appropriate size of active endocytosis (27). As a cell system, we used the established murine macrophage cell line J774A.1 with a mean single cell area of 495 ± 22 μm2 (all numbers shown are the means ± standard error of the mean, unless otherwise stated). We incubated the microplastic particles (nonfluorescent plain polystyrene without any functionalization such as opsonization or carboxylation) in fresh water from an artificial pond or salt water from a marine aquarium (both of which were inhabited by diverse animal, plant, and microbial communities; fig. S1). Microplastic particles incubated in ultrapure water (pristine particles) under sterile conditions with no biomolecules present were used as negative controls, and IgG-opsonized microplastic particles, known to induce receptor-mediated phagocytosis (14), were used as positive controls. Although some biomolecules are adsorbed by surfaces within seconds (21), we incubated the microplastic particles for 2 and 4 weeks to allow the formation of a substantial eco-corona, resembling conditions a microplastic particle is exposed to in nature. For the internalization experiments, 23,000 ± 1000 cells were cultured on glass coverslips, and 29,000 ± 700 microplastic particles were added and allowed to sediment onto the cells for 1 hour on ice. Afterward, the cells and the sedimented particles were incubated for 2 hours. After this incubation, unattached particles were washed off using buffer, and the cells were preserved with paraformaldehyde (PFA). Filamentous actin, as being part of the cytoskeleton and intimately involved in the process of the internalization of particulate matter in the used size (15), was fluorescently labeled to distinguish between internalized and attached microplastic particles: Only those microplastic particles that were fully surrounded by fluorescently labeled actin were considered to have been internalized (Fig. 1 and fig. S2). With our experimental approach, we can show that nonfluorescent microplastic particles exposed to the environment are fully surrounded by cellular material and are therefore unambiguously internalized by the cells. To verify that these particles were indeed the used incubated polystyrene microplastic particles and not contaminants (e.g., dust of a similar size and shape), single Raman spectra were acquired from 10 randomly chosen particles from each treatment and incubation time. All particles were confirmed to be the used polystyrene particles (fig. S3).
Fig. 1. Images of particle-cell interactions of microplastic particles exposed to fresh water for 2 weeks.
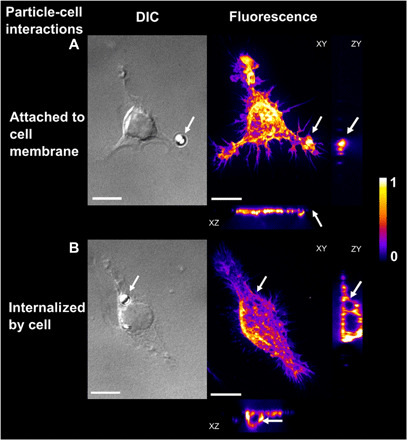
DIC: Differential interference contrast microscopy images of particle-cell interactions. Fluorescence: Spinning disc confocal images of the cells with fluorescently labeled filamentous actin (false color image, maximum intensity projection showing arbitrary units). XY, YZ, and XZ projections of three-dimensional confocal images allow the differentiation of microplastic particles (A) attached to cell membranes or (B) internalized microplastic particles. Arrows indicate microplastic particle position. Scale bars, 10 μm.
Our results show that microplastic particles exposed to fresh water or salt water for either incubation time (2 or 4 weeks) attached to and became internalized by cells approximately 10 times more often than pristine microplastic particles. The nonparametric Kruskal-Wallis test was used to investigate differences between all tested treatments: P < 0.001 for 2 (2w) and 4 (4w) weeks of incubation. A Games-Howell post hoc test was conducted for pairwise analysis (e.g., FW versus SW). Particle-cell interactions (PCI): FW2w versus UW2w P < 0.01, FW4w versus UW4w P < 0.001, SW2w versus UW2w P < 0.05, SW4w versus UW4w P ≤ 0.001; internalized: FW2w versus UW2w P < 0.01, FW4w versus UW4w P < 0.01, SW2w versus UW2w P < 0.01, SW4w versus UW4w P ≤ 0.001 (Fig. 2). IgG-opsonized microplastic particles attached to and were internalized by cells more often than microplastic particles exposed to fresh water and salt water by a factor of approximately 10, and more often than pristine microplastic particles by a factor of approximately 100 (PCI and internalized: IgG2w&4w versus FW/SW/UW2&4w P ≤ 0.001). Furthermore, between 2 and 4 weeks of incubation, we detected significant increases in the numbers of attached microplastic particles exposed to salt water and internalized microplastic particles exposed to fresh water or salt water (for pairwise comparisons of the incubation times within one treatment, a nonparametric Mann-Whitney U test was performed: PCI: SW2w versus SW4w P < 0.01; internalized: SW2w versus SW4w P < 0.01, FW2w versus FW4w P < 0.05; Fig. 2). We did not observe a statistically significant difference between microplastic particles incubated in fresh water and salt water for either incubation time (summary statistical analysis, see table S2).
Fig. 2. Combined results of particle-cell interactions and internalized microplastic particles.
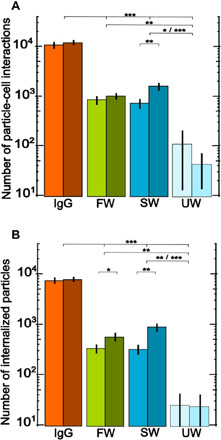
(A) Numbers of particle-cell interactions and (B) numbers of internalized microplastic particles following 2 (light color) or 4 (dark color) weeks of incubation (all numbers indicate the means ± SE, table S1; IgG, positive control; FW, particles exposed to fresh water; SW, particles exposed to salt water; UW, negative control, pristine particles from ultrapure water). The numbers of particle-cell interactions and internalized microplastic particles were standardized to coverslips with 23,000 cells to which 29,000 particles were added. Because the data span almost three orders of magnitude, the ordinate is scaled logarithmically. The IgG treatment differs highly significant from all other treatments for either incubation time. Fresh water and salt water differ significantly from ultrapure water (except where specified otherwise, a Kruskal-Wallis test followed by a Games-Howell post hoc test was conducted to investigate significant differences between treatments, ***P ≤ 0.001, **P ≤ 0.01, *P = 0.05). The salt water treatment shows a significant increase in the numbers of particle-cell interactions and internalized microplastic particles over the incubation time, and fresh water shows a significant increase in the number of internalized microplastic particles during the incubation time (Mann-Whitney U test: **P ≤ 0.01, *P = 0.05).
The increased number of attachments and internalizations observed for the pretreated particles is likely because of the presence of an eco-corona. To determine whether an eco-corona was present, we first investigated the surface morphology of microplastic particles using scanning electron microscopy (SEM). The microplastic particles exposed to fresh water showed heterogeneously distributed irregular surface modifications (Fig. 3A), indicating the adhesion of molecules (28), which might be components of an eco-corona. Those exposed to salt water seemed to be homogeneously covered with structures resembling salt crystals (Fig. 3B), which were likely derived from air drying and which impeded the detection of an eco-corona. In contrast, the pristine microplastic particles had plain surfaces without any visible modifications (Fig. 3C), and IgG-opsonized microplastic particles showed rough but homogeneous surfaces (Fig. 3D), as is characteristic of particles that are opsonized with proteins (28). This finding is supported by the fact that the diameters of these structures were in the same size range (20 to 40 nm) as individual IgG antibodies (29).
Fig. 3. Representative SEM images of microplastic particles after 4 weeks of incubation, with enlarged views of the surface.
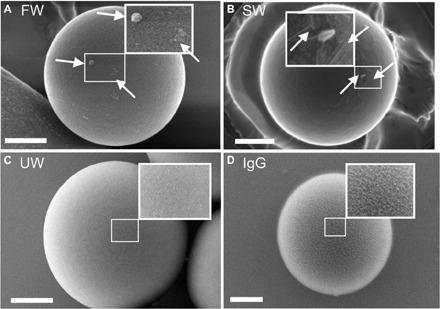
(A) FW: Microplastic particles incubated in fresh water, with an enlarged view of the irregular surface modifications (arrows). (B) SW: Microplastic particles incubated in salt water, with an enlarged view of the irregular surface modifications with small salt crystals (arrows). (C) UW: Microplastic particles incubated in ultrapure water showing a plain surface. (D) IgG: Microplastic particles opsonized with IgG with an enlarged view of its homogeneously rough surface. Scale bars, 1 μm; SEM settings: 2 to 3 kV, InLens/SE2 detector.
Then, to determine whether the surface modifications detected by SEM might represent biomolecules in an eco-corona, we investigated the chemical signatures of these coatings using micro-Raman spectroscopy. We found that the surfaces of particles exposed to fresh water harbored Raman bands, indicating the presence of biomolecules such as carbohydrates (C─O─C band), amino acids (C─N─S band), nucleic acids (PO4 band), lipids (C─H, C─H2 band), and proteins (C─H and C═O band) (30), which are known constituents of an eco-corona (Fig. 4) (3, 23). In contrast, we did not detect Raman bands specific for biomolecules on the pristine microplastic particles. We were not able to detect any chemical signatures specific to biomolecules on the surfaces of microplastic particles exposed to salt water, most likely because of the incrustation with salt crystals that was observed by SEM and bright-field imaging (fig. S4). The very prominent SO42− salt Raman band may have masked other bands specific for biomolecules (fig. S5).
Fig. 4. Raman spectroscopic analysis of the coating of microplastic particles incubated with fresh water.
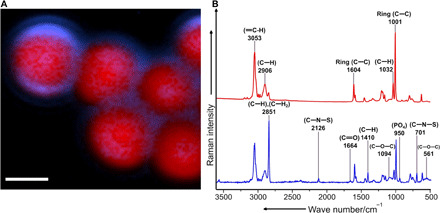
(A) False color Raman image of the microplastic particles (red) and the biomolecules forming a putative eco-corona (blue) on their surfaces, generated from the spectral mapping data. Scale bar, 2 μm. (B) The spectrum in red represents Raman signatures corresponding to the microplastic particles, and the spectrum in blue corresponds to signatures representative of the eco-corona. The Raman vibrational modes associated with biomolecules are mainly the C-S stretching mode (701 cm−1), the PO4 stretching mode (950 cm−1), the C─H bending mode (1410 cm−1), the C═O stretching mode (1664 cm−1), the C─H and C─H2 stretching mode (2851 cm−1), and the Raman band at 2126 cm−1 (C─N─S), together with the stretching mode at 701 cm−1, which could be indicative of the presence of thiocyanate molecules. Spectral signatures such as the ═C─H stretching mode (3053 cm−1), the C─H bending mode (2906 cm−1), the C─C bending mode (1604 cm−1), the C─H bending mode (1032 cm−1), and the C─C ring stretching mode (1001 cm−1) correspond to the PS microplastic particles.
Therefore, we further analyzed the elemental composition of surface coatings of microplastic particles incubated in ultrapure, fresh water, and salt water using x-ray photoelectron spectroscopy (XPS, Fig. 5). Microplastic particles incubated in fresh water and salt water displayed a distinct feature corresponding to nitrogen, which was absent on microplastic particles incubated in ultrapure water (fig. S7). In addition, salt water–incubated microplastic particles showed carbon from either ─O═C─O─ or ─O═C─N─ groups (fig. S8) (31). The origin of this signal is more likely from ─O═C─N─ groups as the presence of nitrogen was unequivocally confirmed on microplastic particles incubated in salt water (Fig. 5B). This is further supported by the nitrogen peak position (400.1 eV) found on both salt water– and fresh water–incubated microplastic particles, which is characteristic of amino acids (31). The presence of a variety of functional groups on microplastic particles incubated in fresh water and salt water was further supported by their broad S 2p and O 1s core spectra (fig. S9). Hence, XPS analysis strongly indicate the presence of biomolecules not only on the surface of fresh water–incubated but also even on the surface of salt water–incubated microplastic particles. Furthermore, the fact that particles incubated in salt water attached to and were internalized by cells as often as particles incubated in fresh water indicates that the two types of particles have similar biomolecular coatings.
Fig. 5. Core-level spectra of microplastic particles incubated in ultrapure water (gray), fresh water (green), and salt water (blue).
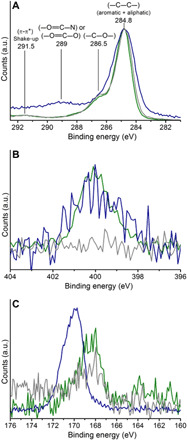
(A) C 1s region showing the characteristic PS signals at 284.8 and 291.5 eV corresponding to carbon from (─C─C─ aliphatic and aromatic) and π-π* shake-up processes, respectively (44). In addition, all the samples display a signal at 286.5 eV, attributable to the carbon bound to oxygen as in alcohol or ether functional groups. Microplastic particles from salt water additionally show a signal at 289 eV, possibly from the carbon in ─O═C─N or ─O═C─O functional groups. (B) N 1s region from all the samples, confirming the presence of nitrogen on the surface of microplastic particles incubated in fresh water and salt water with the maximum at 400.1 eV. (C) S 2p region showing prominent signals at 168.5 and 169.8 eV on the surface of microplastic particles from ultrapure water and fresh water, and salt water, respectively, corresponding to sulfate functional group.
DISCUSSION
Overall, our findings suggest that the coating of the particles with biomolecules enhances the cellular internalization of microplastic particles, indicating that the presence of an eco-corona is an important factor inducing the cellular internalization of microplastics.
It has been shown that some proteins act as opsonins that enhance the internalization of particles into cells, since surface proteins play an important role in nanoparticle-cell interactions (32, 33). For instance, Walkey et al. (25) showed that the internalization of gold nanoparticles by the same murine macrophage cell line as that used in our study is positively correlated with the concentration of proteins adsorbed onto the particle surface. Our results show that the role of surface coatings extends to microplastic particles in the lower micrometer range exposed to fresh water and salt water. Once biomolecules adsorb onto microplastic particle surfaces, they may function as a chemical stimulus for the attachment and internalization of those particles into cells, similar to mechanisms known for specifically functionalized particles. The internalization of foreign materials by macrophages is triggered by membrane receptors such as Fc or scavenger receptors (34). While, for example, Fcγ receptors are highly specific for binding the Fc region of IgG antibodies (15), scavenger receptors are known for their broad range of ligand binding (34). Therefore, the internalization of microplastic particles coated with an eco-corona may occur via scavenger receptor–induced phagocytosis. Consistently, we found that pristine microplastic particles with no eco-corona were very rarely internalized compared with microplastic particles incubated in fresh water and salt water. Although pristine microplastic particles do not have an eco-corona, a coincidental internalization of these particles could be because of membrane ruffling and macropinocytosis, as both processes occur in macrophages (35, 36). Membrane ruffling is intimately linked to the formation of macropinosomes, which can be up to 5 μm in size (32), and therefore, pristine microplastic particles with no chemical stimulus on their surfaces could be nonspecifically internalized together with fluid because of this process.
We show that an eco-corona formed under environmental conditions facilitates cellular internalization of microplastics in a size range frequently found in nature (26). Hence, it may not be pristine plastic particles per se but rather microplastics exposed to the environment that pose a health risk. It enhances the probability of cellular internalization and therefore may pose more of a health risk to organisms that ingest these particles along with their food. The coating with an eco-corona may then lead to a “Trojan horse” effect that particles normally not interacting with membrane receptors become internalized (37). Thus, the generally prevailing assumption that plastic itself behaves as an “inert” material from a toxicological point of view (23, 38) is not valid for microplastic particles when exposed to fresh water and salt water. This is in concordance with the suggestion of Galloway et al. (3) that environmental coating plays an important role in the interaction of microplastics with cells and tissues and therefore in determining their ecological impact.
We anticipate our results to be a starting point for investigations on the cellular mechanisms of microplastic internalization from fresh water and salt water and even terrestrial environments. This will allow us to obtain a comprehensive picture of microplastic internalization by cells, which, in turn, will be indispensable for identifying how microplastic exposure might affect organisms in polluted environments.
MATERIALS AND METHODS
Experimental design
Cell line and cell culture conditions
Murine macrophage J774A.1 cells (DSMZ, Braunschweig, Germany) were cultured as described previously by Keller et al. (39). To maintain suitable cell concentrations, cells were passaged three times per week to an appropriate number and cultured in T-75 culture flasks (Corning, New York, USA). Before the experiments, the cells were scraped off of the culture flask surfaces into the culture media, centrifuged (200g, 2 min, 20°C), and resuspended with 5 ml of cell culture medium in a Falcon tube (Corning, Corning, New York, USA). Then, the cells were counted using a hemocytometer (Neubauer improved, Brand, Wertheim, Germany), seeded on microscope coverslips (diameter, 18 mm; #1, MENZEL GLÄSER, Braunschweig, Germany) in 12-well plates (CellStar, Greiner Bio-One, Frickenhausen, Germany) in 1 ml of cell culture medium, and allowed to adhere onto the coverslips under standard culture conditions (37°C, 5% CO2, humidified) overnight. On each coverslip, 40,000 cells per milliliter were seeded to obtain a mean number of 23,000 cells per coverslip (not all cells adhered to and remained on the coverslips during the experimental procedure).
Microplastic particles and pretreatment conditions
Plain nonfluorescent white polystyrene beads (microplastic particles) with a diameter of 3 μm (Micromod, Rostock, Germany, white particles, micromer plain, Prod. Nr. 01-00-303) were incubated in fresh water or salt water for the environmental samples, or in ultrapure water (Veolia Purelab flex, Veolia, Celle, Germany) to serve as the “pristine microplastic particle” negative control. Freshwater samples were obtained from an artificial outdoor pond, whereas saltwater samples were collected from a marine aquarium facility with a defined salinity of 35 ‰ (fig. S1). Twenty microliters of microplastic particle stock solution (50 mg ml−1) was added to 980 μl of the corresponding water sample (fresh, salt, and ultrapure water) in a glass vial (autosampler vials, 1.2 ml; neoLab, Heidelberg, Germany). The ultrapure water (Veolia Purelab flex, Veolia, Celle, Germany) was filtered (Whatman Puradisc syringe filter, 0.2 μm: GE Healthcare, Freiburg, Germany) under sterile conditions to exclude microbial activity. For each treatment (fresh, salt, and ultrapure water), 10 replicates (glass vials) were prepared. To ensure vital microbial communities within the environmental samples during the incubation process, the corresponding water samples were changed three times per week. Each glass vial was centrifuged (2000g, 20 min, room temperature), and 900 μl of supernatant was replaced by the same amount of new corresponding water samples. To prevent aggregation of the microplastic particles by sedimentation, all samples were placed on a shaker (100 rpm at room temperature) for the whole incubation time. Although we did not add additional surfactant to avoid aggregation of the microplastic particles, only occasionally aggregates were found in all samples. Microplastic particles from fresh, salt, and ultrapure water were collected for the cell experiments after 2 weeks of incubation, while the remaining particles were further incubated under the same conditions for an additional 2 weeks within the same vials.
For the positive control, IgG antibodies (native IgG primary antibodies from mouse serum; Merck Millipore, Darmstadt, Germany) were passively adsorbed onto carboxylated microplastic particles (Micromod, Rostock, Germany, white particles, micromer COOH, Prod. Nr. 01-02-303) according to the protocol described by Keller et al. (39). Opsonization was verified by antibody-antibody labeling with goat anti-mouse IgG cross-adsorbed fluorescent secondary antibody (Dy-Light 488, Thermo Fisher Scientific). The IgG-opsonized microplastic particle stock solution was stored at 4°C for later use as a positive control for assessing internalization.
Quantifying numbers of pretreated, positive, and negative control microplastic particles
The numbers of microplastic particles within each treatment were quantified, because due to the media exchange of microplastic particles incubated in fresh, salt, and ultrapure water, an unknown number of particles were lost. The quantification of the numbers of microplastic particles from fresh, salt, and ultrapure water was conducted in subsamples for both incubation times (2 and 4 weeks). The number of microplastic particles within the IgG treatment was only counted once because those particles were obtained from an unmodified stock solution (no media exchange was performed, and therefore, a loss of IgG particles can be ruled out). To quantify the numbers of microplastic particles for each treatment and incubation time, 100 μl of microplastic particle dilutions [fresh, salt, and ultrapure water:phosphate-buffered saline (PBS) 1:100; IgG stock solution:PBS 1:1000] was added to two sample wells per treatment within a 12-well plate, and the particles were allowed to sediment. For imaging, the 12-well plates were placed onto an inverted microscope (Nikon Eclipse Ti, NIKON, Tokyo, Japan, 20×/0.45 objective), and spatial image series were acquired with an electron-multiplying charge-coupled device (EMCCD) camera (Luca-R, Andor, Belfast, Northern Ireland). For each sample well, three regions of interest (ROIs) (4.23 mm2 each) were imaged, and the numbers of microplastic particles were counted using the Fiji ImageJ cell counter software (40). We tested the distribution of particles within one sample well by microscopy. Heterogeneities of the particle numbers did occur, but we compensated for this by standardizing to 29,000 particles for each treatment for a whole coverslip (245.50 mm2).
Cell experiment
The following experimental procedure was implemented to obtain samples for the quantification of microplastic particles interacting with cells (PCI), the measurement of the area covered by cells on the coverslips, and for investigating the number of internalized microplastic particles from the PCI.
After 2 or 4 weeks of exposure to fresh, salt, and ultrapure water, microplastic particles were added to cells, which were prepared on coverslips in 12-well plates the day before the experiments, as described above. Microplastic particles pretreated with fresh, salt, or ultrapure water and IgG-opsonized microplastic particles were diluted in cell culture medium (fresh, salt, and ultrapure water 1:100; IgG stock solution 1:1000). The 12-well plates containing the prepared cells were placed on ice for 1 hour to reduce cellular activity. From each treatment of the pretreated microplastic particles and the IgG-opsonized particles, 100 μl of each microplastic particle dilution was added to a coverslip. Ten coverslips for each treatment and exposure time were prepared, yielding a total of 80 coverslips. After 1 hour of microplastic particle sedimentation, the well plates were incubated at 37°C (Kelvitron kl BK6160, Heraeus, Hanau, Germany) for 2 hours to activate cells. Then, the coverslips were washed three times with PBS to remove unattached microplastic particles. Cells were fixed using a PBS-PFA solution containing 4% PFA (Sigma Aldrich, St. Louis, Missouri) for 15 min on ice. Then, the coverslips were washed again three times with PBS, and 50 μl of labeling solution was added and allowed to sit for 25 min to label filamentous actin. The labeling solution consisted of Alexa Fluor Phalloidin 488 (Invitrogen, Carlsbad, USA) and dilution buffer, consisting of 98.7% PBS, including 0.3% Triton and 1% bovine serum albumin (AppliChem, Darmstadt, Germany), to a final concentration of 1:25. The coverslips were again washed three times with PBS and transferred to 1 ml of ultrapure water. Last, the coverslips were mounted on glass slides (Servoprax, Wesel, Germany) with Fluoromount-G (SouthernBiotech, Birmingham, Alabama) mounting media and allowed to dry overnight. The next day, the coverslips were fixed to the glass slides with nail polish (fig. S2).
Quantification of PCI and area covered with cells on coverslips
To determine the total number of PCI on each coverslip, five randomly chosen ROIs (0.29 mm2) were selected and imaged by using a DMI 6000 microscope (Leica, Wetzlar, Germany, HCX PL APO 63×/1.30 oil objective) including a spinning disc unit (CSU-X1, Yokogawa, Musashino, Japan) with an EMCCD camera (Evolve 512, Photometrics, Tucson, Arizona, including an additional ×1.2 magnification lens). A differential interference contrast (DIC) microscopy image was acquired to quantify the PCI within the ROIs using the Fiji ImageJ cell counter software. In addition, confocal stacks of fluorescently labeled cells were acquired using a 488-nm laser (50 mW, Sapphire 488, Coherent, Santa Clara, California) at a spinning disc speed of 5000 rpm to excite fluorescence. Axial stacks of the cells were acquired with a vertical distance of 0.2 μm, which is sufficient to oversample the image given the axial resolution of the microscope (41).
The confocal stacks were used to calculate the area covered by cells within an ROI. The area covered by cells was detected using both the DIC and the fluorescence channel simultaneously to obtain robust results. As described previously (42), a local contrast filter was applied to the DIC images to obtain a rough approximation of the cell mask MDIC. The local contrast filter highlights areas in which the difference ∆I between the local intensity maximum and minimum is larger than a given threshold TDIC
A circular filter with a radius of 3 px was chosen. To obtain a mask MF of the area covered by cells in the fluorescence channel, the axial stacks of 25 images IF (index k) were projected to the intensity maximum, and a simple threshold TF was applied to the resulting image
The masks were multiplied to obtain a mask M that highlights areas in which cells were detected in both channels
In the resulting mask, small holes were filled up to a size of 40 μm2, which corresponds to 8% of the average cell size. Next, the mask was smoothed by a Gaussian filter approximation (43) with an SD of 3 px = 0.6 μm to reduce pixel noise at the cell boundary. Objects in the background with an area of less than 80 μm2 were excluded, which appear in M mainly because of noise in the background (42). In this step, care was taken not to exclude any cells (fig. S6). Areas covered by cells within an ROI were extrapolated to a whole coverslip (245.50 mm2). The whole algorithm was implemented in MATLAB 2017b (MathWorks Inc.).
Investigation of internalized microplastic particles
From the same samples used to quantify PCI and areas covered with cells on coverslips, we visually screened each sample for single PCI to distinguish between particles that were only attached to cell membranes or were internalized. All samples were scored blind to exclude personal bias. The same DMI 6000 microscope including a spinning disc unit with a higher magnification (100×/1.40 oil objective) was used. Beginning from a randomly defined starting point, the coverslips were screened in the DIC channel until 100 to 110 PCI were detected or until the whole coverslip was completely screened. Once a PCI was found, a DIC image was taken, and axial stacks of fluorescently labeled cells were acquired (vertical distance of the axial stacks, 0.2 μm). To evaluate internalization of the microplastic particles, each confocal stack of cells with labeled actin filaments was analyzed with Fiji ImageJ (version: 2.0.0-rc-54/1.51 h 2016-09-08) orthogonal views. The microplastic particles used in the experiments were not fluorescent and therefore were not directly visible in the confocal stacks. DIC images were used to mark the particle positions (using the ROI manager in Fiji ImageJ). These positions were then transferred to the confocal stacks, in which internalized particles were visible as spherical black regions within the actin network. Only microplastic particles that were fully surrounded by actin filaments were considered to be internalized. Microplastic particles that were only partly surrounded were considered to be attached to cell membranes.
Combination and standardization of the results
The number of PCI (ROI-PCI experiment, 0.29 mm2) and the number of microplastic particles added (ROI–quantifying numbers of pretreated, positive, and negative control microplastic particles, 4.23 mm2) were extrapolated to a whole coverslip (245.50 mm2). As the areas on the coverslips covered by cells differ slightly between replicates and time points and as the microplastic particle concentrations differed between treatments (because of the loss during media exchange), each coverslip was standardized. A standard coverslip contains CellNumberstandCS = 23,000 cells and ParticleNumberstandCS = 29,000 microplastic particles, which represent the rounded mean values (rounded to the nearest thousand) of the numbers of cells and particles over all treatments and time points. The cell number CellNumberstandCS was determined by calculating the mean area covered by cells on a coverslip (11.33 mm2) divided by the average size of a single cell (495 μm2), which led to 23,000 cells on a standard coverslip. The number of microplastic particles ParticleNumberstandCS was the mean number of microplastic particles added to the coverslips. The standardization was performed with the following equation to obtain comparability between the treatments and incubation times despite slightly varying cell and particle numbers
All PCI determined for a single coverslip (PCICS) were multiplied with the CellNumberstandCS divided by the ratio of the exact area of cells on the coverslip CellAreaCS and the area of a single cell (SingleCellAreaCS) on this specific coverslip and furthermore multiplied with the ParticleNumberstandCS divided by the actual number of microplastic particles counted on this specific coverslip (ParticleNumberCS).
Scanning electron microscopy
To visualize the surface structures of microplastic particles, samples were analyzed using a scanning electron microscope (LEO1530 Zeiss, Oberkochen, Germany; magnification, ×500 to ×50,000, 2 to 3 kV; InLens/SE2 detector). First, each sample of the pretreated and IgG-opsonized microplastic particles was diluted in ultrapure water (1:100), and 10 μl of this dilution was pipetted onto a silicon wafer placed on carbon conductive tabs (Ø 12 mm Plano GmbH, Wetzlar, Germany) fixed to aluminum stubs (Ø 12 mm, Plano GmbH, Wetzlar, Germany). The stubs were then transferred into a desiccator and stored until the images were acquired. Samples were subsequently coated with a 2-nm-thick platinum layer (208HR sputter coater, Cressington, Watford, UK) and analyzed using the scanning electron microscope.
Micro-Raman spectroscopy
All Raman spectroscopic measurements were performed using a micro-Raman spectrometer (WITec Alpha 300 RA+, Ulm, Germany) equipped with a UHTS 300 spectrometer and a back-illuminated Andor Newton 970 EMCCD camera. A frequency-doubled Nd-YAG laser with a wavelength of 532 nm was used as the excitation source. The exciting laser radiation was coupled to a Zeiss microscope through a wavelength-specific single-mode optical fiber. The laser beam was focused onto the sample by means of a 50× long working distance [numerical aperture (NA) = 0.7, lateral resolution ca. 500 nm] and 100× (NA = 0.9, lateral resolution ca. 300 nm) Zeiss objective. The focal length of the spectrometer is 800 mm, and it is equipped with a diffraction grating having a groove density of 600 lines per millimeter to give a spectral resolution of ~3 to 4 cm−1. The laser power used was approximately 5 to 15 mW at the fiber for all measurements. Raman scattered light was detected by a Peltier-cooled complementary metal oxide semiconductor–based CCD with a sensor size of 1600 × 200 px. The instrument was operated by the integrated Witec Control Five software (version 5). All spectra were acquired in the 3600 to 500 cm−1 spectral range. The acquired data were preprocessed using Witec Project Five software (version 5) that allowed compensating for cosmic radiation and other background signals. A true component analysis was also performed using the Witec Project Five software to visualize the spatial distribution of different components within the Raman image (for example, the cell, polystyrene microplastic particles, and mounting media).
Single spectrum acquisition mode with a 50× long working distance objective (NA = 0.7) was used to confirm that the chosen particles from detected PCI showed spectral signatures specific to polystyrene, which showed that the particles were the used microplastic particles and not contaminants of a similar shape and size. For each treatment and incubation time, 10 PCI were investigated per sample. The integration times ranged between 10 and 15 s for single spectrum acquisitions. Second, the chemical components on the surface of the microplastic particles after exposure to fresh water and salt water were investigated. Raman imaging mode was used for these investigations. Therefore, 2 × 20 μl of microplastic particle samples from fresh, salt, and ultrapure water after 4 weeks of incubation and the corresponding water were pipetted onto two marked positions on a glass slide (SERVOPRAX, Wesel, Germany), transferred into a desiccator, and allowed to dry. Raman images were acquired from these samples with the following parameters: 100× objective (NA = 0.9), scan area of 15 × 15 μm, step size of 100 nm, and integration time of 0.5 s/pixel.
X-ray photoelectron spectroscopy
For sample preparation, 2 × 20 μl of microplastic particle solutions from fresh, salt, and ultrapure water after 4 weeks of incubation and the corresponding water were pipetted onto a glass slide (Servoprax, Wesel, Germany). Samples were transferred into a desiccator, and by using a water jet vacuum pump, a slight vacuum was produced and samples were allowed to dry. XPS spectra were measured with the PHI 5000 VersaProbe III system fitted with an Al Kα excitation source (hν = 1486.6 eV) and a dual neutralizer (electron gun and Ar+) at 10−10 mbar pressure. An x-ray source diameter of 100 μm was used to locally excite the samples; the corresponding photoemission with 45° take-off angle was collected at the multichannel analyzer. The survey and the detailed spectra were measured with pass energies of 224 and 69 or 112 eV, respectively. The SD on the reported energy values is ±0.1 eV. The reproducibility of the observed results was confirmed by performing at least three measurements at different places of the samples. The spectra were analyzed with a Multipak software pack, provided by the manufacturer. All emission signals were referenced to adventitious C 1s peak at 284.8 eV. For quantitative analysis of the spectra, Shirley background correction was used. For deconvolution of C 1s and S 2p spectra of salt water– and fresh water–incubated samples, respectively, a combination of Gaussian and Lorentzian functions was used without fixing any constrain on the peak position or FWHM; however, in S 2p spectrum, the multiplet splitting parameters for 2p orbitals were assigned prior to fit the bands. For comparative purposes, the C 1s, N 1s, and S 2p spectra shown in the main text were normalized.
Statistical analysis
Statistical analysis was conducted using R studio software (Version 1.0.143). The data for PCI, as well as for internalized microplastic particles, were tested for normal distribution (Shapiro-Wilk test) and homogeneity of variances (Levene test). A Kruskal-Wallis test with a Games-Howell post hoc test (P-adjust method for multiple nesting: Bonferroni Holm) was conducted to check for differences between treatments. To check for differences within one treatment regarding the incubation time, a Mann-Whitney U test was performed.
Supplementary Material
Acknowledgments
We thank M. Weiss and P. Struntz for the support with the confocal spinning disc microscope. We thank M. Heider for assistance with SEM, and the technicians from the Department of Animal Ecology I and the Biological Physics Group for support with the experiments. We also thank J. Brehm, J. Diller, S. Ritschar, J. Möller, S. Piehl, S. Steibl, S. Keller, K. Berghoff, M. Eisentraut (University of Bayreuth), and G. Griffiths (University of Oslo) for the helpful discussions with the manuscript. Funding: This work was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) project number 391977956–SFB 1357 and DFG INST 91/289-1 FUGG. A.F.R.M.R. was supported by a scholarship of the elite network of Bavaria (BayEFG). W.G. was supported by the German Academic Scholarship Foundation (Studienstiftung des deutschen Volkes). A.F.R.M.R. and W.G. were supported by the University of Bayreuth Graduate School. J.M. and M.T. acknowledge the DFG for the XPS facility at the Keylab-Device engineering, University of Bayreuth. Author contributions: A.F.R.M.R., H.K., and C.L. designed the experiments, and A.F.R.M.R., V.K.B.N., W.G., J.M., H.K. and C.L. wrote the manuscript. A.F.R.M.R. conducted all cell experiments and prepared samples for SEM, micro-Raman spectroscopy, and XPS. V.K.B.N., A.G., and H.S. performed Raman measurements and data analysis. J.M. and M.T. performed XPS measurements and data analysis. W.G. wrote the cell detection algorithm. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eabd1211/DC1
REFERENCES AND NOTES
- 1.Connors K. A., Dyer S. D., Belanger S. E., Advancing the quality of environmental microplastic research. Environ. Toxicol. Chem. 36, 1697–1703 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Conkle J. L., Báez Del Valle C. D., Turner J. W., Are we underestimating microplastic contamination in aquatic environments? Environ. Manag. 61, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Galloway T. S., Cole M., Lewis C., Interactions of microplastic debris throughout the marine ecosystem. Nat. Ecol. Evol. 1, 0116 (2017). [DOI] [PubMed] [Google Scholar]
- 4.D. Laist, Impacts of marine debris: Entanglement of marine life in marine debris including a comprehensive list of species with entanglement and ingestion records, in Marine Debris - Sources, Impacts Solution, J. M. Coe, D. B. Rogers, Eds. (Springer-Verlag, New York, 1997), pp. 99–139. [Google Scholar]
- 5.Desforges J.-P. W., Galbraith M., Ross P. S., Ingestion of microplastics by zooplankton in the Northeast Pacific ocean. Arch. Environ. Contam. Toxicol. 69, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Devriese L. I., van der Meulen M. D., Maes T., Bekaert K., Paul-Pont I., Frère L., Robbens J., Vethaak A. D., Microplastic contamination in brown shrimp (Crangon crangon, Linnaeus 1758) from coastal waters of the Southern North Sea and Channel area. Mar. Pollut. Bull. 98, 179–187 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Browne M. A., Dissanayake A., Galloway T. S., Lowe D. M., Thompson R. C., Ingested microscopic plastic translocates to the circulatory system of the mussel, Mytilus edulis (L). Environ. Sci. Technol. 42, 5026–5031 (2008). [DOI] [PubMed] [Google Scholar]
- 8.von Moos N., Burkhardt-Holm P., Köehler A., Uptake and effects of microplastics on cells and tissue of the blue mussel Mytilus edulis L. after an experimental exposure. Environ. Sci. Technol. 46, 327–335 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Lu Y., Zhang Y., Deng Y., Jiang W., Zhao Y., Geng J., Ding L., Ren H., Uptake and accumulation of polystyrene microplastics in zebrafish (Danio rerio) and toxic effects in liver. Environ. Sci. Technol. 50, 4054–4060 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Delie F., Evaluation of nano- and microparticle uptake by the gastrointestinal tract. Adv. Drug Deliv. Rev. 34, 221–233 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Grainger J. R., Konkel J. E., Zangerle-Murray T., Shaw T. N., Macrophages in gastrointestinal homeostasis and inflammation. Pflugers Arch. 469, 527–539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olivier V., Rivière C., Hindié M., Duval J.-L., Bomila-Koradjim G., Nagel M.-D., Uptake of polystyrene beads bearing functional groups by macrophages and fibroblasts. Colloids Surf. B Biointerfaces 33, 23–31 (2004). [Google Scholar]
- 13.Stock V., Böhmert L., Lisicki E., Block R., Cara-Carmona J., Pack L. K., Selb R., Lichtenstein D., Voss L., Henderson C. J., Zabinsky E., Sieg H., Braeuning A., Lampen A., Uptake and effects of orally ingested polystyrene microplastic particles in vitro and in vivo. Arch. Toxicol. 93, 1817–1833 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Nimmerjahn F., Ravetch J. V., Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Aderem A., Underhill D. M., Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17, 593–623 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Desai M. P., Labhasetwar V., Walter E., Levy R. J., Amidon G. L., The mechanism of uptake of biodegradable microparticles in Caco-2 cells is size dependent. Pharm. Res. 14, 1568–1573 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Yong C. Q. Y., Valiyaveetill S., Tang B. L., Toxicity of microplastics and nanoplastics in mammalian systems. Int. J. Environ. Res. Public Health 17, 1509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudi M., Azadmanesh K., Shokrgozar M. A., Journeay W. S., Laurent S., Effect of nanoparticles on the cell life cycle. Chem. Rev. 111, 3407–3432 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Triebskorn R., Braunbeck T., Grummt T., Hanslik L., Huppertsberg S., Jekel M., Knepper T. P., Krais S., Müller Y. K., Pittroff M., Ruhl A. S., Schmieg H., Schür C., Strobel C., Wagner M., Zumbülte N., Köhler H.-R., Relevance of nano- and microplastics for freshwater ecosystems: A critical review. TrAC Trends Anal. Chem. 110, 375–392 (2019). [Google Scholar]
- 20.Nasser F., Constantinou J., Lynch I., Nanomaterials in the Environment Acquire an “Eco-Corona” Impacting their Toxicity to Daphnia Magna—a Call for Updating Toxicity Testing Policies. Proteomics 20, 1800412 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Loeb G., Neihof R., Marine conditioning films. Adv. Chem. 145, 319–335 (1975). [Google Scholar]
- 22.Monopoli M. P., Åberg C., Salvati A., Dawson K. A., Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 7, 779–786 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Rummel C. D., Jahnke A., Gorokhova E., Kühnel D., Schmitt-Jansen M., Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ. Sci. Technol. Lett. 4, 258–267 (2017). [Google Scholar]
- 24.Smith M. W., Thomast N. W., Jenkinst P. G., Miller N. G. A., Cremaschit D., Portat C., Selective transport of microparticles across Peyer’s patch follicle-associated M cells from mice and rats. Exp. Physiol. 80, 735–743 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Walkey C. D., Olsen J. B., Guo H., Emili A., Chan W. C. W., Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 134, 2139–2147 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Ng K. L., Obbard J. P., Prevalence of microplastics in Singapore’s coastal marine environment. Mar. Pollut. Bull. 52, 761–767 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Tabata Y., Ikada Y., Effect of the size and surface charge of polymer microspheres on their phagocytosis by macrophage. Biomaterials 9, 356–362 (1988). [DOI] [PubMed] [Google Scholar]
- 28.Zhou T., Zhang K., Kamra T., Bülow L., Ye L., Preparation of protein imprinted polymer beads by Pickering emulsion polymerization. J. Mater. Chem. B 3, 1254–1260 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Chen Y., Cai J., Xu Q., Chen Z. W., Atomic force bio-analytics of polymerization and aggregation of phycoerythrin-conjugated immunoglobulin G molecules. Mol. Immunol. 41, 1247–1252 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Movasaghi Z., Rehman S., Rehman I. U., Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42, 493–541 (2007). [Google Scholar]
- 31.Stevens J. S., de Luca A. C., Pelendritis M., Terenghi G., Downes S., Schroeder S. L. M., Quantitative analysis of complex amino acids and RGD peptides by x-ray photoelectron spectroscopy (XPS). Surf. Interface Anal. 45, 1238–1246 (2013). [Google Scholar]
- 32.Norbury C. C., Drinking a lot is good for dendritic cells. Immunology 117, 443–451 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markiewicz M., Kumirska J., Lynch I., Matzke M., Köser J., Bemowsky S., Docter D., Stauber R. H., Westmeier D., Stolte S., Changing environments and biomolecule coronas: Consequences and challenges for the design of environmentally acceptable engineered nanoparticles. Green Chem. 20, 4133–4168 (2018). [Google Scholar]
- 34.Fabriek B. O., Dijkstra C. D., van den Berg T. K., The macrophage scavenger receptor CD163. Immunobiology 210, 153–160 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Champion J. A., Walker A., Mitragotri S., Role of particle size in phagocytosis of polymeric microspheres. Pharm. Res. 25, 1815–1821 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerr M. C., Teasdale R. D., Defining macropinocytosis. Traffic 10, 364–371 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Wang F., Yu L., Monopoli M. P., Sandin P., Mahon E., Salvati A., Dawson K. A., The biomolecular corona is retained during nanoparticle uptake and protects the cells from the damage induced by cationic nanoparticles until degraded in the lysosomes. Nanomed. Nanotechnol. Biol. Med. 9, 1159–1168 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Lithner D., Damberg J., Dave G., Larsson Å., Leachates from plastic consumer products – Screening for toxicity with Daphnia magna. Chemosphere 74, 1195–1200 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Keller S., Berghoff K., Kress H., Phagosomal transport depends strongly on phagosome size. Sci. Rep. 7, 17068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jonkman J. E. N., Swoger J., Kress H., Rohrbach A., Stelzer E. H. K., Resolution in optical microscopy. Methods Enzymol. 360, 416–446 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Berghoff K., Keller S., Gross W., Gebhardt L., Kress H., Application of optical tweezers for biochemical and thermal cell stimulation. Light Robot. Struct. Mediated Nanobiophoton. 2017, 385–410 (2017). [Google Scholar]
- 43.P. Kovesi, Fast almost-Gaussian filtering, in Proceedings of the 2010 Digital Image Computing Techniques and Applications (DICTA 2010) (IEEE, Sydney, Australia, 2010), pp. 121–125. [Google Scholar]
- 44.Girardeaux C., Pireaux J.-J., Analysis of polystyrene (PS) by XPS. Surf. Sci. Spectra. 4, 130–133 (1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/50/eabd1211/DC1


